Project
Inflammatory response time course, HUVEC (Wada, 2009)
Navigation
Downloads
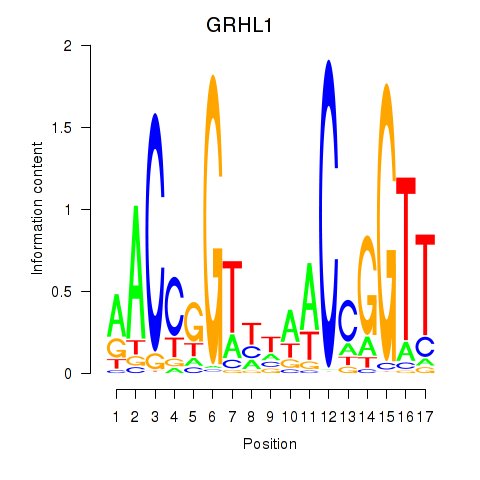
Results for GRHL1
Z-value: 0.53
Transcription factors associated with GRHL1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
GRHL1
|
ENSG00000134317.18 | GRHL1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| GRHL1 | hg38_v1_chr2_+_9951653_9951734 | 0.33 | 1.1e-01 | Click! |
Activity profile of GRHL1 motif
Sorted Z-values of GRHL1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of GRHL1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_+_155666718 | 2.95 |
ENST00000621234.4
ENST00000511108.5 |
GUCY1A1
|
guanylate cyclase 1 soluble subunit alpha 1 |
| chr4_+_155667198 | 2.76 |
ENST00000296518.11
|
GUCY1A1
|
guanylate cyclase 1 soluble subunit alpha 1 |
| chr4_+_155667096 | 2.74 |
ENST00000393832.7
|
GUCY1A1
|
guanylate cyclase 1 soluble subunit alpha 1 |
| chr4_+_155666963 | 2.65 |
ENST00000455639.6
|
GUCY1A1
|
guanylate cyclase 1 soluble subunit alpha 1 |
| chr4_+_155666827 | 1.76 |
ENST00000511507.5
ENST00000506455.6 |
GUCY1A1
|
guanylate cyclase 1 soluble subunit alpha 1 |
| chr9_-_75088198 | 1.10 |
ENST00000376808.8
|
NMRK1
|
nicotinamide riboside kinase 1 |
| chr2_-_208254232 | 1.05 |
ENST00000415913.5
ENST00000415282.5 ENST00000446179.5 |
IDH1
|
isocitrate dehydrogenase (NADP(+)) 1 |
| chr9_+_5450503 | 0.82 |
ENST00000381573.8
ENST00000381577.4 |
CD274
|
CD274 molecule |
| chr3_-_172523460 | 0.69 |
ENST00000420541.6
|
TNFSF10
|
TNF superfamily member 10 |
| chr20_-_25339731 | 0.66 |
ENST00000450393.5
ENST00000491682.5 |
ABHD12
|
abhydrolase domain containing 12, lysophospholipase |
| chr3_-_172523423 | 0.63 |
ENST00000241261.7
|
TNFSF10
|
TNF superfamily member 10 |
| chr19_+_44809053 | 0.61 |
ENST00000611077.5
|
BCAM
|
basal cell adhesion molecule (Lutheran blood group) |
| chr1_+_210232776 | 0.58 |
ENST00000367012.4
|
SERTAD4
|
SERTA domain containing 4 |
| chr19_-_291132 | 0.55 |
ENST00000327790.7
|
PLPP2
|
phospholipid phosphatase 2 |
| chr19_+_10625507 | 0.53 |
ENST00000590857.5
ENST00000588688.5 ENST00000586078.5 ENST00000335757.10 |
SLC44A2
|
solute carrier family 44 member 2 |
| chr3_-_49104457 | 0.47 |
ENST00000635292.1
ENST00000635541.1 ENST00000464962.6 |
QARS1
|
glutaminyl-tRNA synthetase 1 |
| chr3_-_49104745 | 0.47 |
ENST00000635194.1
ENST00000306125.12 ENST00000634602.1 ENST00000414533.5 ENST00000635443.1 ENST00000452739.5 ENST00000635231.1 |
QARS1
|
glutaminyl-tRNA synthetase 1 |
| chr19_+_44809089 | 0.46 |
ENST00000270233.12
ENST00000591520.6 |
BCAM
|
basal cell adhesion molecule (Lutheran blood group) |
| chr12_-_10807286 | 0.44 |
ENST00000240615.3
|
TAS2R8
|
taste 2 receptor member 8 |
| chr19_-_11738882 | 0.43 |
ENST00000586121.1
ENST00000431998.1 ENST00000341191.11 ENST00000440527.1 |
ZNF823
|
zinc finger protein 823 |
| chr1_-_52055156 | 0.41 |
ENST00000371626.9
ENST00000610127.2 |
TXNDC12
|
thioredoxin domain containing 12 |
| chr20_-_62427528 | 0.40 |
ENST00000252998.2
|
RBBP8NL
|
RBBP8 N-terminal like |
| chr7_+_76510516 | 0.40 |
ENST00000257632.9
|
UPK3B
|
uroplakin 3B |
| chr6_-_46921926 | 0.40 |
ENST00000283296.12
|
ADGRF5
|
adhesion G protein-coupled receptor F5 |
| chr7_+_76510608 | 0.39 |
ENST00000394849.1
|
UPK3B
|
uroplakin 3B |
| chr7_+_76510528 | 0.38 |
ENST00000334348.8
|
UPK3B
|
uroplakin 3B |
| chr1_-_24143112 | 0.28 |
ENST00000270800.2
|
IL22RA1
|
interleukin 22 receptor subunit alpha 1 |
| chr18_-_57803307 | 0.28 |
ENST00000648908.2
|
ATP8B1
|
ATPase phospholipid transporting 8B1 |
| chr9_+_75088498 | 0.27 |
ENST00000346234.7
|
OSTF1
|
osteoclast stimulating factor 1 |
| chr6_+_32153441 | 0.25 |
ENST00000414204.5
ENST00000361568.6 ENST00000395523.5 |
PPT2
|
palmitoyl-protein thioesterase 2 |
| chr8_-_129786617 | 0.24 |
ENST00000276708.9
|
GSDMC
|
gasdermin C |
| chr11_-_64166102 | 0.24 |
ENST00000255681.7
ENST00000675777.1 |
MACROD1
|
mono-ADP ribosylhydrolase 1 |
| chr14_+_50533026 | 0.22 |
ENST00000441560.6
|
ATL1
|
atlastin GTPase 1 |
| chr17_-_4560564 | 0.21 |
ENST00000574584.1
ENST00000381550.8 ENST00000301395.7 |
GGT6
|
gamma-glutamyltransferase 6 |
| chr1_-_50960230 | 0.19 |
ENST00000396153.7
|
FAF1
|
Fas associated factor 1 |
| chr11_+_706595 | 0.19 |
ENST00000531348.5
ENST00000530636.5 |
EPS8L2
|
EPS8 like 2 |
| chr2_+_190408324 | 0.18 |
ENST00000417958.5
ENST00000432036.5 ENST00000392328.6 |
MFSD6
|
major facilitator superfamily domain containing 6 |
| chr19_-_43504711 | 0.18 |
ENST00000601646.1
|
PHLDB3
|
pleckstrin homology like domain family B member 3 |
| chr13_+_48232609 | 0.18 |
ENST00000649266.1
|
ITM2B
|
integral membrane protein 2B |
| chr12_+_18261511 | 0.17 |
ENST00000538779.6
ENST00000433979.6 |
PIK3C2G
|
phosphatidylinositol-4-phosphate 3-kinase catalytic subunit type 2 gamma |
| chr1_+_94418435 | 0.15 |
ENST00000647998.2
|
ABCD3
|
ATP binding cassette subfamily D member 3 |
| chr5_+_181040260 | 0.15 |
ENST00000515271.1
ENST00000327705.14 |
BTNL9
|
butyrophilin like 9 |
| chr7_-_102543849 | 0.14 |
ENST00000644544.1
|
UPK3BL2
|
uroplakin 3B like 2 |
| chr11_-_65780917 | 0.14 |
ENST00000532090.3
|
AP5B1
|
adaptor related protein complex 5 subunit beta 1 |
| chr15_+_41838839 | 0.13 |
ENST00000458483.4
|
PLA2G4B
|
phospholipase A2 group IVB |
| chr12_-_3753091 | 0.12 |
ENST00000252322.1
ENST00000440314.7 |
CRACR2A
|
calcium release activated channel regulator 2A |
| chr13_+_48233171 | 0.12 |
ENST00000378549.5
ENST00000647800.2 |
ITM2B
|
integral membrane protein 2B |
| chr6_-_31139063 | 0.12 |
ENST00000259845.5
|
PSORS1C2
|
psoriasis susceptibility 1 candidate 2 |
| chr11_+_65787056 | 0.12 |
ENST00000335987.8
|
OVOL1
|
ovo like transcriptional repressor 1 |
| chr7_-_102642791 | 0.12 |
ENST00000340457.8
|
UPK3BL1
|
uroplakin 3B like 1 |
| chr11_-_67674606 | 0.12 |
ENST00000674110.1
ENST00000349015.7 |
ALDH3B2
|
aldehyde dehydrogenase 3 family member B2 |
| chr11_-_67674725 | 0.10 |
ENST00000525827.6
ENST00000673966.1 ENST00000673873.1 |
ALDH3B2
|
aldehyde dehydrogenase 3 family member B2 |
| chr11_+_119168188 | 0.10 |
ENST00000454811.5
ENST00000409265.8 ENST00000449394.5 |
NLRX1
|
NLR family member X1 |
| chr22_+_18077976 | 0.09 |
ENST00000399744.8
|
PEX26
|
peroxisomal biogenesis factor 26 |
| chr11_+_119168705 | 0.08 |
ENST00000409109.6
ENST00000409991.5 ENST00000292199.6 |
NLRX1
|
NLR family member X1 |
| chr1_-_36397880 | 0.07 |
ENST00000315732.3
|
LSM10
|
LSM10, U7 small nuclear RNA associated |
| chr1_+_40258202 | 0.07 |
ENST00000372759.4
|
ZMPSTE24
|
zinc metallopeptidase STE24 |
| chr1_-_153608136 | 0.07 |
ENST00000368703.6
|
S100A16
|
S100 calcium binding protein A16 |
| chr1_+_43974902 | 0.03 |
ENST00000532642.5
ENST00000236067.8 ENST00000471859.6 ENST00000472174.7 |
ATP6V0B
|
ATPase H+ transporting V0 subunit b |
| chr18_+_9474994 | 0.03 |
ENST00000019317.8
|
RALBP1
|
ralA binding protein 1 |
| chr19_+_11925098 | 0.03 |
ENST00000254321.10
ENST00000591944.1 |
ZNF700
ENSG00000267179.1
|
zinc finger protein 700 novel protein |
| chr19_+_11925062 | 0.03 |
ENST00000622593.4
ENST00000590798.1 |
ZNF700
ENSG00000267179.1
|
zinc finger protein 700 novel protein |
| chr1_+_34782259 | 0.02 |
ENST00000373362.3
|
GJB3
|
gap junction protein beta 3 |
| chr20_-_18057841 | 0.02 |
ENST00000278780.7
|
OVOL2
|
ovo like zinc finger 2 |
| chr14_-_80211472 | 0.02 |
ENST00000557125.1
ENST00000438257.9 ENST00000422005.7 |
DIO2
|
iodothyronine deiodinase 2 |
| chr19_-_17245889 | 0.01 |
ENST00000291442.4
|
NR2F6
|
nuclear receptor subfamily 2 group F member 6 |
| chr5_+_134967901 | 0.01 |
ENST00000282611.8
|
CATSPER3
|
cation channel sperm associated 3 |
| chr14_+_23469681 | 0.01 |
ENST00000408901.8
ENST00000397154.7 ENST00000555128.5 |
NGDN
|
neuroguidin |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:0006097 | glyoxylate cycle(GO:0006097) |
| 0.2 | 0.7 | GO:0046462 | monoacylglycerol metabolic process(GO:0046462) monoacylglycerol catabolic process(GO:0052651) |
| 0.1 | 0.8 | GO:2001181 | positive regulation of interleukin-10 secretion(GO:2001181) |
| 0.1 | 1.3 | GO:0097296 | activation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097296) |
| 0.0 | 0.1 | GO:1901993 | meiotic cell cycle phase transition(GO:0044771) regulation of meiotic cell cycle phase transition(GO:1901993) negative regulation of meiotic cell cycle phase transition(GO:1901994) |
| 0.0 | 0.2 | GO:0015910 | peroxisomal long-chain fatty acid import(GO:0015910) |
| 0.0 | 0.3 | GO:0021650 | vestibulocochlear nerve formation(GO:0021650) |
| 0.0 | 0.4 | GO:0043129 | surfactant homeostasis(GO:0043129) positive regulation of phospholipid biosynthetic process(GO:0071073) |
| 0.0 | 0.5 | GO:0015871 | choline transport(GO:0015871) |
| 0.0 | 0.2 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.0 | 0.3 | GO:0042985 | negative regulation of amyloid precursor protein biosynthetic process(GO:0042985) |
| 0.0 | 1.1 | GO:0009435 | NAD biosynthetic process(GO:0009435) |
| 0.0 | 0.2 | GO:0039536 | negative regulation of RIG-I signaling pathway(GO:0039536) |
| 0.0 | 0.1 | GO:0030327 | prenylated protein catabolic process(GO:0030327) |
| 0.0 | 0.3 | GO:0098734 | macromolecule depalmitoylation(GO:0098734) |
| 0.0 | 0.1 | GO:1901341 | activation of store-operated calcium channel activity(GO:0032237) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.0 | 0.2 | GO:1901748 | leukotriene D4 metabolic process(GO:1901748) leukotriene D4 biosynthetic process(GO:1901750) |
| 0.0 | 0.4 | GO:1902236 | negative regulation of endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway(GO:1902236) |
| 0.0 | 0.4 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.0 | 0.2 | GO:0007253 | cytoplasmic sequestering of NF-kappaB(GO:0007253) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.2 | GO:0031907 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 0.2 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.0 | 0.7 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 0.2 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.1 | GO:0005055 | laminin receptor activity(GO:0005055) |
| 0.3 | 1.0 | GO:0051990 | isocitrate dehydrogenase (NADP+) activity(GO:0004450) (R)-2-hydroxyglutarate dehydrogenase activity(GO:0051990) |
| 0.3 | 1.1 | GO:0050262 | ribosylnicotinamide kinase activity(GO:0050262) ribosylnicotinate kinase activity(GO:0061769) |
| 0.1 | 0.6 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 0.1 | 0.5 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.1 | 0.3 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 0.0 | 0.7 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 1.3 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.1 | GO:0071209 | U7 snRNA binding(GO:0071209) |
| 0.0 | 0.2 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.0 | 0.3 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 0.4 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.0 | 0.4 | GO:0016671 | peptide disulfide oxidoreductase activity(GO:0015037) oxidoreductase activity, acting on a sulfur group of donors, disulfide as acceptor(GO:0016671) |
| 0.0 | 0.3 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.0 | 0.2 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.2 | GO:0004030 | 3-chloroallyl aldehyde dehydrogenase activity(GO:0004028) aldehyde dehydrogenase [NAD(P)+] activity(GO:0004030) |
| 0.0 | 0.1 | GO:0008970 | phosphatidylcholine 1-acylhydrolase activity(GO:0008970) |
| 0.0 | 0.2 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.3 | PID TRAIL PATHWAY | TRAIL signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.3 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 1.0 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 0.8 | REACTOME PD1 SIGNALING | Genes involved in PD-1 signaling |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |