Project
Inflammatory response time course, HUVEC (Wada, 2009)
Navigation
Downloads
Results for GFI1
Z-value: 1.55
Transcription factors associated with GFI1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
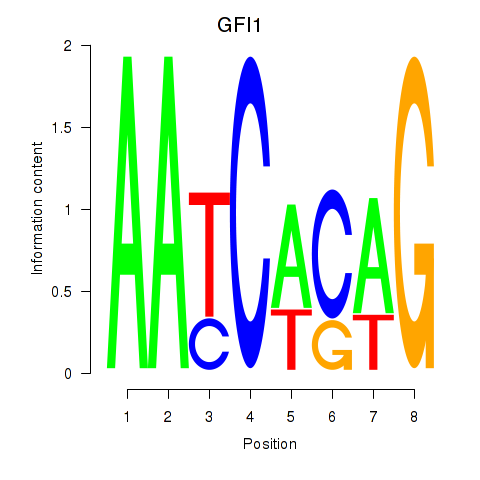
GFI1
|
ENSG00000162676.12 | GFI1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| GFI1 | hg38_v1_chr1_-_92483947_92483960, hg38_v1_chr1_-_92486049_92486104, hg38_v1_chr1_-_92486916_92486932 | -0.05 | 8.1e-01 | Click! |
Activity profile of GFI1 motif
Sorted Z-values of GFI1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of GFI1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_+_41612892 | 7.84 |
ENST00000509454.5
ENST00000396595.7 ENST00000381753.8 |
LIMCH1
|
LIM and calponin homology domains 1 |
| chr1_+_164559173 | 7.81 |
ENST00000420696.7
|
PBX1
|
PBX homeobox 1 |
| chr9_+_2158239 | 7.23 |
ENST00000635133.1
ENST00000634931.1 ENST00000423555.6 ENST00000382185.6 ENST00000302401.8 ENST00000382183.6 ENST00000417599.6 ENST00000382186.6 ENST00000635530.1 ENST00000635388.1 |
SMARCA2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr4_+_41612702 | 7.15 |
ENST00000509277.5
|
LIMCH1
|
LIM and calponin homology domains 1 |
| chr9_+_2158487 | 6.79 |
ENST00000634706.1
ENST00000634338.1 ENST00000635688.1 ENST00000634435.1 |
SMARCA2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr4_+_41360759 | 6.66 |
ENST00000508501.5
ENST00000512946.5 ENST00000313860.11 ENST00000512632.5 ENST00000512820.5 |
LIMCH1
|
LIM and calponin homology domains 1 |
| chr12_-_57818704 | 5.72 |
ENST00000549994.1
|
AVIL
|
advillin |
| chr6_+_135851681 | 5.65 |
ENST00000308191.11
|
PDE7B
|
phosphodiesterase 7B |
| chr1_+_61077219 | 5.11 |
ENST00000407417.7
|
NFIA
|
nuclear factor I A |
| chr18_+_44700796 | 5.10 |
ENST00000677130.1
|
SETBP1
|
SET binding protein 1 |
| chr4_+_42397473 | 4.92 |
ENST00000319234.5
|
SHISA3
|
shisa family member 3 |
| chr3_+_39809602 | 4.34 |
ENST00000302541.11
ENST00000396217.7 |
MYRIP
|
myosin VIIA and Rab interacting protein |
| chr9_+_2159672 | 4.29 |
ENST00000634343.1
|
SMARCA2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr18_-_25352116 | 4.12 |
ENST00000584787.5
ENST00000538137.6 ENST00000361524.8 |
ZNF521
|
zinc finger protein 521 |
| chr15_-_70702273 | 3.94 |
ENST00000558758.5
ENST00000379983.6 ENST00000560441.5 |
UACA
|
uveal autoantigen with coiled-coil domains and ankyrin repeats |
| chr9_-_14321948 | 3.82 |
ENST00000635877.1
ENST00000636432.1 ENST00000646622.1 |
NFIB
|
nuclear factor I B |
| chr17_+_63477052 | 3.82 |
ENST00000290866.10
ENST00000428043.5 |
ACE
|
angiotensin I converting enzyme |
| chr5_-_88883701 | 3.78 |
ENST00000636998.1
|
MEF2C
|
myocyte enhancer factor 2C |
| chr3_+_8501846 | 3.63 |
ENST00000454244.4
|
LMCD1
|
LIM and cysteine rich domains 1 |
| chr2_+_66435116 | 3.63 |
ENST00000272369.14
ENST00000560281.6 |
MEIS1
|
Meis homeobox 1 |
| chr1_+_163069353 | 3.62 |
ENST00000531057.5
ENST00000527809.5 ENST00000367908.8 ENST00000367909.11 |
RGS4
|
regulator of G protein signaling 4 |
| chr4_-_69760596 | 3.60 |
ENST00000510821.1
|
SULT1B1
|
sulfotransferase family 1B member 1 |
| chr15_+_96332432 | 3.51 |
ENST00000559679.1
ENST00000394171.6 |
NR2F2
|
nuclear receptor subfamily 2 group F member 2 |
| chr7_-_120858066 | 3.47 |
ENST00000222747.8
|
TSPAN12
|
tetraspanin 12 |
| chr9_+_2159850 | 3.45 |
ENST00000416751.2
|
SMARCA2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chrX_+_71910818 | 3.31 |
ENST00000633930.1
|
NHSL2
|
NHS like 2 |
| chr9_-_14180779 | 3.20 |
ENST00000380924.1
ENST00000543693.5 |
NFIB
|
nuclear factor I B |
| chr14_-_21536928 | 2.97 |
ENST00000613414.4
|
SALL2
|
spalt like transcription factor 2 |
| chr2_+_66435558 | 2.85 |
ENST00000488550.5
|
MEIS1
|
Meis homeobox 1 |
| chr1_+_162381703 | 2.84 |
ENST00000458626.4
|
C1orf226
|
chromosome 1 open reading frame 226 |
| chr3_+_8501807 | 2.81 |
ENST00000426878.2
ENST00000397386.7 ENST00000415597.5 ENST00000157600.8 |
LMCD1
|
LIM and cysteine rich domains 1 |
| chr7_+_102912983 | 2.78 |
ENST00000339431.9
ENST00000249377.4 |
LRRC17
|
leucine rich repeat containing 17 |
| chr7_-_132576493 | 2.77 |
ENST00000321063.8
|
PLXNA4
|
plexin A4 |
| chr9_-_97697297 | 2.72 |
ENST00000375128.5
|
XPA
|
XPA, DNA damage recognition and repair factor |
| chr6_+_85449584 | 2.63 |
ENST00000369651.7
|
NT5E
|
5'-nucleotidase ecto |
| chr5_-_88883420 | 2.54 |
ENST00000437473.6
|
MEF2C
|
myocyte enhancer factor 2C |
| chr2_+_233195433 | 2.50 |
ENST00000417661.1
|
INPP5D
|
inositol polyphosphate-5-phosphatase D |
| chr3_-_169146595 | 2.49 |
ENST00000468789.5
|
MECOM
|
MDS1 and EVI1 complex locus |
| chr1_+_164559739 | 2.48 |
ENST00000627490.2
|
PBX1
|
PBX homeobox 1 |
| chr5_-_111756245 | 2.46 |
ENST00000447165.6
|
NREP
|
neuronal regeneration related protein |
| chr19_+_36111151 | 2.45 |
ENST00000633214.1
ENST00000585332.3 |
OVOL3
|
ovo like zinc finger 3 |
| chr11_+_5624987 | 2.44 |
ENST00000429814.3
|
TRIM34
|
tripartite motif containing 34 |
| chr14_-_21536884 | 2.41 |
ENST00000546363.5
|
SALL2
|
spalt like transcription factor 2 |
| chr6_-_52994248 | 2.40 |
ENST00000457564.1
ENST00000370960.5 |
GSTA4
|
glutathione S-transferase alpha 4 |
| chr3_-_66038537 | 2.36 |
ENST00000483466.5
|
MAGI1
|
membrane associated guanylate kinase, WW and PDZ domain containing 1 |
| chr3_-_15797930 | 2.22 |
ENST00000683139.1
|
ANKRD28
|
ankyrin repeat domain 28 |
| chr4_-_69760610 | 2.21 |
ENST00000310613.8
|
SULT1B1
|
sulfotransferase family 1B member 1 |
| chr3_-_15798184 | 2.17 |
ENST00000624145.3
|
ANKRD28
|
ankyrin repeat domain 28 |
| chr14_-_21537206 | 2.08 |
ENST00000614342.1
|
SALL2
|
spalt like transcription factor 2 |
| chr5_-_88823763 | 2.05 |
ENST00000635898.1
ENST00000626391.2 ENST00000628656.2 |
MEF2C
|
myocyte enhancer factor 2C |
| chr11_-_83071819 | 2.04 |
ENST00000524635.1
ENST00000526205.5 ENST00000533486.5 ENST00000533276.6 ENST00000527633.6 |
RAB30
|
RAB30, member RAS oncogene family |
| chr7_-_27156646 | 2.03 |
ENST00000242159.5
|
HOXA7
|
homeobox A7 |
| chr1_+_163068775 | 2.02 |
ENST00000421743.6
|
RGS4
|
regulator of G protein signaling 4 |
| chr14_-_22815856 | 1.99 |
ENST00000554758.1
ENST00000397528.8 |
SLC7A7
|
solute carrier family 7 member 7 |
| chr7_-_20786879 | 1.94 |
ENST00000418710.3
ENST00000617581.4 ENST00000361443.4 |
SP8
|
Sp8 transcription factor |
| chr7_+_16753731 | 1.93 |
ENST00000262067.5
|
TSPAN13
|
tetraspanin 13 |
| chr14_-_22815801 | 1.92 |
ENST00000397532.9
|
SLC7A7
|
solute carrier family 7 member 7 |
| chr12_-_58920465 | 1.90 |
ENST00000320743.8
|
LRIG3
|
leucine rich repeats and immunoglobulin like domains 3 |
| chr12_-_104050112 | 1.88 |
ENST00000547583.1
ENST00000546851.1 ENST00000360814.9 |
GLT8D2
|
glycosyltransferase 8 domain containing 2 |
| chr1_+_210232776 | 1.88 |
ENST00000367012.4
|
SERTAD4
|
SERTA domain containing 4 |
| chr18_-_55423757 | 1.85 |
ENST00000675707.1
|
TCF4
|
transcription factor 4 |
| chr9_-_76906090 | 1.85 |
ENST00000376718.8
|
PRUNE2
|
prune homolog 2 with BCH domain |
| chrX_+_139530730 | 1.84 |
ENST00000218099.7
|
F9
|
coagulation factor IX |
| chr22_+_31122923 | 1.80 |
ENST00000620191.4
ENST00000412277.6 ENST00000412985.5 ENST00000331075.10 ENST00000420017.5 ENST00000400294.6 ENST00000405300.5 ENST00000404390.7 |
INPP5J
|
inositol polyphosphate-5-phosphatase J |
| chr14_-_22815421 | 1.79 |
ENST00000674313.1
ENST00000555959.1 |
SLC7A7
|
solute carrier family 7 member 7 |
| chr4_+_107931541 | 1.78 |
ENST00000332884.11
ENST00000508453.1 |
CYP2U1
|
cytochrome P450 family 2 subfamily U member 1 |
| chr21_-_38661694 | 1.77 |
ENST00000417133.6
ENST00000398910.5 ENST00000442448.5 ENST00000429727.6 |
ERG
|
ETS transcription factor ERG |
| chr2_-_157876301 | 1.77 |
ENST00000434821.7
|
ACVR1
|
activin A receptor type 1 |
| chr9_-_76906041 | 1.75 |
ENST00000443509.6
ENST00000428286.5 ENST00000376713.3 |
PRUNE2
|
prune homolog 2 with BCH domain |
| chr4_-_88823306 | 1.73 |
ENST00000395002.6
|
FAM13A
|
family with sequence similarity 13 member A |
| chr17_+_6070361 | 1.72 |
ENST00000317744.10
|
WSCD1
|
WSC domain containing 1 |
| chr6_+_25279359 | 1.71 |
ENST00000329474.7
|
CARMIL1
|
capping protein regulator and myosin 1 linker 1 |
| chr11_-_72080472 | 1.71 |
ENST00000537217.5
ENST00000366394.7 ENST00000358965.10 ENST00000546131.1 ENST00000393695.8 ENST00000543937.5 ENST00000368959.9 ENST00000541641.5 |
NUMA1
|
nuclear mitotic apparatus protein 1 |
| chr16_+_11965193 | 1.71 |
ENST00000053243.6
ENST00000396495.3 |
TNFRSF17
|
TNF receptor superfamily member 17 |
| chr1_-_151909555 | 1.69 |
ENST00000489410.1
|
THEM4
|
thioesterase superfamily member 4 |
| chr2_-_69643152 | 1.68 |
ENST00000606389.7
|
AAK1
|
AP2 associated kinase 1 |
| chr6_-_7910776 | 1.68 |
ENST00000379757.9
|
TXNDC5
|
thioredoxin domain containing 5 |
| chr14_-_106349792 | 1.67 |
ENST00000438142.3
|
IGHV4-31
|
immunoglobulin heavy variable 4-31 |
| chr15_+_96333111 | 1.67 |
ENST00000453270.2
|
NR2F2
|
nuclear receptor subfamily 2 group F member 2 |
| chr2_-_157874976 | 1.60 |
ENST00000682025.1
ENST00000683487.1 ENST00000682300.1 ENST00000683441.1 ENST00000684595.1 ENST00000683426.1 ENST00000683820.1 ENST00000263640.7 |
ACVR1
|
activin A receptor type 1 |
| chr20_+_46709623 | 1.59 |
ENST00000359271.4
|
SLC2A10
|
solute carrier family 2 member 10 |
| chr19_+_4909430 | 1.57 |
ENST00000620565.4
ENST00000613817.4 ENST00000624301.3 ENST00000650932.1 |
UHRF1
|
ubiquitin like with PHD and ring finger domains 1 |
| chr21_-_30372265 | 1.54 |
ENST00000399889.4
|
KRTAP13-2
|
keratin associated protein 13-2 |
| chr18_+_58864866 | 1.53 |
ENST00000588456.5
ENST00000591808.6 ENST00000589481.1 ENST00000591049.1 |
ZNF532
|
zinc finger protein 532 |
| chr8_+_69466617 | 1.53 |
ENST00000525061.5
ENST00000260128.8 ENST00000458141.6 |
SULF1
|
sulfatase 1 |
| chr11_-_72080680 | 1.52 |
ENST00000613205.4
|
NUMA1
|
nuclear mitotic apparatus protein 1 |
| chr2_+_176122712 | 1.52 |
ENST00000249499.8
|
HOXD9
|
homeobox D9 |
| chr19_+_54906140 | 1.51 |
ENST00000291890.9
ENST00000598576.5 ENST00000594765.5 ENST00000350790.9 ENST00000338835.9 ENST00000357397.5 |
NCR1
|
natural cytotoxicity triggering receptor 1 |
| chr3_-_112610262 | 1.51 |
ENST00000479368.1
|
CCDC80
|
coiled-coil domain containing 80 |
| chr1_+_154405326 | 1.50 |
ENST00000368485.8
|
IL6R
|
interleukin 6 receptor |
| chr8_+_117520696 | 1.47 |
ENST00000297347.7
|
MED30
|
mediator complex subunit 30 |
| chr11_+_124115404 | 1.45 |
ENST00000361352.9
ENST00000449321.5 ENST00000392748.5 ENST00000392744.4 ENST00000456829.7 |
VWA5A
|
von Willebrand factor A domain containing 5A |
| chr16_+_82056423 | 1.44 |
ENST00000568090.5
|
HSD17B2
|
hydroxysteroid 17-beta dehydrogenase 2 |
| chr7_-_120858303 | 1.43 |
ENST00000415871.5
ENST00000430985.1 |
TSPAN12
|
tetraspanin 12 |
| chr5_-_91383310 | 1.43 |
ENST00000265138.4
|
ARRDC3
|
arrestin domain containing 3 |
| chr6_-_145964330 | 1.41 |
ENST00000275233.12
ENST00000367505.6 |
SHPRH
|
SNF2 histone linker PHD RING helicase |
| chr16_-_67483541 | 1.39 |
ENST00000290953.3
|
AGRP
|
agouti related neuropeptide |
| chr5_+_41925223 | 1.35 |
ENST00000296812.6
ENST00000281623.8 ENST00000509134.1 |
FBXO4
|
F-box protein 4 |
| chr8_+_94823210 | 1.35 |
ENST00000521860.5
ENST00000523731.6 ENST00000519457.5 ENST00000519053.5 |
INTS8
|
integrator complex subunit 8 |
| chr11_+_73308237 | 1.34 |
ENST00000263674.4
|
ARHGEF17
|
Rho guanine nucleotide exchange factor 17 |
| chr3_-_114624193 | 1.33 |
ENST00000481632.5
|
ZBTB20
|
zinc finger and BTB domain containing 20 |
| chr16_+_4788411 | 1.31 |
ENST00000589327.5
|
SMIM22
|
small integral membrane protein 22 |
| chr17_-_18363451 | 1.31 |
ENST00000354098.7
|
SHMT1
|
serine hydroxymethyltransferase 1 |
| chr7_+_120951116 | 1.31 |
ENST00000431467.1
|
ING3
|
inhibitor of growth family member 3 |
| chr3_+_50269140 | 1.30 |
ENST00000616701.5
ENST00000433753.4 ENST00000611067.4 |
SEMA3B
|
semaphorin 3B |
| chr2_+_188974364 | 1.29 |
ENST00000304636.9
ENST00000317840.9 |
COL3A1
|
collagen type III alpha 1 chain |
| chr1_+_153259684 | 1.29 |
ENST00000368742.4
|
LORICRIN
|
loricrin cornified envelope precursor protein |
| chr3_-_114759115 | 1.29 |
ENST00000471418.5
|
ZBTB20
|
zinc finger and BTB domain containing 20 |
| chr20_-_23086316 | 1.29 |
ENST00000246006.5
|
CD93
|
CD93 molecule |
| chr5_+_150778781 | 1.26 |
ENST00000648745.1
|
SMIM3
|
small integral membrane protein 3 |
| chr5_+_150778733 | 1.26 |
ENST00000526627.2
|
SMIM3
|
small integral membrane protein 3 |
| chr16_+_30378312 | 1.26 |
ENST00000528032.5
ENST00000622647.3 |
ZNF48
|
zinc finger protein 48 |
| chr2_+_232539720 | 1.24 |
ENST00000389492.3
|
CHRNG
|
cholinergic receptor nicotinic gamma subunit |
| chr8_-_6563044 | 1.24 |
ENST00000338312.10
|
ANGPT2
|
angiopoietin 2 |
| chr8_+_67952028 | 1.23 |
ENST00000288368.5
|
PREX2
|
phosphatidylinositol-3,4,5-trisphosphate dependent Rac exchange factor 2 |
| chr15_+_41838839 | 1.21 |
ENST00000458483.4
|
PLA2G4B
|
phospholipase A2 group IVB |
| chr10_+_47322450 | 1.21 |
ENST00000581492.3
|
GDF2
|
growth differentiation factor 2 |
| chr1_+_243256034 | 1.19 |
ENST00000366541.8
|
SDCCAG8
|
SHH signaling and ciliogenesis regulator SDCCAG8 |
| chr1_-_46668394 | 1.19 |
ENST00000371937.8
ENST00000329231.8 |
ATPAF1
|
ATP synthase mitochondrial F1 complex assembly factor 1 |
| chr11_-_83034195 | 1.19 |
ENST00000531021.5
|
RAB30
|
RAB30, member RAS oncogene family |
| chr11_-_95231046 | 1.18 |
ENST00000416495.6
ENST00000536441.7 |
SESN3
|
sestrin 3 |
| chr5_+_58583068 | 1.18 |
ENST00000282878.6
|
RAB3C
|
RAB3C, member RAS oncogene family |
| chr3_+_113838772 | 1.17 |
ENST00000358160.9
|
GRAMD1C
|
GRAM domain containing 1C |
| chr1_-_46668317 | 1.17 |
ENST00000574428.5
|
ATPAF1
|
ATP synthase mitochondrial F1 complex assembly factor 1 |
| chr3_-_114758940 | 1.17 |
ENST00000464560.5
|
ZBTB20
|
zinc finger and BTB domain containing 20 |
| chr1_+_150364136 | 1.16 |
ENST00000369068.5
|
RPRD2
|
regulation of nuclear pre-mRNA domain containing 2 |
| chr6_+_28124596 | 1.16 |
ENST00000340487.5
|
ZSCAN16
|
zinc finger and SCAN domain containing 16 |
| chr2_+_1414382 | 1.15 |
ENST00000423320.5
ENST00000346956.7 ENST00000382198.5 |
TPO
|
thyroid peroxidase |
| chr18_-_55589770 | 1.13 |
ENST00000565018.6
ENST00000636400.2 |
TCF4
|
transcription factor 4 |
| chr19_+_54965252 | 1.13 |
ENST00000543010.5
ENST00000391721.8 ENST00000339757.11 |
NLRP2
|
NLR family pyrin domain containing 2 |
| chr6_-_145964084 | 1.10 |
ENST00000438092.6
ENST00000629427.2 |
SHPRH
|
SNF2 histone linker PHD RING helicase |
| chr8_-_6563238 | 1.07 |
ENST00000629816.3
ENST00000523120.2 |
ANGPT2
|
angiopoietin 2 |
| chr10_-_63269057 | 1.07 |
ENST00000542921.5
|
JMJD1C
|
jumonji domain containing 1C |
| chr17_+_35844077 | 1.06 |
ENST00000604694.1
|
TAF15
|
TATA-box binding protein associated factor 15 |
| chr5_-_35230332 | 1.05 |
ENST00000504500.5
|
PRLR
|
prolactin receptor |
| chr1_+_154405193 | 1.04 |
ENST00000622330.4
ENST00000344086.8 |
IL6R
|
interleukin 6 receptor |
| chr12_-_117968223 | 1.04 |
ENST00000425217.5
|
KSR2
|
kinase suppressor of ras 2 |
| chr18_-_55589836 | 1.03 |
ENST00000537578.5
ENST00000564403.6 |
TCF4
|
transcription factor 4 |
| chr7_-_38909174 | 1.03 |
ENST00000395969.6
ENST00000310301.9 ENST00000414632.5 |
VPS41
|
VPS41 subunit of HOPS complex |
| chr12_-_14951106 | 1.03 |
ENST00000541644.5
ENST00000545895.5 |
ARHGDIB
|
Rho GDP dissociation inhibitor beta |
| chr2_-_159616442 | 1.03 |
ENST00000541068.6
ENST00000392783.7 ENST00000392782.5 |
BAZ2B
|
bromodomain adjacent to zinc finger domain 2B |
| chr1_-_147760592 | 1.02 |
ENST00000579774.3
|
GJA5
|
gap junction protein alpha 5 |
| chr2_-_46617020 | 1.01 |
ENST00000474980.1
ENST00000281382.11 ENST00000306465.8 |
PIGF
|
phosphatidylinositol glycan anchor biosynthesis class F |
| chr8_-_6563409 | 1.01 |
ENST00000325203.9
|
ANGPT2
|
angiopoietin 2 |
| chr18_-_55589795 | 1.01 |
ENST00000568740.5
ENST00000629387.2 |
TCF4
|
transcription factor 4 |
| chr1_-_161069962 | 1.00 |
ENST00000368015.1
|
ARHGAP30
|
Rho GTPase activating protein 30 |
| chr3_-_138132381 | 1.00 |
ENST00000236709.4
|
A4GNT
|
alpha-1,4-N-acetylglucosaminyltransferase |
| chr12_+_48119323 | 0.99 |
ENST00000551339.6
ENST00000629846.2 ENST00000359794.11 ENST00000548345.5 |
PFKM
|
phosphofructokinase, muscle |
| chr1_-_60073750 | 0.99 |
ENST00000371201.3
|
C1orf87
|
chromosome 1 open reading frame 87 |
| chr21_-_46228751 | 0.99 |
ENST00000450351.1
ENST00000397728.8 ENST00000522411.5 ENST00000356396.8 ENST00000457828.6 |
LSS
|
lanosterol synthase |
| chr7_-_130441136 | 0.99 |
ENST00000675596.1
ENST00000676312.1 |
CEP41
|
centrosomal protein 41 |
| chr14_-_20305932 | 0.98 |
ENST00000258821.8
ENST00000553828.1 |
TTC5
|
tetratricopeptide repeat domain 5 |
| chr12_+_48119787 | 0.98 |
ENST00000551804.5
|
PFKM
|
phosphofructokinase, muscle |
| chr1_-_161069857 | 0.97 |
ENST00000368013.8
|
ARHGAP30
|
Rho GTPase activating protein 30 |
| chrX_+_85243983 | 0.97 |
ENST00000674551.1
|
ZNF711
|
zinc finger protein 711 |
| chr18_+_3448456 | 0.97 |
ENST00000549780.5
|
TGIF1
|
TGFB induced factor homeobox 1 |
| chr7_-_92134417 | 0.96 |
ENST00000003100.13
|
CYP51A1
|
cytochrome P450 family 51 subfamily A member 1 |
| chr6_+_78867524 | 0.96 |
ENST00000369940.7
|
IRAK1BP1
|
interleukin 1 receptor associated kinase 1 binding protein 1 |
| chr19_-_39540144 | 0.96 |
ENST00000390658.4
|
EID2
|
EP300 interacting inhibitor of differentiation 2 |
| chr1_-_46668454 | 0.94 |
ENST00000576409.5
|
ATPAF1
|
ATP synthase mitochondrial F1 complex assembly factor 1 |
| chr1_-_60073733 | 0.93 |
ENST00000450089.6
|
C1orf87
|
chromosome 1 open reading frame 87 |
| chr6_+_6588708 | 0.93 |
ENST00000230568.5
|
LY86
|
lymphocyte antigen 86 |
| chr11_-_65780917 | 0.91 |
ENST00000532090.3
|
AP5B1
|
adaptor related protein complex 5 subunit beta 1 |
| chr20_-_35411963 | 0.91 |
ENST00000349714.9
ENST00000438533.5 ENST00000359226.6 ENST00000374384.6 ENST00000374385.10 ENST00000424405.5 ENST00000397554.5 ENST00000374380.6 |
UQCC1
|
ubiquinol-cytochrome c reductase complex assembly factor 1 |
| chr1_+_150364621 | 0.90 |
ENST00000401000.8
|
RPRD2
|
regulation of nuclear pre-mRNA domain containing 2 |
| chrX_-_134658450 | 0.90 |
ENST00000359237.9
|
PLAC1
|
placenta enriched 1 |
| chr1_-_151909501 | 0.89 |
ENST00000368814.8
|
THEM4
|
thioesterase superfamily member 4 |
| chrX_+_85244032 | 0.89 |
ENST00000373165.7
|
ZNF711
|
zinc finger protein 711 |
| chr6_-_36757380 | 0.89 |
ENST00000393189.2
|
CPNE5
|
copine 5 |
| chr19_+_54630497 | 0.88 |
ENST00000396332.8
ENST00000427581.6 |
LILRB1
|
leukocyte immunoglobulin like receptor B1 |
| chr1_-_161069666 | 0.88 |
ENST00000368016.7
|
ARHGAP30
|
Rho GTPase activating protein 30 |
| chr6_-_32953017 | 0.88 |
ENST00000395305.7
ENST00000374843.9 ENST00000395303.7 ENST00000429234.1 |
HLA-DMA
ENSG00000248993.1
|
major histocompatibility complex, class II, DM alpha novel protein |
| chr6_-_79234713 | 0.88 |
ENST00000620514.1
|
HMGN3
|
high mobility group nucleosomal binding domain 3 |
| chr10_+_96129707 | 0.87 |
ENST00000316045.9
|
ZNF518A
|
zinc finger protein 518A |
| chr7_-_92134737 | 0.87 |
ENST00000450723.5
|
CYP51A1
|
cytochrome P450 family 51 subfamily A member 1 |
| chr17_+_1762052 | 0.87 |
ENST00000254722.9
ENST00000576406.5 ENST00000571149.5 |
SERPINF1
|
serpin family F member 1 |
| chr2_+_232539683 | 0.87 |
ENST00000651502.1
|
CHRNG
|
cholinergic receptor nicotinic gamma subunit |
| chr3_+_141738263 | 0.87 |
ENST00000480908.1
ENST00000393000.3 ENST00000273480.4 |
RNF7
|
ring finger protein 7 |
| chr6_-_106629472 | 0.87 |
ENST00000369063.8
|
RTN4IP1
|
reticulon 4 interacting protein 1 |
| chr2_+_46617180 | 0.86 |
ENST00000238892.4
|
CRIPT
|
CXXC repeat containing interactor of PDZ3 domain |
| chr6_+_138773747 | 0.86 |
ENST00000617445.5
|
CCDC28A
|
coiled-coil domain containing 28A |
| chr19_+_15949008 | 0.85 |
ENST00000322107.1
|
OR10H4
|
olfactory receptor family 10 subfamily H member 4 |
| chr4_+_87608529 | 0.84 |
ENST00000651931.1
|
DSPP
|
dentin sialophosphoprotein |
| chr3_+_111674654 | 0.84 |
ENST00000636933.1
ENST00000393934.7 ENST00000477665.2 |
PLCXD2
|
phosphatidylinositol specific phospholipase C X domain containing 2 |
| chr17_-_18363504 | 0.83 |
ENST00000583780.1
ENST00000316694.8 ENST00000352886.10 |
SHMT1
|
serine hydroxymethyltransferase 1 |
| chr10_-_100519829 | 0.83 |
ENST00000370345.8
|
SEC31B
|
SEC31 homolog B, COPII coat complex component |
| chr6_+_80106655 | 0.82 |
ENST00000320393.9
|
BCKDHB
|
branched chain keto acid dehydrogenase E1 subunit beta |
| chr1_+_15341744 | 0.81 |
ENST00000444385.5
|
FHAD1
|
forkhead associated phosphopeptide binding domain 1 |
| chr15_-_23687290 | 0.81 |
ENST00000649030.2
|
NDN
|
necdin, MAGE family member |
| chr1_+_162632454 | 0.81 |
ENST00000367921.8
ENST00000367922.7 |
DDR2
|
discoidin domain receptor tyrosine kinase 2 |
| chr2_+_201257980 | 0.80 |
ENST00000447616.5
ENST00000358485.8 ENST00000392266.7 |
CASP8
|
caspase 8 |
| chr1_+_168280872 | 0.79 |
ENST00000367821.8
|
TBX19
|
T-box transcription factor 19 |
| chr14_+_22105305 | 0.79 |
ENST00000390453.1
|
TRAV24
|
T cell receptor alpha variable 24 |
| chr17_-_59106801 | 0.78 |
ENST00000393065.6
ENST00000262294.12 ENST00000393066.7 |
TRIM37
|
tripartite motif containing 37 |
| chr11_+_22668101 | 0.78 |
ENST00000630668.2
ENST00000278187.7 |
GAS2
|
growth arrest specific 2 |
| chr10_+_134465 | 0.77 |
ENST00000439456.5
ENST00000397962.8 ENST00000309776.8 ENST00000397959.7 |
ZMYND11
|
zinc finger MYND-type containing 11 |
| chr2_-_85561513 | 0.77 |
ENST00000430215.7
|
GGCX
|
gamma-glutamyl carboxylase |
| chr17_+_63622406 | 0.77 |
ENST00000579585.5
ENST00000361733.8 ENST00000584573.5 ENST00000361357.7 |
MAP3K3
|
mitogen-activated protein kinase kinase kinase 3 |
| chr1_-_48400826 | 0.77 |
ENST00000371841.1
|
SPATA6
|
spermatogenesis associated 6 |
| chr19_+_40597168 | 0.77 |
ENST00000308370.11
|
LTBP4
|
latent transforming growth factor beta binding protein 4 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 5.2 | GO:0009956 | radial pattern formation(GO:0009956) |
| 1.1 | 3.4 | GO:0061445 | endocardial cell fate commitment(GO:0060957) endocardial cushion cell fate commitment(GO:0061445) |
| 1.1 | 3.3 | GO:0050928 | negative regulation of positive chemotaxis(GO:0050928) |
| 1.1 | 3.2 | GO:0032888 | regulation of spindle elongation(GO:0032887) regulation of mitotic spindle elongation(GO:0032888) anastral spindle assembly(GO:0055048) protein localization to spindle pole body(GO:0071988) regulation of protein localization to spindle pole body(GO:1902363) positive regulation of protein localization to spindle pole body(GO:1902365) positive regulation of mitotic spindle elongation(GO:1902846) |
| 1.1 | 3.2 | GO:0021793 | chemorepulsion of branchiomotor axon(GO:0021793) |
| 1.0 | 8.4 | GO:0003138 | primary heart field specification(GO:0003138) sinoatrial valve development(GO:0003172) sinoatrial valve morphogenesis(GO:0003185) |
| 1.0 | 3.8 | GO:2000170 | positive regulation of peptidyl-cysteine S-nitrosylation(GO:2000170) |
| 0.9 | 6.4 | GO:0000821 | regulation of arginine metabolic process(GO:0000821) |
| 0.6 | 7.0 | GO:2000795 | negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.6 | 2.5 | GO:0002384 | hepatic immune response(GO:0002384) |
| 0.6 | 2.5 | GO:0045659 | regulation of neutrophil differentiation(GO:0045658) negative regulation of neutrophil differentiation(GO:0045659) |
| 0.6 | 1.7 | GO:0051695 | actin filament uncapping(GO:0051695) |
| 0.5 | 3.3 | GO:0033615 | mitochondrial proton-transporting ATP synthase complex assembly(GO:0033615) |
| 0.5 | 1.5 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) |
| 0.5 | 1.4 | GO:0061011 | hepatic duct development(GO:0061011) |
| 0.5 | 1.9 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.4 | 2.6 | GO:0006196 | AMP catabolic process(GO:0006196) |
| 0.4 | 2.0 | GO:0093001 | glycolysis from storage polysaccharide through glucose-1-phosphate(GO:0093001) |
| 0.4 | 1.4 | GO:0035548 | gamma-delta T cell activation involved in immune response(GO:0002290) negative regulation of interferon-beta secretion(GO:0035548) regulation of gamma-delta T cell activation involved in immune response(GO:2001191) positive regulation of gamma-delta T cell activation involved in immune response(GO:2001193) |
| 0.4 | 2.1 | GO:0019264 | glycine biosynthetic process from serine(GO:0019264) |
| 0.3 | 1.0 | GO:0002503 | peptide antigen assembly with MHC class II protein complex(GO:0002503) |
| 0.3 | 1.0 | GO:0003193 | pulmonary valve formation(GO:0003193) foramen ovale closure(GO:0035922) regulation of bundle of His cell action potential(GO:0098905) |
| 0.3 | 6.5 | GO:0060044 | negative regulation of cardiac muscle cell proliferation(GO:0060044) |
| 0.3 | 11.6 | GO:0035162 | embryonic hemopoiesis(GO:0035162) |
| 0.3 | 4.3 | GO:0030050 | vesicle transport along actin filament(GO:0030050) |
| 0.3 | 2.8 | GO:0048539 | bone marrow development(GO:0048539) |
| 0.3 | 1.8 | GO:0003199 | endocardial cushion to mesenchymal transition involved in heart valve formation(GO:0003199) positive regulation of blood vessel remodeling(GO:2000504) |
| 0.3 | 1.4 | GO:0071879 | positive regulation of adrenergic receptor signaling pathway(GO:0071879) |
| 0.2 | 6.2 | GO:0009812 | flavonoid metabolic process(GO:0009812) |
| 0.2 | 0.7 | GO:0035526 | retrograde transport, plasma membrane to Golgi(GO:0035526) |
| 0.2 | 1.0 | GO:0007181 | transforming growth factor beta receptor complex assembly(GO:0007181) |
| 0.2 | 0.7 | GO:0035752 | lysosomal lumen pH elevation(GO:0035752) |
| 0.2 | 0.9 | GO:0090214 | spongiotrophoblast layer developmental growth(GO:0090214) |
| 0.2 | 1.1 | GO:0006127 | glycerophosphate shuttle(GO:0006127) |
| 0.2 | 6.4 | GO:0070886 | positive regulation of calcineurin-NFAT signaling cascade(GO:0070886) |
| 0.2 | 1.8 | GO:0007598 | blood coagulation, extrinsic pathway(GO:0007598) |
| 0.2 | 0.8 | GO:0039506 | modulation by virus of host molecular function(GO:0039506) suppression by virus of host molecular function(GO:0039507) suppression by virus of host catalytic activity(GO:0039513) modulation by virus of host catalytic activity(GO:0039516) suppression by virus of host cysteine-type endopeptidase activity involved in apoptotic process(GO:0039650) negative regulation by symbiont of host catalytic activity(GO:0052053) negative regulation by symbiont of host molecular function(GO:0052056) modulation by symbiont of host catalytic activity(GO:0052148) |
| 0.2 | 1.8 | GO:0097267 | omega-hydroxylase P450 pathway(GO:0097267) |
| 0.2 | 0.6 | GO:1904845 | response to L-glutamine(GO:1904844) cellular response to L-glutamine(GO:1904845) |
| 0.2 | 0.8 | GO:0042361 | menaquinone catabolic process(GO:0042361) vitamin K catabolic process(GO:0042377) |
| 0.2 | 0.8 | GO:0009236 | cobalamin biosynthetic process(GO:0009236) |
| 0.2 | 0.9 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 0.2 | 1.3 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.2 | 5.5 | GO:0072189 | ureter development(GO:0072189) |
| 0.2 | 1.0 | GO:0071461 | cellular response to redox state(GO:0071461) |
| 0.2 | 2.0 | GO:0045656 | negative regulation of keratinocyte differentiation(GO:0045617) negative regulation of monocyte differentiation(GO:0045656) |
| 0.2 | 1.6 | GO:0090308 | regulation of methylation-dependent chromatin silencing(GO:0090308) |
| 0.1 | 0.4 | GO:0035552 | oxidative single-stranded DNA demethylation(GO:0035552) |
| 0.1 | 0.5 | GO:0000711 | meiotic DNA repair synthesis(GO:0000711) |
| 0.1 | 1.4 | GO:0035726 | common myeloid progenitor cell proliferation(GO:0035726) |
| 0.1 | 1.0 | GO:1902774 | late endosome to lysosome transport(GO:1902774) |
| 0.1 | 2.7 | GO:0070914 | UV-damage excision repair(GO:0070914) |
| 0.1 | 0.9 | GO:0071279 | negative regulation of epithelial cell proliferation involved in prostate gland development(GO:0060770) cellular response to cobalt ion(GO:0071279) |
| 0.1 | 1.0 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.1 | 1.2 | GO:0035148 | tube formation(GO:0035148) |
| 0.1 | 1.0 | GO:0031666 | positive regulation of lipopolysaccharide-mediated signaling pathway(GO:0031666) |
| 0.1 | 0.3 | GO:2000502 | negative regulation of natural killer cell chemotaxis(GO:2000502) |
| 0.1 | 5.1 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.1 | 1.4 | GO:2000253 | adult feeding behavior(GO:0008343) positive regulation of feeding behavior(GO:2000253) |
| 0.1 | 0.4 | GO:0021691 | cerebellar Purkinje cell layer maturation(GO:0021691) |
| 0.1 | 21.1 | GO:0007286 | spermatid development(GO:0007286) |
| 0.1 | 0.5 | GO:1901164 | negative regulation of trophoblast cell migration(GO:1901164) |
| 0.1 | 0.8 | GO:0038031 | non-canonical Wnt signaling pathway via JNK cascade(GO:0038031) |
| 0.1 | 0.4 | GO:0090235 | regulation of metaphase plate congression(GO:0090235) |
| 0.1 | 0.5 | GO:0018243 | protein O-linked glycosylation via threonine(GO:0018243) |
| 0.1 | 1.3 | GO:0071313 | cellular response to caffeine(GO:0071313) |
| 0.1 | 0.4 | GO:1900217 | pronephric field specification(GO:0039003) pattern specification involved in pronephros development(GO:0039017) thyroid-stimulating hormone secretion(GO:0070460) kidney field specification(GO:0072004) metanephric nephron tubule formation(GO:0072289) regulation of mesenchymal cell apoptotic process involved in metanephric nephron morphogenesis(GO:0072304) negative regulation of mesenchymal cell apoptotic process involved in metanephric nephron morphogenesis(GO:0072305) mesenchymal stem cell maintenance involved in metanephric nephron morphogenesis(GO:0072309) apoptotic process involved in metanephric collecting duct development(GO:1900204) apoptotic process involved in metanephric nephron tubule development(GO:1900205) regulation of apoptotic process involved in metanephric collecting duct development(GO:1900214) negative regulation of apoptotic process involved in metanephric collecting duct development(GO:1900215) regulation of apoptotic process involved in metanephric nephron tubule development(GO:1900217) negative regulation of apoptotic process involved in metanephric nephron tubule development(GO:1900218) mesenchymal cell apoptotic process involved in metanephric nephron morphogenesis(GO:1901147) regulation of metanephric DCT cell differentiation(GO:2000592) positive regulation of metanephric DCT cell differentiation(GO:2000594) regulation of thyroid-stimulating hormone secretion(GO:2000612) |
| 0.1 | 0.9 | GO:1902897 | regulation of postsynaptic density protein 95 clustering(GO:1902897) |
| 0.1 | 0.7 | GO:0015798 | myo-inositol transport(GO:0015798) |
| 0.1 | 2.2 | GO:0031115 | negative regulation of microtubule polymerization(GO:0031115) |
| 0.1 | 0.5 | GO:0034196 | acylglycerol transport(GO:0034196) triglyceride transport(GO:0034197) |
| 0.1 | 0.4 | GO:2000213 | nitrogen catabolite regulation of transcription from RNA polymerase II promoter(GO:0001079) nitrogen catabolite activation of transcription from RNA polymerase II promoter(GO:0001080) regulation of urea metabolic process(GO:0034255) intracellular bile acid receptor signaling pathway(GO:0038185) interleukin-17 secretion(GO:0072615) nitrogen catabolite regulation of transcription(GO:0090293) nitrogen catabolite activation of transcription(GO:0090294) regulation of nitrogen cycle metabolic process(GO:1903314) positive regulation of glutamate metabolic process(GO:2000213) regulation of ammonia assimilation cycle(GO:2001248) positive regulation of ammonia assimilation cycle(GO:2001250) |
| 0.1 | 0.3 | GO:0021658 | rhombomere morphogenesis(GO:0021593) rhombomere 3 morphogenesis(GO:0021658) |
| 0.1 | 0.7 | GO:0070842 | aggresome assembly(GO:0070842) |
| 0.1 | 1.4 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.1 | 0.9 | GO:0017062 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) |
| 0.1 | 5.4 | GO:0051693 | actin filament capping(GO:0051693) |
| 0.1 | 0.3 | GO:0042427 | serotonin biosynthetic process(GO:0042427) |
| 0.1 | 2.4 | GO:1901685 | glutathione derivative metabolic process(GO:1901685) glutathione derivative biosynthetic process(GO:1901687) |
| 0.1 | 3.1 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.1 | 0.8 | GO:2000427 | positive regulation of apoptotic cell clearance(GO:2000427) |
| 0.1 | 3.0 | GO:0046329 | negative regulation of JNK cascade(GO:0046329) |
| 0.1 | 1.3 | GO:0045603 | positive regulation of endothelial cell differentiation(GO:0045603) |
| 0.1 | 0.3 | GO:0035910 | ascending aorta development(GO:0035905) ascending aorta morphogenesis(GO:0035910) |
| 0.1 | 0.2 | GO:1903565 | negative regulation of protein localization to cilium(GO:1903565) regulation of protein localization to ciliary membrane(GO:1903567) negative regulation of protein localization to ciliary membrane(GO:1903568) |
| 0.1 | 0.5 | GO:1902459 | positive regulation of stem cell population maintenance(GO:1902459) |
| 0.1 | 2.4 | GO:2000369 | regulation of clathrin-mediated endocytosis(GO:2000369) |
| 0.1 | 0.3 | GO:0061760 | antifungal innate immune response(GO:0061760) |
| 0.1 | 0.1 | GO:0002581 | negative regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002581) |
| 0.1 | 0.6 | GO:0061588 | calcium activated phospholipid scrambling(GO:0061588) calcium activated phosphatidylcholine scrambling(GO:0061590) calcium activated galactosylceramide scrambling(GO:0061591) |
| 0.1 | 0.4 | GO:0006701 | progesterone biosynthetic process(GO:0006701) |
| 0.1 | 0.9 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.1 | 0.6 | GO:0006689 | ganglioside catabolic process(GO:0006689) |
| 0.1 | 0.2 | GO:0002025 | vasodilation by norepinephrine-epinephrine involved in regulation of systemic arterial blood pressure(GO:0002025) |
| 0.1 | 1.3 | GO:0036148 | phosphatidylglycerol acyl-chain remodeling(GO:0036148) |
| 0.1 | 2.1 | GO:0009954 | proximal/distal pattern formation(GO:0009954) |
| 0.1 | 0.8 | GO:0017187 | peptidyl-glutamic acid carboxylation(GO:0017187) protein carboxylation(GO:0018214) |
| 0.1 | 0.3 | GO:0060011 | Sertoli cell proliferation(GO:0060011) |
| 0.1 | 0.3 | GO:0033088 | negative regulation of immature T cell proliferation in thymus(GO:0033088) |
| 0.1 | 1.1 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.1 | 0.5 | GO:0030043 | actin filament fragmentation(GO:0030043) |
| 0.1 | 0.2 | GO:0010752 | regulation of cGMP-mediated signaling(GO:0010752) regulation of B cell chemotaxis(GO:2000537) positive regulation of B cell chemotaxis(GO:2000538) |
| 0.0 | 1.6 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 1.3 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.0 | 0.9 | GO:0010839 | negative regulation of keratinocyte proliferation(GO:0010839) |
| 0.0 | 0.8 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) |
| 0.0 | 0.3 | GO:0006842 | tricarboxylic acid transport(GO:0006842) citrate transport(GO:0015746) |
| 0.0 | 1.6 | GO:1904659 | hexose transmembrane transport(GO:0035428) glucose transmembrane transport(GO:1904659) |
| 0.0 | 2.1 | GO:0043277 | apoptotic cell clearance(GO:0043277) |
| 0.0 | 0.9 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.0 | 0.2 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.0 | 1.0 | GO:0016254 | preassembly of GPI anchor in ER membrane(GO:0016254) |
| 0.0 | 0.5 | GO:0005980 | glycogen catabolic process(GO:0005980) |
| 0.0 | 0.3 | GO:0000480 | endonucleolytic cleavage in 5'-ETS of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000480) |
| 0.0 | 0.3 | GO:0060478 | acrosomal vesicle exocytosis(GO:0060478) |
| 0.0 | 0.1 | GO:0006015 | 5-phosphoribose 1-diphosphate biosynthetic process(GO:0006015) 5-phosphoribose 1-diphosphate metabolic process(GO:0046391) |
| 0.0 | 0.4 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.0 | 0.2 | GO:0030576 | Cajal body organization(GO:0030576) |
| 0.0 | 0.3 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.0 | 0.8 | GO:1900745 | positive regulation of p38MAPK cascade(GO:1900745) |
| 0.0 | 0.7 | GO:0051823 | regulation of synapse structural plasticity(GO:0051823) |
| 0.0 | 0.4 | GO:0042670 | retinal cone cell differentiation(GO:0042670) retinal cone cell development(GO:0046549) |
| 0.0 | 0.1 | GO:0019072 | viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 0.0 | 0.8 | GO:0002693 | positive regulation of cellular extravasation(GO:0002693) |
| 0.0 | 2.0 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
| 0.0 | 1.8 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.0 | 1.6 | GO:0070988 | demethylation(GO:0070988) |
| 0.0 | 3.1 | GO:0048207 | vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) |
| 0.0 | 1.9 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.0 | 10.5 | GO:0031032 | actomyosin structure organization(GO:0031032) |
| 0.0 | 2.6 | GO:1902108 | regulation of mitochondrial membrane permeability involved in apoptotic process(GO:1902108) |
| 0.0 | 2.4 | GO:0070206 | protein trimerization(GO:0070206) |
| 0.0 | 1.5 | GO:0032007 | negative regulation of TOR signaling(GO:0032007) |
| 0.0 | 0.3 | GO:0000733 | DNA strand renaturation(GO:0000733) |
| 0.0 | 0.3 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.0 | 7.9 | GO:0065004 | protein-DNA complex assembly(GO:0065004) |
| 0.0 | 0.5 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.0 | 0.3 | GO:1900029 | positive regulation of ruffle assembly(GO:1900029) |
| 0.0 | 1.3 | GO:0050919 | negative chemotaxis(GO:0050919) |
| 0.0 | 2.3 | GO:0000271 | polysaccharide biosynthetic process(GO:0000271) |
| 0.0 | 0.1 | GO:0061051 | positive regulation of cell growth involved in cardiac muscle cell development(GO:0061051) |
| 0.0 | 0.1 | GO:0015742 | alpha-ketoglutarate transport(GO:0015742) |
| 0.0 | 0.1 | GO:1903936 | cellular response to sodium arsenite(GO:1903936) |
| 0.0 | 1.0 | GO:0042269 | regulation of natural killer cell mediated cytotoxicity(GO:0042269) |
| 0.0 | 0.2 | GO:0071340 | skeletal muscle acetylcholine-gated channel clustering(GO:0071340) |
| 0.0 | 0.5 | GO:0030252 | growth hormone secretion(GO:0030252) |
| 0.0 | 0.3 | GO:0002097 | tRNA wobble base modification(GO:0002097) |
| 0.0 | 2.2 | GO:1901222 | regulation of NIK/NF-kappaB signaling(GO:1901222) |
| 0.0 | 0.3 | GO:1902894 | negative regulation of pri-miRNA transcription from RNA polymerase II promoter(GO:1902894) |
| 0.0 | 0.8 | GO:0044458 | motile cilium assembly(GO:0044458) |
| 0.0 | 1.6 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.0 | 0.4 | GO:0040015 | negative regulation of multicellular organism growth(GO:0040015) |
| 0.0 | 0.4 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.0 | 0.2 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.0 | 0.8 | GO:0048011 | neurotrophin TRK receptor signaling pathway(GO:0048011) |
| 0.0 | 0.2 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.0 | 0.6 | GO:0045907 | positive regulation of vasoconstriction(GO:0045907) |
| 0.0 | 0.6 | GO:0048305 | immunoglobulin secretion(GO:0048305) |
| 0.0 | 0.1 | GO:0019557 | histidine catabolic process to glutamate and formamide(GO:0019556) histidine catabolic process to glutamate and formate(GO:0019557) formamide metabolic process(GO:0043606) |
| 0.0 | 0.1 | GO:0060746 | maternal behavior(GO:0042711) parental behavior(GO:0060746) |
| 0.0 | 1.6 | GO:0001578 | microtubule bundle formation(GO:0001578) |
| 0.0 | 0.3 | GO:0031065 | positive regulation of histone deacetylation(GO:0031065) |
| 0.0 | 1.3 | GO:0042795 | snRNA transcription(GO:0009301) snRNA transcription from RNA polymerase II promoter(GO:0042795) |
| 0.0 | 0.8 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.0 | 8.3 | GO:0051056 | regulation of small GTPase mediated signal transduction(GO:0051056) |
| 0.0 | 0.4 | GO:0032802 | low-density lipoprotein particle receptor catabolic process(GO:0032802) |
| 0.0 | 0.1 | GO:2000507 | positive regulation of energy homeostasis(GO:2000507) |
| 0.0 | 0.0 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.0 | 1.8 | GO:0043280 | positive regulation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0043280) |
| 0.0 | 2.8 | GO:0021915 | neural tube development(GO:0021915) |
| 0.0 | 1.5 | GO:0010811 | positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.0 | 0.2 | GO:0030321 | transepithelial chloride transport(GO:0030321) |
| 0.0 | 0.1 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.0 | 0.1 | GO:0086028 | bundle of His cell to Purkinje myocyte signaling(GO:0086028) bundle of His cell action potential(GO:0086043) |
| 0.0 | 0.2 | GO:0050966 | detection of mechanical stimulus involved in sensory perception of pain(GO:0050966) |
| 0.0 | 0.3 | GO:0016024 | CDP-diacylglycerol biosynthetic process(GO:0016024) |
| 0.0 | 0.3 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 0.0 | 0.1 | GO:0042335 | cuticle development(GO:0042335) |
| 0.0 | 0.3 | GO:0033141 | positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) |
| 0.0 | 0.3 | GO:0007342 | fusion of sperm to egg plasma membrane(GO:0007342) |
| 0.0 | 0.7 | GO:0035065 | regulation of histone acetylation(GO:0035065) |
| 0.0 | 0.7 | GO:0050909 | sensory perception of taste(GO:0050909) |
| 0.0 | 0.2 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.0 | 0.8 | GO:0061178 | regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061178) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 3.2 | GO:0055028 | cortical microtubule(GO:0055028) |
| 0.7 | 3.4 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.6 | 21.8 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.5 | 2.5 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.3 | 2.7 | GO:0000110 | nucleotide-excision repair factor 1 complex(GO:0000110) |
| 0.3 | 2.0 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.3 | 7.0 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.3 | 0.8 | GO:0097224 | sperm connecting piece(GO:0097224) |
| 0.2 | 3.2 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.2 | 1.0 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.2 | 1.4 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 0.2 | 1.6 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.2 | 6.5 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.2 | 1.1 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.2 | 1.1 | GO:0031417 | NatC complex(GO:0031417) |
| 0.2 | 1.5 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 0.2 | 4.0 | GO:0031045 | dense core granule(GO:0031045) |
| 0.2 | 0.5 | GO:0071148 | TEAD-1-YAP complex(GO:0071148) |
| 0.1 | 0.6 | GO:0005889 | hydrogen:potassium-exchanging ATPase complex(GO:0005889) |
| 0.1 | 1.7 | GO:0044354 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.1 | 1.1 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.1 | 1.2 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.1 | 0.9 | GO:0000308 | cytoplasmic cyclin-dependent protein kinase holoenzyme complex(GO:0000308) |
| 0.1 | 0.8 | GO:0070436 | Grb2-EGFR complex(GO:0070436) |
| 0.1 | 2.5 | GO:0000786 | nucleosome(GO:0000786) |
| 0.1 | 0.2 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.1 | 0.3 | GO:0070701 | mucus layer(GO:0070701) |
| 0.1 | 1.3 | GO:0005583 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.1 | 1.3 | GO:0032039 | integrator complex(GO:0032039) |
| 0.1 | 0.5 | GO:0000801 | central element(GO:0000801) |
| 0.1 | 0.9 | GO:0043203 | axon hillock(GO:0043203) |
| 0.1 | 1.2 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.1 | 2.0 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.1 | 1.2 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.1 | 0.3 | GO:0002079 | inner acrosomal membrane(GO:0002079) outer acrosomal membrane(GO:0002081) |
| 0.1 | 0.4 | GO:0036021 | endolysosome lumen(GO:0036021) |
| 0.1 | 0.3 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.1 | 3.8 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 0.5 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.0 | 0.3 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.0 | 1.1 | GO:0005652 | nuclear lamina(GO:0005652) |
| 0.0 | 0.2 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.0 | 6.9 | GO:0032993 | protein-DNA complex(GO:0032993) |
| 0.0 | 2.1 | GO:0016591 | DNA-directed RNA polymerase II, holoenzyme(GO:0016591) |
| 0.0 | 8.9 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.0 | 2.7 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.0 | 0.8 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) |
| 0.0 | 1.5 | GO:0005657 | replication fork(GO:0005657) |
| 0.0 | 1.5 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.6 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 2.8 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 0.8 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.0 | 0.6 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.4 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.0 | 0.5 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.0 | 0.1 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 0.0 | 1.2 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 3.6 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 0.6 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.0 | 0.6 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 0.5 | GO:0036020 | endolysosome membrane(GO:0036020) |
| 0.0 | 0.1 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 0.0 | 0.4 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.0 | 0.7 | GO:0097228 | sperm principal piece(GO:0097228) |
| 0.0 | 0.2 | GO:0097504 | Gemini of coiled bodies(GO:0097504) |
| 0.0 | 1.7 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.0 | 1.7 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.0 | 1.1 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 1.3 | GO:0101003 | ficolin-1-rich granule membrane(GO:0101003) |
| 0.0 | 0.5 | GO:0031904 | endosome lumen(GO:0031904) |
| 0.0 | 1.3 | GO:0048475 | membrane coat(GO:0030117) coated membrane(GO:0048475) |
| 0.0 | 0.1 | GO:0005688 | U6 snRNP(GO:0005688) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.5 | GO:0070119 | ciliary neurotrophic factor binding(GO:0070119) |
| 0.8 | 3.8 | GO:0008240 | tripeptidyl-peptidase activity(GO:0008240) peptidyl-dipeptidase activity(GO:0008241) bradykinin receptor binding(GO:0031711) metallodipeptidase activity(GO:0070573) |
| 0.5 | 2.1 | GO:0070905 | serine binding(GO:0070905) |
| 0.5 | 3.4 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.5 | 1.4 | GO:0047006 | 17-alpha,20-alpha-dihydroxypregn-4-en-3-one dehydrogenase activity(GO:0047006) |
| 0.5 | 1.4 | GO:0003863 | alpha-ketoacid dehydrogenase activity(GO:0003826) 3-methyl-2-oxobutanoate dehydrogenase (2-methylpropanoyl-transferring) activity(GO:0003863) |
| 0.4 | 8.4 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.4 | 4.3 | GO:0034485 | phosphatidylinositol-3,4,5-trisphosphate 5-phosphatase activity(GO:0034485) |
| 0.4 | 1.1 | GO:0032090 | Pyrin domain binding(GO:0032090) |
| 0.4 | 1.4 | GO:0030107 | HLA-A specific inhibitory MHC class I receptor activity(GO:0030107) |
| 0.3 | 5.7 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.3 | 1.6 | GO:0031493 | nucleosomal histone binding(GO:0031493) |
| 0.3 | 1.5 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.3 | 5.5 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.3 | 1.1 | GO:0052590 | sn-glycerol-3-phosphate:ubiquinone oxidoreductase activity(GO:0052590) sn-glycerol-3-phosphate:ubiquinone-8 oxidoreductase activity(GO:0052591) |
| 0.3 | 21.1 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.3 | 1.0 | GO:0086020 | gap junction channel activity involved in SA node cell-atrial cardiac muscle cell electrical coupling(GO:0086020) |
| 0.3 | 1.0 | GO:0004307 | ethanolaminephosphotransferase activity(GO:0004307) |
| 0.2 | 3.2 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.2 | 2.0 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.2 | 0.7 | GO:0036487 | nitric-oxide synthase inhibitor activity(GO:0036487) |
| 0.2 | 5.0 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.2 | 0.9 | GO:0004925 | prolactin receptor activity(GO:0004925) |
| 0.2 | 1.1 | GO:0004447 | iodide peroxidase activity(GO:0004447) |
| 0.2 | 0.9 | GO:0005163 | nerve growth factor receptor binding(GO:0005163) |
| 0.2 | 1.8 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.2 | 0.6 | GO:0032428 | sphingolipid activator protein activity(GO:0030290) beta-N-acetylgalactosaminidase activity(GO:0032428) |
| 0.2 | 0.5 | GO:0017129 | triglyceride binding(GO:0017129) |
| 0.2 | 4.9 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.2 | 0.5 | GO:0051377 | mannose-ethanolamine phosphotransferase activity(GO:0051377) |
| 0.2 | 1.7 | GO:0008970 | phosphatidylcholine 1-acylhydrolase activity(GO:0008970) |
| 0.2 | 3.2 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.2 | 2.6 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.1 | 0.4 | GO:0035514 | DNA demethylase activity(GO:0035514) |
| 0.1 | 5.3 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.1 | 4.1 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.1 | 0.8 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.1 | 0.8 | GO:0097199 | cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097199) |
| 0.1 | 0.5 | GO:0016524 | latrotoxin receptor activity(GO:0016524) |
| 0.1 | 2.6 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.1 | 1.7 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.1 | 1.6 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.1 | 1.0 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.1 | 0.3 | GO:0004967 | glucagon receptor activity(GO:0004967) |
| 0.1 | 0.9 | GO:0019788 | NEDD8 transferase activity(GO:0019788) |
| 0.1 | 1.3 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.1 | 1.5 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.1 | 0.4 | GO:1902122 | chenodeoxycholic acid binding(GO:1902122) |
| 0.1 | 2.1 | GO:0003756 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.1 | 3.6 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.1 | 0.8 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.1 | 2.0 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.1 | 0.3 | GO:0017002 | activin-activated receptor activity(GO:0017002) |
| 0.1 | 1.6 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.1 | 1.9 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.1 | 0.2 | GO:0004939 | beta-adrenergic receptor activity(GO:0004939) |
| 0.1 | 0.3 | GO:0004522 | ribonuclease A activity(GO:0004522) |
| 0.1 | 1.3 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.1 | 0.3 | GO:0043125 | ErbB-3 class receptor binding(GO:0043125) |
| 0.1 | 1.1 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.1 | 1.2 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.1 | 0.7 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 0.5 | GO:0035251 | UDP-glucosyltransferase activity(GO:0035251) |
| 0.0 | 0.5 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.0 | 1.0 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.0 | 2.4 | GO:0016709 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen(GO:0016709) |
| 0.0 | 0.4 | GO:0004996 | thyroid-stimulating hormone receptor activity(GO:0004996) |
| 0.0 | 0.7 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.0 | 2.5 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 0.3 | GO:0015137 | citrate transmembrane transporter activity(GO:0015137) tricarboxylic acid transmembrane transporter activity(GO:0015142) |
| 0.0 | 4.4 | GO:0017022 | myosin binding(GO:0017022) |
| 0.0 | 0.4 | GO:0099580 | ion antiporter activity involved in regulation of postsynaptic membrane potential(GO:0099580) |
| 0.0 | 0.3 | GO:0016714 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced pteridine as one donor, and incorporation of one atom of oxygen(GO:0016714) |
| 0.0 | 1.1 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.6 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 0.8 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.0 | 0.6 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.0 | 0.9 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) |
| 0.0 | 0.5 | GO:0030249 | cyclase regulator activity(GO:0010851) guanylate cyclase regulator activity(GO:0030249) |
| 0.0 | 0.9 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 18.4 | GO:0001077 | transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) |
| 0.0 | 0.1 | GO:0016503 | pheromone receptor activity(GO:0016503) |
| 0.0 | 1.2 | GO:0023026 | MHC class II protein complex binding(GO:0023026) |
| 0.0 | 1.4 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 0.6 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 1.5 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.2 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.0 | 3.3 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.0 | 1.3 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.0 | 2.1 | GO:0051393 | alpha-actinin binding(GO:0051393) |
| 0.0 | 1.0 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 2.3 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
| 0.0 | 1.4 | GO:0031690 | adrenergic receptor binding(GO:0031690) |
| 0.0 | 0.3 | GO:0036310 | annealing helicase activity(GO:0036310) |
| 0.0 | 1.1 | GO:0016866 | intramolecular transferase activity(GO:0016866) |
| 0.0 | 1.9 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 0.8 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.0 | 2.2 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 0.2 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.0 | 0.6 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 2.7 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.0 | 0.8 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.0 | 0.5 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 0.2 | GO:0004340 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.0 | 0.8 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 15.3 | GO:0003779 | actin binding(GO:0003779) |
| 0.0 | 1.2 | GO:0097110 | scaffold protein binding(GO:0097110) |
| 0.0 | 0.7 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.1 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.0 | 0.1 | GO:0061752 | telomeric repeat-containing RNA binding(GO:0061752) |
| 0.0 | 0.2 | GO:0016929 | SUMO-specific protease activity(GO:0016929) |
| 0.0 | 0.3 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.8 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 2.1 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 4.9 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 0.2 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.0 | 0.3 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.0 | 0.5 | GO:0030159 | receptor signaling complex scaffold activity(GO:0030159) |
| 0.0 | 0.3 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.0 | 0.1 | GO:0004983 | neuropeptide Y receptor activity(GO:0004983) |
| 0.0 | 0.7 | GO:0016765 | transferase activity, transferring alkyl or aryl (other than methyl) groups(GO:0016765) |
| 0.0 | 0.1 | GO:0099589 | serotonin receptor activity(GO:0099589) |
| 0.0 | 0.2 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.0 | 0.1 | GO:0016812 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amides(GO:0016812) |
| 0.0 | 0.2 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 0.0 | 0.0 | GO:0072590 | N-acetyl-L-aspartate-L-glutamate ligase activity(GO:0072590) |
| 0.0 | 0.1 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 0.1 | GO:0043008 | ATP-dependent protein binding(GO:0043008) |
| 0.0 | 0.1 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.0 | 0.1 | GO:0023024 | MHC class I protein complex binding(GO:0023024) |
| 0.0 | 5.4 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 0.3 | GO:0030247 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.0 | 0.1 | GO:0004075 | biotin carboxylase activity(GO:0004075) |
| 0.0 | 1.0 | GO:0008375 | acetylglucosaminyltransferase activity(GO:0008375) |
| 0.0 | 0.3 | GO:0001848 | complement binding(GO:0001848) |
| 0.0 | 0.1 | GO:0031404 | chloride ion binding(GO:0031404) |
| 0.0 | 1.1 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.1 | GO:0035197 | siRNA binding(GO:0035197) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 1.4 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.2 | 20.5 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.2 | 17.0 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.2 | 3.4 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.2 | 6.4 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.1 | 8.4 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.1 | 10.4 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.1 | 2.5 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.1 | 1.6 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.1 | 7.0 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.1 | 2.6 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 2.0 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 3.8 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 1.5 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.0 | 0.8 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 5.7 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.7 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.0 | 1.3 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.0 | 0.8 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.0 | 0.8 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.0 | 1.1 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 0.4 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 1.0 | PID ENDOTHELIN PATHWAY | Endothelins |
| 0.0 | 0.8 | ST INTEGRIN SIGNALING PATHWAY | Integrin Signaling Pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 5.5 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.2 | 7.0 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.2 | 2.9 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.2 | 2.6 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.1 | 11.4 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.1 | 2.5 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.1 | 2.6 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.1 | 2.6 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.1 | 5.5 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.1 | 3.3 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.1 | 2.0 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.1 | 2.7 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.1 | 1.6 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.1 | 2.9 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.1 | 1.5 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.1 | 5.0 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.1 | 1.5 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.1 | 0.7 | REACTOME POST TRANSLATIONAL MODIFICATION SYNTHESIS OF GPI ANCHORED PROTEINS | Genes involved in Post-translational modification: synthesis of GPI-anchored proteins |
| 0.1 | 2.4 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 1.6 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 1.8 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 5.1 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 0.8 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 0.9 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 0.8 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.5 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.0 | 2.4 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 4.5 | REACTOME G ALPHA S SIGNALLING EVENTS | Genes involved in G alpha (s) signalling events |
| 0.0 | 1.7 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 1.8 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.0 | 0.4 | REACTOME G PROTEIN ACTIVATION | Genes involved in G-protein activation |
| 0.0 | 6.8 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.8 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.4 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 1.2 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.0 | 0.5 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.0 | 0.5 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 0.0 | 0.7 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 0.6 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.0 | 0.6 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 0.4 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 0.6 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.6 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 0.1 | REACTOME GABA B RECEPTOR ACTIVATION | Genes involved in GABA B receptor activation |
| 0.0 | 0.6 | REACTOME GLUCOSE METABOLISM | Genes involved in Glucose metabolism |
| 0.0 | 0.3 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 0.4 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.0 | 0.4 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 0.2 | REACTOME OPSINS | Genes involved in Opsins |
| 0.0 | 1.5 | REACTOME RECRUITMENT OF MITOTIC CENTROSOME PROTEINS AND COMPLEXES | Genes involved in Recruitment of mitotic centrosome proteins and complexes |
| 0.0 | 0.5 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.4 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 0.5 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 0.3 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.0 | 1.0 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 1.9 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.0 | 0.0 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.0 | 0.4 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.0 | 1.2 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |