Project
Inflammatory response time course, HUVEC (Wada, 2009)
Navigation
Downloads
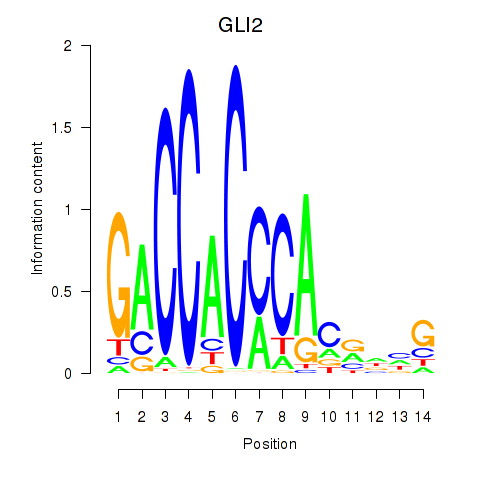
Results for GLI2
Z-value: 0.79
Transcription factors associated with GLI2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
GLI2
|
ENSG00000074047.22 | GLI2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| GLI2 | hg38_v1_chr2_+_120735848_120735899 | 0.42 | 3.6e-02 | Click! |
Activity profile of GLI2 motif
Sorted Z-values of GLI2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of GLI2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_-_7874361 | 1.41 |
ENST00000618550.5
|
PRR36
|
proline rich 36 |
| chr4_-_148444674 | 1.09 |
ENST00000344721.8
|
NR3C2
|
nuclear receptor subfamily 3 group C member 2 |
| chr1_-_205813177 | 1.01 |
ENST00000367137.4
|
SLC41A1
|
solute carrier family 41 member 1 |
| chr2_+_205683109 | 0.83 |
ENST00000357118.8
ENST00000272849.7 ENST00000412873.2 |
NRP2
|
neuropilin 2 |
| chr2_+_11155498 | 0.79 |
ENST00000402361.5
ENST00000428481.1 |
SLC66A3
|
solute carrier family 66 member 3 |
| chr21_-_26843012 | 0.69 |
ENST00000517777.6
|
ADAMTS1
|
ADAM metallopeptidase with thrombospondin type 1 motif 1 |
| chr21_-_26843063 | 0.67 |
ENST00000678221.1
|
ADAMTS1
|
ADAM metallopeptidase with thrombospondin type 1 motif 1 |
| chr19_-_10380454 | 0.67 |
ENST00000530829.1
ENST00000529370.5 |
TYK2
|
tyrosine kinase 2 |
| chrX_-_109733181 | 0.66 |
ENST00000673016.1
|
ACSL4
|
acyl-CoA synthetase long chain family member 4 |
| chr2_+_240625237 | 0.63 |
ENST00000407714.1
|
GPR35
|
G protein-coupled receptor 35 |
| chr17_+_43006740 | 0.62 |
ENST00000438323.2
ENST00000415816.7 |
IFI35
|
interferon induced protein 35 |
| chr7_-_151814819 | 0.59 |
ENST00000392801.6
ENST00000652707.1 ENST00000651378.1 |
PRKAG2
|
protein kinase AMP-activated non-catalytic subunit gamma 2 |
| chr5_-_73565602 | 0.59 |
ENST00000296785.8
|
ANKRA2
|
ankyrin repeat family A member 2 |
| chr3_-_47578832 | 0.58 |
ENST00000264723.9
ENST00000610462.1 |
CSPG5
|
chondroitin sulfate proteoglycan 5 |
| chr16_-_30010972 | 0.56 |
ENST00000565273.5
ENST00000567332.6 ENST00000350119.9 |
DOC2A
|
double C2 domain alpha |
| chr12_-_120250145 | 0.55 |
ENST00000458477.6
|
PXN
|
paxillin |
| chr1_-_16237146 | 0.55 |
ENST00000375599.8
|
CPLANE2
|
ciliogenesis and planar polarity effector 2 |
| chrX_-_109733220 | 0.55 |
ENST00000672282.1
ENST00000340800.7 |
ACSL4
|
acyl-CoA synthetase long chain family member 4 |
| chrX_-_109733249 | 0.53 |
ENST00000469796.7
ENST00000672401.1 ENST00000671846.1 |
ACSL4
|
acyl-CoA synthetase long chain family member 4 |
| chrX_-_109733292 | 0.53 |
ENST00000682031.1
ENST00000502391.6 ENST00000508092.5 ENST00000348502.10 |
ACSL4
|
acyl-CoA synthetase long chain family member 4 |
| chr18_-_48950960 | 0.50 |
ENST00000262158.8
|
SMAD7
|
SMAD family member 7 |
| chr8_+_42152946 | 0.49 |
ENST00000518421.5
ENST00000174653.3 ENST00000396926.8 ENST00000521280.5 ENST00000522288.5 |
AP3M2
|
adaptor related protein complex 3 subunit mu 2 |
| chr16_+_75222609 | 0.49 |
ENST00000495583.1
|
CTRB1
|
chymotrypsinogen B1 |
| chr14_+_94110728 | 0.48 |
ENST00000616764.4
ENST00000618863.1 ENST00000611954.4 ENST00000618200.4 ENST00000621160.4 ENST00000555819.5 ENST00000620396.4 ENST00000612813.4 ENST00000620066.1 |
IFI27
|
interferon alpha inducible protein 27 |
| chr20_-_31722854 | 0.48 |
ENST00000307677.5
|
BCL2L1
|
BCL2 like 1 |
| chr14_+_73950489 | 0.47 |
ENST00000554320.1
|
COQ6
|
coenzyme Q6, monooxygenase |
| chr17_-_61927968 | 0.44 |
ENST00000646954.1
ENST00000251334.7 ENST00000647009.1 |
INTS2
|
integrator complex subunit 2 |
| chr17_-_61928005 | 0.44 |
ENST00000444766.7
|
INTS2
|
integrator complex subunit 2 |
| chr11_+_123430259 | 0.44 |
ENST00000533341.3
ENST00000635736.2 |
GRAMD1B
|
GRAM domain containing 1B |
| chr1_-_44843240 | 0.43 |
ENST00000372192.4
|
PTCH2
|
patched 2 |
| chr12_+_4269771 | 0.42 |
ENST00000676411.1
|
CCND2
|
cyclin D2 |
| chr1_+_1001002 | 0.42 |
ENST00000624697.4
ENST00000624652.1 |
ISG15
|
ISG15 ubiquitin like modifier |
| chr11_+_35180342 | 0.42 |
ENST00000639002.1
|
CD44
|
CD44 molecule (Indian blood group) |
| chr18_-_28177934 | 0.41 |
ENST00000676445.1
|
CDH2
|
cadherin 2 |
| chr19_+_44942268 | 0.41 |
ENST00000591600.1
|
APOC4
|
apolipoprotein C4 |
| chr17_-_82101850 | 0.41 |
ENST00000583053.1
|
CCDC57
|
coiled-coil domain containing 57 |
| chr17_+_79034185 | 0.41 |
ENST00000581774.5
|
C1QTNF1
|
C1q and TNF related 1 |
| chr4_+_94207845 | 0.41 |
ENST00000457823.6
ENST00000354268.9 |
SMARCAD1
|
SWI/SNF-related, matrix-associated actin-dependent regulator of chromatin, subfamily a, containing DEAD/H box 1 |
| chr22_+_39077264 | 0.41 |
ENST00000407997.4
|
APOBEC3G
|
apolipoprotein B mRNA editing enzyme catalytic subunit 3G |
| chr11_-_19240936 | 0.39 |
ENST00000250024.9
|
E2F8
|
E2F transcription factor 8 |
| chr9_+_4490388 | 0.39 |
ENST00000262352.8
|
SLC1A1
|
solute carrier family 1 member 1 |
| chr3_+_44584953 | 0.38 |
ENST00000441021.5
ENST00000322734.2 |
ZNF660
|
zinc finger protein 660 |
| chr17_+_60600178 | 0.38 |
ENST00000629650.2
ENST00000305921.8 |
PPM1D
|
protein phosphatase, Mg2+/Mn2+ dependent 1D |
| chr19_+_43580544 | 0.38 |
ENST00000562255.5
ENST00000569031.6 |
PINLYP
|
phospholipase A2 inhibitor and LY6/PLAUR domain containing |
| chr6_-_43369636 | 0.38 |
ENST00000361428.3
|
ZNF318
|
zinc finger protein 318 |
| chr5_+_132073782 | 0.38 |
ENST00000296871.4
|
CSF2
|
colony stimulating factor 2 |
| chr15_+_58771280 | 0.37 |
ENST00000559228.6
ENST00000450403.3 |
MINDY2
|
MINDY lysine 48 deubiquitinase 2 |
| chr5_-_90409720 | 0.37 |
ENST00000522864.5
ENST00000283122.8 ENST00000522083.5 ENST00000522565.5 ENST00000522842.1 |
CETN3
|
centrin 3 |
| chrX_-_31178220 | 0.37 |
ENST00000681026.1
|
DMD
|
dystrophin |
| chr4_+_185209577 | 0.36 |
ENST00000652585.1
|
SNX25
|
sorting nexin 25 |
| chr16_+_50153248 | 0.36 |
ENST00000561678.7
|
TENT4B
|
terminal nucleotidyltransferase 4B |
| chr17_+_76726948 | 0.36 |
ENST00000586200.1
|
METTL23
|
methyltransferase like 23 |
| chr16_-_53503192 | 0.36 |
ENST00000568596.5
ENST00000394657.12 ENST00000570004.5 ENST00000564497.1 ENST00000300245.8 |
AKTIP
|
AKT interacting protein |
| chr1_-_243255170 | 0.35 |
ENST00000366542.6
|
CEP170
|
centrosomal protein 170 |
| chrX_-_31178149 | 0.35 |
ENST00000679437.1
|
DMD
|
dystrophin |
| chr20_+_49982969 | 0.34 |
ENST00000244050.3
|
SNAI1
|
snail family transcriptional repressor 1 |
| chr8_+_232416 | 0.34 |
ENST00000518414.5
ENST00000521270.5 ENST00000518320.6 ENST00000398612.3 |
ZNF596
|
zinc finger protein 596 |
| chr16_+_2830179 | 0.34 |
ENST00000570670.5
|
ZG16B
|
zymogen granule protein 16B |
| chr8_-_66613229 | 0.33 |
ENST00000517885.5
|
MYBL1
|
MYB proto-oncogene like 1 |
| chr1_-_243255320 | 0.33 |
ENST00000366544.5
ENST00000366543.5 |
CEP170
|
centrosomal protein 170 |
| chr16_+_2830295 | 0.33 |
ENST00000571723.1
|
ZG16B
|
zymogen granule protein 16B |
| chr11_+_117232725 | 0.33 |
ENST00000531287.5
ENST00000531452.5 |
RNF214
|
ring finger protein 214 |
| chr16_+_53130921 | 0.33 |
ENST00000564845.5
|
CHD9
|
chromodomain helicase DNA binding protein 9 |
| chr10_+_48684859 | 0.32 |
ENST00000360890.6
ENST00000325239.11 |
WDFY4
|
WDFY family member 4 |
| chr7_-_151814668 | 0.32 |
ENST00000651764.1
|
PRKAG2
|
protein kinase AMP-activated non-catalytic subunit gamma 2 |
| chr10_-_103452384 | 0.32 |
ENST00000369788.7
|
CALHM2
|
calcium homeostasis modulator family member 2 |
| chr3_-_165196689 | 0.32 |
ENST00000241274.3
|
SLITRK3
|
SLIT and NTRK like family member 3 |
| chr12_-_110445540 | 0.31 |
ENST00000547365.1
|
ARPC3
|
actin related protein 2/3 complex subunit 3 |
| chr1_+_70411180 | 0.31 |
ENST00000411986.6
|
CTH
|
cystathionine gamma-lyase |
| chr14_-_52069228 | 0.30 |
ENST00000617139.4
|
NID2
|
nidogen 2 |
| chr19_-_6481769 | 0.30 |
ENST00000381480.7
ENST00000543576.5 ENST00000590173.5 |
DENND1C
|
DENN domain containing 1C |
| chrX_+_7147819 | 0.30 |
ENST00000660000.2
|
STS
|
steroid sulfatase |
| chrX_-_7148118 | 0.30 |
ENST00000486446.3
ENST00000381077.10 ENST00000412827.6 ENST00000424830.6 |
PUDP
|
pseudouridine 5'-phosphatase |
| chr16_-_74666839 | 0.30 |
ENST00000576652.1
ENST00000572337.5 ENST00000571750.5 ENST00000572990.5 ENST00000361070.9 |
RFWD3
|
ring finger and WD repeat domain 3 |
| chr19_+_1275508 | 0.29 |
ENST00000409293.6
|
FAM174C
|
family with sequence similarity 174 member C |
| chr1_+_70411241 | 0.29 |
ENST00000370938.8
ENST00000346806.2 |
CTH
|
cystathionine gamma-lyase |
| chr6_-_52577012 | 0.29 |
ENST00000182527.4
|
TRAM2
|
translocation associated membrane protein 2 |
| chr7_-_100436425 | 0.29 |
ENST00000292330.3
|
PPP1R35
|
protein phosphatase 1 regulatory subunit 35 |
| chrX_-_153673678 | 0.29 |
ENST00000370150.5
|
PNCK
|
pregnancy up-regulated nonubiquitous CaM kinase |
| chr7_-_101627513 | 0.29 |
ENST00000642629.2
|
MYL10
|
myosin light chain 10 |
| chr3_-_10292901 | 0.29 |
ENST00000430179.5
ENST00000287656.11 ENST00000437422.6 ENST00000439975.6 ENST00000446937.2 ENST00000449238.6 ENST00000457360.5 |
GHRL
|
ghrelin and obestatin prepropeptide |
| chr16_+_53054973 | 0.28 |
ENST00000447540.6
ENST00000615216.4 ENST00000566029.5 |
CHD9
|
chromodomain helicase DNA binding protein 9 |
| chr19_+_1908258 | 0.28 |
ENST00000411971.5
ENST00000588907.2 |
SCAMP4
|
secretory carrier membrane protein 4 |
| chrX_-_153673778 | 0.28 |
ENST00000340888.8
|
PNCK
|
pregnancy up-regulated nonubiquitous CaM kinase |
| chr11_+_117232625 | 0.28 |
ENST00000534428.5
ENST00000300650.9 |
RNF214
|
ring finger protein 214 |
| chr2_-_96116565 | 0.28 |
ENST00000620793.2
|
ADRA2B
|
adrenoceptor alpha 2B |
| chr17_+_41232447 | 0.28 |
ENST00000411528.4
|
KRTAP9-3
|
keratin associated protein 9-3 |
| chr14_-_52069039 | 0.27 |
ENST00000216286.10
|
NID2
|
nidogen 2 |
| chr1_-_41021712 | 0.27 |
ENST00000302946.12
ENST00000372613.6 ENST00000439569.2 |
SLFNL1
|
schlafen like 1 |
| chr1_-_41021743 | 0.27 |
ENST00000372611.5
|
SLFNL1
|
schlafen like 1 |
| chr15_+_97960692 | 0.27 |
ENST00000268042.7
|
ARRDC4
|
arrestin domain containing 4 |
| chr12_+_57459782 | 0.27 |
ENST00000228682.7
|
GLI1
|
GLI family zinc finger 1 |
| chrX_+_152917830 | 0.27 |
ENST00000318529.12
|
ZNF185
|
zinc finger protein 185 with LIM domain |
| chr3_+_88059231 | 0.27 |
ENST00000636215.2
|
ZNF654
|
zinc finger protein 654 |
| chr10_+_70478761 | 0.27 |
ENST00000263563.7
|
PALD1
|
phosphatase domain containing paladin 1 |
| chr4_+_94207596 | 0.27 |
ENST00000359052.8
|
SMARCAD1
|
SWI/SNF-related, matrix-associated actin-dependent regulator of chromatin, subfamily a, containing DEAD/H box 1 |
| chr5_+_90474848 | 0.27 |
ENST00000651687.1
|
POLR3G
|
RNA polymerase III subunit G |
| chr7_-_113086815 | 0.27 |
ENST00000424100.2
|
GPR85
|
G protein-coupled receptor 85 |
| chr7_+_1530684 | 0.27 |
ENST00000343242.9
|
MAFK
|
MAF bZIP transcription factor K |
| chr16_-_75266781 | 0.27 |
ENST00000420641.7
|
BCAR1
|
BCAR1 scaffold protein, Cas family member |
| chr20_-_52191751 | 0.26 |
ENST00000346617.8
ENST00000371515.8 ENST00000371518.6 ENST00000216923.5 |
ZFP64
|
ZFP64 zinc finger protein |
| chr3_-_10292933 | 0.26 |
ENST00000429122.1
ENST00000425479.1 ENST00000335542.13 |
GHRL
|
ghrelin and obestatin prepropeptide |
| chr5_-_59276109 | 0.26 |
ENST00000503258.5
|
PDE4D
|
phosphodiesterase 4D |
| chr2_+_127423265 | 0.26 |
ENST00000402125.2
|
PROC
|
protein C, inactivator of coagulation factors Va and VIIIa |
| chr15_+_49155750 | 0.26 |
ENST00000327171.7
ENST00000560654.5 |
GALK2
|
galactokinase 2 |
| chr2_-_171433950 | 0.26 |
ENST00000375258.9
ENST00000442541.1 ENST00000392599.6 |
METTL8
|
methyltransferase like 8 |
| chr22_+_20021069 | 0.26 |
ENST00000420290.6
ENST00000432883.5 ENST00000434570.6 ENST00000447208.6 ENST00000456048.5 ENST00000398042.6 ENST00000450664.5 ENST00000327374.9 |
TANGO2
|
transport and golgi organization 2 homolog |
| chr11_+_18394586 | 0.25 |
ENST00000227157.8
ENST00000478970.6 ENST00000495052.5 |
LDHA
|
lactate dehydrogenase A |
| chr1_+_154993581 | 0.25 |
ENST00000392487.1
|
LENEP
|
lens epithelial protein |
| chr6_+_1609890 | 0.25 |
ENST00000645831.2
|
FOXC1
|
forkhead box C1 |
| chr9_-_90642855 | 0.25 |
ENST00000637905.1
|
DIRAS2
|
DIRAS family GTPase 2 |
| chr2_+_233060295 | 0.25 |
ENST00000445964.6
|
INPP5D
|
inositol polyphosphate-5-phosphatase D |
| chrX_+_108091752 | 0.25 |
ENST00000457035.5
ENST00000372232.8 |
ATG4A
|
autophagy related 4A cysteine peptidase |
| chr22_-_20268285 | 0.25 |
ENST00000043402.8
|
RTN4R
|
reticulon 4 receptor |
| chrX_+_108091665 | 0.25 |
ENST00000345734.7
|
ATG4A
|
autophagy related 4A cysteine peptidase |
| chr5_+_161848536 | 0.25 |
ENST00000519621.2
ENST00000636573.1 |
GABRA1
|
gamma-aminobutyric acid type A receptor subunit alpha1 |
| chr11_+_18394552 | 0.25 |
ENST00000543445.5
ENST00000430553.6 ENST00000396222.6 ENST00000535451.5 ENST00000422447.8 |
LDHA
|
lactate dehydrogenase A |
| chr16_+_68085420 | 0.25 |
ENST00000349223.9
|
NFATC3
|
nuclear factor of activated T cells 3 |
| chr12_+_57460127 | 0.25 |
ENST00000532291.5
ENST00000543426.5 ENST00000546141.5 |
GLI1
|
GLI family zinc finger 1 |
| chr2_+_200440649 | 0.25 |
ENST00000366118.2
|
SPATS2L
|
spermatogenesis associated serine rich 2 like |
| chr14_+_93927358 | 0.25 |
ENST00000557000.2
|
FAM181A
|
family with sequence similarity 181 member A |
| chr5_-_10761156 | 0.24 |
ENST00000432074.2
ENST00000230895.11 |
DAP
|
death associated protein |
| chr5_+_90474879 | 0.24 |
ENST00000504930.5
ENST00000514483.5 |
POLR3G
|
RNA polymerase III subunit G |
| chr5_+_149345430 | 0.24 |
ENST00000513661.5
ENST00000329271.8 ENST00000416916.2 |
GRPEL2
|
GrpE like 2, mitochondrial |
| chr14_-_63641407 | 0.24 |
ENST00000267522.7
|
WDR89
|
WD repeat domain 89 |
| chr6_+_26204552 | 0.24 |
ENST00000615164.2
|
H4C5
|
H4 clustered histone 5 |
| chr19_-_54280498 | 0.24 |
ENST00000391746.5
|
LILRB2
|
leukocyte immunoglobulin like receptor B2 |
| chr15_+_79311137 | 0.24 |
ENST00000424155.6
ENST00000536821.5 |
TMED3
|
transmembrane p24 trafficking protein 3 |
| chr7_+_116672187 | 0.23 |
ENST00000318493.11
ENST00000397752.8 |
MET
|
MET proto-oncogene, receptor tyrosine kinase |
| chr5_+_98773651 | 0.23 |
ENST00000513185.3
|
RGMB
|
repulsive guidance molecule BMP co-receptor b |
| chr9_+_33264848 | 0.23 |
ENST00000419016.6
|
CHMP5
|
charged multivesicular body protein 5 |
| chr2_+_64988469 | 0.23 |
ENST00000531327.5
|
SLC1A4
|
solute carrier family 1 member 4 |
| chr6_+_16129077 | 0.23 |
ENST00000356840.8
ENST00000349606.4 |
MYLIP
|
myosin regulatory light chain interacting protein |
| chr9_+_33265013 | 0.23 |
ENST00000223500.9
|
CHMP5
|
charged multivesicular body protein 5 |
| chr6_+_3849350 | 0.23 |
ENST00000648326.1
ENST00000380274.2 |
FAM50B
|
family with sequence similarity 50 member B |
| chr13_+_32315071 | 0.23 |
ENST00000544455.6
|
BRCA2
|
BRCA2 DNA repair associated |
| chr20_-_63254429 | 0.23 |
ENST00000370316.8
|
NKAIN4
|
sodium/potassium transporting ATPase interacting 4 |
| chr2_-_240820205 | 0.23 |
ENST00000647731.1
ENST00000648680.1 ENST00000649306.1 ENST00000648129.1 ENST00000498729.9 ENST00000320389.12 ENST00000648364.1 ENST00000647885.1 ENST00000404283.9 ENST00000649096.1 |
KIF1A
|
kinesin family member 1A |
| chr7_+_64903247 | 0.23 |
ENST00000476120.1
|
ZNF273
|
zinc finger protein 273 |
| chr2_-_100142575 | 0.23 |
ENST00000317233.8
ENST00000672204.1 ENST00000416492.5 ENST00000672857.1 ENST00000672756.2 |
AFF3
|
AF4/FMR2 family member 3 |
| chr2_+_233059838 | 0.23 |
ENST00000359570.9
|
INPP5D
|
inositol polyphosphate-5-phosphatase D |
| chr12_-_30754927 | 0.22 |
ENST00000541765.5
ENST00000537108.5 |
CAPRIN2
|
caprin family member 2 |
| chr20_-_31723491 | 0.22 |
ENST00000676582.1
ENST00000422920.2 |
BCL2L1
|
BCL2 like 1 |
| chr14_+_105419938 | 0.22 |
ENST00000405646.5
|
MTA1
|
metastasis associated 1 |
| chr14_+_20999255 | 0.22 |
ENST00000554422.5
ENST00000298681.5 |
SLC39A2
|
solute carrier family 39 member 2 |
| chr17_+_7580442 | 0.22 |
ENST00000584180.1
|
CD68
|
CD68 molecule |
| chrX_+_12791353 | 0.22 |
ENST00000380663.7
ENST00000398491.6 ENST00000380668.10 ENST00000489404.5 |
PRPS2
|
phosphoribosyl pyrophosphate synthetase 2 |
| chr9_+_17579059 | 0.22 |
ENST00000380607.5
|
SH3GL2
|
SH3 domain containing GRB2 like 2, endophilin A1 |
| chr14_+_102922639 | 0.22 |
ENST00000299155.10
|
AMN
|
amnion associated transmembrane protein |
| chr10_+_74151232 | 0.22 |
ENST00000478611.2
ENST00000673352.1 |
ADK
|
adenosine kinase |
| chr20_+_62642492 | 0.22 |
ENST00000217159.6
|
SLCO4A1
|
solute carrier organic anion transporter family member 4A1 |
| chr12_-_47705971 | 0.22 |
ENST00000380650.4
|
RPAP3
|
RNA polymerase II associated protein 3 |
| chr12_-_92929236 | 0.22 |
ENST00000322349.13
|
EEA1
|
early endosome antigen 1 |
| chr10_-_112183698 | 0.22 |
ENST00000369425.5
ENST00000348367.9 |
GPAM
|
glycerol-3-phosphate acyltransferase, mitochondrial |
| chr7_+_142424942 | 0.21 |
ENST00000426318.2
|
TRBV10-2
|
T cell receptor beta variable 10-2 |
| chr12_+_52274610 | 0.21 |
ENST00000423955.7
|
KRT86
|
keratin 86 |
| chr5_+_83077559 | 0.21 |
ENST00000511817.1
|
XRCC4
|
X-ray repair cross complementing 4 |
| chr9_+_4839761 | 0.21 |
ENST00000448872.6
ENST00000441844.2 |
RCL1
|
RNA terminal phosphate cyclase like 1 |
| chr2_+_108786738 | 0.21 |
ENST00000412964.6
ENST00000295124.9 |
CCDC138
|
coiled-coil domain containing 138 |
| chr19_-_23758877 | 0.21 |
ENST00000402377.3
|
ZNF681
|
zinc finger protein 681 |
| chr11_+_95789965 | 0.21 |
ENST00000537677.5
|
CEP57
|
centrosomal protein 57 |
| chr7_+_100373458 | 0.21 |
ENST00000453419.5
ENST00000198536.7 ENST00000394000.6 ENST00000350573.2 |
PILRA
|
paired immunoglobin like type 2 receptor alpha |
| chr2_-_68467272 | 0.21 |
ENST00000377957.4
|
FBXO48
|
F-box protein 48 |
| chr19_-_10928512 | 0.21 |
ENST00000588347.5
|
YIPF2
|
Yip1 domain family member 2 |
| chr5_+_83077498 | 0.21 |
ENST00000282268.7
ENST00000338635.10 ENST00000396027.9 |
XRCC4
|
X-ray repair cross complementing 4 |
| chr5_+_161848314 | 0.21 |
ENST00000437025.6
|
GABRA1
|
gamma-aminobutyric acid type A receptor subunit alpha1 |
| chr12_-_94616061 | 0.21 |
ENST00000551457.1
|
TMCC3
|
transmembrane and coiled-coil domain family 3 |
| chr7_-_138755892 | 0.21 |
ENST00000644341.1
ENST00000478480.2 |
ATP6V0A4
|
ATPase H+ transporting V0 subunit a4 |
| chr19_+_17527232 | 0.21 |
ENST00000601861.5
|
NIBAN3
|
niban apoptosis regulator 3 |
| chr8_+_26291758 | 0.21 |
ENST00000522535.5
ENST00000665949.1 |
PPP2R2A
|
protein phosphatase 2 regulatory subunit Balpha |
| chr12_-_47705990 | 0.21 |
ENST00000432584.7
ENST00000005386.8 |
RPAP3
|
RNA polymerase II associated protein 3 |
| chr19_-_17334844 | 0.21 |
ENST00000159087.7
|
ANO8
|
anoctamin 8 |
| chr7_-_92245795 | 0.20 |
ENST00000412043.6
ENST00000430102.5 ENST00000425073.1 ENST00000394505.7 ENST00000394503.6 ENST00000454017.5 ENST00000440209.5 ENST00000413688.5 ENST00000452773.5 ENST00000433016.5 ENST00000422347.5 ENST00000458493.5 ENST00000425919.5 ENST00000650585.1 |
KRIT1
ENSG00000285953.1
|
KRIT1 ankyrin repeat containing novel protein |
| chr15_-_78234513 | 0.20 |
ENST00000558130.1
ENST00000258873.9 |
ACSBG1
|
acyl-CoA synthetase bubblegum family member 1 |
| chr14_-_105601728 | 0.20 |
ENST00000641420.1
ENST00000390541.2 |
IGHE
|
immunoglobulin heavy constant epsilon |
| chr16_-_90008988 | 0.20 |
ENST00000568662.2
|
DBNDD1
|
dysbindin domain containing 1 |
| chr19_-_39320839 | 0.20 |
ENST00000248668.5
|
LRFN1
|
leucine rich repeat and fibronectin type III domain containing 1 |
| chr1_+_231338242 | 0.20 |
ENST00000008440.9
ENST00000295050.12 |
SPRTN
|
SprT-like N-terminal domain |
| chr1_-_21622509 | 0.20 |
ENST00000374761.6
|
RAP1GAP
|
RAP1 GTPase activating protein |
| chr12_+_120496075 | 0.20 |
ENST00000548214.1
ENST00000392508.2 |
DYNLL1
|
dynein light chain LC8-type 1 |
| chr11_+_68008542 | 0.20 |
ENST00000614849.4
|
ALDH3B1
|
aldehyde dehydrogenase 3 family member B1 |
| chr7_+_100428782 | 0.20 |
ENST00000414441.5
|
MEPCE
|
methylphosphate capping enzyme |
| chr3_-_185821092 | 0.20 |
ENST00000421047.3
|
IGF2BP2
|
insulin like growth factor 2 mRNA binding protein 2 |
| chr6_+_87472925 | 0.20 |
ENST00000369556.7
ENST00000369557.9 ENST00000369552.9 |
SLC35A1
|
solute carrier family 35 member A1 |
| chr12_-_81758641 | 0.19 |
ENST00000552948.5
ENST00000548586.5 |
PPFIA2
|
PTPRF interacting protein alpha 2 |
| chr9_-_21350956 | 0.19 |
ENST00000259555.5
|
IFNA6
|
interferon alpha 6 |
| chr5_+_162067764 | 0.19 |
ENST00000639213.2
ENST00000414552.6 |
GABRG2
|
gamma-aminobutyric acid type A receptor subunit gamma2 |
| chr5_-_132777371 | 0.19 |
ENST00000620483.4
|
SEPTIN8
|
septin 8 |
| chr1_+_107056656 | 0.19 |
ENST00000370078.2
|
PRMT6
|
protein arginine methyltransferase 6 |
| chr13_-_40666600 | 0.19 |
ENST00000379561.6
|
FOXO1
|
forkhead box O1 |
| chr16_+_88382950 | 0.19 |
ENST00000565624.3
|
ZNF469
|
zinc finger protein 469 |
| chr5_+_162067858 | 0.19 |
ENST00000361925.9
|
GABRG2
|
gamma-aminobutyric acid type A receptor subunit gamma2 |
| chr14_+_95876385 | 0.19 |
ENST00000504119.1
|
TUNAR
|
TCL1 upstream neural differentiation-associated RNA |
| chr9_+_40223317 | 0.19 |
ENST00000611747.1
ENST00000377601.3 ENST00000654136.1 ENST00000659103.1 |
ANKRD20A2P
ENSG00000240240.9
|
ankyrin repeat domain 20 family member A2, pseudogene novel transcript |
| chr9_+_92947516 | 0.19 |
ENST00000375482.8
|
FGD3
|
FYVE, RhoGEF and PH domain containing 3 |
| chr7_-_111562455 | 0.19 |
ENST00000452895.5
ENST00000405709.7 ENST00000452753.1 ENST00000331762.7 |
IMMP2L
|
inner mitochondrial membrane peptidase subunit 2 |
| chr2_-_200963633 | 0.19 |
ENST00000234296.7
|
ORC2
|
origin recognition complex subunit 2 |
| chrX_-_15314543 | 0.19 |
ENST00000344384.8
|
ASB11
|
ankyrin repeat and SOCS box containing 11 |
| chr6_-_134052594 | 0.19 |
ENST00000275230.6
|
SLC2A12
|
solute carrier family 2 member 12 |
| chr19_-_1848452 | 0.19 |
ENST00000170168.9
|
REXO1
|
RNA exonuclease 1 homolog |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.3 | GO:0060136 | embryonic process involved in female pregnancy(GO:0060136) |
| 0.2 | 0.5 | GO:2000506 | negative regulation of circadian sleep/wake cycle, REM sleep(GO:0042322) negative regulation of energy homeostasis(GO:2000506) |
| 0.2 | 0.5 | GO:0060032 | notochord regression(GO:0060032) |
| 0.2 | 0.6 | GO:0006175 | adenosine salvage(GO:0006169) dATP biosynthetic process(GO:0006175) |
| 0.1 | 0.4 | GO:0044537 | regulation of circulating fibrinogen levels(GO:0044537) |
| 0.1 | 0.4 | GO:0032877 | positive regulation of DNA endoreduplication(GO:0032877) |
| 0.1 | 1.0 | GO:0010961 | cellular magnesium ion homeostasis(GO:0010961) |
| 0.1 | 0.6 | GO:0018352 | protein-pyridoxal-5-phosphate linkage(GO:0018352) |
| 0.1 | 0.5 | GO:0045659 | regulation of neutrophil differentiation(GO:0045658) negative regulation of neutrophil differentiation(GO:0045659) |
| 0.1 | 0.7 | GO:0051970 | negative regulation of transmission of nerve impulse(GO:0051970) |
| 0.1 | 0.8 | GO:0097490 | sympathetic neuron projection extension(GO:0097490) sympathetic neuron projection guidance(GO:0097491) |
| 0.1 | 0.3 | GO:0032811 | negative regulation of epinephrine secretion(GO:0032811) |
| 0.1 | 0.3 | GO:0071484 | cellular response to light intensity(GO:0071484) cellular response to high light intensity(GO:0071486) retinal rod cell apoptotic process(GO:0097473) retinal cell apoptotic process(GO:1990009) |
| 0.1 | 0.4 | GO:0006550 | isoleucine catabolic process(GO:0006550) |
| 0.1 | 0.7 | GO:0060154 | cellular process regulating host cell cycle in response to virus(GO:0060154) |
| 0.1 | 0.5 | GO:0051697 | protein delipidation(GO:0051697) |
| 0.1 | 0.7 | GO:0060373 | regulation of ventricular cardiac muscle cell membrane depolarization(GO:0060373) |
| 0.1 | 0.3 | GO:1901491 | negative regulation of lymphangiogenesis(GO:1901491) |
| 0.1 | 0.4 | GO:1902498 | regulation of protein autoubiquitination(GO:1902498) |
| 0.1 | 1.4 | GO:0001542 | ovulation from ovarian follicle(GO:0001542) |
| 0.1 | 0.3 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.1 | 0.4 | GO:0070383 | DNA cytosine deamination(GO:0070383) |
| 0.1 | 0.5 | GO:0002774 | Fc receptor mediated inhibitory signaling pathway(GO:0002774) |
| 0.1 | 0.2 | GO:0061300 | cerebellum vasculature development(GO:0061300) |
| 0.1 | 0.2 | GO:0002752 | cell surface pattern recognition receptor signaling pathway(GO:0002752) |
| 0.1 | 0.2 | GO:1902617 | response to fluoride(GO:1902617) |
| 0.1 | 0.6 | GO:0010890 | positive regulation of sequestering of triglyceride(GO:0010890) |
| 0.1 | 0.6 | GO:0000101 | sulfur amino acid transport(GO:0000101) |
| 0.1 | 0.2 | GO:0060903 | positive regulation of meiosis I(GO:0060903) |
| 0.1 | 0.4 | GO:2000860 | positive regulation of mineralocorticoid secretion(GO:2000857) positive regulation of aldosterone secretion(GO:2000860) |
| 0.1 | 0.3 | GO:0045048 | protein insertion into ER membrane(GO:0045048) |
| 0.1 | 0.7 | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.1 | 0.3 | GO:0032747 | positive regulation of interleukin-23 production(GO:0032747) |
| 0.1 | 0.6 | GO:0009957 | epidermal cell fate specification(GO:0009957) |
| 0.1 | 0.8 | GO:0034472 | snRNA 3'-end processing(GO:0034472) |
| 0.1 | 0.2 | GO:0003095 | pressure natriuresis(GO:0003095) |
| 0.1 | 0.2 | GO:0033606 | B cell receptor transport within lipid bilayer(GO:0032595) B cell receptor transport into membrane raft(GO:0032597) protein transport out of membrane raft(GO:0032599) chemokine receptor transport out of membrane raft(GO:0032600) negative regulation of transforming growth factor beta3 production(GO:0032913) chemokine receptor transport within lipid bilayer(GO:0033606) |
| 0.1 | 0.2 | GO:0070105 | positive regulation of interleukin-6-mediated signaling pathway(GO:0070105) |
| 0.0 | 0.1 | GO:0010847 | regulation of chromatin assembly(GO:0010847) |
| 0.0 | 0.1 | GO:0051097 | negative regulation of helicase activity(GO:0051097) |
| 0.0 | 0.2 | GO:0018199 | peptidyl-glutamine modification(GO:0018199) |
| 0.0 | 0.4 | GO:1902897 | regulation of postsynaptic density protein 95 clustering(GO:1902897) |
| 0.0 | 0.2 | GO:1990426 | homologous recombination-dependent replication fork processing(GO:1990426) |
| 0.0 | 1.2 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.0 | 0.1 | GO:0070318 | positive regulation of G0 to G1 transition(GO:0070318) |
| 0.0 | 0.2 | GO:0051582 | positive regulation of neurotransmitter uptake(GO:0051582) |
| 0.0 | 0.2 | GO:0033594 | response to hydroxyisoflavone(GO:0033594) |
| 0.0 | 0.2 | GO:0046391 | 5-phosphoribose 1-diphosphate biosynthetic process(GO:0006015) 5-phosphoribose 1-diphosphate metabolic process(GO:0046391) |
| 0.0 | 0.1 | GO:0018171 | peptidyl-cysteine oxidation(GO:0018171) |
| 0.0 | 0.4 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.0 | 0.3 | GO:0010989 | negative regulation of low-density lipoprotein particle clearance(GO:0010989) |
| 0.0 | 0.1 | GO:1905051 | regulation of base-excision repair(GO:1905051) positive regulation of base-excision repair(GO:1905053) |
| 0.0 | 0.4 | GO:0010529 | regulation of transposition(GO:0010528) negative regulation of transposition(GO:0010529) |
| 0.0 | 0.1 | GO:0046110 | xanthine metabolic process(GO:0046110) |
| 0.0 | 0.7 | GO:0070933 | histone H4 deacetylation(GO:0070933) |
| 0.0 | 0.9 | GO:0006853 | carnitine shuttle(GO:0006853) |
| 0.0 | 0.2 | GO:0097045 | phosphatidylserine exposure on blood platelet(GO:0097045) |
| 0.0 | 0.4 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.0 | 0.5 | GO:1904903 | ESCRT complex disassembly(GO:1904896) ESCRT III complex disassembly(GO:1904903) |
| 0.0 | 0.1 | GO:0072365 | regulation of cellular ketone metabolic process by negative regulation of transcription from RNA polymerase II promoter(GO:0072365) |
| 0.0 | 0.4 | GO:2000582 | regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.0 | 0.2 | GO:0006404 | RNA import into nucleus(GO:0006404) |
| 0.0 | 0.1 | GO:0006478 | peptidyl-tyrosine sulfation(GO:0006478) |
| 0.0 | 0.1 | GO:1902683 | regulation of receptor localization to synapse(GO:1902683) |
| 0.0 | 0.1 | GO:1901143 | insulin catabolic process(GO:1901143) |
| 0.0 | 0.9 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.0 | 0.4 | GO:1900625 | regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 0.0 | 0.2 | GO:0008050 | female courtship behavior(GO:0008050) |
| 0.0 | 0.1 | GO:1903976 | Schwann cell migration(GO:0036135) regulation of Schwann cell migration(GO:1900147) negative regulation of glial cell migration(GO:1903976) |
| 0.0 | 0.2 | GO:0000480 | endonucleolytic cleavage in 5'-ETS of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000480) |
| 0.0 | 0.1 | GO:0042323 | negative regulation of circadian sleep/wake cycle, non-REM sleep(GO:0042323) negative regulation of mucus secretion(GO:0070256) |
| 0.0 | 0.3 | GO:1901299 | negative regulation of hydrogen peroxide-mediated programmed cell death(GO:1901299) |
| 0.0 | 0.1 | GO:1901526 | positive regulation of macromitophagy(GO:1901526) positive regulation of mitophagy in response to mitochondrial depolarization(GO:1904925) |
| 0.0 | 0.9 | GO:0009235 | cobalamin metabolic process(GO:0009235) |
| 0.0 | 0.1 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.0 | 0.3 | GO:0019919 | peptidyl-arginine methylation, to asymmetrical-dimethyl arginine(GO:0019919) |
| 0.0 | 0.2 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) |
| 0.0 | 0.6 | GO:0007172 | signal complex assembly(GO:0007172) |
| 0.0 | 0.5 | GO:0006089 | lactate metabolic process(GO:0006089) |
| 0.0 | 0.1 | GO:2000664 | positive regulation of interleukin-5 secretion(GO:2000664) |
| 0.0 | 0.1 | GO:0038155 | interleukin-23-mediated signaling pathway(GO:0038155) |
| 0.0 | 0.1 | GO:1903377 | negative regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903377) |
| 0.0 | 0.1 | GO:0071442 | positive regulation of histone H3-K14 acetylation(GO:0071442) |
| 0.0 | 0.1 | GO:0098838 | reduced folate transmembrane transport(GO:0098838) |
| 0.0 | 0.1 | GO:0071529 | cementum mineralization(GO:0071529) |
| 0.0 | 0.1 | GO:0036515 | serotonergic neuron axon guidance(GO:0036515) |
| 0.0 | 0.1 | GO:0090273 | regulation of somatostatin secretion(GO:0090273) positive regulation of somatostatin secretion(GO:0090274) |
| 0.0 | 0.1 | GO:0045085 | negative regulation of interleukin-2 biosynthetic process(GO:0045085) |
| 0.0 | 0.3 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.0 | 0.1 | GO:0098528 | skeletal muscle fiber differentiation(GO:0098528) |
| 0.0 | 0.1 | GO:0060672 | epithelial cell differentiation involved in embryonic placenta development(GO:0060671) epithelial cell morphogenesis involved in placental branching(GO:0060672) |
| 0.0 | 0.1 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.0 | 0.2 | GO:0040031 | snRNA modification(GO:0040031) |
| 0.0 | 0.1 | GO:1903568 | negative regulation of protein localization to cilium(GO:1903565) regulation of protein localization to ciliary membrane(GO:1903567) negative regulation of protein localization to ciliary membrane(GO:1903568) |
| 0.0 | 0.1 | GO:2001045 | closure of optic fissure(GO:0061386) negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.0 | 0.1 | GO:0070682 | proteasome regulatory particle assembly(GO:0070682) |
| 0.0 | 0.3 | GO:0046541 | saliva secretion(GO:0046541) |
| 0.0 | 0.2 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.0 | 0.1 | GO:0070370 | heat acclimation(GO:0010286) cellular heat acclimation(GO:0070370) |
| 0.0 | 0.1 | GO:0002143 | tRNA wobble position uridine thiolation(GO:0002143) |
| 0.0 | 0.1 | GO:0032237 | activation of store-operated calcium channel activity(GO:0032237) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.0 | 0.1 | GO:0001555 | oocyte growth(GO:0001555) |
| 0.0 | 0.1 | GO:0010968 | regulation of microtubule nucleation(GO:0010968) |
| 0.0 | 0.1 | GO:1903233 | regulation of calcium ion-dependent exocytosis of neurotransmitter(GO:1903233) |
| 0.0 | 0.1 | GO:0000430 | regulation of transcription from RNA polymerase II promoter by glucose(GO:0000430) positive regulation of transcription from RNA polymerase II promoter by glucose(GO:0000432) late viral transcription(GO:0019086) |
| 0.0 | 0.4 | GO:0075713 | establishment of integrated proviral latency(GO:0075713) |
| 0.0 | 0.1 | GO:0019858 | cytosine metabolic process(GO:0019858) |
| 0.0 | 0.1 | GO:0043321 | regulation of natural killer cell degranulation(GO:0043321) positive regulation of natural killer cell degranulation(GO:0043323) |
| 0.0 | 0.1 | GO:0046900 | tetrahydrofolylpolyglutamate metabolic process(GO:0046900) |
| 0.0 | 0.1 | GO:0019556 | histidine catabolic process to glutamate and formamide(GO:0019556) histidine catabolic process to glutamate and formate(GO:0019557) formamide metabolic process(GO:0043606) |
| 0.0 | 0.6 | GO:0030208 | dermatan sulfate biosynthetic process(GO:0030208) |
| 0.0 | 0.1 | GO:0006121 | mitochondrial electron transport, succinate to ubiquinone(GO:0006121) |
| 0.0 | 0.1 | GO:0002351 | serotonin production involved in inflammatory response(GO:0002351) serotonin secretion involved in inflammatory response(GO:0002442) serotonin secretion by platelet(GO:0002554) |
| 0.0 | 0.2 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.0 | 0.2 | GO:0003138 | primary heart field specification(GO:0003138) sinoatrial valve development(GO:0003172) sinoatrial valve morphogenesis(GO:0003185) |
| 0.0 | 0.1 | GO:0003366 | cell-matrix adhesion involved in ameboidal cell migration(GO:0003366) diapedesis(GO:0050904) |
| 0.0 | 0.4 | GO:0071481 | cellular response to X-ray(GO:0071481) |
| 0.0 | 0.1 | GO:0071072 | negative regulation of phospholipid biosynthetic process(GO:0071072) |
| 0.0 | 0.3 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.0 | 0.2 | GO:0015712 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.0 | 0.1 | GO:0038169 | hormone-mediated apoptotic signaling pathway(GO:0008628) somatostatin receptor signaling pathway(GO:0038169) somatostatin signaling pathway(GO:0038170) |
| 0.0 | 0.1 | GO:0019254 | carnitine metabolic process, CoA-linked(GO:0019254) |
| 0.0 | 0.3 | GO:0023041 | neuronal signal transduction(GO:0023041) |
| 0.0 | 0.5 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.0 | 0.1 | GO:2001287 | negative regulation of caveolin-mediated endocytosis(GO:2001287) |
| 0.0 | 0.2 | GO:0072592 | oxygen metabolic process(GO:0072592) |
| 0.0 | 0.2 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 0.0 | 0.2 | GO:2000786 | positive regulation of autophagosome assembly(GO:2000786) |
| 0.0 | 0.4 | GO:0035729 | cellular response to hepatocyte growth factor stimulus(GO:0035729) |
| 0.0 | 0.1 | GO:0071460 | cellular response to cell-matrix adhesion(GO:0071460) |
| 0.0 | 0.1 | GO:0010902 | positive regulation of very-low-density lipoprotein particle remodeling(GO:0010902) |
| 0.0 | 0.1 | GO:0043126 | regulation of 1-phosphatidylinositol 4-kinase activity(GO:0043126) positive regulation of 1-phosphatidylinositol 4-kinase activity(GO:0043128) |
| 0.0 | 0.0 | GO:1900019 | regulation of protein kinase C activity(GO:1900019) positive regulation of protein kinase C activity(GO:1900020) |
| 0.0 | 0.2 | GO:0048102 | autophagic cell death(GO:0048102) |
| 0.0 | 0.5 | GO:0099514 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.0 | 0.0 | GO:0035281 | pre-miRNA export from nucleus(GO:0035281) |
| 0.0 | 0.8 | GO:0071711 | basement membrane organization(GO:0071711) |
| 0.0 | 0.1 | GO:0051182 | coenzyme transport(GO:0051182) |
| 0.0 | 0.3 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.0 | 0.2 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.0 | 0.1 | GO:0032534 | regulation of microvillus assembly(GO:0032534) |
| 0.0 | 0.1 | GO:0061357 | positive regulation of Wnt protein secretion(GO:0061357) |
| 0.0 | 0.1 | GO:0070269 | pyroptosis(GO:0070269) |
| 0.0 | 0.3 | GO:0033141 | positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) |
| 0.0 | 0.2 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 0.0 | 0.0 | GO:1904479 | negative regulation of intestinal absorption(GO:1904479) |
| 0.0 | 0.1 | GO:0036072 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.0 | 0.2 | GO:0015747 | urate transport(GO:0015747) |
| 0.0 | 0.2 | GO:0010459 | negative regulation of heart rate(GO:0010459) |
| 0.0 | 0.1 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.0 | 0.2 | GO:1902018 | negative regulation of cilium assembly(GO:1902018) |
| 0.0 | 0.2 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 0.1 | GO:0022614 | membrane to membrane docking(GO:0022614) |
| 0.0 | 0.2 | GO:0015781 | pyrimidine nucleotide-sugar transport(GO:0015781) |
| 0.0 | 0.1 | GO:1904751 | positive regulation of protein localization to nucleolus(GO:1904751) |
| 0.0 | 0.1 | GO:0007217 | tachykinin receptor signaling pathway(GO:0007217) |
| 0.0 | 0.0 | GO:0002818 | intracellular defense response(GO:0002818) |
| 0.0 | 0.2 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.0 | 0.4 | GO:0034453 | microtubule anchoring(GO:0034453) |
| 0.0 | 0.0 | GO:0090271 | positive regulation of fibroblast growth factor production(GO:0090271) |
| 0.0 | 0.4 | GO:0031297 | replication fork processing(GO:0031297) |
| 0.0 | 0.2 | GO:0046185 | aldehyde catabolic process(GO:0046185) |
| 0.0 | 0.1 | GO:0014901 | regulation of satellite cell activation involved in skeletal muscle regeneration(GO:0014717) satellite cell activation involved in skeletal muscle regeneration(GO:0014901) |
| 0.0 | 0.1 | GO:0046689 | response to mercury ion(GO:0046689) |
| 0.0 | 0.1 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 0.0 | 0.1 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.0 | 0.1 | GO:0045653 | negative regulation of megakaryocyte differentiation(GO:0045653) |
| 0.0 | 0.2 | GO:0046940 | nucleoside monophosphate phosphorylation(GO:0046940) |
| 0.0 | 0.1 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.0 | 0.1 | GO:1904565 | response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904565) cellular response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904566) |
| 0.0 | 0.3 | GO:0006012 | galactose metabolic process(GO:0006012) |
| 0.0 | 0.0 | GO:2000182 | regulation of progesterone biosynthetic process(GO:2000182) |
| 0.0 | 0.1 | GO:0061740 | protein targeting to lysosome involved in chaperone-mediated autophagy(GO:0061740) protein targeting to vacuole involved in autophagy(GO:0071211) |
| 0.0 | 0.1 | GO:2000323 | negative regulation of glucocorticoid receptor signaling pathway(GO:2000323) |
| 0.0 | 0.0 | GO:0071336 | submandibular salivary gland formation(GO:0060661) hair follicle cell proliferation(GO:0071335) regulation of hair follicle cell proliferation(GO:0071336) positive regulation of hair follicle cell proliferation(GO:0071338) |
| 0.0 | 0.1 | GO:1903412 | response to bile acid(GO:1903412) |
| 0.0 | 0.0 | GO:1904529 | regulation of actin filament binding(GO:1904529) regulation of actin binding(GO:1904616) |
| 0.0 | 0.2 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.0 | 0.2 | GO:1904321 | response to forskolin(GO:1904321) cellular response to forskolin(GO:1904322) |
| 0.0 | 0.5 | GO:0006359 | regulation of transcription from RNA polymerase III promoter(GO:0006359) |
| 0.0 | 0.2 | GO:0008340 | determination of adult lifespan(GO:0008340) |
| 0.0 | 0.3 | GO:0071447 | cellular response to hydroperoxide(GO:0071447) |
| 0.0 | 0.2 | GO:1902894 | negative regulation of pri-miRNA transcription from RNA polymerase II promoter(GO:1902894) |
| 0.0 | 0.1 | GO:0050915 | sensory perception of sour taste(GO:0050915) |
| 0.0 | 0.6 | GO:0060338 | regulation of type I interferon-mediated signaling pathway(GO:0060338) |
| 0.0 | 0.5 | GO:0046825 | regulation of protein export from nucleus(GO:0046825) |
| 0.0 | 0.0 | GO:0048073 | regulation of eye pigmentation(GO:0048073) |
| 0.0 | 0.1 | GO:2000354 | regulation of ovarian follicle development(GO:2000354) |
| 0.0 | 0.0 | GO:1902269 | positive regulation of polyamine transmembrane transport(GO:1902269) |
| 0.0 | 0.2 | GO:0071493 | cellular response to UV-B(GO:0071493) |
| 0.0 | 0.1 | GO:0010908 | regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) positive regulation of proteoglycan biosynthetic process(GO:1902730) |
| 0.0 | 0.1 | GO:0044111 | development involved in symbiotic interaction(GO:0044111) |
| 0.0 | 1.1 | GO:0001895 | retina homeostasis(GO:0001895) |
| 0.0 | 0.1 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.0 | 0.0 | GO:0032077 | positive regulation of deoxyribonuclease activity(GO:0032077) positive regulation of endodeoxyribonuclease activity(GO:0032079) |
| 0.0 | 0.2 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.0 | 0.1 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.0 | 0.0 | GO:0030573 | bile acid catabolic process(GO:0030573) |
| 0.0 | 0.2 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 0.2 | GO:1903830 | magnesium ion transmembrane transport(GO:1903830) |
| 0.0 | 0.1 | GO:1905232 | cellular response to L-glutamate(GO:1905232) |
| 0.0 | 0.2 | GO:0035988 | chondrocyte proliferation(GO:0035988) |
| 0.0 | 0.1 | GO:0021520 | spinal cord motor neuron cell fate specification(GO:0021520) |
| 0.0 | 0.1 | GO:0051177 | meiotic sister chromatid cohesion(GO:0051177) |
| 0.0 | 0.2 | GO:1903608 | protein localization to cytoplasmic stress granule(GO:1903608) |
| 0.0 | 0.1 | GO:0032487 | regulation of Rap protein signal transduction(GO:0032487) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.3 | GO:0044233 | ER-mitochondrion membrane contact site(GO:0044233) |
| 0.1 | 0.6 | GO:0097129 | cyclin D2-CDK4 complex(GO:0097129) |
| 0.1 | 0.4 | GO:0030895 | apolipoprotein B mRNA editing enzyme complex(GO:0030895) |
| 0.1 | 0.6 | GO:1990393 | 3M complex(GO:1990393) |
| 0.1 | 0.4 | GO:0032807 | DNA ligase IV complex(GO:0032807) |
| 0.1 | 0.2 | GO:0033593 | BRCA2-MAGE-D1 complex(GO:0033593) |
| 0.1 | 0.2 | GO:0000939 | condensed chromosome inner kinetochore(GO:0000939) |
| 0.1 | 0.4 | GO:0070695 | FHF complex(GO:0070695) |
| 0.1 | 0.8 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.1 | 0.2 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 0.1 | 0.9 | GO:0032039 | integrator complex(GO:0032039) |
| 0.1 | 0.7 | GO:0016013 | syntrophin complex(GO:0016013) |
| 0.0 | 0.2 | GO:0071817 | MMXD complex(GO:0071817) |
| 0.0 | 0.4 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.0 | 0.9 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.9 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.5 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.0 | 0.2 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.0 | 0.4 | GO:0035692 | macrophage migration inhibitory factor receptor complex(GO:0035692) |
| 0.0 | 0.1 | GO:0045323 | interleukin-1 receptor complex(GO:0045323) |
| 0.0 | 0.2 | GO:0001405 | presequence translocase-associated import motor(GO:0001405) |
| 0.0 | 0.2 | GO:0031501 | mannosyltransferase complex(GO:0031501) |
| 0.0 | 0.1 | GO:0038039 | G-protein coupled receptor heterodimeric complex(GO:0038039) |
| 0.0 | 0.1 | GO:0017102 | methionyl glutamyl tRNA synthetase complex(GO:0017102) |
| 0.0 | 0.4 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.0 | 0.3 | GO:0043203 | axon hillock(GO:0043203) |
| 0.0 | 0.1 | GO:0072536 | interleukin-23 receptor complex(GO:0072536) |
| 0.0 | 0.1 | GO:0034677 | integrin alpha7-beta1 complex(GO:0034677) |
| 0.0 | 1.1 | GO:1902710 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.0 | 0.2 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.0 | 0.1 | GO:1990667 | PCSK9-AnxA2 complex(GO:1990667) |
| 0.0 | 0.1 | GO:0097637 | intrinsic component of autophagosome membrane(GO:0097636) integral component of autophagosome membrane(GO:0097637) |
| 0.0 | 0.2 | GO:0045179 | apical cortex(GO:0045179) |
| 0.0 | 0.1 | GO:0005602 | complement component C1 complex(GO:0005602) |
| 0.0 | 0.1 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.0 | 0.1 | GO:0032279 | asymmetric synapse(GO:0032279) |
| 0.0 | 0.2 | GO:0097165 | nuclear stress granule(GO:0097165) |
| 0.0 | 0.2 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.0 | 0.2 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.0 | 0.2 | GO:0005879 | axonemal microtubule(GO:0005879) |
| 0.0 | 0.1 | GO:0034669 | integrin alpha4-beta7 complex(GO:0034669) |
| 0.0 | 0.4 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 0.1 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.0 | 0.2 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.0 | 0.2 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.0 | 0.9 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 0.1 | GO:0071797 | LUBAC complex(GO:0071797) Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.0 | 0.3 | GO:0001520 | outer dense fiber(GO:0001520) |
| 0.0 | 0.5 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.0 | 0.1 | GO:0045257 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.0 | 0.1 | GO:0098536 | deuterosome(GO:0098536) |
| 0.0 | 0.2 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.0 | 0.1 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.0 | 0.0 | GO:0031085 | BLOC-3 complex(GO:0031085) |
| 0.0 | 0.4 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 0.1 | GO:0016589 | NURF complex(GO:0016589) |
| 0.0 | 0.4 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.4 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 0.1 | GO:0070761 | pre-snoRNP complex(GO:0070761) |
| 0.0 | 0.2 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.0 | 0.3 | GO:0042599 | lamellar body(GO:0042599) |
| 0.0 | 0.7 | GO:0030660 | Golgi-associated vesicle membrane(GO:0030660) |
| 0.0 | 0.1 | GO:0032021 | NELF complex(GO:0032021) |
| 0.0 | 0.1 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 0.0 | 0.1 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.0 | 0.0 | GO:0002945 | cyclin K-CDK13 complex(GO:0002945) |
| 0.0 | 0.1 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.0 | 0.1 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 0.0 | 1.5 | GO:0005604 | basement membrane(GO:0005604) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 2.3 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 0.2 | 0.5 | GO:0016608 | growth hormone-releasing hormone activity(GO:0016608) |
| 0.2 | 0.6 | GO:0004001 | adenosine kinase activity(GO:0004001) |
| 0.1 | 0.6 | GO:0047844 | deoxycytidine deaminase activity(GO:0047844) |
| 0.1 | 0.6 | GO:0097108 | smoothened binding(GO:0005119) hedgehog family protein binding(GO:0097108) |
| 0.1 | 1.1 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.1 | 0.3 | GO:0004938 | alpha2-adrenergic receptor activity(GO:0004938) |
| 0.1 | 0.3 | GO:0033858 | N-acetylgalactosamine kinase activity(GO:0033858) |
| 0.1 | 0.5 | GO:0016314 | phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase activity(GO:0016314) |
| 0.1 | 0.5 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.1 | 0.4 | GO:0052655 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.1 | 0.5 | GO:0032396 | inhibitory MHC class I receptor activity(GO:0032396) |
| 0.1 | 0.8 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.1 | 0.4 | GO:0016807 | cysteine-type carboxypeptidase activity(GO:0016807) cysteine-type exopeptidase activity(GO:0070004) |
| 0.1 | 0.2 | GO:0004584 | dolichyl-phosphate-mannose-glycolipid alpha-mannosyltransferase activity(GO:0004584) |
| 0.1 | 1.2 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.1 | 0.6 | GO:0000099 | sulfur amino acid transmembrane transporter activity(GO:0000099) |
| 0.1 | 0.2 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.1 | 0.7 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.1 | 0.2 | GO:0003963 | RNA-3'-phosphate cyclase activity(GO:0003963) |
| 0.1 | 0.2 | GO:0004773 | steryl-sulfatase activity(GO:0004773) |
| 0.0 | 0.5 | GO:0022851 | GABA-gated chloride ion channel activity(GO:0022851) |
| 0.0 | 0.1 | GO:0033842 | N-acetyl-beta-glucosaminyl-glycoprotein 4-beta-N-acetylgalactosaminyltransferase activity(GO:0033842) |
| 0.0 | 0.2 | GO:0035241 | protein-arginine omega-N monomethyltransferase activity(GO:0035241) |
| 0.0 | 0.2 | GO:0033906 | hyaluronoglucuronidase activity(GO:0033906) |
| 0.0 | 0.8 | GO:0034713 | type I transforming growth factor beta receptor binding(GO:0034713) |
| 0.0 | 0.2 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) |
| 0.0 | 1.3 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 0.1 | GO:0031626 | beta-endorphin binding(GO:0031626) |
| 0.0 | 0.7 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) |
| 0.0 | 0.5 | GO:0005131 | growth hormone receptor binding(GO:0005131) |
| 0.0 | 0.5 | GO:0031386 | protein tag(GO:0031386) |
| 0.0 | 0.2 | GO:0004473 | malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) |
| 0.0 | 0.2 | GO:0086062 | voltage-gated sodium channel activity involved in Purkinje myocyte action potential(GO:0086062) |
| 0.0 | 0.2 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.0 | 0.2 | GO:0005502 | 11-cis retinal binding(GO:0005502) |
| 0.0 | 0.2 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.0 | 0.2 | GO:0018685 | alkane 1-monooxygenase activity(GO:0018685) leukotriene-B4 20-monooxygenase activity(GO:0050051) |
| 0.0 | 0.1 | GO:0008476 | protein-tyrosine sulfotransferase activity(GO:0008476) |
| 0.0 | 0.2 | GO:0070513 | death domain binding(GO:0070513) |
| 0.0 | 0.1 | GO:0090541 | MIT domain binding(GO:0090541) |
| 0.0 | 0.6 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.0 | 0.2 | GO:0046899 | nucleoside triphosphate adenylate kinase activity(GO:0046899) |
| 0.0 | 0.4 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.0 | 0.2 | GO:0015526 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 0.0 | 0.4 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.0 | 0.9 | GO:0071949 | FAD binding(GO:0071949) |
| 0.0 | 0.2 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.0 | 0.1 | GO:0102008 | cytosolic dipeptidase activity(GO:0102008) |
| 0.0 | 0.2 | GO:0071535 | RING-like zinc finger domain binding(GO:0071535) |
| 0.0 | 0.1 | GO:0042020 | interleukin-23 binding(GO:0042019) interleukin-23 receptor activity(GO:0042020) |
| 0.0 | 0.3 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.0 | 0.2 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.0 | 0.1 | GO:0008518 | reduced folate carrier activity(GO:0008518) |
| 0.0 | 0.1 | GO:0051185 | coenzyme transporter activity(GO:0051185) |
| 0.0 | 0.1 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.0 | 0.1 | GO:0032795 | heterotrimeric G-protein binding(GO:0032795) |
| 0.0 | 0.4 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.0 | 0.1 | GO:0008240 | tripeptidyl-peptidase activity(GO:0008240) |
| 0.0 | 0.2 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.0 | 0.1 | GO:0008177 | succinate dehydrogenase (ubiquinone) activity(GO:0008177) |
| 0.0 | 0.3 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.0 | 0.1 | GO:0004667 | prostaglandin-D synthase activity(GO:0004667) |
| 0.0 | 0.7 | GO:0050750 | low-density lipoprotein particle receptor binding(GO:0050750) |
| 0.0 | 0.1 | GO:0004031 | aldehyde oxidase activity(GO:0004031) |
| 0.0 | 0.1 | GO:0044020 | histone methyltransferase activity (H4-R3 specific)(GO:0044020) |
| 0.0 | 0.1 | GO:0047192 | 1-alkylglycerophosphocholine O-acetyltransferase activity(GO:0047192) |
| 0.0 | 0.2 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.0 | 0.3 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.0 | 0.1 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.0 | 0.1 | GO:0017161 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) inositol-1,3,4-trisphosphate 4-phosphatase activity(GO:0017161) phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) inositol-3,4-bisphosphate 4-phosphatase activity(GO:0052828) |
| 0.0 | 0.4 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 0.2 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.0 | 0.4 | GO:0005402 | sugar:proton symporter activity(GO:0005351) cation:sugar symporter activity(GO:0005402) |
| 0.0 | 0.4 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 0.1 | GO:0016262 | protein N-acetylglucosaminyltransferase activity(GO:0016262) |
| 0.0 | 0.1 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.0 | 0.1 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.0 | 0.0 | GO:0032090 | Pyrin domain binding(GO:0032090) |
| 0.0 | 0.2 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.0 | 0.4 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.0 | 0.2 | GO:0015143 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.0 | 0.1 | GO:0070653 | high-density lipoprotein particle receptor binding(GO:0070653) |
| 0.0 | 0.0 | GO:0004772 | sterol O-acyltransferase activity(GO:0004772) cholesterol O-acyltransferase activity(GO:0034736) |
| 0.0 | 0.1 | GO:0043426 | MRF binding(GO:0043426) |
| 0.0 | 0.5 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 0.2 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.0 | 0.1 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.0 | 0.2 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.0 | 0.2 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 0.3 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.0 | 0.1 | GO:0016413 | O-acetyltransferase activity(GO:0016413) |
| 0.0 | 0.8 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 0.1 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.0 | 0.3 | GO:0005024 | transforming growth factor beta-activated receptor activity(GO:0005024) |
| 0.0 | 0.4 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 0.1 | GO:0000702 | oxidized base lesion DNA N-glycosylase activity(GO:0000702) |
| 0.0 | 0.2 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.0 | 0.1 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.0 | 0.1 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.0 | 0.2 | GO:0016755 | transferase activity, transferring amino-acyl groups(GO:0016755) |
| 0.0 | 0.1 | GO:0008853 | exodeoxyribonuclease III activity(GO:0008853) |
| 0.0 | 0.5 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 0.1 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.0 | 0.2 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.0 | 0.1 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.0 | 0.1 | GO:0004999 | vasoactive intestinal polypeptide receptor activity(GO:0004999) |
| 0.0 | 0.2 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.0 | 0.0 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 0.0 | 0.1 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.0 | 0.4 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.0 | 0.1 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.0 | 0.0 | GO:0017089 | glycolipid transporter activity(GO:0017089) |
| 0.0 | 0.1 | GO:0044736 | acid-sensing ion channel activity(GO:0044736) |
| 0.0 | 0.0 | GO:0035242 | histone-arginine N-methyltransferase activity(GO:0008469) protein-arginine omega-N asymmetric methyltransferase activity(GO:0035242) |
| 0.0 | 0.4 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.3 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.0 | 0.2 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.1 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.0 | 0.1 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.0 | 0.1 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.0 | 0.4 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.0 | 0.2 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.0 | 0.1 | GO:0098639 | collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.0 | 0.1 | GO:1990405 | protein antigen binding(GO:1990405) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.0 | 0.6 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 1.4 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 0.7 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 0.6 | ST INTERLEUKIN 4 PATHWAY | Interleukin 4 (IL-4) Pathway |
| 0.0 | 0.6 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.4 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 0.9 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 0.1 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 0.4 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 0.9 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.2 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 0.4 | REACTOME INTEGRATION OF PROVIRUS | Genes involved in Integration of provirus |
| 0.0 | 0.9 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 0.4 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.0 | 1.2 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.0 | 0.6 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.5 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 1.2 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 0.5 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.0 | 0.6 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 0.5 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.0 | 0.5 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.0 | 0.7 | REACTOME INFLAMMASOMES | Genes involved in Inflammasomes |
| 0.0 | 0.3 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 0.7 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 0.4 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.0 | 0.5 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 1.5 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 0.1 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 0.2 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.0 | 0.2 | REACTOME OPSINS | Genes involved in Opsins |
| 0.0 | 0.8 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.4 | REACTOME PYRUVATE METABOLISM | Genes involved in Pyruvate metabolism |
| 0.0 | 0.4 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 0.3 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.0 | 0.5 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.3 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 0.2 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.0 | 0.5 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.0 | 0.1 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.0 | 0.4 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.0 | 0.2 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.0 | 0.4 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |