Project
Inflammatory response time course, HUVEC (Wada, 2009)
Navigation
Downloads
Results for FOXJ2
Z-value: 0.61
Transcription factors associated with FOXJ2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
FOXJ2
|
ENSG00000065970.9 | FOXJ2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| FOXJ2 | hg38_v1_chr12_+_8032692_8032744 | -0.47 | 1.8e-02 | Click! |
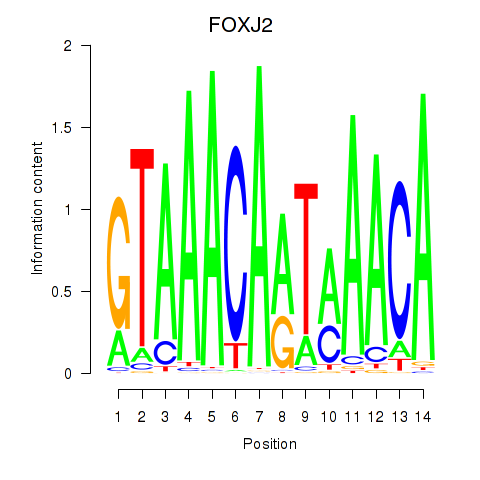
Activity profile of FOXJ2 motif
Sorted Z-values of FOXJ2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of FOXJ2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr15_+_67138001 | 3.81 |
ENST00000439724.7
|
SMAD3
|
SMAD family member 3 |
| chr8_+_103371490 | 1.75 |
ENST00000330295.10
ENST00000415886.2 |
CTHRC1
|
collagen triple helix repeat containing 1 |
| chr2_+_161160299 | 1.69 |
ENST00000440506.5
ENST00000429217.5 ENST00000406287.5 ENST00000402568.5 |
TANK
|
TRAF family member associated NFKB activator |
| chr21_-_34526850 | 1.54 |
ENST00000481448.5
ENST00000381132.6 |
RCAN1
|
regulator of calcineurin 1 |
| chr11_+_35180279 | 1.11 |
ENST00000531873.5
|
CD44
|
CD44 molecule (Indian blood group) |
| chr9_+_5510492 | 1.02 |
ENST00000397747.5
|
PDCD1LG2
|
programmed cell death 1 ligand 2 |
| chr2_+_161160420 | 0.96 |
ENST00000392749.7
ENST00000405852.5 |
TANK
|
TRAF family member associated NFKB activator |
| chr21_-_34526815 | 0.92 |
ENST00000492600.1
|
RCAN1
|
regulator of calcineurin 1 |
| chr1_-_220089818 | 0.87 |
ENST00000498791.6
ENST00000480959.6 |
BPNT1
|
3'(2'), 5'-bisphosphate nucleotidase 1 |
| chr11_+_35180342 | 0.85 |
ENST00000639002.1
|
CD44
|
CD44 molecule (Indian blood group) |
| chr16_-_58546702 | 0.81 |
ENST00000567133.1
|
CNOT1
|
CCR4-NOT transcription complex subunit 1 |
| chr21_-_26051023 | 0.75 |
ENST00000415997.1
|
APP
|
amyloid beta precursor protein |
| chr9_-_131740056 | 0.74 |
ENST00000372195.5
ENST00000683357.1 |
RAPGEF1
|
Rap guanine nucleotide exchange factor 1 |
| chr2_+_201116143 | 0.69 |
ENST00000443227.5
ENST00000309955.8 ENST00000341222.10 ENST00000341582.10 |
CFLAR
|
CASP8 and FADD like apoptosis regulator |
| chr19_+_9087061 | 0.55 |
ENST00000641627.1
|
OR1M1
|
olfactory receptor family 1 subfamily M member 1 |
| chr6_+_30571393 | 0.54 |
ENST00000376545.7
ENST00000441867.6 ENST00000468958.1 ENST00000326195.13 |
ABCF1
|
ATP binding cassette subfamily F member 1 |
| chr17_+_27631148 | 0.52 |
ENST00000313648.10
ENST00000395473.7 ENST00000577392.5 ENST00000584661.5 |
LGALS9
|
galectin 9 |
| chr9_+_100429511 | 0.50 |
ENST00000613183.1
|
MSANTD3
|
Myb/SANT DNA binding domain containing 3 |
| chrX_+_109537118 | 0.50 |
ENST00000372103.1
|
NXT2
|
nuclear transport factor 2 like export factor 2 |
| chr11_-_57322197 | 0.48 |
ENST00000532437.1
|
TNKS1BP1
|
tankyrase 1 binding protein 1 |
| chr6_+_31494881 | 0.46 |
ENST00000538442.5
|
MICB
|
MHC class I polypeptide-related sequence B |
| chr19_-_7058640 | 0.44 |
ENST00000333843.8
|
MBD3L3
|
methyl-CpG binding domain protein 3 like 3 |
| chr7_-_102642791 | 0.44 |
ENST00000340457.8
|
UPK3BL1
|
uroplakin 3B like 1 |
| chr8_+_38728186 | 0.44 |
ENST00000519416.5
ENST00000520615.5 |
TACC1
|
transforming acidic coiled-coil containing protein 1 |
| chr4_+_87975829 | 0.42 |
ENST00000614857.5
|
SPP1
|
secreted phosphoprotein 1 |
| chr7_-_115968302 | 0.42 |
ENST00000457268.5
|
TFEC
|
transcription factor EC |
| chr19_+_7030578 | 0.41 |
ENST00000329753.5
|
MBD3L5
|
methyl-CpG binding domain protein 3 like 5 |
| chr7_+_23598144 | 0.40 |
ENST00000410069.1
|
CCDC126
|
coiled-coil domain containing 126 |
| chr19_-_7040179 | 0.40 |
ENST00000381394.9
|
MBD3L4
|
methyl-CpG binding domain protein 3 like 4 |
| chr17_+_68515399 | 0.40 |
ENST00000588188.6
|
PRKAR1A
|
protein kinase cAMP-dependent type I regulatory subunit alpha |
| chr15_+_73873604 | 0.40 |
ENST00000535547.6
ENST00000562056.1 |
TBC1D21
|
TBC1 domain family member 21 |
| chr11_-_82845734 | 0.39 |
ENST00000681883.1
ENST00000680040.1 ENST00000681432.1 |
PRCP
|
prolylcarboxypeptidase |
| chr9_-_96655280 | 0.39 |
ENST00000446045.1
ENST00000375234.8 |
PRXL2C
|
peroxiredoxin like 2C |
| chr3_+_157436842 | 0.38 |
ENST00000295927.4
|
PTX3
|
pentraxin 3 |
| chrX_-_6535118 | 0.37 |
ENST00000381089.7
ENST00000612369.4 ENST00000398729.1 |
VCX3A
|
variable charge X-linked 3A |
| chr6_+_10887036 | 0.37 |
ENST00000283141.11
|
SYCP2L
|
synaptonemal complex protein 2 like |
| chr1_+_160796098 | 0.36 |
ENST00000392203.8
|
LY9
|
lymphocyte antigen 9 |
| chr8_+_100158029 | 0.36 |
ENST00000251809.4
|
SPAG1
|
sperm associated antigen 1 |
| chr1_+_160796070 | 0.35 |
ENST00000368037.9
|
LY9
|
lymphocyte antigen 9 |
| chr1_+_150149819 | 0.35 |
ENST00000369124.5
|
PLEKHO1
|
pleckstrin homology domain containing O1 |
| chr12_-_10098940 | 0.35 |
ENST00000420265.2
|
CLEC1A
|
C-type lectin domain family 1 member A |
| chr7_-_33100886 | 0.35 |
ENST00000448915.1
|
RP9
|
RP9 pre-mRNA splicing factor |
| chr10_+_116545907 | 0.35 |
ENST00000369221.2
|
PNLIP
|
pancreatic lipase |
| chr8_-_33567118 | 0.34 |
ENST00000256257.2
|
RNF122
|
ring finger protein 122 |
| chr4_+_67558719 | 0.34 |
ENST00000265404.7
ENST00000396225.1 |
STAP1
|
signal transducing adaptor family member 1 |
| chr20_-_43726989 | 0.34 |
ENST00000373003.2
|
GTSF1L
|
gametocyte specific factor 1 like |
| chr11_-_82846128 | 0.33 |
ENST00000679809.1
ENST00000680186.1 ENST00000681592.1 |
PRCP
|
prolylcarboxypeptidase |
| chr10_+_84230660 | 0.32 |
ENST00000652073.1
|
RGR
|
retinal G protein coupled receptor |
| chr14_+_88005128 | 0.32 |
ENST00000267549.5
|
GPR65
|
G protein-coupled receptor 65 |
| chr9_-_127916978 | 0.31 |
ENST00000361444.3
ENST00000335791.10 |
ST6GALNAC4
|
ST6 N-acetylgalactosaminide alpha-2,6-sialyltransferase 4 |
| chr12_-_52680398 | 0.31 |
ENST00000252244.3
|
KRT1
|
keratin 1 |
| chr15_+_66505289 | 0.31 |
ENST00000565627.5
ENST00000564179.5 ENST00000307897.10 |
ZWILCH
|
zwilch kinetochore protein |
| chr2_+_100974849 | 0.31 |
ENST00000450763.1
|
NPAS2
|
neuronal PAS domain protein 2 |
| chr15_+_73873557 | 0.30 |
ENST00000300504.7
|
TBC1D21
|
TBC1 domain family member 21 |
| chr2_+_209653171 | 0.30 |
ENST00000447185.5
|
MAP2
|
microtubule associated protein 2 |
| chr1_-_72100930 | 0.29 |
ENST00000306821.3
|
NEGR1
|
neuronal growth regulator 1 |
| chr12_-_10098977 | 0.29 |
ENST00000315330.8
ENST00000457018.6 |
CLEC1A
|
C-type lectin domain family 1 member A |
| chr15_+_66504959 | 0.29 |
ENST00000535141.6
ENST00000613446.4 ENST00000446801.6 |
ZWILCH
|
zwilch kinetochore protein |
| chr6_-_167157980 | 0.28 |
ENST00000366834.2
|
GPR31
|
G protein-coupled receptor 31 |
| chr1_+_44746401 | 0.28 |
ENST00000372217.5
|
KIF2C
|
kinesin family member 2C |
| chr19_+_54497879 | 0.27 |
ENST00000412608.5
ENST00000610651.1 |
LAIR2
|
leukocyte associated immunoglobulin like receptor 2 |
| chr16_-_30429207 | 0.26 |
ENST00000568434.1
|
DCTPP1
|
dCTP pyrophosphatase 1 |
| chr3_-_190449782 | 0.26 |
ENST00000354905.3
|
TMEM207
|
transmembrane protein 207 |
| chr17_-_58417521 | 0.26 |
ENST00000584437.5
ENST00000407977.7 |
RNF43
|
ring finger protein 43 |
| chr17_-_41168219 | 0.26 |
ENST00000391356.4
|
KRTAP4-3
|
keratin associated protein 4-3 |
| chr10_-_25062279 | 0.25 |
ENST00000615958.4
|
ENKUR
|
enkurin, TRPC channel interacting protein |
| chr9_-_21141832 | 0.25 |
ENST00000380229.4
|
IFNW1
|
interferon omega 1 |
| chr17_+_39461468 | 0.25 |
ENST00000447079.6
ENST00000584632.5 |
CDK12
|
cyclin dependent kinase 12 |
| chr11_+_63369779 | 0.24 |
ENST00000279178.4
|
SLC22A9
|
solute carrier family 22 member 9 |
| chrX_+_109535775 | 0.24 |
ENST00000218004.5
|
NXT2
|
nuclear transport factor 2 like export factor 2 |
| chr1_-_43389768 | 0.24 |
ENST00000372455.4
ENST00000372457.9 ENST00000290663.10 |
MED8
|
mediator complex subunit 8 |
| chr17_-_58417547 | 0.24 |
ENST00000577716.5
|
RNF43
|
ring finger protein 43 |
| chr10_-_72626111 | 0.24 |
ENST00000604238.2
ENST00000642044.1 ENST00000635239.1 ENST00000398761.8 |
MICU1
|
mitochondrial calcium uptake 1 |
| chr16_+_57413861 | 0.23 |
ENST00000616880.1
|
CCL17
|
C-C motif chemokine ligand 17 |
| chr21_-_36079382 | 0.23 |
ENST00000399201.5
|
SETD4
|
SET domain containing 4 |
| chr19_+_11547840 | 0.23 |
ENST00000588935.1
|
CNN1
|
calponin 1 |
| chr1_-_27626229 | 0.22 |
ENST00000399173.5
|
FGR
|
FGR proto-oncogene, Src family tyrosine kinase |
| chr12_+_108129276 | 0.22 |
ENST00000547525.6
|
WSCD2
|
WSC domain containing 2 |
| chr7_+_99828010 | 0.22 |
ENST00000631161.2
ENST00000354829.7 ENST00000342499.8 ENST00000417625.5 ENST00000415413.5 ENST00000444905.5 ENST00000222382.5 ENST00000312017.9 |
CYP3A43
|
cytochrome P450 family 3 subfamily A member 43 |
| chr4_-_47981535 | 0.21 |
ENST00000402813.9
|
CNGA1
|
cyclic nucleotide gated channel subunit alpha 1 |
| chr15_-_55588937 | 0.21 |
ENST00000302000.10
|
PYGO1
|
pygopus family PHD finger 1 |
| chr3_+_49007365 | 0.21 |
ENST00000608424.6
ENST00000438660.5 ENST00000415265.6 |
WDR6
|
WD repeat domain 6 |
| chr3_-_44510602 | 0.21 |
ENST00000436261.6
|
ZNF852
|
zinc finger protein 852 |
| chr1_+_162069768 | 0.20 |
ENST00000530878.5
|
NOS1AP
|
nitric oxide synthase 1 adaptor protein |
| chr3_+_179148341 | 0.20 |
ENST00000263967.4
|
PIK3CA
|
phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha |
| chr9_+_105662457 | 0.19 |
ENST00000334077.6
|
TAL2
|
TAL bHLH transcription factor 2 |
| chr16_-_18896874 | 0.19 |
ENST00000565324.5
ENST00000561947.5 |
SMG1
|
SMG1 nonsense mediated mRNA decay associated PI3K related kinase |
| chr3_+_57890011 | 0.19 |
ENST00000494088.6
ENST00000438794.5 |
SLMAP
|
sarcolemma associated protein |
| chr1_+_162069674 | 0.19 |
ENST00000361897.10
|
NOS1AP
|
nitric oxide synthase 1 adaptor protein |
| chr11_+_64318091 | 0.19 |
ENST00000265462.9
ENST00000352435.8 ENST00000347941.4 |
PRDX5
|
peroxiredoxin 5 |
| chr15_-_34336749 | 0.19 |
ENST00000397707.6
ENST00000560611.5 |
SLC12A6
|
solute carrier family 12 member 6 |
| chr1_+_50105666 | 0.18 |
ENST00000651347.1
|
ELAVL4
|
ELAV like RNA binding protein 4 |
| chr8_+_30387064 | 0.18 |
ENST00000523115.5
ENST00000519647.5 |
RBPMS
|
RNA binding protein, mRNA processing factor |
| chr19_+_54115726 | 0.18 |
ENST00000445811.5
ENST00000321030.9 ENST00000445124.5 ENST00000447810.5 |
PRPF31
|
pre-mRNA processing factor 31 |
| chr9_+_113444725 | 0.18 |
ENST00000374140.6
|
RGS3
|
regulator of G protein signaling 3 |
| chr1_+_35869750 | 0.18 |
ENST00000373206.5
|
AGO1
|
argonaute RISC component 1 |
| chr10_+_59176600 | 0.18 |
ENST00000373880.9
|
PHYHIPL
|
phytanoyl-CoA 2-hydroxylase interacting protein like |
| chr11_+_27055215 | 0.17 |
ENST00000525090.1
|
BBOX1
|
gamma-butyrobetaine hydroxylase 1 |
| chr1_+_224183197 | 0.17 |
ENST00000323699.9
|
DEGS1
|
delta 4-desaturase, sphingolipid 1 |
| chr2_-_85414039 | 0.17 |
ENST00000447219.6
ENST00000409670.5 ENST00000409724.5 |
CAPG
|
capping actin protein, gelsolin like |
| chr11_-_5516690 | 0.17 |
ENST00000380184.2
|
UBQLNL
|
ubiquilin like |
| chr1_+_43389889 | 0.16 |
ENST00000562955.2
ENST00000634258.3 |
SZT2
|
SZT2 subunit of KICSTOR complex |
| chr3_+_128726151 | 0.16 |
ENST00000675342.1
ENST00000493186.6 ENST00000674748.1 ENST00000265062.8 ENST00000482525.5 |
RAB7A
|
RAB7A, member RAS oncogene family |
| chr12_+_54497712 | 0.15 |
ENST00000293373.11
|
NCKAP1L
|
NCK associated protein 1 like |
| chr11_-_16356538 | 0.15 |
ENST00000683767.1
|
SOX6
|
SRY-box transcription factor 6 |
| chr4_+_95051671 | 0.15 |
ENST00000440890.7
|
BMPR1B
|
bone morphogenetic protein receptor type 1B |
| chr17_+_75633420 | 0.14 |
ENST00000375215.3
|
SMIM5
|
small integral membrane protein 5 |
| chr11_-_125680869 | 0.13 |
ENST00000527795.1
|
ACRV1
|
acrosomal vesicle protein 1 |
| chr1_+_239719542 | 0.13 |
ENST00000448020.1
|
CHRM3
|
cholinergic receptor muscarinic 3 |
| chrX_+_55717796 | 0.13 |
ENST00000262850.7
|
RRAGB
|
Ras related GTP binding B |
| chr2_-_206086057 | 0.13 |
ENST00000403263.6
|
INO80D
|
INO80 complex subunit D |
| chr15_-_55196608 | 0.13 |
ENST00000677989.1
ENST00000562895.2 ENST00000569386.2 |
RSL24D1
|
ribosomal L24 domain containing 1 |
| chr3_+_63443076 | 0.13 |
ENST00000295894.9
|
SYNPR
|
synaptoporin |
| chr1_-_27374816 | 0.13 |
ENST00000270879.9
ENST00000354982.2 |
FCN3
|
ficolin 3 |
| chr1_-_217076889 | 0.13 |
ENST00000493748.5
ENST00000463665.5 |
ESRRG
|
estrogen related receptor gamma |
| chr10_+_103245887 | 0.13 |
ENST00000441178.2
|
RPEL1
|
ribulose-5-phosphate-3-epimerase like 1 |
| chr4_-_119322128 | 0.13 |
ENST00000274024.4
|
FABP2
|
fatty acid binding protein 2 |
| chr17_+_17782108 | 0.12 |
ENST00000395774.1
|
RAI1
|
retinoic acid induced 1 |
| chr17_+_28719972 | 0.12 |
ENST00000422514.7
|
RPL23A
|
ribosomal protein L23a |
| chr6_-_130222813 | 0.12 |
ENST00000437477.6
ENST00000439090.7 |
SAMD3
|
sterile alpha motif domain containing 3 |
| chr12_-_8662619 | 0.12 |
ENST00000544889.1
ENST00000543369.5 |
MFAP5
|
microfibril associated protein 5 |
| chr3_+_172039556 | 0.12 |
ENST00000415807.7
ENST00000421757.5 |
FNDC3B
|
fibronectin type III domain containing 3B |
| chr7_+_133253064 | 0.11 |
ENST00000393161.6
ENST00000253861.5 |
EXOC4
|
exocyst complex component 4 |
| chr12_-_8662703 | 0.11 |
ENST00000535336.5
|
MFAP5
|
microfibril associated protein 5 |
| chr4_-_41748713 | 0.11 |
ENST00000226382.4
|
PHOX2B
|
paired like homeobox 2B |
| chr3_-_57199938 | 0.11 |
ENST00000473921.2
ENST00000295934.8 |
HESX1
|
HESX homeobox 1 |
| chr7_+_8433602 | 0.11 |
ENST00000405863.6
|
NXPH1
|
neurexophilin 1 |
| chrX_-_80809854 | 0.11 |
ENST00000373275.5
|
BRWD3
|
bromodomain and WD repeat domain containing 3 |
| chr4_-_122621011 | 0.11 |
ENST00000611104.2
ENST00000648588.1 |
IL21
|
interleukin 21 |
| chr15_+_64136330 | 0.11 |
ENST00000560861.1
|
SNX1
|
sorting nexin 1 |
| chr12_-_102480552 | 0.11 |
ENST00000337514.11
ENST00000307046.8 |
IGF1
|
insulin like growth factor 1 |
| chr3_+_128726130 | 0.11 |
ENST00000676425.1
ENST00000675864.1 |
RAB7A
|
RAB7A, member RAS oncogene family |
| chr1_+_160796157 | 0.11 |
ENST00000263285.11
ENST00000368039.2 |
LY9
|
lymphocyte antigen 9 |
| chr10_-_104232301 | 0.11 |
ENST00000369720.6
ENST00000369719.2 ENST00000278064.7 ENST00000357060.8 |
CFAP43
|
cilia and flagella associated protein 43 |
| chr1_-_152325232 | 0.10 |
ENST00000368799.2
|
FLG
|
filaggrin |
| chr6_-_26123910 | 0.10 |
ENST00000314332.5
ENST00000396984.1 |
H2BC4
|
H2B clustered histone 4 |
| chr16_+_3018390 | 0.10 |
ENST00000573001.5
|
TNFRSF12A
|
TNF receptor superfamily member 12A |
| chr11_-_57649894 | 0.10 |
ENST00000534810.3
ENST00000300022.8 |
YPEL4
|
yippee like 4 |
| chr14_+_36657560 | 0.10 |
ENST00000402703.6
|
PAX9
|
paired box 9 |
| chr20_+_34194569 | 0.10 |
ENST00000568305.5
|
ASIP
|
agouti signaling protein |
| chr17_+_4788926 | 0.10 |
ENST00000331264.8
|
GLTPD2
|
glycolipid transfer protein domain containing 2 |
| chr3_-_149221811 | 0.10 |
ENST00000455472.3
ENST00000264613.11 |
CP
|
ceruloplasmin |
| chr9_+_5890872 | 0.10 |
ENST00000381477.8
ENST00000381476.5 ENST00000381471.1 |
MLANA
|
melan-A |
| chr11_-_57649932 | 0.09 |
ENST00000524669.5
|
YPEL4
|
yippee like 4 |
| chr6_+_33454543 | 0.09 |
ENST00000621915.1
ENST00000395064.3 |
ZBTB9
|
zinc finger and BTB domain containing 9 |
| chr9_+_130693756 | 0.09 |
ENST00000546165.5
ENST00000372352.7 ENST00000372358.10 ENST00000372351.7 ENST00000372350.7 ENST00000495699.2 |
EXOSC2
|
exosome component 2 |
| chr6_+_29457155 | 0.09 |
ENST00000377133.6
ENST00000377136.5 |
OR2H1
|
olfactory receptor family 2 subfamily H member 1 |
| chr7_-_56106444 | 0.09 |
ENST00000395422.4
|
CHCHD2
|
coiled-coil-helix-coiled-coil-helix domain containing 2 |
| chr3_+_49007062 | 0.09 |
ENST00000395474.7
ENST00000610967.4 ENST00000429900.6 |
WDR6
|
WD repeat domain 6 |
| chrX_+_55717733 | 0.08 |
ENST00000414239.5
ENST00000374941.9 |
RRAGB
|
Ras related GTP binding B |
| chr10_+_116621306 | 0.08 |
ENST00000611850.4
ENST00000591655.3 |
PNLIPRP2
|
pancreatic lipase related protein 2 (gene/pseudogene) |
| chr6_-_24666591 | 0.08 |
ENST00000341060.3
|
TDP2
|
tyrosyl-DNA phosphodiesterase 2 |
| chr5_-_64768619 | 0.08 |
ENST00000513458.9
|
SREK1IP1
|
SREK1 interacting protein 1 |
| chr6_-_52763473 | 0.08 |
ENST00000493422.3
|
GSTA2
|
glutathione S-transferase alpha 2 |
| chr7_+_73827737 | 0.08 |
ENST00000435050.1
|
CLDN4
|
claudin 4 |
| chr3_-_109337572 | 0.07 |
ENST00000335658.6
|
DPPA4
|
developmental pluripotency associated 4 |
| chr17_-_28406160 | 0.07 |
ENST00000618626.1
ENST00000612814.5 |
SLC46A1
|
solute carrier family 46 member 1 |
| chr9_-_26892732 | 0.07 |
ENST00000333916.8
ENST00000520187.1 |
CAAP1
|
caspase activity and apoptosis inhibitor 1 |
| chr3_+_51983438 | 0.06 |
ENST00000476351.5
ENST00000476854.5 ENST00000494103.5 ENST00000404366.7 ENST00000635797.1 ENST00000636358.2 ENST00000469863.1 |
ACY1
|
aminoacylase 1 |
| chr11_+_61430100 | 0.06 |
ENST00000542074.1
ENST00000534878.5 ENST00000301761.7 ENST00000537782.5 ENST00000543265.1 |
SDHAF2
|
succinate dehydrogenase complex assembly factor 2 |
| chr2_-_74392025 | 0.06 |
ENST00000440727.1
ENST00000409240.5 |
DCTN1
|
dynactin subunit 1 |
| chr14_+_90397019 | 0.06 |
ENST00000447653.8
ENST00000356978.9 ENST00000626705.2 |
CALM1
|
calmodulin 1 |
| chr15_-_80252205 | 0.06 |
ENST00000560778.3
|
CTXND1
|
cortexin domain containing 1 |
| chrX_-_53422170 | 0.05 |
ENST00000675504.1
|
SMC1A
|
structural maintenance of chromosomes 1A |
| chr18_+_3447562 | 0.05 |
ENST00000618001.4
|
TGIF1
|
TGFB induced factor homeobox 1 |
| chr6_+_31114793 | 0.05 |
ENST00000259881.10
|
PSORS1C1
|
psoriasis susceptibility 1 candidate 1 |
| chr2_-_89160329 | 0.05 |
ENST00000390256.2
|
IGKV6-21
|
immunoglobulin kappa variable 6-21 (non-functional) |
| chr1_-_37554277 | 0.05 |
ENST00000296215.8
|
SNIP1
|
Smad nuclear interacting protein 1 |
| chr13_-_37598750 | 0.05 |
ENST00000379743.8
ENST00000379742.4 ENST00000379749.8 ENST00000379747.9 ENST00000541179.5 ENST00000541481.5 |
POSTN
|
periostin |
| chr13_-_46142834 | 0.05 |
ENST00000674665.1
|
LCP1
|
lymphocyte cytosolic protein 1 |
| chr11_+_120210991 | 0.05 |
ENST00000328965.9
|
OAF
|
out at first homolog |
| chrX_+_80335504 | 0.05 |
ENST00000538312.5
|
TENT5D
|
terminal nucleotidyltransferase 5D |
| chr1_-_179877790 | 0.04 |
ENST00000495650.1
ENST00000367612.7 ENST00000482587.5 ENST00000609928.6 |
TOR1AIP2
|
torsin 1A interacting protein 2 |
| chr19_-_55363243 | 0.04 |
ENST00000424985.3
|
FAM71E2
|
family with sequence similarity 71 member E2 |
| chr8_+_47961028 | 0.04 |
ENST00000650216.1
|
MCM4
|
minichromosome maintenance complex component 4 |
| chr4_-_67883987 | 0.04 |
ENST00000283916.11
|
TMPRSS11D
|
transmembrane serine protease 11D |
| chr6_+_10747753 | 0.04 |
ENST00000612333.4
|
TMEM14B
|
transmembrane protein 14B |
| chr17_-_41184895 | 0.04 |
ENST00000620667.1
ENST00000398472.2 |
KRTAP4-1
|
keratin associated protein 4-1 |
| chr1_-_216805367 | 0.04 |
ENST00000360012.7
|
ESRRG
|
estrogen related receptor gamma |
| chr12_-_68159732 | 0.04 |
ENST00000229135.4
|
IFNG
|
interferon gamma |
| chr4_-_38804783 | 0.04 |
ENST00000308979.7
ENST00000505940.1 ENST00000515861.5 |
TLR1
|
toll like receptor 1 |
| chr6_+_10747765 | 0.04 |
ENST00000461342.5
ENST00000475942.5 ENST00000379542.10 ENST00000379530.7 ENST00000473276.5 ENST00000481240.5 ENST00000467317.5 |
TMEM14B
|
transmembrane protein 14B |
| chr17_-_81947233 | 0.03 |
ENST00000409745.2
|
MYADML2
|
myeloid associated differentiation marker like 2 |
| chr12_+_21526287 | 0.03 |
ENST00000256969.7
|
SPX
|
spexin hormone |
| chr14_+_37795308 | 0.03 |
ENST00000267368.11
ENST00000382320.4 |
TTC6
|
tetratricopeptide repeat domain 6 |
| chr1_+_32274111 | 0.03 |
ENST00000619559.4
ENST00000461712.6 ENST00000373562.7 ENST00000477031.6 ENST00000373557.6 ENST00000333070.4 |
LCK
|
LCK proto-oncogene, Src family tyrosine kinase |
| chrX_+_8464830 | 0.03 |
ENST00000453306.4
ENST00000381032.6 ENST00000444481.3 |
VCX3B
|
variable charge X-linked 3B |
| chr2_-_24123251 | 0.03 |
ENST00000313213.5
ENST00000436622.1 |
PFN4
|
profilin family member 4 |
| chr7_-_101211672 | 0.02 |
ENST00000454310.5
|
PLOD3
|
procollagen-lysine,2-oxoglutarate 5-dioxygenase 3 |
| chr19_+_49363074 | 0.02 |
ENST00000597873.5
|
DKKL1
|
dickkopf like acrosomal protein 1 |
| chr17_+_28719995 | 0.02 |
ENST00000496182.5
|
RPL23A
|
ribosomal protein L23a |
| chr2_+_231056845 | 0.01 |
ENST00000677230.1
ENST00000677259.1 ENST00000677180.1 ENST00000409643.6 ENST00000619128.5 ENST00000678679.1 ENST00000676740.1 ENST00000308696.11 ENST00000440838.5 ENST00000373635.9 |
PSMD1
|
proteasome 26S subunit, non-ATPase 1 |
| chr3_+_26694499 | 0.01 |
ENST00000456208.2
|
LRRC3B
|
leucine rich repeat containing 3B |
| chr19_+_13023958 | 0.01 |
ENST00000587760.5
ENST00000585575.5 |
NFIX
|
nuclear factor I X |
| chr17_-_66229380 | 0.01 |
ENST00000205948.11
|
APOH
|
apolipoprotein H |
| chr4_-_159035226 | 0.01 |
ENST00000434826.3
|
C4orf45
|
chromosome 4 open reading frame 45 |
| chr20_+_37744630 | 0.01 |
ENST00000373473.5
|
CTNNBL1
|
catenin beta like 1 |
| chr2_+_90021567 | 0.01 |
ENST00000436451.2
|
IGKV6D-21
|
immunoglobulin kappa variable 6D-21 (non-functional) |
| chr12_+_27710795 | 0.01 |
ENST00000081029.8
ENST00000538315.5 ENST00000542791.1 |
MRPS35
|
mitochondrial ribosomal protein S35 |
| chr17_+_28720224 | 0.00 |
ENST00000394938.8
ENST00000394935.7 ENST00000355731.8 |
RPL23A
|
ribosomal protein L23a |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 3.8 | GO:0061767 | negative regulation of lung blood pressure(GO:0061767) |
| 0.4 | 2.6 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 0.2 | 1.7 | GO:0043932 | ossification involved in bone remodeling(GO:0043932) |
| 0.2 | 0.6 | GO:0032831 | regulation of CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation(GO:0032829) positive regulation of CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation(GO:0032831) |
| 0.2 | 2.0 | GO:1900625 | regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 0.1 | 0.4 | GO:1903947 | positive regulation of voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization(GO:1903762) positive regulation of ventricular cardiac muscle cell action potential(GO:1903947) positive regulation of membrane repolarization during ventricular cardiac muscle cell action potential(GO:1905026) positive regulation of membrane repolarization during cardiac muscle cell action potential(GO:1905033) |
| 0.1 | 0.4 | GO:1903015 | regulation of exo-alpha-sialidase activity(GO:1903015) |
| 0.1 | 0.8 | GO:0048669 | microglia differentiation(GO:0014004) microglia development(GO:0014005) collateral sprouting in absence of injury(GO:0048669) smooth endoplasmic reticulum calcium ion homeostasis(GO:0051563) cellular response to norepinephrine stimulus(GO:0071874) |
| 0.1 | 0.5 | GO:0038018 | Wnt receptor catabolic process(GO:0038018) |
| 0.1 | 0.3 | GO:1903973 | negative regulation of macrophage colony-stimulating factor signaling pathway(GO:1902227) negative regulation of response to macrophage colony-stimulating factor(GO:1903970) negative regulation of cellular response to macrophage colony-stimulating factor stimulus(GO:1903973) |
| 0.1 | 0.4 | GO:2000863 | positive regulation of estrogen secretion(GO:2000863) positive regulation of estradiol secretion(GO:2000866) |
| 0.1 | 1.4 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.1 | 0.7 | GO:0002353 | kinin cascade(GO:0002254) plasma kallikrein-kinin cascade(GO:0002353) |
| 0.1 | 0.3 | GO:0007174 | epidermal growth factor catabolic process(GO:0007174) |
| 0.1 | 0.8 | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.1 | 0.2 | GO:2000653 | regulation of genetic imprinting(GO:2000653) |
| 0.1 | 0.7 | GO:1903943 | regulation of hepatocyte apoptotic process(GO:1903943) negative regulation of hepatocyte apoptotic process(GO:1903944) |
| 0.1 | 0.5 | GO:0050689 | negative regulation of defense response to virus by host(GO:0050689) |
| 0.1 | 0.3 | GO:0009149 | pyrimidine nucleoside triphosphate catabolic process(GO:0009149) pyrimidine deoxyribonucleoside triphosphate catabolic process(GO:0009213) |
| 0.1 | 0.2 | GO:0043376 | regulation of CD8-positive, alpha-beta T cell differentiation(GO:0043376) |
| 0.0 | 2.4 | GO:0070884 | regulation of calcineurin-NFAT signaling cascade(GO:0070884) |
| 0.0 | 0.2 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.0 | 1.3 | GO:0006346 | methylation-dependent chromatin silencing(GO:0006346) |
| 0.0 | 0.9 | GO:0032740 | positive regulation of interleukin-17 production(GO:0032740) |
| 0.0 | 0.2 | GO:0016480 | negative regulation of transcription from RNA polymerase III promoter(GO:0016480) |
| 0.0 | 0.1 | GO:0003358 | noradrenergic neuron development(GO:0003358) |
| 0.0 | 0.1 | GO:0021780 | oligodendrocyte cell fate specification(GO:0021778) oligodendrocyte cell fate commitment(GO:0021779) glial cell fate specification(GO:0021780) |
| 0.0 | 0.7 | GO:0038180 | nerve growth factor signaling pathway(GO:0038180) |
| 0.0 | 0.3 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.0 | 0.4 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.0 | 0.2 | GO:0071477 | hypotonic salinity response(GO:0042539) cellular hypotonic salinity response(GO:0071477) |
| 0.0 | 0.1 | GO:1902626 | assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.0 | 0.2 | GO:0048254 | snoRNA localization(GO:0048254) |
| 0.0 | 0.2 | GO:1990253 | positive regulation of TORC1 signaling(GO:1904263) cellular response to leucine starvation(GO:1990253) |
| 0.0 | 0.2 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.0 | 0.3 | GO:0051451 | myoblast migration(GO:0051451) |
| 0.0 | 0.2 | GO:0090625 | mRNA cleavage involved in gene silencing by siRNA(GO:0090625) |
| 0.0 | 0.1 | GO:1902731 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) negative regulation of chondrocyte proliferation(GO:1902731) |
| 0.0 | 0.1 | GO:0019376 | galactolipid catabolic process(GO:0019376) |
| 0.0 | 0.1 | GO:0040030 | regulation of molecular function, epigenetic(GO:0040030) |
| 0.0 | 0.0 | GO:0070340 | detection of triacyl bacterial lipopeptide(GO:0042495) detection of bacterial lipopeptide(GO:0070340) |
| 0.0 | 0.1 | GO:0000459 | exonucleolytic trimming involved in rRNA processing(GO:0000459) exonucleolytic trimming to generate mature 3'-end of 5.8S rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000467) CUT catabolic process(GO:0071034) nuclear polyadenylation-dependent tRNA catabolic process(GO:0071038) CUT metabolic process(GO:0071043) |
| 0.0 | 0.4 | GO:0061365 | positive regulation of triglyceride lipase activity(GO:0061365) |
| 0.0 | 0.1 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) |
| 0.0 | 0.1 | GO:0034553 | mitochondrial electron transport, succinate to ubiquinone(GO:0006121) respiratory chain complex II assembly(GO:0034552) mitochondrial respiratory chain complex II assembly(GO:0034553) mitochondrial respiratory chain complex II biogenesis(GO:0097032) |
| 0.0 | 0.1 | GO:0009052 | pentose-phosphate shunt, non-oxidative branch(GO:0009052) pentose catabolic process(GO:0019323) |
| 0.0 | 0.2 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.0 | 0.3 | GO:0051775 | response to redox state(GO:0051775) |
| 0.0 | 0.1 | GO:1901668 | regulation of superoxide dismutase activity(GO:1901668) |
| 0.0 | 0.2 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.0 | 0.0 | GO:1990523 | bone regeneration(GO:1990523) |
| 0.0 | 0.2 | GO:0043306 | positive regulation of mast cell activation involved in immune response(GO:0033008) positive regulation of mast cell degranulation(GO:0043306) |
| 0.0 | 0.3 | GO:0010447 | response to acidic pH(GO:0010447) |
| 0.0 | 0.2 | GO:0060391 | positive regulation of SMAD protein import into nucleus(GO:0060391) |
| 0.0 | 0.5 | GO:0031954 | positive regulation of protein autophosphorylation(GO:0031954) |
| 0.0 | 0.2 | GO:1904706 | negative regulation of vascular smooth muscle cell proliferation(GO:1904706) |
| 0.0 | 0.1 | GO:0051958 | methotrexate transport(GO:0051958) |
| 0.0 | 0.1 | GO:0048341 | paraxial mesoderm formation(GO:0048341) |
| 0.0 | 0.3 | GO:0033141 | positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) |
| 0.0 | 0.2 | GO:0071803 | positive regulation of podosome assembly(GO:0071803) |
| 0.0 | 0.2 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 3.8 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.2 | 2.0 | GO:0035692 | macrophage migration inhibitory factor receptor complex(GO:0035692) |
| 0.1 | 0.6 | GO:1990423 | RZZ complex(GO:1990423) |
| 0.1 | 0.3 | GO:0036195 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 0.1 | 0.3 | GO:0032419 | extrinsic component of lysosome membrane(GO:0032419) |
| 0.1 | 0.8 | GO:1990761 | growth cone lamellipodium(GO:1990761) |
| 0.1 | 0.4 | GO:1902937 | inward rectifier potassium channel complex(GO:1902937) |
| 0.1 | 0.8 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.0 | 0.2 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.0 | 0.2 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.0 | 0.3 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.0 | 0.2 | GO:1990131 | Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.0 | 0.5 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.0 | 0.7 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) |
| 0.0 | 0.3 | GO:0005785 | signal recognition particle receptor complex(GO:0005785) |
| 0.0 | 0.2 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) |
| 0.0 | 0.4 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.1 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.0 | 0.1 | GO:0032279 | asymmetric synapse(GO:0032279) |
| 0.0 | 0.1 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.0 | 0.0 | GO:0035354 | Toll-like receptor 1-Toll-like receptor 2 protein complex(GO:0035354) |
| 0.0 | 0.2 | GO:0005687 | U4 snRNP(GO:0005687) |
| 0.0 | 0.1 | GO:0030905 | retromer, tubulation complex(GO:0030905) |
| 0.0 | 0.2 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.0 | 0.2 | GO:0005685 | U1 snRNP(GO:0005685) |
| 0.0 | 0.2 | GO:0035068 | micro-ribonucleoprotein complex(GO:0035068) |
| 0.0 | 0.1 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.0 | 1.3 | GO:0005581 | collagen trimer(GO:0005581) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 3.8 | GO:0031962 | mineralocorticoid receptor binding(GO:0031962) |
| 0.4 | 2.6 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.3 | 2.5 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.1 | 0.5 | GO:0048030 | disaccharide binding(GO:0048030) |
| 0.1 | 0.8 | GO:0051425 | PTB domain binding(GO:0051425) |
| 0.1 | 0.5 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.1 | 0.4 | GO:0030144 | alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase activity(GO:0030144) |
| 0.1 | 0.2 | GO:0008336 | gamma-butyrobetaine dioxygenase activity(GO:0008336) |
| 0.1 | 0.2 | GO:0042284 | sphingolipid delta-4 desaturase activity(GO:0042284) |
| 0.1 | 0.7 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.0 | 2.2 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 2.0 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.3 | GO:0005157 | macrophage colony-stimulating factor receptor binding(GO:0005157) |
| 0.0 | 0.4 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.0 | 0.5 | GO:0046703 | natural killer cell lectin-like receptor binding(GO:0046703) |
| 0.0 | 0.2 | GO:0034988 | Fc-gamma receptor I complex binding(GO:0034988) |
| 0.0 | 0.1 | GO:0004750 | ribulose-phosphate 3-epimerase activity(GO:0004750) |
| 0.0 | 0.8 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 0.2 | GO:0035614 | snRNA stem-loop binding(GO:0035614) |
| 0.0 | 0.2 | GO:0030622 | U4atac snRNA binding(GO:0030622) |
| 0.0 | 0.3 | GO:0019237 | centromeric DNA binding(GO:0019237) |
| 0.0 | 0.3 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.0 | 1.3 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.4 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.0 | 0.7 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.0 | 0.1 | GO:0008441 | 3'(2'),5'-bisphosphate nucleotidase activity(GO:0008441) |
| 0.0 | 0.1 | GO:0070259 | tyrosyl-DNA phosphodiesterase activity(GO:0070259) |
| 0.0 | 0.3 | GO:0032552 | deoxyribonucleotide binding(GO:0032552) |
| 0.0 | 0.7 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.0 | 0.1 | GO:1990460 | leptin receptor binding(GO:1990460) |
| 0.0 | 0.2 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.0 | 0.4 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.0 | 0.2 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.2 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 0.0 | 0.2 | GO:0005221 | intracellular cyclic nucleotide activated cation channel activity(GO:0005221) cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.0 | 0.1 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.0 | 0.4 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.0 | 0.3 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.0 | 0.2 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 0.3 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.4 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.0 | 0.2 | GO:0022820 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.0 | 0.2 | GO:0015245 | fatty acid transporter activity(GO:0015245) |
| 0.0 | 0.1 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 0.0 | GO:0035663 | Toll-like receptor 2 binding(GO:0035663) |
| 0.0 | 0.1 | GO:0031779 | melanocortin receptor binding(GO:0031779) type 3 melanocortin receptor binding(GO:0031781) type 4 melanocortin receptor binding(GO:0031782) |
| 0.0 | 0.1 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.0 | 0.2 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.0 | 0.1 | GO:0015350 | methotrexate transporter activity(GO:0015350) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.8 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.1 | 2.0 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 1.7 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 2.1 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 0.9 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.0 | 0.7 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 1.0 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 0.3 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 0.4 | PID AVB3 OPN PATHWAY | Osteopontin-mediated events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.8 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.1 | 2.0 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.1 | 2.1 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.0 | 0.8 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 1.1 | REACTOME PD1 SIGNALING | Genes involved in PD-1 signaling |
| 0.0 | 0.7 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.0 | 0.7 | REACTOME ARMS MEDIATED ACTIVATION | Genes involved in ARMS-mediated activation |
| 0.0 | 0.7 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 0.2 | REACTOME OPSINS | Genes involved in Opsins |
| 0.0 | 0.5 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 0.4 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |