Project
Inflammatory response time course, HUVEC (Wada, 2009)
Navigation
Downloads
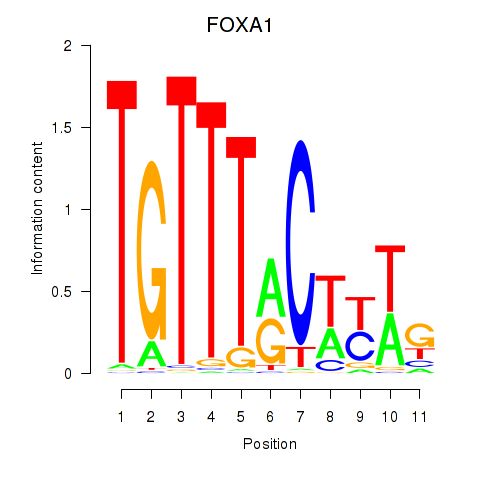
Results for FOXA1
Z-value: 0.70
Transcription factors associated with FOXA1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
FOXA1
|
ENSG00000129514.8 | FOXA1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| FOXA1 | hg38_v1_chr14_-_37595224_37595281 | 0.12 | 5.7e-01 | Click! |
Activity profile of FOXA1 motif
Sorted Z-values of FOXA1 motif
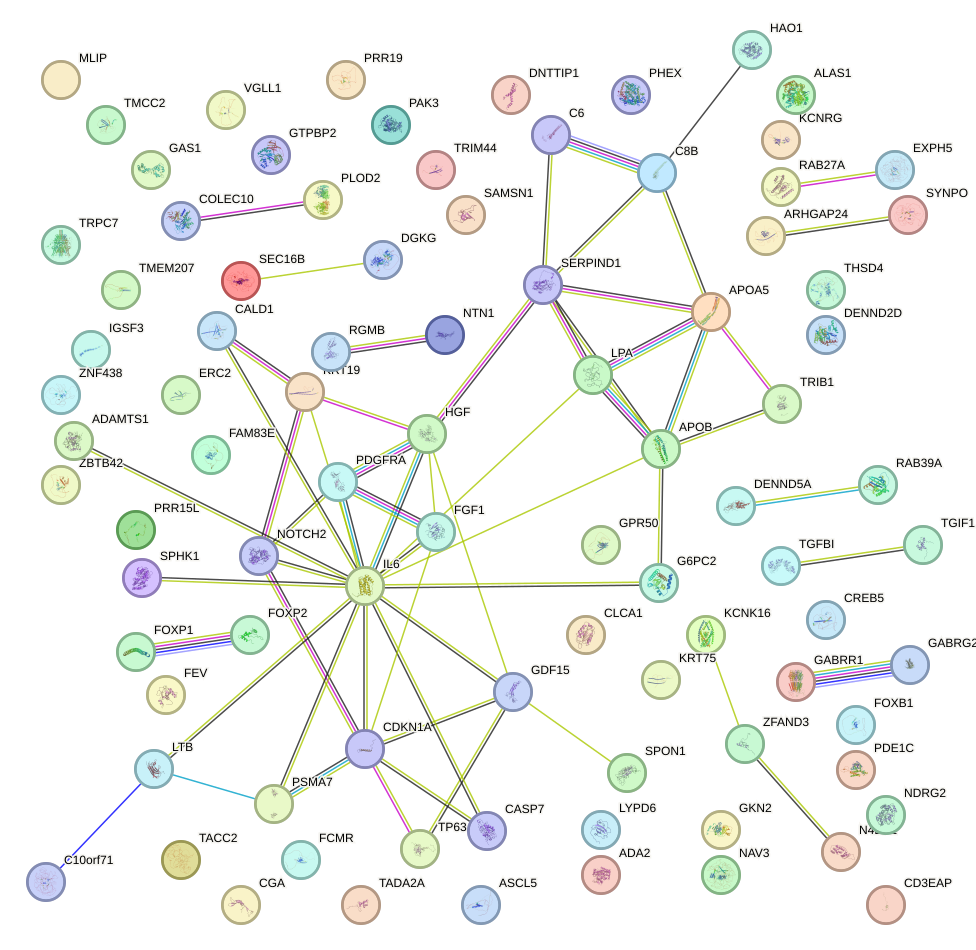
Network of associatons between targets according to the STRING database.
First level regulatory network of FOXA1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_+_77830886 | 2.71 |
ENST00000397909.7
ENST00000549464.5 |
NAV3
|
neuron navigator 3 |
| chr2_+_149330506 | 2.06 |
ENST00000334166.9
|
LYPD6
|
LY6/PLAUR domain containing 6 |
| chr5_+_136058849 | 2.03 |
ENST00000508076.5
|
TGFBI
|
transforming growth factor beta induced |
| chr22_+_20774092 | 1.57 |
ENST00000215727.10
|
SERPIND1
|
serpin family D member 1 |
| chr19_+_18386150 | 1.55 |
ENST00000252809.3
|
GDF15
|
growth differentiation factor 15 |
| chr6_-_31582415 | 1.53 |
ENST00000429299.3
ENST00000446745.2 |
LTB
|
lymphotoxin beta |
| chr17_+_76376581 | 1.49 |
ENST00000591651.5
ENST00000545180.5 |
SPHK1
|
sphingosine kinase 1 |
| chr1_-_120100688 | 1.33 |
ENST00000652264.1
|
NOTCH2
|
notch receptor 2 |
| chr1_-_201127184 | 1.12 |
ENST00000449188.3
|
ASCL5
|
achaete-scute family bHLH transcription factor 5 |
| chr10_+_89332484 | 1.07 |
ENST00000371811.4
ENST00000680037.1 ENST00000679583.1 ENST00000679897.1 |
IFIT3
|
interferon induced protein with tetratricopeptide repeats 3 |
| chr10_+_113709261 | 1.07 |
ENST00000672138.1
ENST00000452490.3 |
CASP7
|
caspase 7 |
| chr5_+_150601060 | 1.05 |
ENST00000394243.5
|
SYNPO
|
synaptopodin |
| chr1_-_111200633 | 0.98 |
ENST00000357640.9
|
DENND2D
|
DENN domain containing 2D |
| chr3_-_146161167 | 0.97 |
ENST00000360060.7
ENST00000282903.10 |
PLOD2
|
procollagen-lysine,2-oxoglutarate 5-dioxygenase 2 |
| chr8_+_119067239 | 0.97 |
ENST00000332843.3
|
COLEC10
|
collectin subfamily member 10 |
| chr2_-_144431001 | 0.95 |
ENST00000636413.1
|
ZEB2
|
zinc finger E-box binding homeobox 2 |
| chr8_+_32647080 | 0.94 |
ENST00000520502.7
ENST00000523041.2 ENST00000650819.1 |
NRG1
|
neuregulin 1 |
| chr6_+_36676489 | 0.90 |
ENST00000448526.6
|
CDKN1A
|
cyclin dependent kinase inhibitor 1A |
| chr1_+_205256189 | 0.88 |
ENST00000329800.7
|
TMCC2
|
transmembrane and coiled-coil domain family 2 |
| chr2_-_144516397 | 0.87 |
ENST00000638128.1
|
ZEB2
|
zinc finger E-box binding homeobox 2 |
| chr2_-_144430934 | 0.85 |
ENST00000638087.1
ENST00000638007.1 |
ZEB2
|
zinc finger E-box binding homeobox 2 |
| chr4_+_54229261 | 0.84 |
ENST00000508170.5
ENST00000512143.1 ENST00000257290.10 |
PDGFRA
|
platelet derived growth factor receptor alpha |
| chr2_-_144516154 | 0.83 |
ENST00000637304.1
|
ZEB2
|
zinc finger E-box binding homeobox 2 |
| chr11_-_9265302 | 0.82 |
ENST00000328194.8
|
DENND5A
|
DENN domain containing 5A |
| chr15_-_55249029 | 0.79 |
ENST00000566877.5
|
RAB27A
|
RAB27A, member RAS oncogene family |
| chr15_+_71096941 | 0.76 |
ENST00000355327.7
|
THSD4
|
thrombospondin type 1 domain containing 4 |
| chr18_+_3449413 | 0.75 |
ENST00000549253.5
|
TGIF1
|
TGFB induced factor homeobox 1 |
| chr6_+_116399395 | 0.74 |
ENST00000644499.1
|
ENSG00000285446.1
|
novel protein |
| chr6_-_87095059 | 0.73 |
ENST00000369582.6
ENST00000610310.3 ENST00000630630.2 ENST00000627148.3 ENST00000625577.1 |
CGA
|
glycoprotein hormones, alpha polypeptide |
| chr22_-_17219424 | 0.72 |
ENST00000649540.1
ENST00000399837.8 ENST00000543038.1 |
ADA2
|
adenosine deaminase 2 |
| chr3_-_71583713 | 0.69 |
ENST00000649528.3
ENST00000471386.3 ENST00000493089.7 |
FOXP1
|
forkhead box P1 |
| chr14_+_104801082 | 0.67 |
ENST00000342537.8
|
ZBTB42
|
zinc finger and BTB domain containing 42 |
| chr22_-_17219571 | 0.66 |
ENST00000610390.4
|
ADA2
|
adenosine deaminase 2 |
| chr6_+_54018992 | 0.64 |
ENST00000509997.5
|
MLIP
|
muscular LMNA interacting protein |
| chr8_+_125430333 | 0.61 |
ENST00000311922.4
|
TRIB1
|
tribbles pseudokinase 1 |
| chr3_+_148730100 | 0.61 |
ENST00000474935.5
ENST00000475347.5 ENST00000461609.1 |
AGTR1
|
angiotensin II receptor type 1 |
| chr15_+_60004305 | 0.61 |
ENST00000396057.6
|
FOXB1
|
forkhead box B1 |
| chr21_-_14658812 | 0.60 |
ENST00000647101.1
|
SAMSN1
|
SAM domain, SH3 domain and nuclear localization signals 1 |
| chr5_+_162067764 | 0.57 |
ENST00000639213.2
ENST00000414552.6 |
GABRG2
|
gamma-aminobutyric acid type A receptor subunit gamma2 |
| chr6_+_37929959 | 0.57 |
ENST00000373389.5
|
ZFAND3
|
zinc finger AN1-type containing 3 |
| chr5_+_98769273 | 0.56 |
ENST00000308234.11
|
RGMB
|
repulsive guidance molecule BMP co-receptor b |
| chr5_+_162067500 | 0.56 |
ENST00000639384.1
ENST00000640985.1 ENST00000638772.1 |
GABRG2
|
gamma-aminobutyric acid type A receptor subunit gamma2 |
| chr5_+_162067858 | 0.55 |
ENST00000361925.9
|
GABRG2
|
gamma-aminobutyric acid type A receptor subunit gamma2 |
| chr6_-_89217339 | 0.54 |
ENST00000454853.7
|
GABRR1
|
gamma-aminobutyric acid type A receptor subunit rho1 |
| chr11_-_108593738 | 0.52 |
ENST00000525344.5
ENST00000265843.9 |
EXPH5
|
exophilin 5 |
| chr6_+_54018910 | 0.52 |
ENST00000514921.5
ENST00000274897.9 ENST00000370877.6 |
MLIP
|
muscular LMNA interacting protein |
| chr8_+_32646838 | 0.52 |
ENST00000651333.1
ENST00000652592.1 |
NRG1
|
neuregulin 1 |
| chr5_+_162067458 | 0.50 |
ENST00000639975.1
ENST00000639111.2 ENST00000639683.1 |
GABRG2
|
gamma-aminobutyric acid type A receptor subunit gamma2 |
| chr5_+_162068031 | 0.49 |
ENST00000356592.8
|
GABRG2
|
gamma-aminobutyric acid type A receptor subunit gamma2 |
| chr3_+_52211442 | 0.49 |
ENST00000459884.1
|
ALAS1
|
5'-aminolevulinate synthase 1 |
| chr11_+_107928394 | 0.47 |
ENST00000320578.3
|
RAB39A
|
RAB39A, member RAS oncogene family |
| chr7_-_32299287 | 0.47 |
ENST00000396193.5
|
PDE1C
|
phosphodiesterase 1C |
| chr11_+_35662739 | 0.45 |
ENST00000299413.7
|
TRIM44
|
tripartite motif containing 44 |
| chr16_-_87936529 | 0.43 |
ENST00000649794.3
ENST00000649158.1 ENST00000648177.1 |
CA5A
|
carbonic anhydrase 5A |
| chr6_-_43629222 | 0.43 |
ENST00000307126.10
|
GTPBP2
|
GTP binding protein 2 |
| chr17_+_9021501 | 0.43 |
ENST00000173229.7
|
NTN1
|
netrin 1 |
| chr4_+_164754116 | 0.43 |
ENST00000507311.1
|
SMIM31
|
small integral membrane protein 31 |
| chr2_+_202073282 | 0.43 |
ENST00000459709.5
|
KIAA2012
|
KIAA2012 |
| chr7_+_114414809 | 0.43 |
ENST00000350908.9
|
FOXP2
|
forkhead box P2 |
| chr14_-_56805648 | 0.42 |
ENST00000554788.5
ENST00000554845.1 ENST00000408990.8 |
OTX2
|
orthodenticle homeobox 2 |
| chr2_+_202073249 | 0.41 |
ENST00000498697.3
|
KIAA2012
|
KIAA2012 |
| chr1_-_248748327 | 0.41 |
ENST00000651827.1
|
ENSG00000286015.1
|
novel protein |
| chr2_-_68952880 | 0.41 |
ENST00000481498.1
ENST00000328895.9 |
GKN2
|
gastrokine 2 |
| chr1_-_116667668 | 0.40 |
ENST00000369486.8
ENST00000369483.5 |
IGSF3
|
immunoglobulin superfamily member 3 |
| chr3_+_189631373 | 0.40 |
ENST00000264731.8
ENST00000418709.6 ENST00000320472.9 ENST00000392460.7 ENST00000440651.6 |
TP63
|
tumor protein p63 |
| chr11_-_116792386 | 0.38 |
ENST00000433069.2
ENST00000542499.5 |
APOA5
|
apolipoprotein A5 |
| chr9_-_86947496 | 0.38 |
ENST00000298743.9
|
GAS1
|
growth arrest specific 1 |
| chr5_-_41213505 | 0.37 |
ENST00000337836.10
ENST00000433294.1 |
C6
|
complement C6 |
| chr1_+_86468902 | 0.37 |
ENST00000394711.2
|
CLCA1
|
chloride channel accessory 1 |
| chr12_-_52434363 | 0.36 |
ENST00000252245.6
|
KRT75
|
keratin 75 |
| chr2_-_218985176 | 0.36 |
ENST00000295727.2
|
FEV
|
FEV transcription factor, ETS family member |
| chr13_+_50015438 | 0.35 |
ENST00000312942.2
|
KCNRG
|
potassium channel regulator |
| chr13_+_50015254 | 0.35 |
ENST00000360473.8
|
KCNRG
|
potassium channel regulator |
| chr17_-_28368012 | 0.35 |
ENST00000555059.2
|
ENSG00000273171.1
|
novel protein, readthrough between VTN and SEBOX |
| chr4_+_85475131 | 0.35 |
ENST00000395184.6
|
ARHGAP24
|
Rho GTPase activating protein 24 |
| chr16_-_48610150 | 0.35 |
ENST00000262384.4
|
N4BP1
|
NEDD4 binding protein 1 |
| chr1_-_56966006 | 0.34 |
ENST00000371237.9
|
C8B
|
complement C8 beta chain |
| chr21_-_26843012 | 0.34 |
ENST00000517777.6
|
ADAMTS1
|
ADAM metallopeptidase with thrombospondin type 1 motif 1 |
| chr2_+_168901290 | 0.34 |
ENST00000429379.2
ENST00000375363.8 ENST00000421979.1 |
G6PC2
|
glucose-6-phosphatase catalytic subunit 2 |
| chrX_+_151176582 | 0.34 |
ENST00000218316.4
|
GPR50
|
G protein-coupled receptor 50 |
| chr3_-_186362223 | 0.34 |
ENST00000265022.8
|
DGKG
|
diacylglycerol kinase gamma |
| chr17_-_41528293 | 0.34 |
ENST00000455635.1
ENST00000361566.7 |
KRT19
|
keratin 19 |
| chr7_-_81770122 | 0.33 |
ENST00000423064.7
|
HGF
|
hepatocyte growth factor |
| chr4_-_163613505 | 0.33 |
ENST00000339875.9
|
MARCHF1
|
membrane associated ring-CH-type finger 1 |
| chr1_-_206921867 | 0.33 |
ENST00000628511.2
ENST00000367091.8 |
FCMR
|
Fc fragment of IgM receptor |
| chr14_-_21025490 | 0.33 |
ENST00000553442.5
ENST00000555869.5 ENST00000556457.5 ENST00000397844.6 ENST00000554415.5 ENST00000556974.5 ENST00000554419.5 ENST00000298687.9 ENST00000397858.5 ENST00000360463.7 ENST00000350792.7 ENST00000397847.6 |
NDRG2
|
NDRG family member 2 |
| chr1_-_206921987 | 0.32 |
ENST00000530505.1
ENST00000442471.4 |
FCMR
|
Fc fragment of IgM receptor |
| chr6_-_160664270 | 0.32 |
ENST00000316300.10
|
LPA
|
lipoprotein(a) |
| chr7_+_28412511 | 0.32 |
ENST00000357727.7
|
CREB5
|
cAMP responsive element binding protein 5 |
| chr17_-_50468871 | 0.32 |
ENST00000508540.6
ENST00000258969.4 |
CHAD
|
chondroadherin |
| chrX_+_136532205 | 0.31 |
ENST00000370634.8
|
VGLL1
|
vestigial like family member 1 |
| chr1_-_177970213 | 0.31 |
ENST00000464631.6
|
SEC16B
|
SEC16 homolog B, endoplasmic reticulum export factor |
| chr2_-_21044063 | 0.31 |
ENST00000233242.5
|
APOB
|
apolipoprotein B |
| chr16_+_7332839 | 0.31 |
ENST00000355637.9
|
RBFOX1
|
RNA binding fox-1 homolog 1 |
| chr11_+_13962676 | 0.31 |
ENST00000576479.4
|
SPON1
|
spondin 1 |
| chr3_-_190449782 | 0.30 |
ENST00000354905.3
|
TMEM207
|
transmembrane protein 207 |
| chr21_-_26843063 | 0.30 |
ENST00000678221.1
|
ADAMTS1
|
ADAM metallopeptidase with thrombospondin type 1 motif 1 |
| chr5_+_96663010 | 0.30 |
ENST00000506811.5
ENST00000514055.5 ENST00000508608.6 |
CAST
|
calpastatin |
| chr10_+_122163590 | 0.29 |
ENST00000368999.5
|
TACC2
|
transforming acidic coiled-coil containing protein 2 |
| chrX_+_22032301 | 0.29 |
ENST00000379374.5
|
PHEX
|
phosphate regulating endopeptidase homolog X-linked |
| chr7_+_134891566 | 0.29 |
ENST00000424922.5
ENST00000495522.1 |
CALD1
|
caldesmon 1 |
| chr6_-_39322968 | 0.29 |
ENST00000507712.5
|
KCNK16
|
potassium two pore domain channel subfamily K member 16 |
| chr20_-_7940444 | 0.28 |
ENST00000378789.4
|
HAO1
|
hydroxyacid oxidase 1 |
| chr3_-_71583683 | 0.28 |
ENST00000649631.1
ENST00000648718.1 |
FOXP1
|
forkhead box P1 |
| chr19_-_48615050 | 0.28 |
ENST00000263266.4
|
FAM83E
|
family with sequence similarity 83 member E |
| chr22_-_40533808 | 0.28 |
ENST00000422851.1
ENST00000651694.1 ENST00000652095.2 |
MRTFA
|
myocardin related transcription factor A |
| chr4_+_164754045 | 0.28 |
ENST00000515485.5
|
SMIM31
|
small integral membrane protein 31 |
| chr16_+_7332744 | 0.28 |
ENST00000436368.6
ENST00000311745.9 ENST00000340209.8 ENST00000620507.4 |
RBFOX1
|
RNA binding fox-1 homolog 1 |
| chr17_-_47957824 | 0.27 |
ENST00000300557.3
|
PRR15L
|
proline rich 15 like |
| chr5_-_142686079 | 0.27 |
ENST00000337706.7
|
FGF1
|
fibroblast growth factor 1 |
| chr12_+_120978686 | 0.26 |
ENST00000541395.5
ENST00000544413.2 |
HNF1A
|
HNF1 homeobox A |
| chr1_-_56966133 | 0.26 |
ENST00000535057.5
ENST00000543257.5 |
C8B
|
complement C8 beta chain |
| chr10_-_31031911 | 0.26 |
ENST00000375311.1
ENST00000413025.5 ENST00000436087.6 ENST00000452305.5 ENST00000442986.5 |
ZNF438
|
zinc finger protein 438 |
| chr19_+_42302098 | 0.26 |
ENST00000598490.1
ENST00000341747.8 |
PRR19
|
proline rich 19 |
| chr10_+_122163672 | 0.26 |
ENST00000369004.7
ENST00000260733.7 |
TACC2
|
transforming acidic coiled-coil containing protein 2 |
| chr20_+_45791930 | 0.26 |
ENST00000372622.8
|
DNTTIP1
|
deoxynucleotidyltransferase terminal interacting protein 1 |
| chr10_+_49299159 | 0.26 |
ENST00000374144.8
|
C10orf71
|
chromosome 10 open reading frame 71 |
| chr12_+_10505890 | 0.26 |
ENST00000538173.1
|
EIF2S3B
|
eukaryotic translation initiation factor 2 subunit gamma B |
| chr11_+_31812307 | 0.25 |
ENST00000643436.1
ENST00000646959.1 ENST00000645942.1 ENST00000530348.5 |
PAUPAR
ENSG00000285283.1
|
PAX6 upstream antisense RNA novel protein |
| chr5_-_136365476 | 0.25 |
ENST00000378459.7
ENST00000502753.4 ENST00000513104.6 ENST00000352189.8 |
TRPC7
|
transient receptor potential cation channel subfamily C member 7 |
| chr7_+_130266847 | 0.24 |
ENST00000222481.9
|
CPA2
|
carboxypeptidase A2 |
| chr21_-_42350987 | 0.24 |
ENST00000291526.5
|
TFF2
|
trefoil factor 2 |
| chr1_+_204870831 | 0.24 |
ENST00000404076.5
ENST00000539706.6 |
NFASC
|
neurofascin |
| chr3_+_108602776 | 0.24 |
ENST00000497905.5
ENST00000463306.1 |
DZIP3
|
DAZ interacting zinc finger protein 3 |
| chrX_+_1268807 | 0.24 |
ENST00000381524.8
ENST00000381529.9 ENST00000412290.6 |
CSF2RA
|
colony stimulating factor 2 receptor subunit alpha |
| chr13_-_46142834 | 0.24 |
ENST00000674665.1
|
LCP1
|
lymphocyte cytosolic protein 1 |
| chr14_-_31457495 | 0.24 |
ENST00000310850.9
|
DTD2
|
D-aminoacyl-tRNA deacylase 2 |
| chr6_+_52420992 | 0.24 |
ENST00000636954.1
ENST00000636566.1 ENST00000638075.1 |
EFHC1
|
EF-hand domain containing 1 |
| chr3_+_160677152 | 0.23 |
ENST00000320767.4
|
ARL14
|
ADP ribosylation factor like GTPase 14 |
| chr6_+_10528326 | 0.23 |
ENST00000379597.7
|
GCNT2
|
glucosaminyl (N-acetyl) transferase 2 (I blood group) |
| chr19_+_38390055 | 0.23 |
ENST00000587947.5
ENST00000338502.8 |
SPRED3
|
sprouty related EVH1 domain containing 3 |
| chr9_-_14722725 | 0.22 |
ENST00000380911.4
|
CER1
|
cerberus 1, DAN family BMP antagonist |
| chr12_+_120978537 | 0.22 |
ENST00000257555.11
ENST00000400024.6 |
HNF1A
|
HNF1 homeobox A |
| chr11_+_66258467 | 0.22 |
ENST00000394066.6
|
KLC2
|
kinesin light chain 2 |
| chr4_+_67558719 | 0.22 |
ENST00000265404.7
ENST00000396225.1 |
STAP1
|
signal transducing adaptor family member 1 |
| chr9_-_120477354 | 0.21 |
ENST00000416449.5
|
CDK5RAP2
|
CDK5 regulatory subunit associated protein 2 |
| chr12_-_21334858 | 0.21 |
ENST00000445053.1
ENST00000458504.5 ENST00000422327.5 ENST00000683939.1 |
SLCO1A2
|
solute carrier organic anion transporter family member 1A2 |
| chr7_-_100100716 | 0.21 |
ENST00000354230.7
ENST00000425308.5 |
MCM7
|
minichromosome maintenance complex component 7 |
| chr4_+_164877164 | 0.21 |
ENST00000507152.6
ENST00000515275.1 |
APELA
|
apelin receptor early endogenous ligand |
| chr14_+_36657560 | 0.21 |
ENST00000402703.6
|
PAX9
|
paired box 9 |
| chr10_+_18400562 | 0.21 |
ENST00000377315.5
ENST00000650685.1 |
CACNB2
|
calcium voltage-gated channel auxiliary subunit beta 2 |
| chr10_+_102776237 | 0.21 |
ENST00000369889.5
|
WBP1L
|
WW domain binding protein 1 like |
| chr2_+_151409878 | 0.20 |
ENST00000453091.6
ENST00000428287.6 ENST00000444746.7 ENST00000243326.9 ENST00000414861.6 |
RIF1
|
replication timing regulatory factor 1 |
| chr19_-_45406327 | 0.20 |
ENST00000593226.5
ENST00000418234.6 |
PPP1R13L
|
protein phosphatase 1 regulatory subunit 13 like |
| chr15_+_84237586 | 0.20 |
ENST00000512109.1
|
GOLGA6L4
|
golgin A6 family like 4 |
| chr11_-_107712049 | 0.19 |
ENST00000305991.3
|
SLN
|
sarcolipin |
| chr1_-_246566238 | 0.19 |
ENST00000366514.5
|
TFB2M
|
transcription factor B2, mitochondrial |
| chr2_+_151410090 | 0.19 |
ENST00000430328.6
|
RIF1
|
replication timing regulatory factor 1 |
| chr10_+_116590956 | 0.19 |
ENST00000358834.9
ENST00000528052.5 |
PNLIPRP1
|
pancreatic lipase related protein 1 |
| chr13_-_85799400 | 0.19 |
ENST00000647374.2
|
SLITRK6
|
SLIT and NTRK like family member 6 |
| chr2_-_49154433 | 0.18 |
ENST00000454032.5
ENST00000304421.8 |
FSHR
|
follicle stimulating hormone receptor |
| chr14_-_31457417 | 0.18 |
ENST00000356180.4
|
DTD2
|
D-aminoacyl-tRNA deacylase 2 |
| chr11_+_119334511 | 0.18 |
ENST00000311413.5
|
RNF26
|
ring finger protein 26 |
| chr6_-_132659178 | 0.18 |
ENST00000275216.3
|
TAAR1
|
trace amine associated receptor 1 |
| chr7_+_134745460 | 0.18 |
ENST00000436461.6
|
CALD1
|
caldesmon 1 |
| chr5_-_149379286 | 0.18 |
ENST00000261796.4
|
IL17B
|
interleukin 17B |
| chr6_-_89315291 | 0.18 |
ENST00000402938.4
|
GABRR2
|
gamma-aminobutyric acid type A receptor subunit rho2 |
| chr2_-_49154507 | 0.17 |
ENST00000406846.7
|
FSHR
|
follicle stimulating hormone receptor |
| chr18_+_3449620 | 0.17 |
ENST00000405385.7
|
TGIF1
|
TGFB induced factor homeobox 1 |
| chr2_+_17541157 | 0.17 |
ENST00000406397.1
|
VSNL1
|
visinin like 1 |
| chr14_-_37595224 | 0.16 |
ENST00000250448.5
|
FOXA1
|
forkhead box A1 |
| chr11_+_121102666 | 0.16 |
ENST00000264037.2
|
TECTA
|
tectorin alpha |
| chrX_-_48958348 | 0.16 |
ENST00000428668.2
|
OTUD5
|
OTU deubiquitinase 5 |
| chr14_+_36661852 | 0.16 |
ENST00000361487.7
|
PAX9
|
paired box 9 |
| chr1_-_244860376 | 0.16 |
ENST00000638716.1
|
HNRNPU
|
heterogeneous nuclear ribonucleoprotein U |
| chr14_+_23185316 | 0.16 |
ENST00000399910.5
ENST00000492621.5 |
RNF212B
|
ring finger protein 212B |
| chr6_-_107824294 | 0.16 |
ENST00000369020.8
ENST00000369022.6 |
SCML4
|
Scm polycomb group protein like 4 |
| chr14_-_99271485 | 0.15 |
ENST00000345514.2
ENST00000443726.2 |
BCL11B
|
BAF chromatin remodeling complex subunit BCL11B |
| chr1_-_26913964 | 0.15 |
ENST00000254227.4
|
NR0B2
|
nuclear receptor subfamily 0 group B member 2 |
| chr2_-_213151590 | 0.15 |
ENST00000374319.8
ENST00000457361.5 ENST00000451136.6 ENST00000434687.6 |
IKZF2
|
IKAROS family zinc finger 2 |
| chr14_-_99272184 | 0.15 |
ENST00000357195.8
|
BCL11B
|
BAF chromatin remodeling complex subunit BCL11B |
| chr5_+_40841308 | 0.15 |
ENST00000381677.4
ENST00000254691.10 |
CARD6
|
caspase recruitment domain family member 6 |
| chr3_+_98353854 | 0.15 |
ENST00000354924.2
|
OR5K4
|
olfactory receptor family 5 subfamily K member 4 |
| chr3_+_186996444 | 0.14 |
ENST00000676633.1
|
ST6GAL1
|
ST6 beta-galactoside alpha-2,6-sialyltransferase 1 |
| chr1_+_50105666 | 0.14 |
ENST00000651347.1
|
ELAVL4
|
ELAV like RNA binding protein 4 |
| chr7_-_107803215 | 0.14 |
ENST00000340010.10
ENST00000453332.1 |
SLC26A3
|
solute carrier family 26 member 3 |
| chr8_+_86866415 | 0.14 |
ENST00000518476.6
|
CNBD1
|
cyclic nucleotide binding domain containing 1 |
| chr14_+_22829879 | 0.14 |
ENST00000355151.9
ENST00000397496.7 ENST00000555345.5 ENST00000432849.7 ENST00000553711.5 ENST00000556465.5 ENST00000397505.2 ENST00000557221.1 ENST00000556840.5 ENST00000555536.1 |
MRPL52
|
mitochondrial ribosomal protein L52 |
| chr1_-_178871022 | 0.14 |
ENST00000367629.1
|
ANGPTL1
|
angiopoietin like 1 |
| chr1_-_11047225 | 0.14 |
ENST00000400898.3
ENST00000400897.8 |
MASP2
|
mannan binding lectin serine peptidase 2 |
| chr9_+_137230757 | 0.14 |
ENST00000673865.1
ENST00000538474.5 ENST00000673835.1 ENST00000673953.1 ENST00000361134.2 |
SLC34A3
|
solute carrier family 34 member 3 |
| chr5_-_138338325 | 0.14 |
ENST00000510119.1
ENST00000513970.5 |
CDC25C
|
cell division cycle 25C |
| chr1_-_152360004 | 0.14 |
ENST00000388718.5
|
FLG2
|
filaggrin family member 2 |
| chrX_-_15384402 | 0.13 |
ENST00000297904.4
|
VEGFD
|
vascular endothelial growth factor D |
| chr21_-_42315336 | 0.13 |
ENST00000398431.2
ENST00000518498.3 |
TFF3
|
trefoil factor 3 |
| chr7_+_112423137 | 0.13 |
ENST00000005558.8
ENST00000621379.4 |
IFRD1
|
interferon related developmental regulator 1 |
| chr10_-_79949098 | 0.13 |
ENST00000372292.8
|
SFTPD
|
surfactant protein D |
| chr1_+_74235377 | 0.13 |
ENST00000326637.8
|
TNNI3K
|
TNNI3 interacting kinase |
| chrX_-_15600953 | 0.13 |
ENST00000679212.1
ENST00000679278.1 ENST00000678046.1 ENST00000252519.8 |
ACE2
|
angiotensin I converting enzyme 2 |
| chr1_+_43389874 | 0.13 |
ENST00000372450.8
|
SZT2
|
SZT2 subunit of KICSTOR complex |
| chr2_+_172085499 | 0.12 |
ENST00000361725.5
ENST00000341900.6 |
DLX1
|
distal-less homeobox 1 |
| chr22_-_45008938 | 0.12 |
ENST00000420689.1
ENST00000403565.5 ENST00000396103.7 |
PHF21B
|
PHD finger protein 21B |
| chr5_+_96662969 | 0.11 |
ENST00000514845.5
ENST00000675663.1 |
CAST
|
calpastatin |
| chr1_-_43389768 | 0.11 |
ENST00000372455.4
ENST00000372457.9 ENST00000290663.10 |
MED8
|
mediator complex subunit 8 |
| chr14_-_69797232 | 0.11 |
ENST00000216540.5
|
SLC10A1
|
solute carrier family 10 member 1 |
| chr20_-_57266606 | 0.11 |
ENST00000450594.6
ENST00000395863.8 |
BMP7
|
bone morphogenetic protein 7 |
| chr5_+_96936071 | 0.11 |
ENST00000231368.10
|
LNPEP
|
leucyl and cystinyl aminopeptidase |
| chr19_-_48390847 | 0.11 |
ENST00000597017.5
|
KDELR1
|
KDEL endoplasmic reticulum protein retention receptor 1 |
| chr10_-_79560386 | 0.11 |
ENST00000372327.9
ENST00000417041.1 ENST00000640627.1 ENST00000372325.7 |
SFTPA2
|
surfactant protein A2 |
| chr1_-_94925759 | 0.11 |
ENST00000415017.1
ENST00000545882.5 |
CNN3
|
calponin 3 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.5 | GO:0046521 | sphingoid catabolic process(GO:0046521) |
| 0.5 | 4.1 | GO:0061373 | mammillary body development(GO:0021767) mammillary axonal complex development(GO:0061373) |
| 0.3 | 0.8 | GO:0072276 | metanephric glomerulus morphogenesis(GO:0072275) metanephric glomerulus vasculature morphogenesis(GO:0072276) metanephric glomerular capillary formation(GO:0072277) |
| 0.3 | 1.1 | GO:0098886 | modification of dendritic spine(GO:0098886) |
| 0.3 | 1.6 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.2 | 1.0 | GO:0046946 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 0.2 | 0.6 | GO:0045643 | regulation of eosinophil differentiation(GO:0045643) positive regulation of eosinophil differentiation(GO:0045645) |
| 0.2 | 1.4 | GO:0006154 | adenosine catabolic process(GO:0006154) inosine biosynthetic process(GO:0046103) |
| 0.2 | 0.5 | GO:0021757 | caudate nucleus development(GO:0021757) putamen development(GO:0021758) |
| 0.2 | 1.5 | GO:0045084 | positive regulation of interleukin-12 biosynthetic process(GO:0045084) |
| 0.2 | 0.5 | GO:0035565 | regulation of pronephros size(GO:0035565) |
| 0.2 | 0.6 | GO:0086097 | phospholipase C-activating angiotensin-activated signaling pathway(GO:0086097) |
| 0.1 | 1.1 | GO:0072733 | response to staurosporine(GO:0072733) cellular response to staurosporine(GO:0072734) |
| 0.1 | 0.8 | GO:1903435 | positive regulation of constitutive secretory pathway(GO:1903435) |
| 0.1 | 0.4 | GO:0001545 | primary ovarian follicle growth(GO:0001545) |
| 0.1 | 1.5 | GO:0021840 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) ERBB3 signaling pathway(GO:0038129) |
| 0.1 | 0.3 | GO:1900155 | regulation of bone trabecula formation(GO:1900154) negative regulation of bone trabecula formation(GO:1900155) |
| 0.1 | 0.3 | GO:0097535 | lymphoid lineage cell migration(GO:0097534) lymphoid lineage cell migration into thymus(GO:0097535) |
| 0.1 | 0.4 | GO:0007499 | ectoderm and mesoderm interaction(GO:0007499) |
| 0.1 | 0.9 | GO:0060574 | intestinal epithelial cell maturation(GO:0060574) |
| 0.1 | 2.7 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.1 | 0.4 | GO:0010902 | positive regulation of very-low-density lipoprotein particle remodeling(GO:0010902) |
| 0.1 | 0.4 | GO:2000259 | positive regulation of complement activation(GO:0045917) positive regulation of protein activation cascade(GO:2000259) |
| 0.1 | 1.3 | GO:0002315 | marginal zone B cell differentiation(GO:0002315) |
| 0.1 | 0.4 | GO:2001034 | positive regulation of double-strand break repair via nonhomologous end joining(GO:2001034) |
| 0.1 | 0.3 | GO:1904383 | response to sodium phosphate(GO:1904383) |
| 0.1 | 0.2 | GO:1903973 | negative regulation of macrophage colony-stimulating factor signaling pathway(GO:1902227) negative regulation of response to macrophage colony-stimulating factor(GO:1903970) negative regulation of cellular response to macrophage colony-stimulating factor stimulus(GO:1903973) |
| 0.1 | 0.2 | GO:0090133 | mesendoderm migration(GO:0090133) cell migration involved in mesendoderm migration(GO:0090134) |
| 0.1 | 1.1 | GO:0035457 | cellular response to interferon-alpha(GO:0035457) |
| 0.1 | 1.1 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.1 | 0.1 | GO:0007518 | myoblast fate determination(GO:0007518) |
| 0.1 | 0.2 | GO:1901877 | regulation of calcium ion binding(GO:1901876) negative regulation of calcium ion binding(GO:1901877) |
| 0.1 | 0.8 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.1 | 0.2 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.1 | 1.0 | GO:0032625 | interleukin-21 production(GO:0032625) interleukin-21 secretion(GO:0072619) |
| 0.1 | 0.2 | GO:1900108 | negative regulation of nodal signaling pathway(GO:1900108) |
| 0.1 | 0.4 | GO:0060665 | regulation of branching involved in salivary gland morphogenesis by mesenchymal-epithelial signaling(GO:0060665) |
| 0.1 | 0.2 | GO:0060738 | epithelial-mesenchymal signaling involved in prostate gland development(GO:0060738) |
| 0.0 | 1.6 | GO:1901741 | positive regulation of myoblast fusion(GO:1901741) |
| 0.0 | 0.4 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.0 | 0.4 | GO:0009441 | glycolate metabolic process(GO:0009441) |
| 0.0 | 0.2 | GO:1905069 | neural fold elevation formation(GO:0021502) nephrogenic mesenchyme morphogenesis(GO:0072134) allantois development(GO:1905069) |
| 0.0 | 0.3 | GO:0006642 | triglyceride mobilization(GO:0006642) |
| 0.0 | 0.2 | GO:0010966 | regulation of phosphate transport(GO:0010966) |
| 0.0 | 0.1 | GO:0021893 | regulation of transcription from RNA polymerase II promoter involved in forebrain neuron fate commitment(GO:0021882) cerebral cortex GABAergic interneuron fate commitment(GO:0021893) positive regulation of neural retina development(GO:0061075) positive regulation of retina development in camera-type eye(GO:1902868) positive regulation of amacrine cell differentiation(GO:1902871) |
| 0.0 | 0.7 | GO:1902260 | negative regulation of delayed rectifier potassium channel activity(GO:1902260) |
| 0.0 | 0.3 | GO:1901509 | regulation of endothelial tube morphogenesis(GO:1901509) |
| 0.0 | 0.6 | GO:0001542 | ovulation from ovarian follicle(GO:0001542) |
| 0.0 | 2.1 | GO:2000272 | negative regulation of receptor activity(GO:2000272) |
| 0.0 | 0.5 | GO:0071985 | multivesicular body sorting pathway(GO:0071985) |
| 0.0 | 0.1 | GO:0045085 | negative regulation of interleukin-2 biosynthetic process(GO:0045085) |
| 0.0 | 0.5 | GO:0090385 | phagosome-lysosome fusion(GO:0090385) |
| 0.0 | 0.4 | GO:0032808 | lacrimal gland development(GO:0032808) |
| 0.0 | 0.6 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.0 | 0.1 | GO:0035638 | patched ligand maturation(GO:0007225) signal maturation(GO:0035638) |
| 0.0 | 0.2 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.0 | 0.4 | GO:2000543 | positive regulation of gastrulation(GO:2000543) |
| 0.0 | 0.2 | GO:0002175 | protein localization to paranode region of axon(GO:0002175) |
| 0.0 | 0.5 | GO:0006782 | protoporphyrinogen IX biosynthetic process(GO:0006782) |
| 0.0 | 0.2 | GO:0099566 | regulation of postsynaptic cytosolic calcium ion concentration(GO:0099566) |
| 0.0 | 0.4 | GO:0006450 | regulation of translational fidelity(GO:0006450) |
| 0.0 | 0.1 | GO:0010897 | negative regulation of triglyceride catabolic process(GO:0010897) secretory granule localization(GO:0032252) |
| 0.0 | 0.4 | GO:0060628 | regulation of ER to Golgi vesicle-mediated transport(GO:0060628) |
| 0.0 | 0.2 | GO:1904879 | positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1904879) |
| 0.0 | 0.7 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.0 | 0.1 | GO:1903598 | positive regulation of gap junction assembly(GO:1903598) |
| 0.0 | 0.1 | GO:0072502 | cellular phosphate ion homeostasis(GO:0030643) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.0 | 0.3 | GO:0090361 | platelet-derived growth factor production(GO:0090360) regulation of platelet-derived growth factor production(GO:0090361) |
| 0.0 | 0.3 | GO:0034374 | low-density lipoprotein particle remodeling(GO:0034374) |
| 0.0 | 0.1 | GO:0007181 | transforming growth factor beta receptor complex assembly(GO:0007181) |
| 0.0 | 0.1 | GO:1901668 | regulation of superoxide dismutase activity(GO:1901668) |
| 0.0 | 0.2 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.0 | 0.3 | GO:0036066 | protein O-linked fucosylation(GO:0036066) |
| 0.0 | 0.4 | GO:2000675 | negative regulation of type B pancreatic cell apoptotic process(GO:2000675) |
| 0.0 | 0.2 | GO:0046600 | negative regulation of centriole replication(GO:0046600) |
| 0.0 | 0.1 | GO:0010193 | response to ozone(GO:0010193) |
| 0.0 | 0.1 | GO:1902164 | mRNA localization resulting in posttranscriptional regulation of gene expression(GO:0010609) platelet alpha granule organization(GO:0070889) regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902162) positive regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902164) |
| 0.0 | 0.2 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.0 | 0.1 | GO:0060754 | positive regulation of mast cell chemotaxis(GO:0060754) |
| 0.0 | 0.8 | GO:0050982 | detection of mechanical stimulus(GO:0050982) |
| 0.0 | 0.2 | GO:0006828 | manganese ion transport(GO:0006828) |
| 0.0 | 0.7 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.0 | 0.2 | GO:0071803 | positive regulation of podosome assembly(GO:0071803) |
| 0.0 | 0.9 | GO:0042982 | amyloid precursor protein metabolic process(GO:0042982) |
| 0.0 | 0.1 | GO:0097056 | selenocysteinyl-tRNA(Sec) biosynthetic process(GO:0097056) |
| 0.0 | 0.2 | GO:1900017 | positive regulation of cytokine production involved in inflammatory response(GO:1900017) |
| 0.0 | 0.1 | GO:0060081 | membrane hyperpolarization(GO:0060081) |
| 0.0 | 0.2 | GO:0007039 | protein catabolic process in the vacuole(GO:0007039) |
| 0.0 | 0.6 | GO:0050732 | negative regulation of peptidyl-tyrosine phosphorylation(GO:0050732) |
| 0.0 | 0.4 | GO:0042481 | regulation of odontogenesis(GO:0042481) |
| 0.0 | 0.1 | GO:0002480 | antigen processing and presentation of exogenous peptide antigen via MHC class I, TAP-independent(GO:0002480) |
| 0.0 | 0.1 | GO:0070863 | positive regulation of protein exit from endoplasmic reticulum(GO:0070863) |
| 0.0 | 0.2 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.0 | 0.1 | GO:0035992 | tendon cell differentiation(GO:0035990) tendon formation(GO:0035992) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.1 | GO:0097444 | spine apparatus(GO:0097444) |
| 0.2 | 0.9 | GO:0070557 | PCNA-p21 complex(GO:0070557) |
| 0.1 | 1.0 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.1 | 3.4 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.1 | 0.2 | GO:0016938 | kinesin I complex(GO:0016938) |
| 0.1 | 0.8 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.1 | 0.2 | GO:0038038 | G-protein coupled receptor homodimeric complex(GO:0038038) |
| 0.0 | 0.2 | GO:0097454 | Schwann cell microvillus(GO:0097454) |
| 0.0 | 2.3 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 0.3 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 0.0 | 0.4 | GO:0001940 | female pronucleus(GO:0001939) male pronucleus(GO:0001940) |
| 0.0 | 0.3 | GO:1990357 | terminal web(GO:1990357) |
| 0.0 | 0.1 | GO:0005944 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) |
| 0.0 | 0.2 | GO:0000836 | ER ubiquitin ligase complex(GO:0000835) Hrd1p ubiquitin ligase complex(GO:0000836) |
| 0.0 | 0.8 | GO:0043205 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.0 | 0.5 | GO:0030478 | actin cap(GO:0030478) |
| 0.0 | 1.5 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 1.0 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.0 | 0.8 | GO:0030904 | retromer complex(GO:0030904) |
| 0.0 | 0.1 | GO:0031905 | early endosome lumen(GO:0031905) |
| 0.0 | 3.1 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 0.4 | GO:0042627 | chylomicron(GO:0042627) |
| 0.0 | 0.9 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 0.1 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.0 | 0.2 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 0.1 | GO:0035867 | alphav-beta3 integrin-IGF-1-IGF1R complex(GO:0035867) |
| 0.0 | 1.4 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 1.2 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.2 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.1 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 0.3 | 0.8 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 0.2 | 1.0 | GO:0033823 | procollagen-lysine 5-dioxygenase activity(GO:0008475) procollagen glucosyltransferase activity(GO:0033823) |
| 0.2 | 1.5 | GO:0017050 | sphinganine kinase activity(GO:0008481) D-erythro-sphingosine kinase activity(GO:0017050) |
| 0.2 | 0.6 | GO:0001596 | angiotensin type I receptor activity(GO:0001596) |
| 0.2 | 0.9 | GO:0019912 | cyclin-dependent protein kinase activating kinase activity(GO:0019912) |
| 0.2 | 2.7 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) |
| 0.1 | 1.3 | GO:0038049 | transcription factor activity, ligand-activated RNA polymerase II transcription factor binding(GO:0038049) |
| 0.1 | 1.4 | GO:0031685 | adenosine receptor binding(GO:0031685) |
| 0.1 | 0.4 | GO:0004963 | follicle-stimulating hormone receptor activity(GO:0004963) |
| 0.1 | 0.3 | GO:0050309 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 0.1 | 0.3 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.1 | 0.4 | GO:0051499 | D-aminoacyl-tRNA deacylase activity(GO:0051499) D-tyrosyl-tRNA(Tyr) deacylase activity(GO:0051500) |
| 0.1 | 0.7 | GO:0035473 | lipase binding(GO:0035473) |
| 0.1 | 0.5 | GO:0003870 | 5-aminolevulinate synthase activity(GO:0003870) N-succinyltransferase activity(GO:0016749) |
| 0.1 | 0.3 | GO:0052852 | (S)-2-hydroxy-acid oxidase activity(GO:0003973) very-long-chain-(S)-2-hydroxy-acid oxidase activity(GO:0052852) long-chain-(S)-2-hydroxy-long-chain-acid oxidase activity(GO:0052853) medium-chain-(S)-2-hydroxy-acid oxidase activity(GO:0052854) |
| 0.1 | 1.5 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.1 | 3.4 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.0 | 1.0 | GO:0005537 | mannose binding(GO:0005537) |
| 0.0 | 0.4 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.0 | 0.2 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.0 | 0.1 | GO:0005046 | KDEL sequence binding(GO:0005046) |
| 0.0 | 1.1 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.0 | 0.1 | GO:0001855 | complement component C4b binding(GO:0001855) |
| 0.0 | 0.6 | GO:0055106 | ubiquitin-protein transferase regulator activity(GO:0055106) |
| 0.0 | 0.2 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.0 | 0.4 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.0 | 0.2 | GO:0005157 | macrophage colony-stimulating factor receptor binding(GO:0005157) |
| 0.0 | 0.9 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.0 | 1.7 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.0 | 0.8 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 1.3 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.2 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.0 | 1.8 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.2 | GO:0099583 | neurotransmitter receptor activity involved in regulation of postsynaptic cytosolic calcium ion concentration(GO:0099583) |
| 0.0 | 0.1 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.0 | 0.1 | GO:0004967 | glucagon receptor activity(GO:0004967) |
| 0.0 | 0.7 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.0 | 0.4 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.2 | GO:0036122 | BMP binding(GO:0036122) |
| 0.0 | 0.1 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.0 | 0.1 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.0 | 0.2 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.1 | GO:0008241 | peptidyl-dipeptidase activity(GO:0008241) |
| 0.0 | 1.9 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 0.5 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 0.1 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.0 | 0.3 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.4 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 0.3 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.0 | 0.2 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.0 | 0.2 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.0 | 1.5 | GO:0050681 | androgen receptor binding(GO:0050681) |
| 0.0 | 0.5 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.0 | 0.1 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.3 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.0 | 0.2 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.0 | 0.2 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.0 | 0.1 | GO:0015321 | sodium-dependent phosphate transmembrane transporter activity(GO:0015321) |
| 0.0 | 0.2 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.0 | 0.4 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.0 | 0.3 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.0 | 0.1 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 0.0 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 0.0 | 0.0 | GO:0000773 | phosphatidyl-N-methylethanolamine N-methyltransferase activity(GO:0000773) phosphatidylethanolamine N-methyltransferase activity(GO:0004608) phosphatidyl-N-dimethylethanolamine N-methyltransferase activity(GO:0080101) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 3.5 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.0 | 1.5 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 1.1 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 2.0 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 0.8 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.0 | 1.0 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 1.1 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.3 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.0 | 1.3 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 0.8 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 0.6 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.3 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.1 | 1.1 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.1 | 3.4 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |
| 0.1 | 1.0 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.1 | 0.7 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.0 | 2.1 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 1.5 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 1.5 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 1.1 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 0.7 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 0.4 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.0 | 0.7 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 1.1 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.0 | 0.3 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 0.9 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 0.3 | REACTOME LIPOPROTEIN METABOLISM | Genes involved in Lipoprotein metabolism |
| 0.0 | 0.4 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.5 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.3 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 0.0 | 0.5 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 1.0 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.2 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.0 | 0.1 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.0 | 1.1 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |