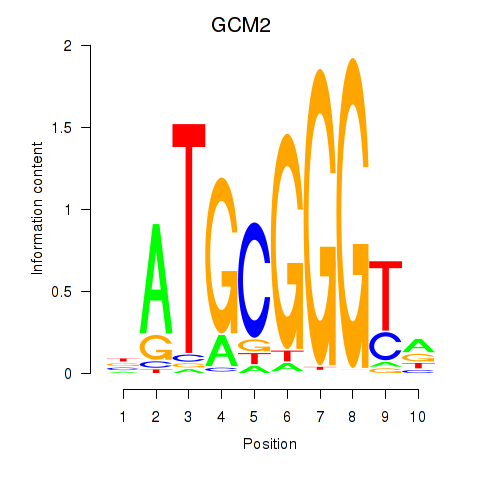
|
chr8_-_92103217
Show fit
|
1.70 |
ENST00000615601.4
ENST00000523629.5
|
RUNX1T1
|
RUNX1 partner transcriptional co-repressor 1
|
|
chr11_+_7576408
Show fit
|
1.48 |
ENST00000533792.5
|
PPFIBP2
|
PPFIA binding protein 2
|
|
chr1_+_14924100
Show fit
|
1.14 |
ENST00000361144.9
|
KAZN
|
kazrin, periplakin interacting protein
|
|
chr11_+_7513966
Show fit
|
1.14 |
ENST00000299492.9
|
PPFIBP2
|
PPFIA binding protein 2
|
|
chr15_-_92655712
Show fit
|
1.10 |
ENST00000327355.6
|
FAM174B
|
family with sequence similarity 174 member B
|
|
chr1_+_99646025
Show fit
|
0.98 |
ENST00000263174.9
ENST00000605497.5
ENST00000615664.1
|
PALMD
|
palmdelphin
|
|
chr1_+_26111139
Show fit
|
0.94 |
ENST00000619836.4
ENST00000444713.5
|
PDIK1L
|
PDLIM1 interacting kinase 1 like
|
|
chr5_+_123512214
Show fit
|
0.79 |
ENST00000511130.6
ENST00000512718.7
|
CSNK1G3
|
casein kinase 1 gamma 3
|
|
chr1_-_39901996
Show fit
|
0.78 |
ENST00000397332.2
|
MYCL
|
MYCL proto-oncogene, bHLH transcription factor
|
|
chr6_-_31897200
Show fit
|
0.73 |
ENST00000395728.7
ENST00000375528.8
|
EHMT2
|
euchromatic histone lysine methyltransferase 2
|
|
chr12_-_49828394
Show fit
|
0.72 |
ENST00000335999.7
|
NCKAP5L
|
NCK associated protein 5 like
|
|
chr14_-_105168753
Show fit
|
0.70 |
ENST00000331782.8
ENST00000347004.2
|
JAG2
|
jagged canonical Notch ligand 2
|
|
chr14_+_64504574
Show fit
|
0.67 |
ENST00000358738.3
|
ZBTB1
|
zinc finger and BTB domain containing 1
|
|
chr17_-_27793716
Show fit
|
0.67 |
ENST00000646938.1
|
NOS2
|
nitric oxide synthase 2
|
|
chr5_+_177133005
Show fit
|
0.64 |
ENST00000510954.5
ENST00000354179.8
|
NSD1
|
nuclear receptor binding SET domain protein 1
|
|
chr13_+_31846713
Show fit
|
0.63 |
ENST00000645780.1
|
FRY
|
FRY microtubule binding protein
|
|
chr6_+_7107941
Show fit
|
0.62 |
ENST00000379938.7
ENST00000467782.5
ENST00000334984.10
ENST00000349384.10
|
RREB1
|
ras responsive element binding protein 1
|
|
chr17_+_6070361
Show fit
|
0.59 |
ENST00000317744.10
|
WSCD1
|
WSC domain containing 1
|
|
chr9_-_127755243
Show fit
|
0.57 |
ENST00000629203.2
ENST00000420366.5
|
SH2D3C
|
SH2 domain containing 3C
|
|
chr1_-_39901861
Show fit
|
0.56 |
ENST00000372816.3
ENST00000372815.1
|
MYCL
|
MYCL proto-oncogene, bHLH transcription factor
|
|
chr7_-_101165114
Show fit
|
0.54 |
ENST00000445482.2
|
VGF
|
VGF nerve growth factor inducible
|
|
chr7_+_12687625
Show fit
|
0.54 |
ENST00000651779.1
ENST00000404894.1
|
ARL4A
|
ADP ribosylation factor like GTPase 4A
|
|
chr19_-_38869921
Show fit
|
0.53 |
ENST00000593809.1
ENST00000593424.5
|
RINL
|
Ras and Rab interactor like
|
|
chr12_+_57522801
Show fit
|
0.53 |
ENST00000355673.8
ENST00000546632.1
ENST00000549623.1
|
MBD6
|
methyl-CpG binding domain protein 6
|
|
chr19_+_35000275
Show fit
|
0.50 |
ENST00000317991.10
ENST00000680623.1
|
GRAMD1A
|
GRAM domain containing 1A
|
|
chr13_-_98577131
Show fit
|
0.49 |
ENST00000397517.6
|
STK24
|
serine/threonine kinase 24
|
|
chr10_+_92689946
Show fit
|
0.49 |
ENST00000282728.10
|
HHEX
|
hematopoietically expressed homeobox
|
|
chr17_-_67245165
Show fit
|
0.48 |
ENST00000580168.5
ENST00000358691.10
|
HELZ
|
helicase with zinc finger
|
|
chr12_+_55997180
Show fit
|
0.47 |
ENST00000356124.8
ENST00000266971.8
ENST00000394115.6
ENST00000547586.5
ENST00000552258.5
ENST00000548274.5
ENST00000546833.5
|
SUOX
|
sulfite oxidase
|
|
chr9_+_127612222
Show fit
|
0.46 |
ENST00000637953.1
ENST00000636962.2
|
STXBP1
|
syntaxin binding protein 1
|
|
chr2_-_219543793
Show fit
|
0.43 |
ENST00000243776.11
|
CHPF
|
chondroitin polymerizing factor
|
|
chr12_+_14973020
Show fit
|
0.43 |
ENST00000266395.3
|
PDE6H
|
phosphodiesterase 6H
|
|
chr9_+_70043840
Show fit
|
0.43 |
ENST00000377182.5
|
MAMDC2
|
MAM domain containing 2
|
|
chr6_-_6007511
Show fit
|
0.43 |
ENST00000616243.1
|
NRN1
|
neuritin 1
|
|
chr9_+_127612257
Show fit
|
0.42 |
ENST00000637173.2
ENST00000630492.2
ENST00000627871.2
ENST00000373302.8
ENST00000373299.5
ENST00000650920.1
ENST00000476182.3
|
STXBP1
|
syntaxin binding protein 1
|
|
chr11_+_117232625
Show fit
|
0.42 |
ENST00000534428.5
ENST00000300650.9
|
RNF214
|
ring finger protein 214
|
|
chrX_-_63785510
Show fit
|
0.42 |
ENST00000437457.6
ENST00000374878.5
ENST00000623517.3
|
ARHGEF9
|
Cdc42 guanine nucleotide exchange factor 9
|
|
chr19_+_17470474
Show fit
|
0.42 |
ENST00000598424.5
ENST00000252595.12
|
SLC27A1
|
solute carrier family 27 member 1
|
|
chr5_-_173328407
Show fit
|
0.41 |
ENST00000265087.9
|
STC2
|
stanniocalcin 2
|
|
chr18_-_31684504
Show fit
|
0.41 |
ENST00000383131.3
ENST00000237019.11
ENST00000306851.10
|
B4GALT6
|
beta-1,4-galactosyltransferase 6
|
|
chr2_+_188292771
Show fit
|
0.40 |
ENST00000359135.7
|
GULP1
|
GULP PTB domain containing engulfment adaptor 1
|
|
chr9_-_124500986
Show fit
|
0.40 |
ENST00000373587.3
|
NR5A1
|
nuclear receptor subfamily 5 group A member 1
|
|
chr7_+_121328991
Show fit
|
0.40 |
ENST00000222462.3
|
WNT16
|
Wnt family member 16
|
|
chr14_-_75069478
Show fit
|
0.39 |
ENST00000555463.1
|
ACYP1
|
acylphosphatase 1
|
|
chr9_+_4490388
Show fit
|
0.39 |
ENST00000262352.8
|
SLC1A1
|
solute carrier family 1 member 1
|
|
chr14_-_49586325
Show fit
|
0.39 |
ENST00000557519.1
ENST00000396020.7
ENST00000554075.2
ENST00000245458.11
|
RPS29
|
ribosomal protein S29
|
|
chr5_-_177311882
Show fit
|
0.39 |
ENST00000513169.1
ENST00000423571.6
ENST00000502529.1
ENST00000427908.6
|
MXD3
|
MAX dimerization protein 3
|
|
chr1_+_2050387
Show fit
|
0.39 |
ENST00000378567.8
|
PRKCZ
|
protein kinase C zeta
|
|
chr7_-_101165558
Show fit
|
0.39 |
ENST00000611537.1
ENST00000249330.3
|
VGF
|
VGF nerve growth factor inducible
|
|
chr6_+_130018565
Show fit
|
0.39 |
ENST00000361794.7
ENST00000526087.5
ENST00000533560.5
|
L3MBTL3
|
L3MBTL histone methyl-lysine binding protein 3
|
|
chr17_-_4903088
Show fit
|
0.38 |
ENST00000649488.2
|
CHRNE
|
cholinergic receptor nicotinic epsilon subunit
|
|
chr11_+_66857056
Show fit
|
0.38 |
ENST00000309602.5
ENST00000393952.3
|
LRFN4
|
leucine rich repeat and fibronectin type III domain containing 4
|
|
chr7_+_5879827
Show fit
|
0.38 |
ENST00000416608.5
|
OCM
|
oncomodulin
|
|
chr1_+_156728442
Show fit
|
0.37 |
ENST00000368218.8
ENST00000368216.9
|
RRNAD1
|
ribosomal RNA adenine dimethylase domain containing 1
|
|
chrX_-_63785149
Show fit
|
0.37 |
ENST00000671741.2
ENST00000625116.3
ENST00000624355.1
|
ARHGEF9
|
Cdc42 guanine nucleotide exchange factor 9
|
|
chrX_+_47078380
Show fit
|
0.37 |
ENST00000352078.8
|
RGN
|
regucalcin
|
|
chrX_+_47078434
Show fit
|
0.37 |
ENST00000397180.6
|
RGN
|
regucalcin
|
|
chr11_-_58578096
Show fit
|
0.37 |
ENST00000528954.5
ENST00000528489.1
|
LPXN
|
leupaxin
|
|
chr17_+_1742836
Show fit
|
0.36 |
ENST00000324015.7
ENST00000450523.6
ENST00000453723.5
ENST00000453066.6
ENST00000382061.5
|
SERPINF2
|
serpin family F member 2
|
|
chr19_-_19515542
Show fit
|
0.36 |
ENST00000585580.4
|
TSSK6
|
testis specific serine kinase 6
|
|
chr12_-_43552196
Show fit
|
0.35 |
ENST00000389420.8
|
ADAMTS20
|
ADAM metallopeptidase with thrombospondin type 1 motif 20
|
|
chr12_+_56224318
Show fit
|
0.35 |
ENST00000267023.9
ENST00000380198.6
ENST00000341463.5
|
NABP2
|
nucleic acid binding protein 2
|
|
chr10_+_74824919
Show fit
|
0.34 |
ENST00000648828.1
ENST00000648725.1
|
KAT6B
|
lysine acetyltransferase 6B
|
|
chr5_-_524322
Show fit
|
0.34 |
ENST00000644203.1
ENST00000514375.1
ENST00000264938.8
|
SLC9A3
|
solute carrier family 9 member A3
|
|
chrX_+_47078330
Show fit
|
0.34 |
ENST00000457380.5
|
RGN
|
regucalcin
|
|
chr1_+_14945775
Show fit
|
0.33 |
ENST00000400797.3
|
KAZN
|
kazrin, periplakin interacting protein
|
|
chr22_+_19718390
Show fit
|
0.33 |
ENST00000383045.7
ENST00000438754.6
|
SEPTIN5
|
septin 5
|
|
chr3_+_186666003
Show fit
|
0.33 |
ENST00000232003.5
|
HRG
|
histidine rich glycoprotein
|
|
chr6_+_7107597
Show fit
|
0.32 |
ENST00000379933.7
ENST00000491191.5
ENST00000471433.5
|
RREB1
|
ras responsive element binding protein 1
|
|
chr3_-_86991135
Show fit
|
0.32 |
ENST00000398399.7
|
VGLL3
|
vestigial like family member 3
|
|
chr14_-_64504570
Show fit
|
0.32 |
ENST00000394715.1
|
ZBTB25
|
zinc finger and BTB domain containing 25
|
|
chr4_-_146521891
Show fit
|
0.32 |
ENST00000394059.8
ENST00000502607.1
ENST00000335472.12
ENST00000432059.6
|
SLC10A7
|
solute carrier family 10 member 7
|
|
chr20_-_1002264
Show fit
|
0.32 |
ENST00000400634.2
ENST00000217260.9
|
RSPO4
|
R-spondin 4
|
|
chr11_+_63938971
Show fit
|
0.32 |
ENST00000539656.5
ENST00000377793.9
|
NAA40
|
N-alpha-acetyltransferase 40, NatD catalytic subunit
|
|
chr9_+_134025455
Show fit
|
0.31 |
ENST00000603928.3
ENST00000432807.1
|
BRD3OS
|
BRD3 opposite strand
|
|
chr17_+_6071075
Show fit
|
0.31 |
ENST00000574232.5
ENST00000539421.1
|
WSCD1
|
WSC domain containing 1
|
|
chr9_+_136712531
Show fit
|
0.31 |
ENST00000371692.9
|
DIPK1B
|
divergent protein kinase domain 1B
|
|
chr15_-_74956758
Show fit
|
0.31 |
ENST00000322177.6
|
RPP25
|
ribonuclease P and MRP subunit p25
|
|
chr12_-_48004467
Show fit
|
0.30 |
ENST00000380518.8
|
COL2A1
|
collagen type II alpha 1 chain
|
|
chr7_-_44325421
Show fit
|
0.30 |
ENST00000395747.6
ENST00000347193.8
ENST00000346990.8
ENST00000258682.10
ENST00000353625.8
ENST00000421607.1
ENST00000424197.5
|
CAMK2B
|
calcium/calmodulin dependent protein kinase II beta
|
|
chr5_-_91383310
Show fit
|
0.30 |
ENST00000265138.4
|
ARRDC3
|
arrestin domain containing 3
|
|
chr19_+_17871937
Show fit
|
0.30 |
ENST00000222248.4
|
SLC5A5
|
solute carrier family 5 member 5
|
|
chr13_-_98577094
Show fit
|
0.29 |
ENST00000539966.6
|
STK24
|
serine/threonine kinase 24
|
|
chr2_+_188292814
Show fit
|
0.28 |
ENST00000409580.5
ENST00000409637.7
|
GULP1
|
GULP PTB domain containing engulfment adaptor 1
|
|
chr9_+_125748175
Show fit
|
0.27 |
ENST00000491787.7
ENST00000447726.6
|
PBX3
|
PBX homeobox 3
|
|
chr7_-_44325653
Show fit
|
0.27 |
ENST00000440254.6
|
CAMK2B
|
calcium/calmodulin dependent protein kinase II beta
|
|
chr3_+_155080088
Show fit
|
0.27 |
ENST00000462745.5
|
MME
|
membrane metalloendopeptidase
|
|
chr3_+_23945271
Show fit
|
0.26 |
ENST00000312521.9
|
NR1D2
|
nuclear receptor subfamily 1 group D member 2
|
|
chr7_-_5423543
Show fit
|
0.26 |
ENST00000399537.8
|
TNRC18
|
trinucleotide repeat containing 18
|
|
chr2_+_232406836
Show fit
|
0.26 |
ENST00000295453.8
|
ALPG
|
alkaline phosphatase, germ cell
|
|
chr7_-_8262052
Show fit
|
0.26 |
ENST00000396675.7
ENST00000430867.5
ENST00000402384.8
ENST00000265577.11
|
ICA1
|
islet cell autoantigen 1
|
|
chr17_-_7219813
Show fit
|
0.25 |
ENST00000399510.8
ENST00000648172.8
|
DLG4
|
discs large MAGUK scaffold protein 4
|
|
chr7_-_5423826
Show fit
|
0.25 |
ENST00000430969.6
|
TNRC18
|
trinucleotide repeat containing 18
|
|
chr13_-_20161038
Show fit
|
0.25 |
ENST00000241125.4
|
GJA3
|
gap junction protein alpha 3
|
|
chr7_-_27130182
Show fit
|
0.25 |
ENST00000511914.1
|
HOXA4
|
homeobox A4
|
|
chr1_-_156500763
Show fit
|
0.24 |
ENST00000348159.9
ENST00000489057.1
|
MEF2D
|
myocyte enhancer factor 2D
|
|
chrX_-_154805386
Show fit
|
0.24 |
ENST00000393531.5
ENST00000369534.8
ENST00000453245.5
ENST00000428488.1
ENST00000369531.1
|
MPP1
|
membrane palmitoylated protein 1
|
|
chr19_-_51368979
Show fit
|
0.24 |
ENST00000601435.1
ENST00000291715.5
|
CLDND2
|
claudin domain containing 2
|
|
chr2_-_197310646
Show fit
|
0.24 |
ENST00000647377.1
|
ANKRD44
|
ankyrin repeat domain 44
|
|
chr3_-_19946970
Show fit
|
0.23 |
ENST00000344838.8
|
EFHB
|
EF-hand domain family member B
|
|
chr15_+_40569290
Show fit
|
0.23 |
ENST00000315616.12
ENST00000559271.1
ENST00000616318.1
|
RPUSD2
|
RNA pseudouridine synthase domain containing 2
|
|
chr12_-_51270351
Show fit
|
0.23 |
ENST00000603798.6
|
SMAGP
|
small cell adhesion glycoprotein
|
|
chr19_+_40717091
Show fit
|
0.23 |
ENST00000263370.3
|
ITPKC
|
inositol-trisphosphate 3-kinase C
|
|
chr2_-_44361555
Show fit
|
0.22 |
ENST00000409957.5
|
PREPL
|
prolyl endopeptidase like
|
|
chr7_-_44325490
Show fit
|
0.22 |
ENST00000350811.7
|
CAMK2B
|
calcium/calmodulin dependent protein kinase II beta
|
|
chr4_-_17781613
Show fit
|
0.22 |
ENST00000265018.4
|
FAM184B
|
family with sequence similarity 184 member B
|
|
chr11_+_66638661
Show fit
|
0.21 |
ENST00000396053.9
ENST00000408993.6
|
RBM4
|
RNA binding motif protein 4
|
|
chr22_+_31093358
Show fit
|
0.21 |
ENST00000404574.5
|
SMTN
|
smoothelin
|
|
chr14_-_21098848
Show fit
|
0.21 |
ENST00000556174.5
ENST00000554478.5
ENST00000553980.1
ENST00000421093.6
|
ZNF219
|
zinc finger protein 219
|
|
chr3_-_122022122
Show fit
|
0.21 |
ENST00000393631.5
ENST00000273691.7
ENST00000344209.10
|
ILDR1
|
immunoglobulin like domain containing receptor 1
|
|
chr18_-_14132423
Show fit
|
0.20 |
ENST00000589498.5
ENST00000590202.3
|
ZNF519
|
zinc finger protein 519
|
|
chr19_+_45039040
Show fit
|
0.20 |
ENST00000221455.8
ENST00000391953.8
ENST00000588936.5
|
CLASRP
|
CLK4 associating serine/arginine rich protein
|
|
chrX_+_43654888
Show fit
|
0.20 |
ENST00000542639.5
|
MAOA
|
monoamine oxidase A
|
|
chrX_-_154805516
Show fit
|
0.20 |
ENST00000413259.7
|
MPP1
|
membrane palmitoylated protein 1
|
|
chr3_-_50567711
Show fit
|
0.20 |
ENST00000357203.8
|
C3orf18
|
chromosome 3 open reading frame 18
|
|
chr14_-_77616630
Show fit
|
0.20 |
ENST00000216484.7
|
SPTLC2
|
serine palmitoyltransferase long chain base subunit 2
|
|
chr16_+_28985043
Show fit
|
0.19 |
ENST00000395456.7
ENST00000564277.5
ENST00000630764.2
ENST00000354453.7
|
LAT
|
linker for activation of T cells
|
|
chrX_-_136880715
Show fit
|
0.19 |
ENST00000431446.7
ENST00000320676.11
ENST00000562646.5
|
RBMX
|
RNA binding motif protein X-linked
|
|
chr11_-_74170975
Show fit
|
0.19 |
ENST00000539061.6
ENST00000680645.1
ENST00000334126.12
ENST00000680718.1
|
C2CD3
|
C2 domain containing 3 centriole elongation regulator
|
|
chr5_-_73448769
Show fit
|
0.19 |
ENST00000615637.3
|
FOXD1
|
forkhead box D1
|
|
chr14_-_21098570
Show fit
|
0.19 |
ENST00000360947.8
|
ZNF219
|
zinc finger protein 219
|
|
chr1_+_960549
Show fit
|
0.19 |
ENST00000338591.8
ENST00000622660.1
|
KLHL17
|
kelch like family member 17
|
|
chr11_-_796185
Show fit
|
0.19 |
ENST00000533385.5
ENST00000528936.5
ENST00000629634.2
ENST00000625752.2
ENST00000528606.5
ENST00000320230.9
|
SLC25A22
|
solute carrier family 25 member 22
|
|
chr19_+_45039509
Show fit
|
0.19 |
ENST00000544944.6
|
CLASRP
|
CLK4 associating serine/arginine rich protein
|
|
chr11_+_63681444
Show fit
|
0.18 |
ENST00000341307.6
ENST00000356000.7
ENST00000542238.5
|
RTN3
|
reticulon 3
|
|
chr6_-_32109291
Show fit
|
0.18 |
ENST00000479795.1
ENST00000647633.1
ENST00000644971.2
ENST00000375244.7
|
TNXB
|
tenascin XB
|
|
chr14_+_92513766
Show fit
|
0.18 |
ENST00000216487.12
ENST00000620541.4
ENST00000557762.1
|
RIN3
|
Ras and Rab interactor 3
|
|
chr11_-_74170792
Show fit
|
0.18 |
ENST00000680231.1
ENST00000681143.1
ENST00000679906.1
ENST00000681310.1
ENST00000414160.7
ENST00000538361.2
|
C2CD3
|
C2 domain containing 3 centriole elongation regulator
|
|
chr9_-_69014090
Show fit
|
0.18 |
ENST00000377276.5
|
PRKACG
|
protein kinase cAMP-activated catalytic subunit gamma
|
|
chr11_+_63681483
Show fit
|
0.18 |
ENST00000339997.8
ENST00000540798.5
ENST00000545432.5
ENST00000543552.5
ENST00000377819.10
ENST00000537981.5
|
RTN3
|
reticulon 3
|
|
chr19_-_38388020
Show fit
|
0.17 |
ENST00000334928.11
ENST00000587676.1
|
GGN
|
gametogenetin
|
|
chr3_+_155079847
Show fit
|
0.17 |
ENST00000615825.2
|
MME
|
membrane metalloendopeptidase
|
|
chr1_+_150549734
Show fit
|
0.17 |
ENST00000674043.1
ENST00000674058.1
|
ADAMTSL4
|
ADAMTS like 4
|
|
chr1_-_16431371
Show fit
|
0.17 |
ENST00000612240.1
|
SPATA21
|
spermatogenesis associated 21
|
|
chr20_+_43514426
Show fit
|
0.17 |
ENST00000422861.3
|
L3MBTL1
|
L3MBTL histone methyl-lysine binding protein 1
|
|
chr22_-_29315656
Show fit
|
0.17 |
ENST00000401450.3
ENST00000216101.7
|
RASL10A
|
RAS like family 10 member A
|
|
chr3_+_155079663
Show fit
|
0.17 |
ENST00000460393.6
|
MME
|
membrane metalloendopeptidase
|
|
chr3_-_50567646
Show fit
|
0.16 |
ENST00000426034.5
ENST00000441239.5
|
C3orf18
|
chromosome 3 open reading frame 18
|
|
chr15_-_93089192
Show fit
|
0.16 |
ENST00000329082.11
|
RGMA
|
repulsive guidance molecule BMP co-receptor a
|
|
chr3_+_155079911
Show fit
|
0.16 |
ENST00000675418.2
|
MME
|
membrane metalloendopeptidase
|
|
chr19_-_10565990
Show fit
|
0.16 |
ENST00000539027.5
ENST00000312962.12
ENST00000543682.3
ENST00000652042.1
ENST00000432197.5
|
KRI1
|
KRI1 homolog
|
|
chr16_+_28985251
Show fit
|
0.16 |
ENST00000360872.9
ENST00000566177.5
|
LAT
|
linker for activation of T cells
|
|
chr17_-_3893040
Show fit
|
0.15 |
ENST00000158166.5
ENST00000348335.7
|
CAMKK1
|
calcium/calmodulin dependent protein kinase kinase 1
|
|
chr1_+_150549384
Show fit
|
0.15 |
ENST00000369041.9
ENST00000271643.9
|
ADAMTSL4
|
ADAMTS like 4
|
|
chr19_-_51027662
Show fit
|
0.15 |
ENST00000594768.5
|
KLK11
|
kallikrein related peptidase 11
|
|
chr2_+_178194460
Show fit
|
0.15 |
ENST00000392505.6
ENST00000359685.7
ENST00000357080.8
ENST00000190611.9
ENST00000409045.7
|
OSBPL6
|
oxysterol binding protein like 6
|
|
chr19_-_14206168
Show fit
|
0.15 |
ENST00000361434.7
ENST00000340736.10
|
ADGRL1
|
adhesion G protein-coupled receptor L1
|
|
chr14_+_75069577
Show fit
|
0.15 |
ENST00000238686.8
|
ZC2HC1C
|
zinc finger C2HC-type containing 1C
|
|
chr14_+_75069632
Show fit
|
0.15 |
ENST00000439583.2
ENST00000554763.2
ENST00000524913.3
ENST00000525046.2
ENST00000674086.1
ENST00000526130.2
ENST00000674094.1
ENST00000532198.2
|
ZC2HC1C
|
zinc finger C2HC-type containing 1C
|
|
chr16_+_71358713
Show fit
|
0.15 |
ENST00000349553.9
ENST00000302628.9
ENST00000562305.5
|
CALB2
|
calbindin 2
|
|
chr22_+_19723525
Show fit
|
0.15 |
ENST00000366425.4
|
GP1BB
|
glycoprotein Ib platelet subunit beta
|
|
chr11_-_6320494
Show fit
|
0.15 |
ENST00000303927.4
ENST00000530979.1
|
CAVIN3
|
caveolae associated protein 3
|
|
chr3_+_15206179
Show fit
|
0.15 |
ENST00000253693.7
|
CAPN7
|
calpain 7
|
|
chr11_-_120120880
Show fit
|
0.15 |
ENST00000526881.1
|
TRIM29
|
tripartite motif containing 29
|
|
chr2_-_219571241
Show fit
|
0.14 |
ENST00000373876.5
ENST00000603926.5
ENST00000373873.8
ENST00000289656.3
|
OBSL1
|
obscurin like cytoskeletal adaptor 1
|
|
chr13_-_36131286
Show fit
|
0.14 |
ENST00000255448.8
ENST00000379892.4
|
DCLK1
|
doublecortin like kinase 1
|
|
chr1_-_202807988
Show fit
|
0.14 |
ENST00000648738.1
ENST00000648354.1
|
KDM5B
|
lysine demethylase 5B
|
|
chr17_-_8118489
Show fit
|
0.14 |
ENST00000380149.6
ENST00000448843.7
|
ALOXE3
|
arachidonate lipoxygenase 3
|
|
chr12_-_52618559
Show fit
|
0.14 |
ENST00000305748.7
|
KRT73
|
keratin 73
|
|
chr17_-_46193372
Show fit
|
0.14 |
ENST00000638275.1
|
KANSL1
|
KAT8 regulatory NSL complex subunit 1
|
|
chr19_+_22286433
Show fit
|
0.14 |
ENST00000601693.2
|
ZNF729
|
zinc finger protein 729
|
|
chr17_-_46193412
Show fit
|
0.14 |
ENST00000432791.7
|
KANSL1
|
KAT8 regulatory NSL complex subunit 1
|
|
chr2_-_44361485
Show fit
|
0.14 |
ENST00000438314.1
ENST00000409411.6
ENST00000409936.5
|
PREPL
|
prolyl endopeptidase like
|
|
chr2_-_40512361
Show fit
|
0.14 |
ENST00000403092.5
|
SLC8A1
|
solute carrier family 8 member A1
|
|
chr5_+_100535317
Show fit
|
0.14 |
ENST00000312637.5
|
FAM174A
|
family with sequence similarity 174 member A
|
|
chr11_+_63681573
Show fit
|
0.14 |
ENST00000354497.4
|
RTN3
|
reticulon 3
|
|
chr12_-_51270274
Show fit
|
0.14 |
ENST00000605627.1
|
SMAGP
|
small cell adhesion glycoprotein
|
|
chr18_+_3449413
Show fit
|
0.13 |
ENST00000549253.5
|
TGIF1
|
TGFB induced factor homeobox 1
|
|
chr11_+_5488685
Show fit
|
0.13 |
ENST00000322641.5
|
OR52D1
|
olfactory receptor family 52 subfamily D member 1
|
|
chr7_-_8262668
Show fit
|
0.13 |
ENST00000446305.1
|
ICA1
|
islet cell autoantigen 1
|
|
chr2_+_219544002
Show fit
|
0.13 |
ENST00000421791.1
ENST00000373883.4
ENST00000451952.1
|
TMEM198
|
transmembrane protein 198
|
|
chr17_-_10114546
Show fit
|
0.13 |
ENST00000323816.8
|
GAS7
|
growth arrest specific 7
|
|
chr9_-_21974821
Show fit
|
0.13 |
ENST00000304494.10
ENST00000579122.1
ENST00000498124.1
|
CDKN2A
|
cyclin dependent kinase inhibitor 2A
|
|
chr17_-_46192767
Show fit
|
0.13 |
ENST00000262419.10
|
KANSL1
|
KAT8 regulatory NSL complex subunit 1
|
|
chr16_+_28984795
Show fit
|
0.13 |
ENST00000395461.7
|
LAT
|
linker for activation of T cells
|
|
chr1_+_75724431
Show fit
|
0.12 |
ENST00000541113.6
ENST00000680805.1
|
ACADM
|
acyl-CoA dehydrogenase medium chain
|
|
chr11_+_31816266
Show fit
|
0.12 |
ENST00000644607.1
ENST00000646221.1
ENST00000643671.1
ENST00000643931.1
ENST00000642614.1
ENST00000642818.1
ENST00000645848.1
ENST00000506388.2
ENST00000645824.1
ENST00000532942.5
|
PAUPAR
ENSG00000285283.1
|
PAX6 upstream antisense RNA
novel protein
|
|
chr5_+_177086867
Show fit
|
0.12 |
ENST00000503708.5
ENST00000393648.6
ENST00000514472.1
ENST00000502906.5
ENST00000292408.9
ENST00000510911.5
|
FGFR4
|
fibroblast growth factor receptor 4
|
|
chr5_+_123512087
Show fit
|
0.12 |
ENST00000345990.8
|
CSNK1G3
|
casein kinase 1 gamma 3
|
|
chr14_-_67515153
Show fit
|
0.12 |
ENST00000555994.6
|
TMEM229B
|
transmembrane protein 229B
|
|
chr11_-_5324297
Show fit
|
0.11 |
ENST00000624187.1
|
OR51B2
|
olfactory receptor family 51 subfamily B member 2
|
|
chr19_-_17460893
Show fit
|
0.11 |
ENST00000301944.3
|
NXNL1
|
nucleoredoxin like 1
|
|
chr5_+_134905100
Show fit
|
0.11 |
ENST00000512783.5
ENST00000254908.11
|
PCBD2
|
pterin-4 alpha-carbinolamine dehydratase 2
|
|
chr12_-_16606102
Show fit
|
0.11 |
ENST00000537304.6
|
LMO3
|
LIM domain only 3
|
|
chr11_-_118152775
Show fit
|
0.11 |
ENST00000324727.9
|
SCN4B
|
sodium voltage-gated channel beta subunit 4
|
|
chr17_-_5123102
Show fit
|
0.11 |
ENST00000250076.7
|
ZNF232
|
zinc finger protein 232
|
|
chrX_-_41923547
Show fit
|
0.11 |
ENST00000378163.7
|
CASK
|
calcium/calmodulin dependent serine protein kinase
|
|
chr12_-_16605939
Show fit
|
0.11 |
ENST00000541295.5
ENST00000535535.5
|
LMO3
|
LIM domain only 3
|
|
chr19_+_35292145
Show fit
|
0.11 |
ENST00000595791.5
ENST00000597035.5
ENST00000537831.2
ENST00000392213.8
|
MAG
|
myelin associated glycoprotein
|
|
chr16_+_24845987
Show fit
|
0.11 |
ENST00000565769.5
ENST00000569520.5
|
SLC5A11
|
solute carrier family 5 member 11
|
|
chr19_+_7348930
Show fit
|
0.10 |
ENST00000668164.2
|
ARHGEF18
|
Rho/Rac guanine nucleotide exchange factor 18
|
|
chr12_+_4909895
Show fit
|
0.10 |
ENST00000638821.1
ENST00000382545.5
|
ENSG00000256654.4
KCNA1
|
novel transcript, sense overlapping KCNA1
potassium voltage-gated channel subfamily A member 1
|
|
chr16_-_10182394
Show fit
|
0.10 |
ENST00000330684.4
|
GRIN2A
|
glutamate ionotropic receptor NMDA type subunit 2A
|
|
chr22_-_29555178
Show fit
|
0.10 |
ENST00000418021.1
|
THOC5
|
THO complex 5
|
|
chr19_+_35292125
Show fit
|
0.10 |
ENST00000600291.5
ENST00000361922.8
|
MAG
|
myelin associated glycoprotein
|
|
chr15_+_78872954
Show fit
|
0.10 |
ENST00000558746.5
ENST00000558830.5
ENST00000559345.5
|
MORF4L1
|
mortality factor 4 like 1
|
|
chr17_+_29593118
Show fit
|
0.10 |
ENST00000394859.8
|
ANKRD13B
|
ankyrin repeat domain 13B
|
|
chr10_+_44959780
Show fit
|
0.10 |
ENST00000340258.10
ENST00000427758.5
|
RASSF4
|
Ras association domain family member 4
|
|
chr14_-_77271200
Show fit
|
0.10 |
ENST00000298352.5
|
NGB
|
neuroglobin
|
|
chr2_-_40512423
Show fit
|
0.10 |
ENST00000402441.5
ENST00000448531.1
|
SLC8A1
|
solute carrier family 8 member A1
|