Project
Inflammatory response time course, HUVEC (Wada, 2009)
Navigation
Downloads
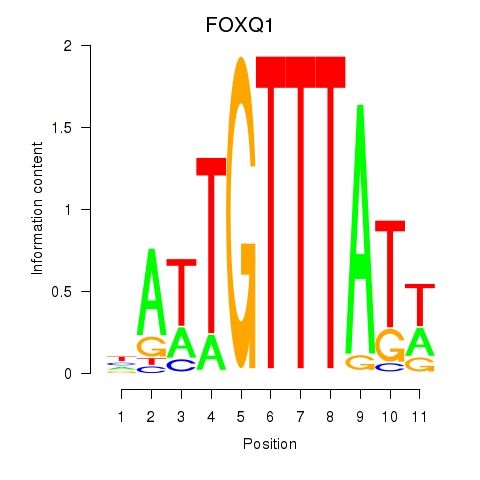
Results for FOXQ1
Z-value: 0.83
Transcription factors associated with FOXQ1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
FOXQ1
|
ENSG00000164379.7 | FOXQ1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| FOXQ1 | hg38_v1_chr6_+_1312090_1312105 | -0.40 | 4.9e-02 | Click! |
Activity profile of FOXQ1 motif
Sorted Z-values of FOXQ1 motif
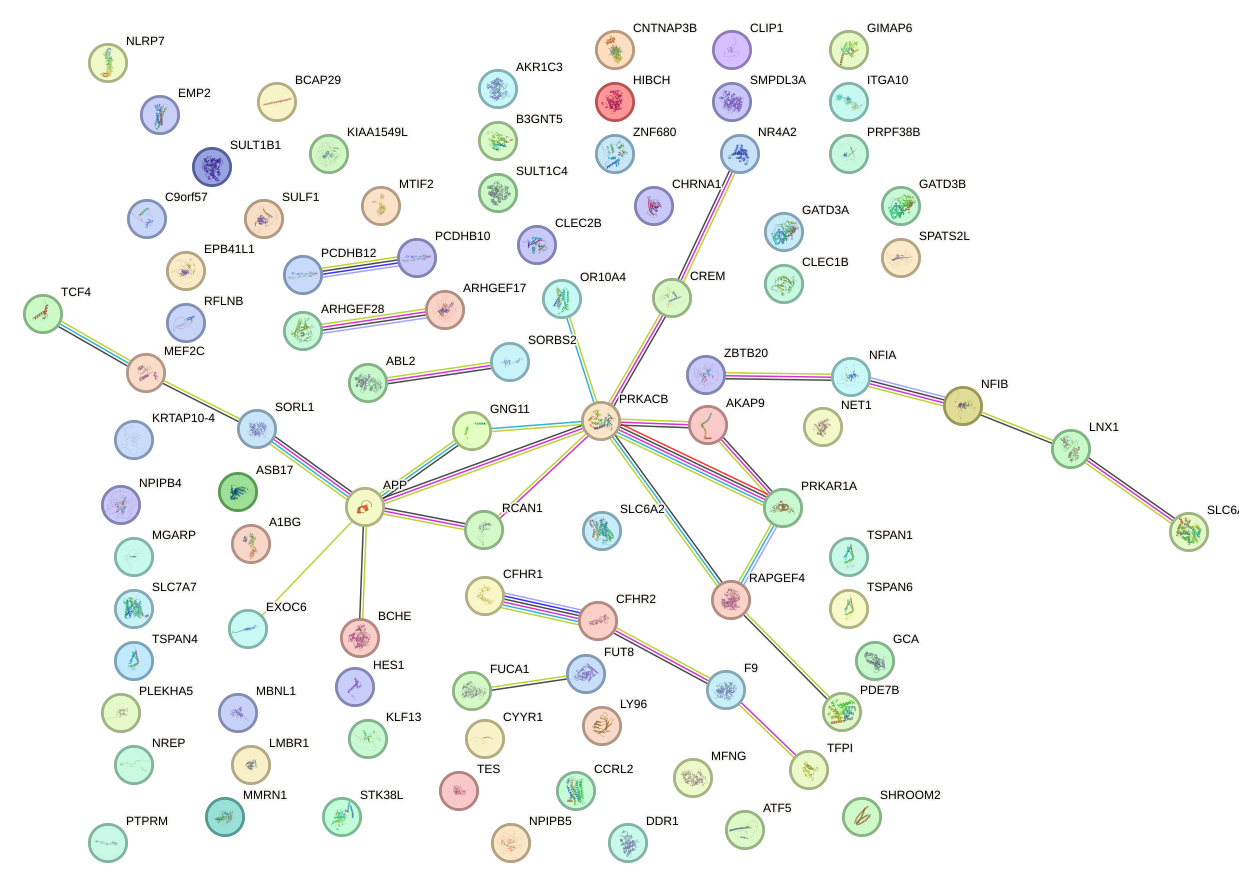
Network of associatons between targets according to the STRING database.
First level regulatory network of FOXQ1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_-_185775271 | 3.90 |
ENST00000430503.5
ENST00000319454.10 ENST00000450341.5 |
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr4_+_41613476 | 3.55 |
ENST00000508466.1
|
LIMCH1
|
LIM and calponin homology domains 1 |
| chr1_+_61077219 | 3.43 |
ENST00000407417.7
|
NFIA
|
nuclear factor I A |
| chr4_-_69760596 | 2.97 |
ENST00000510821.1
|
SULT1B1
|
sulfotransferase family 1B member 1 |
| chr1_+_84164370 | 2.90 |
ENST00000446538.5
ENST00000610703.4 ENST00000370682.7 ENST00000394838.6 ENST00000432111.5 |
PRKACB
|
protein kinase cAMP-activated catalytic subunit beta |
| chr1_+_84164962 | 2.26 |
ENST00000614872.4
ENST00000394839.6 |
PRKACB
|
protein kinase cAMP-activated catalytic subunit beta |
| chr21_-_26573211 | 2.23 |
ENST00000299340.9
ENST00000652641.2 |
CYYR1
|
cysteine and tyrosine rich 1 |
| chr8_-_3409528 | 2.10 |
ENST00000335551.11
|
CSMD1
|
CUB and Sushi multiple domains 1 |
| chr9_-_72060590 | 2.03 |
ENST00000652156.1
|
C9orf57
|
chromosome 9 open reading frame 57 |
| chr2_+_200440649 | 1.95 |
ENST00000366118.2
|
SPATS2L
|
spermatogenesis associated serine rich 2 like |
| chr9_-_72060605 | 1.86 |
ENST00000377024.8
ENST00000651200.2 ENST00000652752.1 |
C9orf57
|
chromosome 9 open reading frame 57 |
| chr4_-_69760610 | 1.82 |
ENST00000310613.8
|
SULT1B1
|
sulfotransferase family 1B member 1 |
| chr4_-_158173042 | 1.77 |
ENST00000592057.1
ENST00000393807.9 |
GASK1B
|
golgi associated kinase 1B |
| chr10_+_35195843 | 1.69 |
ENST00000488741.5
ENST00000474931.5 ENST00000468236.5 ENST00000344351.5 ENST00000490511.1 |
CREM
|
cAMP responsive element modulator |
| chr9_-_41908681 | 1.62 |
ENST00000476961.5
|
CNTNAP3B
|
contactin associated protein family member 3B |
| chr2_-_55269038 | 1.59 |
ENST00000417363.5
ENST00000412530.1 ENST00000366137.6 ENST00000420637.5 |
MTIF2
|
mitochondrial translational initiation factor 2 |
| chr9_-_14300231 | 1.57 |
ENST00000636735.1
|
NFIB
|
nuclear factor I B |
| chr12_+_19205294 | 1.49 |
ENST00000424268.5
|
PLEKHA5
|
pleckstrin homology domain containing A5 |
| chr2_+_108377947 | 1.49 |
ENST00000272452.7
|
SULT1C4
|
sulfotransferase family 1C member 4 |
| chr4_-_158173004 | 1.47 |
ENST00000585682.6
|
GASK1B
|
golgi associated kinase 1B |
| chr3_+_152300135 | 1.46 |
ENST00000465907.6
ENST00000492948.5 ENST00000485509.5 ENST00000464596.5 |
MBNL1
|
muscleblind like splicing regulator 1 |
| chr2_+_162318884 | 1.45 |
ENST00000446271.5
ENST00000429691.6 |
GCA
|
grancalcin |
| chr2_-_156342348 | 1.43 |
ENST00000409572.5
|
NR4A2
|
nuclear receptor subfamily 4 group A member 2 |
| chr1_+_60865259 | 1.33 |
ENST00000371191.5
|
NFIA
|
nuclear factor I A |
| chr1_-_145910031 | 1.32 |
ENST00000369304.8
|
ITGA10
|
integrin subunit alpha 10 |
| chr3_+_46407558 | 1.28 |
ENST00000357392.4
ENST00000400880.3 ENST00000433848.1 |
CCRL2
|
C-C motif chemokine receptor like 2 |
| chr1_-_145910066 | 1.25 |
ENST00000539363.2
|
ITGA10
|
integrin subunit alpha 10 |
| chr21_+_44573724 | 1.20 |
ENST00000622352.3
ENST00000400374.4 ENST00000616689.2 |
KRTAP10-4
|
keratin associated protein 10-4 |
| chr10_+_92834594 | 1.15 |
ENST00000371552.8
|
EXOC6
|
exocyst complex component 6 |
| chr12_+_27244222 | 1.13 |
ENST00000545470.5
ENST00000389032.8 ENST00000540996.5 |
STK38L
|
serine/threonine kinase 38 like |
| chr12_-_9869345 | 1.11 |
ENST00000228438.3
|
CLEC2B
|
C-type lectin domain family 2 member B |
| chr14_-_22819721 | 1.11 |
ENST00000554517.5
ENST00000285850.11 ENST00000397529.6 ENST00000555702.5 |
SLC7A7
|
solute carrier family 7 member 7 |
| chr10_+_35195124 | 1.10 |
ENST00000487763.5
ENST00000473940.5 ENST00000488328.5 ENST00000356917.9 |
CREM
|
cAMP responsive element modulator |
| chrX_+_9912434 | 1.10 |
ENST00000418909.6
|
SHROOM2
|
shroom family member 2 |
| chr20_+_36214373 | 1.04 |
ENST00000432603.1
|
EPB41L1
|
erythrocyte membrane protein band 4.1 like 1 |
| chr5_+_141192330 | 1.02 |
ENST00000239446.6
|
PCDHB10
|
protocadherin beta 10 |
| chr17_+_68525795 | 0.99 |
ENST00000592800.5
|
PRKAR1A
|
protein kinase cAMP-dependent type I regulatory subunit alpha |
| chr22_-_37484505 | 0.95 |
ENST00000442496.1
|
MFNG
|
MFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase |
| chr5_+_73813518 | 0.95 |
ENST00000296799.8
|
ARHGEF28
|
Rho guanine nucleotide exchange factor 28 |
| chr1_+_196819731 | 0.94 |
ENST00000320493.10
ENST00000367424.4 |
CFHR1
|
complement factor H related 1 |
| chr7_+_93921720 | 0.91 |
ENST00000248564.6
|
GNG11
|
G protein subunit gamma 11 |
| chr7_+_92057602 | 0.90 |
ENST00000491695.2
|
AKAP9
|
A-kinase anchoring protein 9 |
| chr7_-_150632333 | 0.90 |
ENST00000493969.2
ENST00000328902.9 |
GIMAP6
|
GTPase, IMAP family member 6 |
| chr2_+_108378176 | 0.90 |
ENST00000409309.3
|
SULT1C4
|
sulfotransferase family 1C member 4 |
| chr10_+_5048748 | 0.90 |
ENST00000602997.5
ENST00000439082.7 |
AKR1C3
|
aldo-keto reductase family 1 member C3 |
| chr3_+_194136138 | 0.88 |
ENST00000232424.4
|
HES1
|
hes family bHLH transcription factor 1 |
| chr2_-_187554473 | 0.86 |
ENST00000453013.5
ENST00000417013.5 |
TFPI
|
tissue factor pathway inhibitor |
| chr2_-_105438503 | 0.85 |
ENST00000393352.7
ENST00000607522.1 |
FHL2
|
four and a half LIM domains 2 |
| chr7_-_156892987 | 0.85 |
ENST00000415428.5
|
LMBR1
|
limb development membrane protein 1 |
| chr3_+_183265302 | 0.84 |
ENST00000465010.1
|
B3GNT5
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 5 |
| chr5_-_111756245 | 0.83 |
ENST00000447165.6
|
NREP
|
neuronal regeneration related protein |
| chr4_-_139280179 | 0.83 |
ENST00000398955.2
|
MGARP
|
mitochondria localized glutamic acid rich protein |
| chr11_+_33376077 | 0.82 |
ENST00000658780.2
|
KIAA1549L
|
KIAA1549 like |
| chr19_+_49930219 | 0.82 |
ENST00000596658.1
|
ATF5
|
activating transcription factor 5 |
| chr4_+_89879532 | 0.78 |
ENST00000394980.5
|
MMRN1
|
multimerin 1 |
| chr17_-_445939 | 0.78 |
ENST00000329099.4
|
RFLNB
|
refilin B |
| chr3_+_184812138 | 0.78 |
ENST00000287546.8
|
VPS8
|
VPS8 subunit of CORVET complex |
| chr3_+_184812159 | 0.77 |
ENST00000625842.3
ENST00000436792.6 ENST00000446204.6 ENST00000422105.5 |
VPS8
|
VPS8 subunit of CORVET complex |
| chr7_+_116222804 | 0.77 |
ENST00000393481.6
|
TES
|
testin LIM domain protein |
| chr2_-_187554351 | 0.77 |
ENST00000437725.5
ENST00000409676.5 ENST00000233156.9 ENST00000339091.8 ENST00000420747.1 |
TFPI
|
tissue factor pathway inhibitor |
| chr3_-_165837412 | 0.77 |
ENST00000479451.5
ENST00000488954.1 ENST00000264381.8 |
BCHE
|
butyrylcholinesterase |
| chr11_+_6876625 | 0.75 |
ENST00000379829.2
|
OR10A4
|
olfactory receptor family 10 subfamily A member 4 |
| chr5_-_88785493 | 0.73 |
ENST00000503554.4
|
MEF2C
|
myocyte enhancer factor 2C |
| chr21_-_26051023 | 0.73 |
ENST00000415997.1
|
APP
|
amyloid beta precursor protein |
| chr3_-_114624193 | 0.68 |
ENST00000481632.5
|
ZBTB20
|
zinc finger and BTB domain containing 20 |
| chr10_+_80413817 | 0.67 |
ENST00000372187.9
|
PRXL2A
|
peroxiredoxin like 2A |
| chr14_+_65411845 | 0.66 |
ENST00000556518.5
ENST00000557164.5 |
FUT8
|
fucosyltransferase 8 |
| chrX_+_139530730 | 0.66 |
ENST00000218099.7
|
F9
|
coagulation factor IX |
| chr1_+_196943738 | 0.66 |
ENST00000367415.8
ENST00000367421.5 ENST00000649283.1 ENST00000476712.6 ENST00000496448.6 ENST00000473386.1 ENST00000649960.1 |
CFHR2
|
complement factor H related 2 |
| chr7_-_64563032 | 0.66 |
ENST00000447137.2
|
ZNF680
|
zinc finger protein 680 |
| chr9_-_13279407 | 0.65 |
ENST00000546205.5
|
MPDZ
|
multiple PDZ domain crumbs cell polarity complex component |
| chr9_-_123268538 | 0.64 |
ENST00000360998.3
ENST00000348403.10 |
STRBP
|
spermatid perinuclear RNA binding protein |
| chr16_-_10559135 | 0.63 |
ENST00000536829.1
|
EMP2
|
epithelial membrane protein 2 |
| chr16_+_22505845 | 0.62 |
ENST00000356156.7
|
NPIPB5
|
nuclear pore complex interacting protein family member B5 |
| chr19_-_58353482 | 0.62 |
ENST00000263100.8
|
A1BG
|
alpha-1-B glycoprotein |
| chr5_-_88823763 | 0.62 |
ENST00000635898.1
ENST00000626391.2 ENST00000628656.2 |
MEF2C
|
myocyte enhancer factor 2C |
| chr11_+_73308237 | 0.62 |
ENST00000263674.4
|
ARHGEF17
|
Rho guanine nucleotide exchange factor 17 |
| chr18_-_55589770 | 0.61 |
ENST00000565018.6
ENST00000636400.2 |
TCF4
|
transcription factor 4 |
| chr21_-_34526815 | 0.61 |
ENST00000492600.1
|
RCAN1
|
regulator of calcineurin 1 |
| chr18_-_55589836 | 0.61 |
ENST00000537578.5
ENST00000564403.6 |
TCF4
|
transcription factor 4 |
| chr9_-_123184233 | 0.61 |
ENST00000447404.6
|
STRBP
|
spermatid perinuclear RNA binding protein |
| chr8_+_69492793 | 0.60 |
ENST00000616868.1
ENST00000419716.7 ENST00000402687.9 |
SULF1
|
sulfatase 1 |
| chr6_+_30888672 | 0.60 |
ENST00000446312.5
|
DDR1
|
discoidin domain receptor tyrosine kinase 1 |
| chr1_+_84164684 | 0.58 |
ENST00000370680.5
|
PRKACB
|
protein kinase cAMP-activated catalytic subunit beta |
| chr19_-_54947492 | 0.58 |
ENST00000592784.5
ENST00000340844.6 |
NLRP7
|
NLR family pyrin domain containing 7 |
| chr14_+_22516273 | 0.58 |
ENST00000390510.1
|
TRAJ27
|
T cell receptor alpha joining 27 |
| chr2_+_172860038 | 0.57 |
ENST00000538974.5
ENST00000540783.5 |
RAPGEF4
|
Rap guanine nucleotide exchange factor 4 |
| chr6_+_135851681 | 0.57 |
ENST00000308191.11
|
PDE7B
|
phosphodiesterase 7B |
| chr9_-_13279564 | 0.56 |
ENST00000541718.5
ENST00000447879.6 |
MPDZ
|
multiple PDZ domain crumbs cell polarity complex component |
| chr6_-_130956371 | 0.56 |
ENST00000639623.1
ENST00000525193.5 ENST00000527659.5 |
EPB41L2
|
erythrocyte membrane protein band 4.1 like 2 |
| chr3_-_185821092 | 0.56 |
ENST00000421047.3
|
IGF2BP2
|
insulin like growth factor 2 mRNA binding protein 2 |
| chr2_-_174764407 | 0.56 |
ENST00000409219.5
ENST00000409542.5 |
CHRNA1
|
cholinergic receptor nicotinic alpha 1 subunit |
| chr2_-_216372432 | 0.56 |
ENST00000273067.5
|
MARCHF4
|
membrane associated ring-CH-type finger 4 |
| chr9_-_13279642 | 0.56 |
ENST00000319217.12
|
MPDZ
|
multiple PDZ domain crumbs cell polarity complex component |
| chr10_+_5412542 | 0.56 |
ENST00000355029.9
|
NET1
|
neuroepithelial cell transforming 1 |
| chr2_+_108621260 | 0.55 |
ENST00000409441.5
|
LIMS1
|
LIM zinc finger domain containing 1 |
| chr18_-_55589795 | 0.55 |
ENST00000568740.5
ENST00000629387.2 |
TCF4
|
transcription factor 4 |
| chr16_+_22490337 | 0.55 |
ENST00000415833.6
|
NPIPB5
|
nuclear pore complex interacting protein family member B5 |
| chr5_+_141208697 | 0.55 |
ENST00000624949.1
ENST00000622978.1 ENST00000239450.4 |
PCDHB12
|
protocadherin beta 12 |
| chr7_+_107583919 | 0.54 |
ENST00000491150.5
|
BCAP29
|
B cell receptor associated protein 29 |
| chrX_-_100636799 | 0.54 |
ENST00000373020.9
|
TSPAN6
|
tetraspanin 6 |
| chr6_+_122789197 | 0.54 |
ENST00000368440.5
|
SMPDL3A
|
sphingomyelin phosphodiesterase acid like 3A |
| chr2_-_190319845 | 0.54 |
ENST00000392332.7
|
HIBCH
|
3-hydroxyisobutyryl-CoA hydrolase |
| chr18_+_7754959 | 0.51 |
ENST00000400053.8
|
PTPRM
|
protein tyrosine phosphatase receptor type M |
| chr12_-_122395422 | 0.51 |
ENST00000540304.6
|
CLIP1
|
CAP-Gly domain containing linker protein 1 |
| chr11_+_121576760 | 0.50 |
ENST00000532694.5
ENST00000534286.5 |
SORL1
|
sortilin related receptor 1 |
| chr10_+_5446601 | 0.50 |
ENST00000449083.5
ENST00000380359.3 |
NET1
|
neuroepithelial cell transforming 1 |
| chr1_-_75932392 | 0.49 |
ENST00000284142.7
|
ASB17
|
ankyrin repeat and SOCS box containing 17 |
| chr8_+_73991345 | 0.49 |
ENST00000284818.7
ENST00000518893.1 |
LY96
|
lymphocyte antigen 96 |
| chr11_+_844406 | 0.49 |
ENST00000397404.5
|
TSPAN4
|
tetraspanin 4 |
| chr1_-_179143044 | 0.49 |
ENST00000504405.5
ENST00000512653.5 ENST00000344730.7 |
ABL2
|
ABL proto-oncogene 2, non-receptor tyrosine kinase |
| chr15_+_31366138 | 0.48 |
ENST00000558844.1
|
KLF13
|
Kruppel like factor 13 |
| chr1_-_23868279 | 0.48 |
ENST00000374479.4
|
FUCA1
|
alpha-L-fucosidase 1 |
| chr3_-_114759115 | 0.48 |
ENST00000471418.5
|
ZBTB20
|
zinc finger and BTB domain containing 20 |
| chr6_+_156776020 | 0.47 |
ENST00000346085.10
|
ARID1B
|
AT-rich interaction domain 1B |
| chr21_-_34526850 | 0.47 |
ENST00000481448.5
ENST00000381132.6 |
RCAN1
|
regulator of calcineurin 1 |
| chr7_+_135148041 | 0.45 |
ENST00000275767.3
|
TMEM140
|
transmembrane protein 140 |
| chr1_+_162632454 | 0.45 |
ENST00000367921.8
ENST00000367922.7 |
DDR2
|
discoidin domain receptor tyrosine kinase 2 |
| chr3_+_136930469 | 0.45 |
ENST00000469404.1
ENST00000467911.1 |
NCK1
|
NCK adaptor protein 1 |
| chr1_+_213988501 | 0.44 |
ENST00000261454.8
ENST00000435016.2 |
PROX1
|
prospero homeobox 1 |
| chr2_+_201183120 | 0.42 |
ENST00000272879.9
ENST00000286186.11 ENST00000374650.7 ENST00000346817.9 ENST00000313728.11 ENST00000448480.1 |
CASP10
|
caspase 10 |
| chr5_+_73626158 | 0.41 |
ENST00000296794.10
ENST00000545377.5 ENST00000509848.5 ENST00000513042.7 |
ARHGEF28
|
Rho guanine nucleotide exchange factor 28 |
| chr6_+_38715288 | 0.41 |
ENST00000327475.11
|
DNAH8
|
dynein axonemal heavy chain 8 |
| chr4_+_85475167 | 0.41 |
ENST00000503995.5
|
ARHGAP24
|
Rho GTPase activating protein 24 |
| chrX_+_10158448 | 0.41 |
ENST00000380829.5
ENST00000421085.7 ENST00000674669.1 ENST00000454850.1 |
CLCN4
|
chloride voltage-gated channel 4 |
| chr10_+_114938187 | 0.40 |
ENST00000298746.5
|
TRUB1
|
TruB pseudouridine synthase family member 1 |
| chr9_-_20382461 | 0.40 |
ENST00000380321.5
ENST00000629733.3 |
MLLT3
|
MLLT3 super elongation complex subunit |
| chr1_+_63523490 | 0.40 |
ENST00000371088.5
|
EFCAB7
|
EF-hand calcium binding domain 7 |
| chr5_+_141199555 | 0.40 |
ENST00000624887.1
ENST00000354757.5 |
PCDHB11
|
protocadherin beta 11 |
| chr11_+_7088991 | 0.40 |
ENST00000306904.7
|
RBMXL2
|
RBMX like 2 |
| chr2_+_172928165 | 0.40 |
ENST00000535187.5
|
RAPGEF4
|
Rap guanine nucleotide exchange factor 4 |
| chr4_+_85827745 | 0.40 |
ENST00000509300.5
|
ARHGAP24
|
Rho GTPase activating protein 24 |
| chr17_-_48613468 | 0.39 |
ENST00000498634.2
|
HOXB8
|
homeobox B8 |
| chr4_+_85827891 | 0.39 |
ENST00000514229.5
|
ARHGAP24
|
Rho GTPase activating protein 24 |
| chr7_+_143288215 | 0.39 |
ENST00000619992.4
ENST00000310447.10 |
CASP2
|
caspase 2 |
| chr13_-_77919390 | 0.39 |
ENST00000475537.2
ENST00000646605.1 |
EDNRB
|
endothelin receptor type B |
| chr18_+_63777773 | 0.39 |
ENST00000447428.5
ENST00000546027.5 |
SERPINB7
|
serpin family B member 7 |
| chr12_-_102197827 | 0.39 |
ENST00000329406.5
|
PMCH
|
pro-melanin concentrating hormone |
| chr20_-_63956382 | 0.39 |
ENST00000358711.7
ENST00000354216.11 |
UCKL1
|
uridine-cytidine kinase 1 like 1 |
| chr3_+_159273235 | 0.38 |
ENST00000638749.1
|
IQCJ-SCHIP1
|
IQCJ-SCHIP1 readthrough |
| chr17_+_35844077 | 0.38 |
ENST00000604694.1
|
TAF15
|
TATA-box binding protein associated factor 15 |
| chr20_-_61998132 | 0.38 |
ENST00000474089.5
|
TAF4
|
TATA-box binding protein associated factor 4 |
| chr12_-_10388976 | 0.37 |
ENST00000540818.5
|
KLRK1
|
killer cell lectin like receptor K1 |
| chr7_+_80638662 | 0.37 |
ENST00000394788.7
|
CD36
|
CD36 molecule |
| chr2_-_174764436 | 0.37 |
ENST00000409323.1
ENST00000261007.9 ENST00000348749.9 ENST00000672640.1 |
CHRNA1
|
cholinergic receptor nicotinic alpha 1 subunit |
| chr1_-_93614091 | 0.37 |
ENST00000370247.7
|
BCAR3
|
BCAR3 adaptor protein, NSP family member |
| chr12_-_47079859 | 0.37 |
ENST00000266581.4
|
AMIGO2
|
adhesion molecule with Ig like domain 2 |
| chrX_+_106611930 | 0.36 |
ENST00000372544.6
ENST00000372548.9 |
RADX
|
RPA1 related single stranded DNA binding protein, X-linked |
| chr14_+_30577752 | 0.36 |
ENST00000547532.5
ENST00000555429.1 |
G2E3
|
G2/M-phase specific E3 ubiquitin protein ligase |
| chr4_-_110198579 | 0.36 |
ENST00000302274.8
|
ELOVL6
|
ELOVL fatty acid elongase 6 |
| chr8_+_74339566 | 0.36 |
ENST00000675376.1
|
GDAP1
|
ganglioside induced differentiation associated protein 1 |
| chr18_-_12656716 | 0.36 |
ENST00000462226.1
ENST00000497844.6 ENST00000309836.9 ENST00000453447.6 |
SPIRE1
|
spire type actin nucleation factor 1 |
| chr1_-_216805367 | 0.36 |
ENST00000360012.7
|
ESRRG
|
estrogen related receptor gamma |
| chr3_-_134250831 | 0.36 |
ENST00000623711.4
|
RYK
|
receptor like tyrosine kinase |
| chr9_-_96383675 | 0.35 |
ENST00000375257.2
ENST00000375259.9 ENST00000253270.13 |
SLC35D2
|
solute carrier family 35 member D2 |
| chr1_+_220748297 | 0.34 |
ENST00000366913.8
|
MTARC2
|
mitochondrial amidoxime reducing component 2 |
| chr6_+_87472925 | 0.34 |
ENST00000369556.7
ENST00000369557.9 ENST00000369552.9 |
SLC35A1
|
solute carrier family 35 member A1 |
| chr3_+_172754457 | 0.34 |
ENST00000441497.6
|
ECT2
|
epithelial cell transforming 2 |
| chr1_-_53940100 | 0.34 |
ENST00000371376.1
|
HSPB11
|
heat shock protein family B (small) member 11 |
| chr10_+_68560317 | 0.34 |
ENST00000373644.5
|
TET1
|
tet methylcytosine dioxygenase 1 |
| chr7_-_156893150 | 0.33 |
ENST00000353442.10
|
LMBR1
|
limb development membrane protein 1 |
| chr4_-_110198650 | 0.33 |
ENST00000394607.7
|
ELOVL6
|
ELOVL fatty acid elongase 6 |
| chr6_+_41637005 | 0.33 |
ENST00000419164.6
ENST00000373051.6 |
MDFI
|
MyoD family inhibitor |
| chr10_+_21524627 | 0.33 |
ENST00000651097.1
|
MLLT10
|
MLLT10 histone lysine methyltransferase DOT1L cofactor |
| chr7_-_139716980 | 0.33 |
ENST00000342645.7
|
HIPK2
|
homeodomain interacting protein kinase 2 |
| chr15_-_55365231 | 0.32 |
ENST00000568543.1
|
CCPG1
|
cell cycle progression 1 |
| chr11_+_65027402 | 0.32 |
ENST00000377244.8
ENST00000534637.5 ENST00000524831.5 |
SNX15
|
sorting nexin 15 |
| chr17_+_30970984 | 0.32 |
ENST00000443677.6
ENST00000324689.8 ENST00000328381.10 ENST00000535306.6 ENST00000580444.2 |
RNF135
|
ring finger protein 135 |
| chr11_+_112175526 | 0.32 |
ENST00000532612.5
ENST00000438022.5 |
BCO2
|
beta-carotene oxygenase 2 |
| chr3_+_107377433 | 0.31 |
ENST00000261058.3
|
CCDC54
|
coiled-coil domain containing 54 |
| chr10_+_70404129 | 0.31 |
ENST00000373218.5
|
EIF4EBP2
|
eukaryotic translation initiation factor 4E binding protein 2 |
| chr12_-_8650529 | 0.31 |
ENST00000543467.5
|
MFAP5
|
microfibril associated protein 5 |
| chr15_+_41256907 | 0.31 |
ENST00000560965.1
|
CHP1
|
calcineurin like EF-hand protein 1 |
| chr4_-_139302460 | 0.31 |
ENST00000394223.2
ENST00000676245.1 |
NDUFC1
|
NADH:ubiquinone oxidoreductase subunit C1 |
| chr10_+_21524670 | 0.30 |
ENST00000631589.1
|
MLLT10
|
MLLT10 histone lysine methyltransferase DOT1L cofactor |
| chr17_+_19282979 | 0.30 |
ENST00000395626.5
ENST00000571254.1 |
EPN2
|
epsin 2 |
| chr14_-_106639589 | 0.30 |
ENST00000390630.3
|
IGHV4-61
|
immunoglobulin heavy variable 4-61 |
| chr18_-_33136075 | 0.30 |
ENST00000581852.5
|
CCDC178
|
coiled-coil domain containing 178 |
| chr2_-_8837589 | 0.29 |
ENST00000319688.5
ENST00000489024.5 ENST00000256707.8 ENST00000473731.5 |
KIDINS220
|
kinase D interacting substrate 220 |
| chr1_+_95151377 | 0.29 |
ENST00000604203.1
|
TLCD4-RWDD3
|
TLCD4-RWDD3 readthrough |
| chr4_+_39182497 | 0.28 |
ENST00000509560.5
ENST00000512112.5 ENST00000399820.8 ENST00000506503.1 |
WDR19
|
WD repeat domain 19 |
| chr19_-_9219589 | 0.28 |
ENST00000641244.1
ENST00000641669.1 |
OR7D4
|
olfactory receptor family 7 subfamily D member 4 |
| chr6_+_116369837 | 0.27 |
ENST00000645988.1
|
DSE
|
dermatan sulfate epimerase |
| chr11_-_107457801 | 0.27 |
ENST00000282251.9
|
CWF19L2
|
CWF19 like cell cycle control factor 2 |
| chr7_+_1688119 | 0.27 |
ENST00000424383.4
|
ELFN1
|
extracellular leucine rich repeat and fibronectin type III domain containing 1 |
| chr6_+_42929127 | 0.26 |
ENST00000394142.7
|
CNPY3
|
canopy FGF signaling regulator 3 |
| chr1_+_151766655 | 0.26 |
ENST00000400999.7
|
OAZ3
|
ornithine decarboxylase antizyme 3 |
| chr5_+_55853464 | 0.26 |
ENST00000652039.1
|
IL31RA
|
interleukin 31 receptor A |
| chr11_+_6926417 | 0.26 |
ENST00000610573.4
ENST00000278319.10 |
ZNF215
|
zinc finger protein 215 |
| chr5_+_119476530 | 0.26 |
ENST00000645099.1
ENST00000513628.5 |
HSD17B4
|
hydroxysteroid 17-beta dehydrogenase 4 |
| chr7_+_134843884 | 0.25 |
ENST00000445569.6
|
CALD1
|
caldesmon 1 |
| chr18_-_24272179 | 0.25 |
ENST00000399443.7
|
OSBPL1A
|
oxysterol binding protein like 1A |
| chr2_+_206275002 | 0.25 |
ENST00000650097.1
ENST00000649525.1 ENST00000649998.1 |
ZDBF2
|
zinc finger DBF-type containing 2 |
| chr15_+_42404842 | 0.25 |
ENST00000673928.1
|
CAPN3
|
calpain 3 |
| chr2_+_234050679 | 0.25 |
ENST00000373368.5
ENST00000168148.8 |
SPP2
|
secreted phosphoprotein 2 |
| chr12_-_120325936 | 0.25 |
ENST00000549767.1
|
PLA2G1B
|
phospholipase A2 group IB |
| chr6_-_116829037 | 0.24 |
ENST00000368549.7
ENST00000530250.1 ENST00000310357.8 |
GPRC6A
|
G protein-coupled receptor class C group 6 member A |
| chr11_+_112176364 | 0.24 |
ENST00000526088.5
ENST00000532593.5 ENST00000531169.5 |
BCO2
|
beta-carotene oxygenase 2 |
| chr5_+_141245384 | 0.24 |
ENST00000623671.1
ENST00000231173.6 |
PCDHB15
|
protocadherin beta 15 |
| chr2_-_144517663 | 0.23 |
ENST00000427902.5
ENST00000462355.2 ENST00000470879.5 ENST00000409487.7 ENST00000435831.5 ENST00000630572.2 |
ZEB2
|
zinc finger E-box binding homeobox 2 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 5.7 | GO:0097338 | response to clozapine(GO:0097338) |
| 0.5 | 1.6 | GO:0070124 | mitochondrial translational initiation(GO:0070124) |
| 0.5 | 1.4 | GO:0021538 | epithalamus development(GO:0021538) habenula development(GO:0021986) |
| 0.4 | 0.8 | GO:0014016 | neuroblast differentiation(GO:0014016) |
| 0.3 | 0.9 | GO:0006714 | sesquiterpenoid metabolic process(GO:0006714) sesquiterpenoid catabolic process(GO:0016107) farnesol metabolic process(GO:0016487) farnesol catabolic process(GO:0016488) regulation of testosterone biosynthetic process(GO:2000224) |
| 0.3 | 0.9 | GO:0021558 | trochlear nerve development(GO:0021558) auditory receptor cell fate determination(GO:0042668) negative regulation of auditory receptor cell differentiation(GO:0045608) regulation of timing of neuron differentiation(GO:0060164) negative regulation of pro-B cell differentiation(GO:2000974) |
| 0.3 | 0.8 | GO:0061182 | negative regulation of chondrocyte development(GO:0061182) |
| 0.3 | 6.5 | GO:0006068 | ethanol catabolic process(GO:0006068) |
| 0.3 | 2.3 | GO:0007598 | blood coagulation, extrinsic pathway(GO:0007598) |
| 0.2 | 0.7 | GO:0036071 | N-glycan fucosylation(GO:0036071) |
| 0.2 | 0.8 | GO:0055014 | atrial cardiac muscle cell differentiation(GO:0055011) atrial cardiac muscle cell development(GO:0055014) |
| 0.2 | 0.6 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) |
| 0.2 | 0.6 | GO:0046247 | tetraterpenoid metabolic process(GO:0016108) carotenoid metabolic process(GO:0016116) carotene catabolic process(GO:0016121) xanthophyll metabolic process(GO:0016122) terpene catabolic process(GO:0046247) |
| 0.2 | 1.3 | GO:0003172 | primary heart field specification(GO:0003138) sinoatrial valve development(GO:0003172) sinoatrial valve morphogenesis(GO:0003185) |
| 0.2 | 0.5 | GO:1902769 | regulation of choline O-acetyltransferase activity(GO:1902769) positive regulation of choline O-acetyltransferase activity(GO:1902771) negative regulation of tau-protein kinase activity(GO:1902948) positive regulation of early endosome to recycling endosome transport(GO:1902955) negative regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902960) negative regulation of neurofibrillary tangle assembly(GO:1902997) negative regulation of aspartic-type peptidase activity(GO:1905246) |
| 0.2 | 0.8 | GO:0097210 | response to gonadotropin-releasing hormone(GO:0097210) cellular response to gonadotropin-releasing hormone(GO:0097211) |
| 0.2 | 1.1 | GO:0000821 | regulation of arginine metabolic process(GO:0000821) |
| 0.1 | 0.4 | GO:1903676 | regulation of cap-dependent translational initiation(GO:1903674) positive regulation of cap-dependent translational initiation(GO:1903676) |
| 0.1 | 0.4 | GO:0002194 | hepatocyte cell migration(GO:0002194) otic placode formation(GO:0043049) branching involved in pancreas morphogenesis(GO:0061114) acinar cell differentiation(GO:0090425) positive regulation of forebrain neuron differentiation(GO:2000979) |
| 0.1 | 1.6 | GO:2000795 | negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.1 | 4.5 | GO:0072189 | ureter development(GO:0072189) |
| 0.1 | 0.4 | GO:0001554 | luteolysis(GO:0001554) |
| 0.1 | 1.5 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.1 | 0.5 | GO:0044861 | protein transport into plasma membrane raft(GO:0044861) |
| 0.1 | 0.4 | GO:2000502 | negative regulation of natural killer cell chemotaxis(GO:2000502) |
| 0.1 | 0.7 | GO:0071874 | microglia differentiation(GO:0014004) microglia development(GO:0014005) collateral sprouting in absence of injury(GO:0048669) smooth endoplasmic reticulum calcium ion homeostasis(GO:0051563) cellular response to norepinephrine stimulus(GO:0071874) |
| 0.1 | 0.6 | GO:0035645 | posterior midgut development(GO:0007497) enteric smooth muscle cell differentiation(GO:0035645) |
| 0.1 | 0.9 | GO:0048630 | skeletal muscle tissue growth(GO:0048630) |
| 0.1 | 3.9 | GO:0061049 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.1 | 0.5 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.1 | 0.5 | GO:0006574 | valine catabolic process(GO:0006574) |
| 0.1 | 0.4 | GO:0036518 | chemorepulsion of dopaminergic neuron axon(GO:0036518) |
| 0.1 | 0.3 | GO:0015847 | putrescine transport(GO:0015847) |
| 0.1 | 0.3 | GO:0036111 | very long-chain fatty-acyl-CoA metabolic process(GO:0036111) |
| 0.1 | 1.1 | GO:0045176 | apical protein localization(GO:0045176) |
| 0.1 | 0.4 | GO:0090362 | positive regulation of platelet-derived growth factor production(GO:0090362) |
| 0.1 | 0.3 | GO:0070885 | negative regulation of calcineurin-NFAT signaling cascade(GO:0070885) |
| 0.1 | 0.4 | GO:1901350 | cell-cell signaling involved in cell-cell junction organization(GO:1901350) |
| 0.1 | 0.2 | GO:0090310 | negative regulation of methylation-dependent chromatin silencing(GO:0090310) |
| 0.1 | 0.2 | GO:0000964 | mitochondrial RNA 5'-end processing(GO:0000964) |
| 0.1 | 0.2 | GO:1904845 | response to L-glutamine(GO:1904844) cellular response to L-glutamine(GO:1904845) |
| 0.1 | 0.6 | GO:0070836 | caveola assembly(GO:0070836) |
| 0.1 | 0.5 | GO:0032497 | detection of lipopolysaccharide(GO:0032497) |
| 0.1 | 0.9 | GO:0098909 | regulation of cardiac muscle cell action potential involved in regulation of contraction(GO:0098909) |
| 0.1 | 0.6 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.1 | 1.0 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.1 | 0.2 | GO:0035526 | retrograde transport, plasma membrane to Golgi(GO:0035526) |
| 0.1 | 0.7 | GO:0019368 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.1 | 1.0 | GO:0002315 | marginal zone B cell differentiation(GO:0002315) |
| 0.1 | 0.5 | GO:0097026 | dendritic cell dendrite assembly(GO:0097026) |
| 0.1 | 0.4 | GO:1990481 | mRNA pseudouridine synthesis(GO:1990481) |
| 0.1 | 0.7 | GO:0015781 | pyrimidine nucleotide-sugar transport(GO:0015781) |
| 0.1 | 0.2 | GO:0044313 | protein K6-linked deubiquitination(GO:0044313) |
| 0.1 | 0.3 | GO:0060830 | ciliary receptor clustering involved in smoothened signaling pathway(GO:0060830) |
| 0.1 | 0.8 | GO:0051451 | myoblast migration(GO:0051451) |
| 0.1 | 0.1 | GO:1990535 | neuron projection maintenance(GO:1990535) |
| 0.1 | 0.4 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.1 | 0.8 | GO:0021891 | olfactory bulb interneuron development(GO:0021891) |
| 0.0 | 0.2 | GO:0044240 | multicellular organism lipid catabolic process(GO:0044240) |
| 0.0 | 0.2 | GO:0090131 | mesenchyme migration(GO:0090131) |
| 0.0 | 3.2 | GO:0048384 | retinoic acid receptor signaling pathway(GO:0048384) |
| 0.0 | 0.4 | GO:0044211 | CTP salvage(GO:0044211) |
| 0.0 | 0.5 | GO:0016139 | glycoside catabolic process(GO:0016139) |
| 0.0 | 0.1 | GO:0050787 | antibiotic metabolic process(GO:0016999) detoxification of mercury ion(GO:0050787) |
| 0.0 | 0.1 | GO:0036451 | cap mRNA methylation(GO:0036451) |
| 0.0 | 0.1 | GO:0035262 | gonad morphogenesis(GO:0035262) |
| 0.0 | 0.2 | GO:1904977 | lymphatic endothelial cell migration(GO:1904977) |
| 0.0 | 0.4 | GO:0072564 | response to linoleic acid(GO:0070543) blood microparticle formation(GO:0072564) regulation of blood microparticle formation(GO:2000332) positive regulation of blood microparticle formation(GO:2000334) |
| 0.0 | 0.5 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.0 | 0.2 | GO:1990637 | response to prolactin(GO:1990637) |
| 0.0 | 0.5 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.0 | 0.2 | GO:0044314 | protein K27-linked ubiquitination(GO:0044314) |
| 0.0 | 0.4 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) |
| 0.0 | 0.6 | GO:1901223 | negative regulation of NIK/NF-kappaB signaling(GO:1901223) |
| 0.0 | 0.3 | GO:0030578 | PML body organization(GO:0030578) |
| 0.0 | 0.3 | GO:2001106 | regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.0 | 0.2 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.0 | 0.2 | GO:0006196 | AMP catabolic process(GO:0006196) |
| 0.0 | 0.2 | GO:0009624 | response to nematode(GO:0009624) |
| 0.0 | 0.1 | GO:0060723 | spongiotrophoblast cell proliferation(GO:0060720) regulation of spongiotrophoblast cell proliferation(GO:0060721) cell proliferation involved in embryonic placenta development(GO:0060722) regulation of cell proliferation involved in embryonic placenta development(GO:0060723) |
| 0.0 | 1.1 | GO:0070884 | regulation of calcineurin-NFAT signaling cascade(GO:0070884) |
| 0.0 | 0.3 | GO:0070649 | formin-nucleated actin cable assembly(GO:0070649) |
| 0.0 | 0.2 | GO:1902748 | positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.0 | 1.3 | GO:0007638 | mechanosensory behavior(GO:0007638) |
| 0.0 | 0.4 | GO:2000096 | segment specification(GO:0007379) positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.0 | 0.1 | GO:2000653 | regulation of genetic imprinting(GO:2000653) |
| 0.0 | 0.6 | GO:1904776 | regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.0 | 0.2 | GO:0035093 | spermatogenesis, exchange of chromosomal proteins(GO:0035093) |
| 0.0 | 0.2 | GO:0014718 | positive regulation of satellite cell activation involved in skeletal muscle regeneration(GO:0014718) |
| 0.0 | 0.1 | GO:0014057 | positive regulation of acetylcholine secretion, neurotransmission(GO:0014057) |
| 0.0 | 0.1 | GO:0016259 | selenocysteine metabolic process(GO:0016259) |
| 0.0 | 0.4 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.0 | 0.4 | GO:0070986 | left/right axis specification(GO:0070986) |
| 0.0 | 0.1 | GO:0019072 | viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 0.0 | 1.0 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.0 | 1.0 | GO:0071377 | cellular response to glucagon stimulus(GO:0071377) |
| 0.0 | 0.1 | GO:0009597 | detection of virus(GO:0009597) |
| 0.0 | 0.3 | GO:0051988 | regulation of attachment of spindle microtubules to kinetochore(GO:0051988) |
| 0.0 | 0.1 | GO:0044278 | cell wall disruption in other organism(GO:0044278) |
| 0.0 | 0.6 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.0 | 0.3 | GO:0060707 | trophoblast giant cell differentiation(GO:0060707) |
| 0.0 | 0.1 | GO:0006127 | glycerophosphate shuttle(GO:0006127) |
| 0.0 | 0.4 | GO:0021516 | dorsal spinal cord development(GO:0021516) |
| 0.0 | 0.2 | GO:0051026 | chiasma assembly(GO:0051026) |
| 0.0 | 1.2 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.0 | 0.5 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.0 | 0.1 | GO:0032700 | negative regulation of interleukin-17 production(GO:0032700) |
| 0.0 | 0.1 | GO:0042997 | negative regulation of Golgi to plasma membrane protein transport(GO:0042997) |
| 0.0 | 1.2 | GO:0042733 | embryonic digit morphogenesis(GO:0042733) |
| 0.0 | 0.1 | GO:0046116 | queuosine biosynthetic process(GO:0008616) queuosine metabolic process(GO:0046116) |
| 0.0 | 0.2 | GO:0015886 | heme transport(GO:0015886) |
| 0.0 | 0.3 | GO:0038180 | nerve growth factor signaling pathway(GO:0038180) |
| 0.0 | 0.2 | GO:0046689 | response to mercury ion(GO:0046689) |
| 0.0 | 0.1 | GO:0051088 | PMA-inducible membrane protein ectodomain proteolysis(GO:0051088) |
| 0.0 | 0.3 | GO:0043031 | T-helper 2 cell cytokine production(GO:0035745) negative regulation of macrophage activation(GO:0043031) |
| 0.0 | 0.2 | GO:0045008 | depyrimidination(GO:0045008) |
| 0.0 | 0.2 | GO:0090023 | positive regulation of neutrophil chemotaxis(GO:0090023) |
| 0.0 | 1.3 | GO:0070098 | chemokine-mediated signaling pathway(GO:0070098) |
| 0.0 | 1.5 | GO:0032091 | negative regulation of protein binding(GO:0032091) |
| 0.0 | 0.6 | GO:0051894 | positive regulation of focal adhesion assembly(GO:0051894) |
| 0.0 | 0.0 | GO:0038193 | thromboxane A2 signaling pathway(GO:0038193) |
| 0.0 | 0.2 | GO:0015939 | pantothenate metabolic process(GO:0015939) |
| 0.0 | 1.0 | GO:0019933 | cAMP-mediated signaling(GO:0019933) |
| 0.0 | 0.3 | GO:0002089 | lens morphogenesis in camera-type eye(GO:0002089) |
| 0.0 | 1.3 | GO:0035113 | embryonic limb morphogenesis(GO:0030326) embryonic appendage morphogenesis(GO:0035113) |
| 0.0 | 0.2 | GO:0098719 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.0 | 0.9 | GO:0032760 | positive regulation of tumor necrosis factor production(GO:0032760) |
| 0.0 | 0.2 | GO:0015866 | ADP transport(GO:0015866) |
| 0.0 | 0.0 | GO:0045897 | positive regulation of transcription during mitosis(GO:0045897) |
| 0.0 | 0.1 | GO:0010968 | regulation of microtubule nucleation(GO:0010968) |
| 0.0 | 0.0 | GO:0018094 | protein polyglycylation(GO:0018094) |
| 0.0 | 0.5 | GO:2000249 | regulation of actin cytoskeleton reorganization(GO:2000249) |
| 0.0 | 1.7 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.0 | 0.1 | GO:0044130 | negative regulation of growth of symbiont in host(GO:0044130) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.6 | GO:0034680 | integrin alpha10-beta1 complex(GO:0034680) |
| 0.3 | 1.5 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.3 | 6.7 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.2 | 0.9 | GO:0044307 | dendritic branch(GO:0044307) |
| 0.1 | 2.0 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.1 | 1.8 | GO:0043220 | Schmidt-Lanterman incisure(GO:0043220) |
| 0.1 | 0.2 | GO:0030678 | mitochondrial ribonuclease P complex(GO:0030678) |
| 0.1 | 1.6 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.1 | 0.3 | GO:0097149 | centralspindlin complex(GO:0097149) |
| 0.1 | 0.5 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.1 | 0.5 | GO:0044354 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.1 | 0.2 | GO:1990876 | cytoplasmic side of nuclear pore(GO:1990876) |
| 0.0 | 0.5 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.0 | 0.2 | GO:0005965 | protein farnesyltransferase complex(GO:0005965) |
| 0.0 | 1.2 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 1.2 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 0.6 | GO:0008091 | spectrin(GO:0008091) |
| 0.0 | 0.9 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 0.1 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.0 | 0.1 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.0 | 0.1 | GO:0097629 | extrinsic component of omegasome membrane(GO:0097629) |
| 0.0 | 0.3 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.0 | 0.4 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 0.4 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) |
| 0.0 | 0.4 | GO:0033276 | transcription factor TFTC complex(GO:0033276) |
| 0.0 | 0.3 | GO:0030478 | actin cap(GO:0030478) |
| 0.0 | 1.5 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 1.0 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 0.1 | GO:0032280 | symmetric synapse(GO:0032280) |
| 0.0 | 0.3 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.0 | 4.1 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 0.1 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.0 | 0.2 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 0.5 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 0.0 | 0.1 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.0 | 1.5 | GO:0035578 | azurophil granule lumen(GO:0035578) |
| 0.0 | 1.0 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 0.3 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 0.1 | GO:0031595 | nuclear proteasome complex(GO:0031595) |
| 0.0 | 0.2 | GO:0032982 | myosin filament(GO:0032982) |
| 0.0 | 0.8 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.1 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.0 | 5.2 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 0.3 | GO:0030128 | clathrin coat of endocytic vesicle(GO:0030128) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.5 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.4 | 5.7 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.3 | 6.5 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.3 | 2.6 | GO:0098639 | collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.3 | 0.9 | GO:0047787 | geranylgeranyl reductase activity(GO:0045550) delta4-3-oxosteroid 5beta-reductase activity(GO:0047787) |
| 0.3 | 0.8 | GO:0047256 | beta-galactosyl-N-acetylglucosaminylgalactosylglucosyl-ceramide beta-1,3-acetylglucosaminyltransferase activity(GO:0008457) lactosylceramide 1,3-N-acetyl-beta-D-glucosaminyltransferase activity(GO:0047256) |
| 0.2 | 1.0 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.2 | 0.7 | GO:0008424 | glycoprotein 6-alpha-L-fucosyltransferase activity(GO:0008424) alpha-(1->6)-fucosyltransferase activity(GO:0046921) |
| 0.2 | 0.8 | GO:0003990 | acetylcholinesterase activity(GO:0003990) |
| 0.2 | 0.5 | GO:0003860 | 3-hydroxyisobutyryl-CoA hydrolase activity(GO:0003860) |
| 0.2 | 1.0 | GO:0038064 | protein tyrosine kinase collagen receptor activity(GO:0038062) collagen receptor activity(GO:0038064) |
| 0.2 | 2.8 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.1 | 4.3 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.1 | 0.6 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.1 | 1.1 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.1 | 0.3 | GO:0004556 | alpha-amylase activity(GO:0004556) |
| 0.1 | 0.3 | GO:0015489 | polyamine transmembrane transporter activity(GO:0015203) putrescine transmembrane transporter activity(GO:0015489) |
| 0.1 | 1.5 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.1 | 1.6 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.1 | 0.3 | GO:0044594 | 3alpha,7alpha,12alpha-trihydroxy-5beta-cholest-24-enoyl-CoA hydratase activity(GO:0033989) 17-beta-hydroxysteroid dehydrogenase (NAD+) activity(GO:0044594) |
| 0.1 | 0.6 | GO:0004962 | endothelin receptor activity(GO:0004962) |
| 0.1 | 1.8 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.1 | 0.8 | GO:0051425 | PTB domain binding(GO:0051425) |
| 0.1 | 1.3 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.1 | 0.4 | GO:0097199 | cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097199) |
| 0.1 | 0.7 | GO:0015165 | pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
| 0.1 | 1.1 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.1 | 0.3 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.1 | 0.3 | GO:0047757 | chondroitin-glucuronate 5-epimerase activity(GO:0047757) |
| 0.1 | 1.1 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.1 | 0.7 | GO:0102337 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.1 | 0.5 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.1 | 1.4 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.1 | 1.0 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.1 | 0.6 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.0 | 0.2 | GO:0000702 | oxidized base lesion DNA N-glycosylase activity(GO:0000702) |
| 0.0 | 0.4 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 0.0 | 0.2 | GO:0004660 | protein farnesyltransferase activity(GO:0004660) |
| 0.0 | 0.5 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 0.4 | GO:1904929 | coreceptor activity involved in Wnt signaling pathway, planar cell polarity pathway(GO:1904929) |
| 0.0 | 0.1 | GO:0004483 | mRNA (nucleoside-2'-O-)-methyltransferase activity(GO:0004483) |
| 0.0 | 0.2 | GO:0016429 | tRNA (adenine-N1-)-methyltransferase activity(GO:0016429) |
| 0.0 | 0.2 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.0 | 1.1 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.0 | 0.4 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.0 | 0.4 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 0.0 | 0.2 | GO:0071987 | WD40-repeat domain binding(GO:0071987) |
| 0.0 | 0.4 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.0 | 0.1 | GO:0004796 | thromboxane-A synthase activity(GO:0004796) 12-hydroxyheptadecatrienoic acid synthase activity(GO:0036134) |
| 0.0 | 0.9 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.0 | 0.5 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.0 | 0.6 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 1.3 | GO:0004950 | G-protein coupled chemoattractant receptor activity(GO:0001637) chemokine receptor activity(GO:0004950) |
| 0.0 | 1.2 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.9 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.0 | 0.7 | GO:0031005 | filamin binding(GO:0031005) |
| 0.0 | 0.4 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.0 | 0.2 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.4 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 0.1 | GO:0052590 | sn-glycerol-3-phosphate:ubiquinone oxidoreductase activity(GO:0052590) sn-glycerol-3-phosphate:ubiquinone-8 oxidoreductase activity(GO:0052591) |
| 0.0 | 0.2 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.0 | 0.2 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.0 | 0.3 | GO:0032395 | MHC class II receptor activity(GO:0032395) |
| 0.0 | 1.0 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.0 | 0.1 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.0 | 0.1 | GO:1990763 | arrestin family protein binding(GO:1990763) |
| 0.0 | 0.5 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.4 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.0 | 0.4 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.4 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.0 | 0.1 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 0.0 | 0.6 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.0 | 0.0 | GO:0055100 | adiponectin binding(GO:0055100) |
| 0.0 | 0.6 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.0 | 0.1 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.0 | 0.1 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.0 | 0.2 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 0.0 | GO:0004960 | thromboxane receptor activity(GO:0004960) thromboxane A2 receptor activity(GO:0004961) |
| 0.0 | 0.1 | GO:0005152 | interleukin-1 receptor antagonist activity(GO:0005152) |
| 0.0 | 2.0 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 0.2 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.2 | GO:0015217 | ATP transmembrane transporter activity(GO:0005347) ADP transmembrane transporter activity(GO:0015217) |
| 0.0 | 0.4 | GO:0008198 | ferrous iron binding(GO:0008198) |
| 0.0 | 0.2 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.0 | 0.4 | GO:0042288 | MHC class I protein binding(GO:0042288) |
| 0.0 | 0.1 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 0.0 | 0.1 | GO:0015194 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.0 | 0.5 | GO:0003785 | actin monomer binding(GO:0003785) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 5.9 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.1 | 3.9 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.1 | 6.3 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.1 | 4.3 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.0 | 1.6 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.0 | 0.8 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 1.0 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 0.9 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.0 | 0.5 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.0 | 1.0 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.0 | 1.3 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 2.0 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 0.7 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 0.8 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.0 | 0.9 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.0 | 1.7 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 0.6 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.0 | 0.4 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.0 | 0.1 | ST DIFFERENTIATION PATHWAY IN PC12 CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
| 0.0 | 0.5 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 0.3 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 0.4 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 6.5 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.2 | 6.7 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.1 | 0.5 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.0 | 0.9 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.0 | 1.9 | REACTOME FORMATION OF FIBRIN CLOT CLOTTING CASCADE | Genes involved in Formation of Fibrin Clot (Clotting Cascade) |
| 0.0 | 1.5 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.0 | 0.8 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.0 | 0.7 | REACTOME NFKB ACTIVATION THROUGH FADD RIP1 PATHWAY MEDIATED BY CASPASE 8 AND10 | Genes involved in NF-kB activation through FADD/RIP-1 pathway mediated by caspase-8 and -10 |
| 0.0 | 1.2 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 2.8 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 1.0 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 1.0 | REACTOME G BETA GAMMA SIGNALLING THROUGH PLC BETA | Genes involved in G beta:gamma signalling through PLC beta |
| 0.0 | 1.0 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 0.7 | REACTOME TRAF6 MEDIATED NFKB ACTIVATION | Genes involved in TRAF6 mediated NF-kB activation |
| 0.0 | 0.3 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.0 | 0.9 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF CAMKII | Genes involved in CREB phosphorylation through the activation of CaMKII |
| 0.0 | 1.2 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 0.7 | REACTOME N GLYCAN ANTENNAE ELONGATION IN THE MEDIAL TRANS GOLGI | Genes involved in N-glycan antennae elongation in the medial/trans-Golgi |
| 0.0 | 0.4 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 2.3 | REACTOME CELL DEATH SIGNALLING VIA NRAGE NRIF AND NADE | Genes involved in Cell death signalling via NRAGE, NRIF and NADE |
| 0.0 | 0.4 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 0.7 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 0.6 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.3 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 0.5 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 0.3 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.0 | 1.0 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.0 | 1.0 | REACTOME TRAFFICKING OF AMPA RECEPTORS | Genes involved in Trafficking of AMPA receptors |
| 0.0 | 1.8 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 1.9 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 0.4 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.0 | 0.2 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.0 | 0.1 | REACTOME SIGNALING BY NOTCH4 | Genes involved in Signaling by NOTCH4 |
| 0.0 | 0.7 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 0.3 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 0.2 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.0 | 0.2 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 0.5 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |