Project
Inflammatory response time course, HUVEC (Wada, 2009)
Navigation
Downloads
Results for FOXA3_FOXC2
Z-value: 0.68
Transcription factors associated with FOXA3_FOXC2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
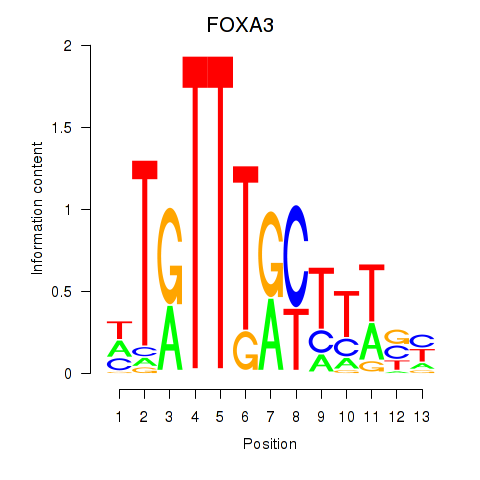
FOXA3
|
ENSG00000170608.3 | FOXA3 |
|
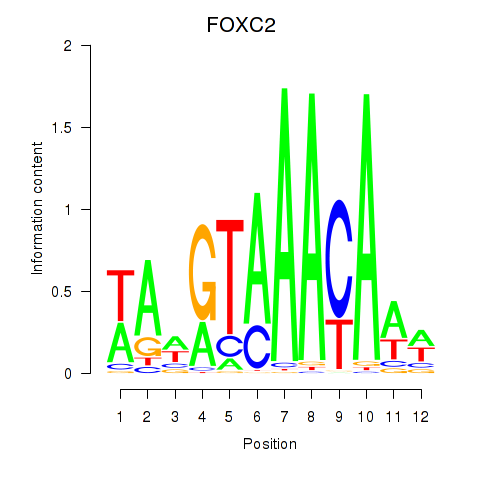
FOXC2
|
ENSG00000176692.8 | FOXC2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| FOXC2 | hg38_v1_chr16_+_86566821_86566836 | -0.52 | 8.3e-03 | Click! |
| FOXA3 | hg38_v1_chr19_+_45864318_45864334 | 0.33 | 1.1e-01 | Click! |
Activity profile of FOXA3_FOXC2 motif
Sorted Z-values of FOXA3_FOXC2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of FOXA3_FOXC2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_+_103372388 | 2.74 |
ENST00000520337.1
|
CTHRC1
|
collagen triple helix repeat containing 1 |
| chr8_+_119067239 | 1.65 |
ENST00000332843.3
|
COLEC10
|
collectin subfamily member 10 |
| chr1_-_120100688 | 1.34 |
ENST00000652264.1
|
NOTCH2
|
notch receptor 2 |
| chr2_-_144516397 | 1.28 |
ENST00000638128.1
|
ZEB2
|
zinc finger E-box binding homeobox 2 |
| chr2_-_144516154 | 1.23 |
ENST00000637304.1
|
ZEB2
|
zinc finger E-box binding homeobox 2 |
| chr2_-_174846405 | 1.22 |
ENST00000409597.5
ENST00000413882.6 |
CHN1
|
chimerin 1 |
| chr1_-_201127184 | 1.14 |
ENST00000449188.3
|
ASCL5
|
achaete-scute family bHLH transcription factor 5 |
| chr17_+_76376581 | 1.09 |
ENST00000591651.5
ENST00000545180.5 |
SPHK1
|
sphingosine kinase 1 |
| chr1_+_205256189 | 1.09 |
ENST00000329800.7
|
TMCC2
|
transmembrane and coiled-coil domain family 2 |
| chr5_+_136058849 | 0.91 |
ENST00000508076.5
|
TGFBI
|
transforming growth factor beta induced |
| chr10_+_69801892 | 0.91 |
ENST00000398978.8
ENST00000645393.2 ENST00000354547.7 ENST00000674121.1 ENST00000673842.1 ENST00000520267.5 |
COL13A1
|
collagen type XIII alpha 1 chain |
| chr11_+_20022550 | 0.91 |
ENST00000533917.5
|
NAV2
|
neuron navigator 2 |
| chr1_+_74235377 | 0.91 |
ENST00000326637.8
|
TNNI3K
|
TNNI3 interacting kinase |
| chr14_+_104801082 | 0.87 |
ENST00000342537.8
|
ZBTB42
|
zinc finger and BTB domain containing 42 |
| chr11_-_108593738 | 0.85 |
ENST00000525344.5
ENST00000265843.9 |
EXPH5
|
exophilin 5 |
| chr22_+_20774092 | 0.85 |
ENST00000215727.10
|
SERPIND1
|
serpin family D member 1 |
| chr8_+_31639755 | 0.84 |
ENST00000520407.5
|
NRG1
|
neuregulin 1 |
| chr6_+_37929959 | 0.81 |
ENST00000373389.5
|
ZFAND3
|
zinc finger AN1-type containing 3 |
| chr4_-_69961007 | 0.80 |
ENST00000353151.3
|
CSN2
|
casein beta |
| chr1_-_206921987 | 0.79 |
ENST00000530505.1
ENST00000442471.4 |
FCMR
|
Fc fragment of IgM receptor |
| chr1_-_206921867 | 0.79 |
ENST00000628511.2
ENST00000367091.8 |
FCMR
|
Fc fragment of IgM receptor |
| chr10_+_122163590 | 0.78 |
ENST00000368999.5
|
TACC2
|
transforming acidic coiled-coil containing protein 2 |
| chr3_+_109136707 | 0.75 |
ENST00000622536.6
|
C3orf85
|
chromosome 3 open reading frame 85 |
| chr10_+_113709261 | 0.74 |
ENST00000672138.1
ENST00000452490.3 |
CASP7
|
caspase 7 |
| chr2_-_174847015 | 0.73 |
ENST00000650938.1
|
CHN1
|
chimerin 1 |
| chr10_+_122163672 | 0.73 |
ENST00000369004.7
ENST00000260733.7 |
TACC2
|
transforming acidic coiled-coil containing protein 2 |
| chr7_+_151341764 | 0.68 |
ENST00000413040.7
ENST00000470229.6 ENST00000568733.6 |
NUB1
|
negative regulator of ubiquitin like proteins 1 |
| chr7_-_116030735 | 0.68 |
ENST00000393485.5
|
TFEC
|
transcription factor EC |
| chr3_-_116444983 | 0.66 |
ENST00000333617.8
|
LSAMP
|
limbic system associated membrane protein |
| chr7_+_28412511 | 0.66 |
ENST00000357727.7
|
CREB5
|
cAMP responsive element binding protein 5 |
| chr7_-_116030750 | 0.65 |
ENST00000265440.12
ENST00000320239.11 |
TFEC
|
transcription factor EC |
| chr11_+_62419025 | 0.64 |
ENST00000278282.3
|
SCGB1A1
|
secretoglobin family 1A member 1 |
| chrX_-_15384402 | 0.64 |
ENST00000297904.4
|
VEGFD
|
vascular endothelial growth factor D |
| chr16_-_48247533 | 0.64 |
ENST00000356608.7
ENST00000569991.1 |
ABCC11
|
ATP binding cassette subfamily C member 11 |
| chr1_-_56966006 | 0.64 |
ENST00000371237.9
|
C8B
|
complement C8 beta chain |
| chr1_+_239719542 | 0.61 |
ENST00000448020.1
|
CHRM3
|
cholinergic receptor muscarinic 3 |
| chr3_+_114294020 | 0.59 |
ENST00000383671.8
|
TIGIT
|
T cell immunoreceptor with Ig and ITIM domains |
| chr8_-_63026179 | 0.58 |
ENST00000677919.1
|
GGH
|
gamma-glutamyl hydrolase |
| chr4_+_168092530 | 0.58 |
ENST00000359299.8
|
ANXA10
|
annexin A10 |
| chr3_-_186362223 | 0.58 |
ENST00000265022.8
|
DGKG
|
diacylglycerol kinase gamma |
| chr1_+_172420681 | 0.57 |
ENST00000367727.9
|
C1orf105
|
chromosome 1 open reading frame 105 |
| chr12_-_39340963 | 0.56 |
ENST00000552961.5
|
KIF21A
|
kinesin family member 21A |
| chr17_-_31314066 | 0.56 |
ENST00000577894.1
|
EVI2B
|
ecotropic viral integration site 2B |
| chr5_+_162067764 | 0.55 |
ENST00000639213.2
ENST00000414552.6 |
GABRG2
|
gamma-aminobutyric acid type A receptor subunit gamma2 |
| chr11_+_27055215 | 0.55 |
ENST00000525090.1
|
BBOX1
|
gamma-butyrobetaine hydroxylase 1 |
| chr1_-_56966133 | 0.53 |
ENST00000535057.5
ENST00000543257.5 |
C8B
|
complement C8 beta chain |
| chr5_+_162067858 | 0.53 |
ENST00000361925.9
|
GABRG2
|
gamma-aminobutyric acid type A receptor subunit gamma2 |
| chr3_+_130850585 | 0.53 |
ENST00000505330.5
ENST00000504381.5 ENST00000507488.6 |
ATP2C1
|
ATPase secretory pathway Ca2+ transporting 1 |
| chr2_+_167248638 | 0.53 |
ENST00000295237.10
|
XIRP2
|
xin actin binding repeat containing 2 |
| chr6_-_87095059 | 0.53 |
ENST00000369582.6
ENST00000610310.3 ENST00000630630.2 ENST00000627148.3 ENST00000625577.1 |
CGA
|
glycoprotein hormones, alpha polypeptide |
| chr5_+_162067500 | 0.52 |
ENST00000639384.1
ENST00000640985.1 ENST00000638772.1 |
GABRG2
|
gamma-aminobutyric acid type A receptor subunit gamma2 |
| chr3_+_148730100 | 0.52 |
ENST00000474935.5
ENST00000475347.5 ENST00000461609.1 |
AGTR1
|
angiotensin II receptor type 1 |
| chr10_+_69088096 | 0.52 |
ENST00000242465.4
|
SRGN
|
serglycin |
| chr12_-_21334858 | 0.50 |
ENST00000445053.1
ENST00000458504.5 ENST00000422327.5 ENST00000683939.1 |
SLCO1A2
|
solute carrier organic anion transporter family member 1A2 |
| chr5_-_116536458 | 0.49 |
ENST00000510263.5
|
SEMA6A
|
semaphorin 6A |
| chr4_-_163613505 | 0.49 |
ENST00000339875.9
|
MARCHF1
|
membrane associated ring-CH-type finger 1 |
| chr11_-_85686123 | 0.49 |
ENST00000316398.5
|
CCDC89
|
coiled-coil domain containing 89 |
| chr5_+_162067458 | 0.48 |
ENST00000639975.1
ENST00000639111.2 ENST00000639683.1 |
GABRG2
|
gamma-aminobutyric acid type A receptor subunit gamma2 |
| chr4_-_152352800 | 0.47 |
ENST00000393956.9
|
FBXW7
|
F-box and WD repeat domain containing 7 |
| chr3_-_108222362 | 0.47 |
ENST00000492106.1
|
IFT57
|
intraflagellar transport 57 |
| chr5_+_162068031 | 0.45 |
ENST00000356592.8
|
GABRG2
|
gamma-aminobutyric acid type A receptor subunit gamma2 |
| chr7_+_120273129 | 0.45 |
ENST00000331113.9
|
KCND2
|
potassium voltage-gated channel subfamily D member 2 |
| chr7_+_130266847 | 0.45 |
ENST00000222481.9
|
CPA2
|
carboxypeptidase A2 |
| chr4_+_164877164 | 0.44 |
ENST00000507152.6
ENST00000515275.1 |
APELA
|
apelin receptor early endogenous ligand |
| chr15_+_70892809 | 0.44 |
ENST00000260382.10
ENST00000560755.5 |
LRRC49
|
leucine rich repeat containing 49 |
| chr16_-_87936529 | 0.44 |
ENST00000649794.3
ENST00000649158.1 ENST00000648177.1 |
CA5A
|
carbonic anhydrase 5A |
| chr12_-_21775581 | 0.44 |
ENST00000537950.1
ENST00000665145.1 |
KCNJ8
|
potassium inwardly rectifying channel subfamily J member 8 |
| chr5_-_59276109 | 0.43 |
ENST00000503258.5
|
PDE4D
|
phosphodiesterase 4D |
| chr5_-_138338325 | 0.42 |
ENST00000510119.1
ENST00000513970.5 |
CDC25C
|
cell division cycle 25C |
| chr2_-_179562576 | 0.41 |
ENST00000336917.9
|
ZNF385B
|
zinc finger protein 385B |
| chr6_+_125203639 | 0.41 |
ENST00000392482.6
|
TPD52L1
|
TPD52 like 1 |
| chr4_-_122456725 | 0.41 |
ENST00000226730.5
|
IL2
|
interleukin 2 |
| chr5_+_98769273 | 0.41 |
ENST00000308234.11
|
RGMB
|
repulsive guidance molecule BMP co-receptor b |
| chr10_+_69801874 | 0.41 |
ENST00000357811.8
|
COL13A1
|
collagen type XIII alpha 1 chain |
| chr7_-_81770122 | 0.41 |
ENST00000423064.7
|
HGF
|
hepatocyte growth factor |
| chr7_+_134891566 | 0.40 |
ENST00000424922.5
ENST00000495522.1 |
CALD1
|
caldesmon 1 |
| chr2_-_207165923 | 0.39 |
ENST00000309446.11
|
KLF7
|
Kruppel like factor 7 |
| chr9_-_14722725 | 0.39 |
ENST00000380911.4
|
CER1
|
cerberus 1, DAN family BMP antagonist |
| chr14_-_21094488 | 0.38 |
ENST00000555270.5
|
ZNF219
|
zinc finger protein 219 |
| chr1_+_50105666 | 0.38 |
ENST00000651347.1
|
ELAVL4
|
ELAV like RNA binding protein 4 |
| chr14_-_56805648 | 0.38 |
ENST00000554788.5
ENST00000554845.1 ENST00000408990.8 |
OTX2
|
orthodenticle homeobox 2 |
| chr18_+_3449413 | 0.38 |
ENST00000549253.5
|
TGIF1
|
TGFB induced factor homeobox 1 |
| chr16_+_56191476 | 0.37 |
ENST00000262493.12
|
GNAO1
|
G protein subunit alpha o1 |
| chr4_+_15427998 | 0.37 |
ENST00000444304.3
|
C1QTNF7
|
C1q and TNF related 7 |
| chr13_-_85799400 | 0.37 |
ENST00000647374.2
|
SLITRK6
|
SLIT and NTRK like family member 6 |
| chr17_+_9021501 | 0.37 |
ENST00000173229.7
|
NTN1
|
netrin 1 |
| chr10_+_69802424 | 0.36 |
ENST00000673802.2
ENST00000517713.5 ENST00000520133.5 ENST00000522165.5 ENST00000673641.2 ENST00000673628.2 |
COL13A1
|
collagen type XIII alpha 1 chain |
| chr13_+_50015438 | 0.35 |
ENST00000312942.2
|
KCNRG
|
potassium channel regulator |
| chr6_+_10528326 | 0.35 |
ENST00000379597.7
|
GCNT2
|
glucosaminyl (N-acetyl) transferase 2 (I blood group) |
| chr10_+_122163426 | 0.34 |
ENST00000360561.7
|
TACC2
|
transforming acidic coiled-coil containing protein 2 |
| chr6_-_160664270 | 0.34 |
ENST00000316300.10
|
LPA
|
lipoprotein(a) |
| chr11_+_13962676 | 0.34 |
ENST00000576479.4
|
SPON1
|
spondin 1 |
| chr5_+_40841308 | 0.34 |
ENST00000381677.4
ENST00000254691.10 |
CARD6
|
caspase recruitment domain family member 6 |
| chr13_+_50015254 | 0.34 |
ENST00000360473.8
|
KCNRG
|
potassium channel regulator |
| chr17_-_19387170 | 0.33 |
ENST00000395592.6
ENST00000299610.5 |
MFAP4
|
microfibril associated protein 4 |
| chr1_-_28058087 | 0.33 |
ENST00000373864.5
|
EYA3
|
EYA transcriptional coactivator and phosphatase 3 |
| chr7_-_141973773 | 0.33 |
ENST00000547270.1
|
TAS2R38
|
taste 2 receptor member 38 |
| chr10_-_116273009 | 0.33 |
ENST00000439649.8
ENST00000369234.5 ENST00000682194.1 ENST00000355422.11 |
GFRA1
|
GDNF family receptor alpha 1 |
| chr7_-_22220226 | 0.32 |
ENST00000420196.5
|
RAPGEF5
|
Rap guanine nucleotide exchange factor 5 |
| chr3_-_71064915 | 0.31 |
ENST00000614176.5
ENST00000485326.7 |
FOXP1
|
forkhead box P1 |
| chr3_-_108222383 | 0.31 |
ENST00000264538.4
|
IFT57
|
intraflagellar transport 57 |
| chr3_-_71064964 | 0.31 |
ENST00000650387.1
|
FOXP1
|
forkhead box P1 |
| chr2_-_174395640 | 0.30 |
ENST00000342016.8
|
CIR1
|
corepressor interacting with RBPJ, CIR1 |
| chr1_+_86468902 | 0.30 |
ENST00000394711.2
|
CLCA1
|
chloride channel accessory 1 |
| chr7_+_112423137 | 0.30 |
ENST00000005558.8
ENST00000621379.4 |
IFRD1
|
interferon related developmental regulator 1 |
| chr11_-_66345066 | 0.29 |
ENST00000359957.8
ENST00000425825.6 |
BRMS1
|
BRMS1 transcriptional repressor and anoikis regulator |
| chr7_+_134745460 | 0.29 |
ENST00000436461.6
|
CALD1
|
caldesmon 1 |
| chr12_-_21775045 | 0.29 |
ENST00000667884.1
|
KCNJ8
|
potassium inwardly rectifying channel subfamily J member 8 |
| chr2_-_219178103 | 0.28 |
ENST00000409789.5
|
CNPPD1
|
cyclin Pas1/PHO80 domain containing 1 |
| chr18_-_28036585 | 0.28 |
ENST00000399380.7
|
CDH2
|
cadherin 2 |
| chr20_-_7940444 | 0.28 |
ENST00000378789.4
|
HAO1
|
hydroxyacid oxidase 1 |
| chr12_-_52814106 | 0.28 |
ENST00000551956.2
|
KRT4
|
keratin 4 |
| chr1_-_167553745 | 0.28 |
ENST00000370509.5
|
CREG1
|
cellular repressor of E1A stimulated genes 1 |
| chr3_+_171843337 | 0.27 |
ENST00000334567.9
ENST00000619900.4 ENST00000450693.1 |
TMEM212
|
transmembrane protein 212 |
| chr1_+_222928415 | 0.27 |
ENST00000284476.7
|
DISP1
|
dispatched RND transporter family member 1 |
| chr3_+_119597874 | 0.27 |
ENST00000488919.5
ENST00000273371.9 ENST00000495992.5 |
PLA1A
|
phospholipase A1 member A |
| chr6_+_36676489 | 0.27 |
ENST00000448526.6
|
CDKN1A
|
cyclin dependent kinase inhibitor 1A |
| chr4_+_183905266 | 0.27 |
ENST00000308497.9
|
STOX2
|
storkhead box 2 |
| chr7_+_123601815 | 0.27 |
ENST00000451215.6
|
ASB15
|
ankyrin repeat and SOCS box containing 15 |
| chr4_-_99144238 | 0.26 |
ENST00000512499.5
ENST00000504125.1 ENST00000505590.5 ENST00000629236.2 ENST00000508393.5 ENST00000265512.12 |
ADH4
|
alcohol dehydrogenase 4 (class II), pi polypeptide |
| chr19_+_42302098 | 0.26 |
ENST00000598490.1
ENST00000341747.8 |
PRR19
|
proline rich 19 |
| chr6_-_43629222 | 0.25 |
ENST00000307126.10
|
GTPBP2
|
GTP binding protein 2 |
| chr4_-_70666492 | 0.25 |
ENST00000254801.9
ENST00000391614.7 |
JCHAIN
|
joining chain of multimeric IgA and IgM |
| chr7_+_123601836 | 0.25 |
ENST00000434204.5
|
ASB15
|
ankyrin repeat and SOCS box containing 15 |
| chr15_-_58749569 | 0.24 |
ENST00000402627.5
ENST00000559053.1 ENST00000260408.8 ENST00000561288.1 ENST00000461408.2 ENST00000439637.5 ENST00000558004.1 |
ADAM10
|
ADAM metallopeptidase domain 10 |
| chr3_-_197183963 | 0.24 |
ENST00000653795.1
|
DLG1
|
discs large MAGUK scaffold protein 1 |
| chr21_-_14658812 | 0.24 |
ENST00000647101.1
|
SAMSN1
|
SAM domain, SH3 domain and nuclear localization signals 1 |
| chr1_-_43389768 | 0.23 |
ENST00000372455.4
ENST00000372457.9 ENST00000290663.10 |
MED8
|
mediator complex subunit 8 |
| chr11_+_119334511 | 0.23 |
ENST00000311413.5
|
RNF26
|
ring finger protein 26 |
| chr5_+_141330494 | 0.23 |
ENST00000517417.3
ENST00000378105.4 |
PCDHGA1
|
protocadherin gamma subfamily A, 1 |
| chr3_-_116445458 | 0.23 |
ENST00000490035.7
|
LSAMP
|
limbic system associated membrane protein |
| chr7_-_138627444 | 0.23 |
ENST00000463557.1
|
SVOPL
|
SVOP like |
| chr16_-_48610150 | 0.23 |
ENST00000262384.4
|
N4BP1
|
NEDD4 binding protein 1 |
| chr17_-_31314040 | 0.23 |
ENST00000330927.5
|
EVI2B
|
ecotropic viral integration site 2B |
| chr6_+_29301701 | 0.23 |
ENST00000641895.1
|
OR14J1
|
olfactory receptor family 14 subfamily J member 1 |
| chr20_+_9069076 | 0.23 |
ENST00000378473.9
|
PLCB4
|
phospholipase C beta 4 |
| chr1_-_150697128 | 0.23 |
ENST00000427665.1
ENST00000271732.8 |
GOLPH3L
|
golgi phosphoprotein 3 like |
| chr10_-_79949098 | 0.23 |
ENST00000372292.8
|
SFTPD
|
surfactant protein D |
| chr7_+_114414809 | 0.23 |
ENST00000350908.9
|
FOXP2
|
forkhead box P2 |
| chr14_+_22829879 | 0.23 |
ENST00000355151.9
ENST00000397496.7 ENST00000555345.5 ENST00000432849.7 ENST00000553711.5 ENST00000556465.5 ENST00000397505.2 ENST00000557221.1 ENST00000556840.5 ENST00000555536.1 |
MRPL52
|
mitochondrial ribosomal protein L52 |
| chr19_+_15049469 | 0.22 |
ENST00000427043.4
|
CASP14
|
caspase 14 |
| chr12_+_92702983 | 0.21 |
ENST00000344636.6
ENST00000544406.2 |
PLEKHG7
|
pleckstrin homology and RhoGEF domain containing G7 |
| chr1_-_152360004 | 0.21 |
ENST00000388718.5
|
FLG2
|
filaggrin family member 2 |
| chr18_+_63907948 | 0.21 |
ENST00000238508.8
|
SERPINB10
|
serpin family B member 10 |
| chrX_-_41665766 | 0.21 |
ENST00000643043.2
ENST00000486402.1 ENST00000646087.2 |
CASK
|
calcium/calmodulin dependent serine protein kinase |
| chr1_+_18631006 | 0.21 |
ENST00000375375.7
|
PAX7
|
paired box 7 |
| chr4_+_164754116 | 0.21 |
ENST00000507311.1
|
SMIM31
|
small integral membrane protein 31 |
| chr14_-_69797232 | 0.21 |
ENST00000216540.5
|
SLC10A1
|
solute carrier family 10 member 1 |
| chr12_-_49187369 | 0.21 |
ENST00000547939.6
|
TUBA1A
|
tubulin alpha 1a |
| chr19_-_45584810 | 0.21 |
ENST00000323060.3
|
OPA3
|
outer mitochondrial membrane lipid metabolism regulator OPA3 |
| chr12_+_92702843 | 0.21 |
ENST00000397833.3
|
PLEKHG7
|
pleckstrin homology and RhoGEF domain containing G7 |
| chr11_-_116837586 | 0.20 |
ENST00000375320.5
ENST00000359492.6 ENST00000375329.6 ENST00000375323.5 ENST00000236850.5 |
APOA1
|
apolipoprotein A1 |
| chr18_+_57352541 | 0.20 |
ENST00000324000.4
|
ST8SIA3
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 3 |
| chr5_-_41213505 | 0.20 |
ENST00000337836.10
ENST00000433294.1 |
C6
|
complement C6 |
| chr1_-_246566238 | 0.20 |
ENST00000366514.5
|
TFB2M
|
transcription factor B2, mitochondrial |
| chr7_+_130207847 | 0.20 |
ENST00000297819.4
|
SSMEM1
|
serine rich single-pass membrane protein 1 |
| chr3_-_197183806 | 0.20 |
ENST00000671246.1
ENST00000660553.1 |
DLG1
|
discs large MAGUK scaffold protein 1 |
| chr20_+_37744630 | 0.20 |
ENST00000373473.5
|
CTNNBL1
|
catenin beta like 1 |
| chr14_-_100569780 | 0.19 |
ENST00000355173.7
|
BEGAIN
|
brain enriched guanylate kinase associated |
| chr15_+_84237586 | 0.19 |
ENST00000512109.1
|
GOLGA6L4
|
golgin A6 family like 4 |
| chr18_+_23453275 | 0.19 |
ENST00000581585.5
ENST00000339486.8 ENST00000577501.5 |
RIOK3
|
RIO kinase 3 |
| chr6_-_25830557 | 0.19 |
ENST00000468082.1
|
SLC17A1
|
solute carrier family 17 member 1 |
| chr2_+_227616998 | 0.19 |
ENST00000641801.1
|
SCYGR4
|
small cysteine and glycine repeat containing 4 |
| chr8_-_23404076 | 0.19 |
ENST00000524168.1
ENST00000389131.8 ENST00000523833.2 ENST00000519243.1 |
LOXL2
|
lysyl oxidase like 2 |
| chr14_-_99272184 | 0.19 |
ENST00000357195.8
|
BCL11B
|
BAF chromatin remodeling complex subunit BCL11B |
| chr4_+_15374541 | 0.19 |
ENST00000382383.7
ENST00000429690.5 |
C1QTNF7
|
C1q and TNF related 7 |
| chr2_+_113127588 | 0.19 |
ENST00000409930.4
|
IL1RN
|
interleukin 1 receptor antagonist |
| chr14_-_25010604 | 0.19 |
ENST00000550887.5
|
STXBP6
|
syntaxin binding protein 6 |
| chr8_-_94436926 | 0.18 |
ENST00000481490.3
|
FSBP
|
fibrinogen silencer binding protein |
| chr6_+_44342684 | 0.18 |
ENST00000288390.2
|
SPATS1
|
spermatogenesis associated serine rich 1 |
| chr10_-_30999469 | 0.18 |
ENST00000538351.6
|
ZNF438
|
zinc finger protein 438 |
| chr3_+_189631373 | 0.18 |
ENST00000264731.8
ENST00000418709.6 ENST00000320472.9 ENST00000392460.7 ENST00000440651.6 |
TP63
|
tumor protein p63 |
| chr4_+_15339818 | 0.18 |
ENST00000397700.6
ENST00000295297.4 |
C1QTNF7
|
C1q and TNF related 7 |
| chr11_-_41459592 | 0.18 |
ENST00000528697.6
ENST00000530763.5 |
LRRC4C
|
leucine rich repeat containing 4C |
| chr8_+_19313685 | 0.18 |
ENST00000265807.8
ENST00000518040.5 |
SH2D4A
|
SH2 domain containing 4A |
| chr6_-_132659178 | 0.18 |
ENST00000275216.3
|
TAAR1
|
trace amine associated receptor 1 |
| chr1_-_167553799 | 0.17 |
ENST00000466652.2
|
CREG1
|
cellular repressor of E1A stimulated genes 1 |
| chr7_+_86643902 | 0.17 |
ENST00000361669.7
|
GRM3
|
glutamate metabotropic receptor 3 |
| chr12_-_18090185 | 0.17 |
ENST00000229002.6
ENST00000538724.6 |
RERGL
|
RERG like |
| chr6_+_44342639 | 0.17 |
ENST00000674044.1
ENST00000515220.5 ENST00000323108.12 |
SPATS1
|
spermatogenesis associated serine rich 1 |
| chr1_+_43389874 | 0.17 |
ENST00000372450.8
|
SZT2
|
SZT2 subunit of KICSTOR complex |
| chr21_-_42350987 | 0.17 |
ENST00000291526.5
|
TFF2
|
trefoil factor 2 |
| chr12_+_120302316 | 0.17 |
ENST00000536460.1
ENST00000202967.4 |
SIRT4
|
sirtuin 4 |
| chrX_+_136532205 | 0.17 |
ENST00000370634.8
|
VGLL1
|
vestigial like family member 1 |
| chr3_-_197183849 | 0.17 |
ENST00000443183.5
|
DLG1
|
discs large MAGUK scaffold protein 1 |
| chr16_+_56191728 | 0.16 |
ENST00000638705.1
ENST00000262494.12 |
GNAO1
|
G protein subunit alpha o1 |
| chr14_-_99271485 | 0.16 |
ENST00000345514.2
ENST00000443726.2 |
BCL11B
|
BAF chromatin remodeling complex subunit BCL11B |
| chr5_+_96663010 | 0.16 |
ENST00000506811.5
ENST00000514055.5 ENST00000508608.6 |
CAST
|
calpastatin |
| chr4_+_87799546 | 0.16 |
ENST00000226284.7
|
IBSP
|
integrin binding sialoprotein |
| chr7_+_134779625 | 0.16 |
ENST00000454108.5
ENST00000361675.7 |
CALD1
|
caldesmon 1 |
| chr9_+_100578071 | 0.16 |
ENST00000307584.6
|
CAVIN4
|
caveolae associated protein 4 |
| chr17_+_7583828 | 0.16 |
ENST00000396501.8
ENST00000250124.11 ENST00000584378.5 ENST00000423172.6 ENST00000579445.5 ENST00000585217.5 ENST00000581380.1 |
MPDU1
|
mannose-P-dolichol utilization defect 1 |
| chrX_+_100584928 | 0.16 |
ENST00000373031.5
|
TNMD
|
tenomodulin |
| chr8_+_91249307 | 0.16 |
ENST00000309536.6
ENST00000276609.8 |
SLC26A7
|
solute carrier family 26 member 7 |
| chr2_+_168802610 | 0.16 |
ENST00000397206.6
ENST00000317647.12 ENST00000397209.6 |
NOSTRIN
|
nitric oxide synthase trafficking |
| chr2_-_21044063 | 0.16 |
ENST00000233242.5
|
APOB
|
apolipoprotein B |
| chr4_+_164754045 | 0.15 |
ENST00000515485.5
|
SMIM31
|
small integral membrane protein 31 |
| chr17_-_39607876 | 0.15 |
ENST00000302584.5
|
NEUROD2
|
neuronal differentiation 2 |
| chr15_+_58138368 | 0.15 |
ENST00000219919.9
ENST00000536493.1 |
AQP9
|
aquaporin 9 |
| chr19_-_45584769 | 0.15 |
ENST00000263275.5
|
OPA3
|
outer mitochondrial membrane lipid metabolism regulator OPA3 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 2.7 | GO:0043932 | ossification involved in bone remodeling(GO:0043932) |
| 0.4 | 1.1 | GO:0046521 | sphingoid catabolic process(GO:0046521) |
| 0.3 | 2.5 | GO:1902748 | positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.3 | 0.8 | GO:1903487 | regulation of lactation(GO:1903487) |
| 0.2 | 0.5 | GO:0033364 | mast cell secretory granule organization(GO:0033364) |
| 0.2 | 0.9 | GO:0003025 | regulation of systemic arterial blood pressure by baroreceptor feedback(GO:0003025) vagus nerve development(GO:0021564) |
| 0.1 | 0.4 | GO:0090133 | mesendoderm migration(GO:0090133) cell migration involved in mesendoderm migration(GO:0090134) |
| 0.1 | 0.6 | GO:0046900 | tetrahydrofolylpolyglutamate metabolic process(GO:0046900) |
| 0.1 | 0.6 | GO:2000638 | regulation of SREBP signaling pathway(GO:2000638) negative regulation of SREBP signaling pathway(GO:2000639) |
| 0.1 | 0.5 | GO:0086097 | phospholipase C-activating angiotensin-activated signaling pathway(GO:0086097) |
| 0.1 | 0.6 | GO:0010193 | response to ozone(GO:0010193) |
| 0.1 | 0.4 | GO:0097534 | lymphoid lineage cell migration(GO:0097534) lymphoid lineage cell migration into thymus(GO:0097535) |
| 0.1 | 0.1 | GO:0021767 | mammillary body development(GO:0021767) mammillary axonal complex development(GO:0061373) |
| 0.1 | 1.7 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.1 | 0.3 | GO:0007518 | myoblast fate determination(GO:0007518) |
| 0.1 | 0.4 | GO:1900108 | negative regulation of nodal signaling pathway(GO:1900108) |
| 0.1 | 2.5 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.1 | 0.7 | GO:0072734 | response to staurosporine(GO:0072733) cellular response to staurosporine(GO:0072734) |
| 0.1 | 0.3 | GO:0035638 | patched ligand maturation(GO:0007225) signal maturation(GO:0035638) |
| 0.1 | 0.5 | GO:0032468 | Golgi calcium ion homeostasis(GO:0032468) |
| 0.1 | 0.4 | GO:0046013 | regulation of T cell homeostatic proliferation(GO:0046013) |
| 0.1 | 1.3 | GO:0002315 | marginal zone B cell differentiation(GO:0002315) |
| 0.1 | 0.6 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) |
| 0.1 | 0.2 | GO:0021758 | caudate nucleus development(GO:0021757) putamen development(GO:0021758) |
| 0.1 | 0.5 | GO:0060665 | regulation of branching involved in salivary gland morphogenesis by mesenchymal-epithelial signaling(GO:0060665) |
| 0.1 | 0.6 | GO:0060754 | positive regulation of mast cell chemotaxis(GO:0060754) |
| 0.1 | 0.2 | GO:2000259 | positive regulation of complement activation(GO:0045917) positive regulation of protein activation cascade(GO:2000259) |
| 0.1 | 0.2 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.1 | 0.8 | GO:0021840 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) ERBB3 signaling pathway(GO:0038129) |
| 0.1 | 0.2 | GO:0045085 | negative regulation of interleukin-2 biosynthetic process(GO:0045085) |
| 0.1 | 0.2 | GO:1903216 | regulation of protein processing involved in protein targeting to mitochondrion(GO:1903216) negative regulation of protein processing involved in protein targeting to mitochondrion(GO:1903217) |
| 0.1 | 1.2 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.1 | 0.2 | GO:0014813 | skeletal muscle satellite cell commitment(GO:0014813) |
| 0.0 | 0.5 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.0 | 0.7 | GO:1990573 | potassium ion import across plasma membrane(GO:1990573) |
| 0.0 | 0.2 | GO:0039534 | negative regulation of MDA-5 signaling pathway(GO:0039534) |
| 0.0 | 0.2 | GO:0051088 | PMA-inducible membrane protein ectodomain proteolysis(GO:0051088) |
| 0.0 | 2.0 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.0 | 0.2 | GO:0007499 | ectoderm and mesoderm interaction(GO:0007499) |
| 0.0 | 0.9 | GO:0086069 | bundle of His cell to Purkinje myocyte communication(GO:0086069) |
| 0.0 | 0.1 | GO:0019836 | hemolysis by symbiont of host erythrocytes(GO:0019836) hemolysis in other organism(GO:0044179) hemolysis in other organism involved in symbiotic interaction(GO:0052331) |
| 0.0 | 0.3 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.0 | 0.4 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.0 | 0.1 | GO:0002305 | gamma-delta intraepithelial T cell differentiation(GO:0002304) CD8-positive, gamma-delta intraepithelial T cell differentiation(GO:0002305) |
| 0.0 | 0.4 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.0 | 0.5 | GO:0021562 | vestibulocochlear nerve development(GO:0021562) |
| 0.0 | 0.2 | GO:0010903 | negative regulation of very-low-density lipoprotein particle remodeling(GO:0010903) |
| 0.0 | 0.6 | GO:1903764 | regulation of potassium ion export across plasma membrane(GO:1903764) |
| 0.0 | 0.9 | GO:0071985 | multivesicular body sorting pathway(GO:0071985) |
| 0.0 | 0.7 | GO:1902260 | negative regulation of delayed rectifier potassium channel activity(GO:1902260) |
| 0.0 | 0.2 | GO:2000297 | negative regulation of synapse maturation(GO:2000297) |
| 0.0 | 0.6 | GO:0072619 | interleukin-21 production(GO:0032625) interleukin-21 secretion(GO:0072619) |
| 0.0 | 0.1 | GO:1904823 | canalicular bile acid transport(GO:0015722) pyrimidine nucleobase transport(GO:0015855) purine nucleobase transmembrane transport(GO:1904823) |
| 0.0 | 0.3 | GO:1901668 | regulation of superoxide dismutase activity(GO:1901668) |
| 0.0 | 0.9 | GO:0035641 | locomotory exploration behavior(GO:0035641) |
| 0.0 | 0.2 | GO:0010966 | regulation of phosphate transport(GO:0010966) |
| 0.0 | 0.1 | GO:0035106 | operant conditioning(GO:0035106) |
| 0.0 | 0.3 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.0 | 0.2 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 0.0 | 0.2 | GO:0035990 | tendon cell differentiation(GO:0035990) tendon formation(GO:0035992) |
| 0.0 | 0.2 | GO:0009441 | glycolate metabolic process(GO:0009441) |
| 0.0 | 0.3 | GO:0060574 | intestinal epithelial cell maturation(GO:0060574) |
| 0.0 | 0.4 | GO:0035860 | glial cell-derived neurotrophic factor receptor signaling pathway(GO:0035860) |
| 0.0 | 0.3 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.0 | 1.7 | GO:0001958 | endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.0 | 0.3 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.0 | 0.3 | GO:0060267 | positive regulation of respiratory burst(GO:0060267) |
| 0.0 | 0.5 | GO:0034374 | low-density lipoprotein particle remodeling(GO:0034374) |
| 0.0 | 0.1 | GO:0021592 | midbrain-hindbrain boundary morphogenesis(GO:0021555) fourth ventricle development(GO:0021592) |
| 0.0 | 0.5 | GO:0045475 | locomotor rhythm(GO:0045475) |
| 0.0 | 0.1 | GO:0014740 | mesodermal cell fate determination(GO:0007500) negative regulation of muscle hyperplasia(GO:0014740) |
| 0.0 | 0.6 | GO:0015865 | purine nucleotide transport(GO:0015865) |
| 0.0 | 0.4 | GO:2000543 | positive regulation of gastrulation(GO:2000543) |
| 0.0 | 0.1 | GO:0097056 | selenocysteinyl-tRNA(Sec) biosynthetic process(GO:0097056) |
| 0.0 | 0.1 | GO:0035799 | ureter maturation(GO:0035799) |
| 0.0 | 0.2 | GO:2000660 | negative regulation of interleukin-1-mediated signaling pathway(GO:2000660) |
| 0.0 | 0.5 | GO:2001224 | positive regulation of neuron migration(GO:2001224) |
| 0.0 | 0.3 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.0 | 0.8 | GO:0044458 | motile cilium assembly(GO:0044458) |
| 0.0 | 0.3 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.0 | 0.1 | GO:0030241 | skeletal muscle myosin thick filament assembly(GO:0030241) |
| 0.0 | 0.2 | GO:0048194 | Golgi vesicle budding(GO:0048194) |
| 0.0 | 0.3 | GO:2000809 | positive regulation of synaptic vesicle clustering(GO:2000809) |
| 0.0 | 0.4 | GO:0007039 | protein catabolic process in the vacuole(GO:0007039) |
| 0.0 | 0.1 | GO:0044537 | regulation of circulating fibrinogen levels(GO:0044537) |
| 0.0 | 0.3 | GO:0036066 | protein O-linked fucosylation(GO:0036066) |
| 0.0 | 0.1 | GO:0036135 | Schwann cell migration(GO:0036135) regulation of Schwann cell migration(GO:1900147) |
| 0.0 | 0.1 | GO:0032277 | negative regulation of gonadotropin secretion(GO:0032277) |
| 0.0 | 0.5 | GO:0032695 | negative regulation of interleukin-12 production(GO:0032695) |
| 0.0 | 0.7 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.0 | 1.6 | GO:0006968 | cellular defense response(GO:0006968) |
| 0.0 | 0.3 | GO:0007196 | adenylate cyclase-inhibiting G-protein coupled glutamate receptor signaling pathway(GO:0007196) |
| 0.0 | 1.1 | GO:0042982 | amyloid precursor protein metabolic process(GO:0042982) |
| 0.0 | 0.6 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.0 | 0.1 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 0.0 | 0.4 | GO:0036150 | phosphatidylserine acyl-chain remodeling(GO:0036150) |
| 0.0 | 0.1 | GO:0002215 | defense response to nematode(GO:0002215) |
| 0.0 | 0.2 | GO:0035542 | regulation of SNARE complex assembly(GO:0035542) |
| 0.0 | 0.1 | GO:0071847 | TNFSF11-mediated signaling pathway(GO:0071847) negative regulation of osteoclast development(GO:2001205) |
| 0.0 | 0.1 | GO:0035948 | positive regulation of gluconeogenesis by positive regulation of transcription from RNA polymerase II promoter(GO:0035948) |
| 0.0 | 0.4 | GO:0061436 | establishment of skin barrier(GO:0061436) |
| 0.0 | 0.1 | GO:0032571 | response to vitamin K(GO:0032571) |
| 0.0 | 0.6 | GO:0050913 | sensory perception of bitter taste(GO:0050913) |
| 0.0 | 0.1 | GO:0097646 | calcitonin family receptor signaling pathway(GO:0097646) amylin receptor signaling pathway(GO:0097647) |
| 0.0 | 0.2 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.0 | 0.1 | GO:0070574 | cadmium ion transport(GO:0015691) cadmium ion transmembrane transport(GO:0070574) |
| 0.0 | 0.1 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 0.0 | 0.2 | GO:2001214 | positive regulation of vasculogenesis(GO:2001214) |
| 0.0 | 0.5 | GO:0043278 | response to isoquinoline alkaloid(GO:0014072) response to morphine(GO:0043278) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.7 | GO:0005600 | collagen type XIII trimer(GO:0005600) transmembrane collagen trimer(GO:0030936) |
| 0.2 | 1.4 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.1 | 0.3 | GO:0071750 | dimeric IgA immunoglobulin complex(GO:0071750) secretory dimeric IgA immunoglobulin complex(GO:0071752) IgM immunoglobulin complex(GO:0071753) IgM immunoglobulin complex, circulating(GO:0071754) pentameric IgM immunoglobulin complex(GO:0071756) |
| 0.1 | 0.6 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.1 | 0.7 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.1 | 0.6 | GO:0032279 | asymmetric synapse(GO:0032279) |
| 0.1 | 1.0 | GO:0030478 | actin cap(GO:0030478) |
| 0.1 | 2.5 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.1 | 0.8 | GO:0044292 | dendrite terminus(GO:0044292) |
| 0.1 | 0.3 | GO:0070557 | PCNA-p21 complex(GO:0070557) |
| 0.1 | 0.4 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 0.0 | 5.4 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.3 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.0 | 0.9 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.3 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.0 | 0.1 | GO:0038038 | G-protein coupled receptor homodimeric complex(GO:0038038) |
| 0.0 | 0.6 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.0 | 0.2 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.0 | 0.2 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 0.2 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.0 | 0.8 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 0.1 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.0 | 0.3 | GO:0042588 | zymogen granule(GO:0042588) |
| 0.0 | 0.1 | GO:0036398 | TCR signalosome(GO:0036398) |
| 0.0 | 0.3 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.1 | GO:1990682 | CSF1-CSF1R complex(GO:1990682) |
| 0.0 | 0.5 | GO:0044298 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.0 | 0.3 | GO:0070822 | Sin3-type complex(GO:0070822) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0008336 | gamma-butyrobetaine dioxygenase activity(GO:0008336) |
| 0.2 | 0.5 | GO:0001596 | angiotensin type I receptor activity(GO:0001596) |
| 0.2 | 1.1 | GO:0017050 | sphinganine kinase activity(GO:0008481) D-erythro-sphingosine kinase activity(GO:0017050) |
| 0.1 | 2.5 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) |
| 0.1 | 1.3 | GO:0038049 | transcription factor activity, ligand-activated RNA polymerase II transcription factor binding(GO:0038049) |
| 0.1 | 0.6 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.1 | 0.7 | GO:0017098 | sulfonylurea receptor binding(GO:0017098) |
| 0.1 | 0.3 | GO:0052854 | (S)-2-hydroxy-acid oxidase activity(GO:0003973) very-long-chain-(S)-2-hydroxy-acid oxidase activity(GO:0052852) long-chain-(S)-2-hydroxy-long-chain-acid oxidase activity(GO:0052853) medium-chain-(S)-2-hydroxy-acid oxidase activity(GO:0052854) |
| 0.1 | 0.9 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.1 | 0.3 | GO:0052739 | phosphatidylserine 1-acylhydrolase activity(GO:0052739) |
| 0.1 | 0.6 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.1 | 1.7 | GO:0005537 | mannose binding(GO:0005537) |
| 0.1 | 0.6 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.1 | 0.4 | GO:0043208 | glycosphingolipid binding(GO:0043208) |
| 0.1 | 0.5 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.1 | 0.2 | GO:0005150 | interleukin-1, Type I receptor binding(GO:0005150) |
| 0.1 | 2.5 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.1 | 0.2 | GO:0001641 | group II metabotropic glutamate receptor activity(GO:0001641) |
| 0.1 | 0.3 | GO:0019912 | cyclin-dependent protein kinase activating kinase activity(GO:0019912) |
| 0.1 | 0.3 | GO:0035276 | ethanol binding(GO:0035276) |
| 0.1 | 0.4 | GO:0016167 | glial cell-derived neurotrophic factor receptor activity(GO:0016167) |
| 0.1 | 0.3 | GO:0019862 | IgA binding(GO:0019862) |
| 0.0 | 0.3 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.0 | 0.1 | GO:0005275 | amine transmembrane transporter activity(GO:0005275) |
| 0.0 | 0.8 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) transmembrane receptor protein tyrosine kinase activator activity(GO:0030297) |
| 0.0 | 0.1 | GO:0016497 | substance K receptor activity(GO:0016497) |
| 0.0 | 0.2 | GO:0047708 | biotinidase activity(GO:0047708) |
| 0.0 | 0.2 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.0 | 1.0 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 0.2 | GO:0070653 | high-density lipoprotein particle receptor binding(GO:0070653) |
| 0.0 | 0.2 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.0 | 0.6 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.0 | 2.4 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.0 | 0.4 | GO:0016015 | morphogen activity(GO:0016015) |
| 0.0 | 0.1 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.0 | 0.5 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.0 | 0.4 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.0 | 0.6 | GO:0043225 | anion transmembrane-transporting ATPase activity(GO:0043225) |
| 0.0 | 1.8 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.1 | GO:0070259 | tyrosyl-DNA phosphodiesterase activity(GO:0070259) |
| 0.0 | 0.6 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.0 | 0.1 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.0 | 0.2 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.0 | 1.0 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.0 | 0.6 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.4 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.0 | 0.3 | GO:0015321 | sodium-dependent phosphate transmembrane transporter activity(GO:0015321) |
| 0.0 | 0.1 | GO:0005011 | macrophage colony-stimulating factor receptor activity(GO:0005011) |
| 0.0 | 0.2 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.0 | 0.2 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.0 | 0.4 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.3 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.0 | 0.3 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 0.2 | GO:0035473 | lipase binding(GO:0035473) |
| 0.0 | 0.2 | GO:0089720 | caspase binding(GO:0089720) |
| 0.0 | 0.2 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.0 | 0.1 | GO:0045145 | single-stranded DNA 5'-3' exodeoxyribonuclease activity(GO:0045145) |
| 0.0 | 0.1 | GO:0051500 | D-aminoacyl-tRNA deacylase activity(GO:0051499) D-tyrosyl-tRNA(Tyr) deacylase activity(GO:0051500) |
| 0.0 | 0.6 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.5 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.0 | 0.4 | GO:0031698 | beta-2 adrenergic receptor binding(GO:0031698) |
| 0.0 | 0.1 | GO:0001010 | transcription factor activity, sequence-specific DNA binding transcription factor recruiting(GO:0001010) |
| 0.0 | 0.1 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.0 | 0.4 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 0.3 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.0 | 0.3 | GO:0015197 | peptide transporter activity(GO:0015197) |
| 0.0 | 0.2 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.0 | 0.1 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.0 | 0.5 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.0 | 0.1 | GO:0099583 | neurotransmitter receptor activity involved in regulation of postsynaptic cytosolic calcium ion concentration(GO:0099583) |
| 0.0 | 0.3 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 0.4 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.0 | 0.3 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.0 | 0.1 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.0 | 0.2 | GO:0016918 | retinal binding(GO:0016918) |
| 0.0 | 0.3 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.0 | 1.4 | GO:0005518 | collagen binding(GO:0005518) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.5 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 1.8 | PID S1P S1P1 PATHWAY | S1P1 pathway |
| 0.0 | 2.2 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.0 | 0.7 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.0 | 0.8 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 2.3 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 1.8 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 0.7 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 0.9 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 0.5 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.0 | 1.0 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 0.5 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.0 | 1.2 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.0 | 0.6 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.2 | REACTOME PLC BETA MEDIATED EVENTS | Genes involved in PLC beta mediated events |
| 0.1 | 1.6 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.1 | 2.5 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.1 | 0.8 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.1 | 0.8 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 0.5 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.0 | 1.7 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.4 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.0 | 1.3 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.0 | 0.4 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.0 | 1.2 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 0.8 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 0.7 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 0.5 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.4 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.0 | 0.4 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.0 | 1.8 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.6 | REACTOME G PROTEIN ACTIVATION | Genes involved in G-protein activation |
| 0.0 | 0.7 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.5 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.8 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.5 | REACTOME LIPOPROTEIN METABOLISM | Genes involved in Lipoprotein metabolism |
| 0.0 | 0.6 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.6 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.7 | REACTOME NUCLEAR SIGNALING BY ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.0 | 0.7 | REACTOME INWARDLY RECTIFYING K CHANNELS | Genes involved in Inwardly rectifying K+ channels |
| 0.0 | 0.4 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.4 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 0.2 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.0 | 0.4 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |