Project
Inflammatory response time course, HUVEC (Wada, 2009)
Navigation
Downloads
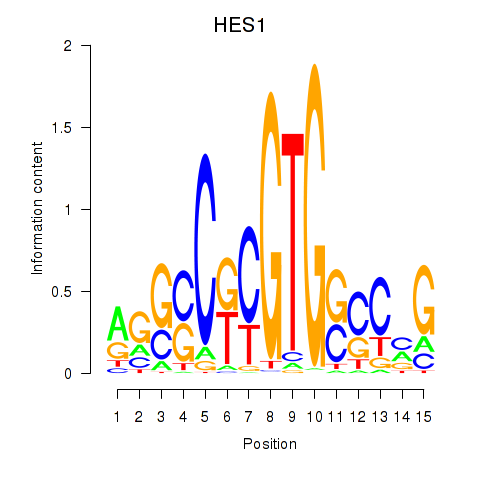
Results for HES1
Z-value: 0.98
Transcription factors associated with HES1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HES1
|
ENSG00000114315.4 | HES1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HES1 | hg38_v1_chr3_+_194136138_194136155 | 0.49 | 1.2e-02 | Click! |
Activity profile of HES1 motif
Sorted Z-values of HES1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of HES1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 1.0 | GO:0003169 | coronary vein morphogenesis(GO:0003169) |
| 0.5 | 2.1 | GO:0021571 | rhombomere 5 development(GO:0021571) central nervous system segmentation(GO:0035283) brain segmentation(GO:0035284) |
| 0.4 | 7.2 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.4 | 3.5 | GO:0089700 | protein kinase D signaling(GO:0089700) |
| 0.4 | 2.6 | GO:1904849 | positive regulation of cell chemotaxis to fibroblast growth factor(GO:1904849) positive regulation of endothelial cell chemotaxis to fibroblast growth factor(GO:2000546) |
| 0.3 | 1.7 | GO:0035948 | N-terminal peptidyl-lysine acetylation(GO:0018076) positive regulation of gluconeogenesis by positive regulation of transcription from RNA polymerase II promoter(GO:0035948) |
| 0.3 | 1.3 | GO:0015910 | peroxisomal long-chain fatty acid import(GO:0015910) |
| 0.3 | 0.9 | GO:0070662 | positive regulation of Fc receptor mediated stimulatory signaling pathway(GO:0060369) mast cell proliferation(GO:0070662) |
| 0.3 | 0.9 | GO:0015920 | lipopolysaccharide transport(GO:0015920) |
| 0.3 | 0.8 | GO:0006212 | uracil catabolic process(GO:0006212) beta-alanine biosynthetic process(GO:0019483) |
| 0.2 | 0.7 | GO:0002086 | diaphragm contraction(GO:0002086) |
| 0.2 | 0.7 | GO:0010932 | macrophage tolerance induction(GO:0010931) regulation of macrophage tolerance induction(GO:0010932) positive regulation of macrophage tolerance induction(GO:0010933) |
| 0.2 | 0.7 | GO:0060957 | endocardial cell fate commitment(GO:0060957) endocardial cushion cell fate commitment(GO:0061445) |
| 0.2 | 0.7 | GO:0042412 | taurine biosynthetic process(GO:0042412) |
| 0.2 | 1.1 | GO:0035645 | enteric smooth muscle cell differentiation(GO:0035645) |
| 0.2 | 0.9 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.2 | 0.7 | GO:0021722 | superior olivary nucleus development(GO:0021718) superior olivary nucleus maturation(GO:0021722) |
| 0.2 | 1.1 | GO:0035992 | tendon cell differentiation(GO:0035990) tendon formation(GO:0035992) |
| 0.2 | 1.5 | GO:0018406 | protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 0.2 | 1.4 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.2 | 1.4 | GO:0003138 | primary heart field specification(GO:0003138) sinoatrial valve development(GO:0003172) sinoatrial valve morphogenesis(GO:0003185) |
| 0.2 | 1.4 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.1 | 0.6 | GO:1904379 | protein localization to cytosolic proteasome complex(GO:1904327) protein localization to cytosolic proteasome complex involved in ERAD pathway(GO:1904379) |
| 0.1 | 0.3 | GO:0090272 | negative regulation of fibroblast growth factor production(GO:0090272) |
| 0.1 | 1.0 | GO:0000101 | sulfur amino acid transport(GO:0000101) |
| 0.1 | 0.7 | GO:0006404 | RNA import into nucleus(GO:0006404) |
| 0.1 | 0.4 | GO:1902490 | regulation of sperm capacitation(GO:1902490) |
| 0.1 | 0.8 | GO:0010748 | regulation of plasma membrane long-chain fatty acid transport(GO:0010746) negative regulation of plasma membrane long-chain fatty acid transport(GO:0010748) |
| 0.1 | 0.4 | GO:0035494 | SNARE complex disassembly(GO:0035494) |
| 0.1 | 0.5 | GO:0061086 | negative regulation of histone H3-K27 methylation(GO:0061086) |
| 0.1 | 0.5 | GO:1904381 | Golgi apparatus mannose trimming(GO:1904381) |
| 0.1 | 0.6 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.1 | 0.9 | GO:0010641 | positive regulation of platelet-derived growth factor receptor signaling pathway(GO:0010641) |
| 0.1 | 1.7 | GO:0021853 | cerebral cortex GABAergic interneuron migration(GO:0021853) interneuron migration(GO:1904936) |
| 0.1 | 1.5 | GO:0014894 | response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.1 | 0.9 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.1 | 3.4 | GO:0072189 | ureter development(GO:0072189) |
| 0.1 | 0.6 | GO:0006668 | sphinganine-1-phosphate metabolic process(GO:0006668) |
| 0.1 | 1.1 | GO:0032328 | alanine transport(GO:0032328) |
| 0.1 | 2.2 | GO:0007351 | tripartite regional subdivision(GO:0007351) anterior/posterior axis specification, embryo(GO:0008595) |
| 0.1 | 1.1 | GO:0032057 | negative regulation of translational initiation in response to stress(GO:0032057) |
| 0.1 | 0.8 | GO:0097338 | response to clozapine(GO:0097338) |
| 0.1 | 0.6 | GO:2000563 | positive regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000563) |
| 0.1 | 0.3 | GO:0090526 | regulation of gluconeogenesis involved in cellular glucose homeostasis(GO:0090526) |
| 0.1 | 2.1 | GO:0035518 | histone H2A monoubiquitination(GO:0035518) |
| 0.1 | 0.5 | GO:2000667 | positive regulation of interleukin-5 secretion(GO:2000664) positive regulation of interleukin-13 secretion(GO:2000667) positive regulation of interleukin-10 secretion(GO:2001181) |
| 0.1 | 0.4 | GO:0060671 | epithelial cell differentiation involved in embryonic placenta development(GO:0060671) epithelial cell morphogenesis involved in placental branching(GO:0060672) |
| 0.1 | 0.6 | GO:0032417 | positive regulation of sodium:proton antiporter activity(GO:0032417) |
| 0.1 | 0.7 | GO:0021678 | third ventricle development(GO:0021678) |
| 0.1 | 0.3 | GO:0035425 | autocrine signaling(GO:0035425) |
| 0.1 | 0.9 | GO:0097012 | cellular response to granulocyte macrophage colony-stimulating factor stimulus(GO:0097011) response to granulocyte macrophage colony-stimulating factor(GO:0097012) |
| 0.1 | 0.6 | GO:0061525 | hindgut development(GO:0061525) |
| 0.1 | 0.4 | GO:0003199 | endocardial cushion to mesenchymal transition involved in heart valve formation(GO:0003199) |
| 0.1 | 0.7 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.1 | 0.6 | GO:0002329 | pre-B cell differentiation(GO:0002329) |
| 0.1 | 0.2 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.1 | 0.9 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.1 | 0.2 | GO:2000583 | regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000583) negative regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000584) |
| 0.1 | 0.2 | GO:0001560 | regulation of cell growth by extracellular stimulus(GO:0001560) |
| 0.1 | 0.8 | GO:2000601 | positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.1 | 0.2 | GO:2000410 | regulation of thymocyte migration(GO:2000410) |
| 0.1 | 0.4 | GO:0006535 | cysteine biosynthetic process from serine(GO:0006535) |
| 0.1 | 0.7 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.1 | 0.4 | GO:2001295 | malonyl-CoA biosynthetic process(GO:2001295) |
| 0.1 | 0.8 | GO:0097267 | omega-hydroxylase P450 pathway(GO:0097267) |
| 0.1 | 0.7 | GO:2001028 | positive regulation of endothelial cell chemotaxis(GO:2001028) |
| 0.1 | 0.2 | GO:0035674 | tricarboxylic acid transmembrane transport(GO:0035674) |
| 0.1 | 1.6 | GO:0097094 | craniofacial suture morphogenesis(GO:0097094) |
| 0.1 | 0.4 | GO:0031291 | Ran protein signal transduction(GO:0031291) |
| 0.1 | 0.2 | GO:0070245 | positive regulation of thymocyte apoptotic process(GO:0070245) |
| 0.1 | 0.3 | GO:0098838 | reduced folate transmembrane transport(GO:0098838) |
| 0.1 | 0.7 | GO:0010571 | positive regulation of nuclear cell cycle DNA replication(GO:0010571) |
| 0.1 | 0.1 | GO:1900194 | negative regulation of oocyte maturation(GO:1900194) |
| 0.1 | 0.7 | GO:0032525 | somite rostral/caudal axis specification(GO:0032525) |
| 0.1 | 0.3 | GO:0006169 | adenosine salvage(GO:0006169) dATP biosynthetic process(GO:0006175) |
| 0.1 | 0.3 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.1 | 0.2 | GO:0036515 | serotonergic neuron axon guidance(GO:0036515) |
| 0.1 | 0.5 | GO:0015798 | myo-inositol transport(GO:0015798) |
| 0.1 | 0.5 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.1 | 0.2 | GO:1902824 | positive regulation of late endosome to lysosome transport(GO:1902824) |
| 0.1 | 3.0 | GO:0032094 | response to food(GO:0032094) |
| 0.1 | 0.5 | GO:0009256 | 10-formyltetrahydrofolate metabolic process(GO:0009256) |
| 0.1 | 0.2 | GO:0018242 | protein O-linked glycosylation via serine(GO:0018242) |
| 0.1 | 1.3 | GO:0002115 | store-operated calcium entry(GO:0002115) |
| 0.1 | 0.2 | GO:1903722 | regulation of centriole elongation(GO:1903722) |
| 0.1 | 0.4 | GO:0006041 | glucosamine metabolic process(GO:0006041) |
| 0.1 | 0.5 | GO:0090219 | negative regulation of lipid kinase activity(GO:0090219) |
| 0.1 | 0.7 | GO:0045176 | apical protein localization(GO:0045176) |
| 0.1 | 0.5 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.1 | 0.2 | GO:0018106 | peptidyl-histidine phosphorylation(GO:0018106) |
| 0.1 | 0.2 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.1 | 0.6 | GO:0046549 | retinal cone cell differentiation(GO:0042670) retinal cone cell development(GO:0046549) |
| 0.1 | 0.6 | GO:0034626 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.1 | 0.5 | GO:0070535 | histone H2A K63-linked ubiquitination(GO:0070535) |
| 0.1 | 0.6 | GO:0017183 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.1 | 1.1 | GO:0036159 | inner dynein arm assembly(GO:0036159) |
| 0.1 | 3.1 | GO:0045599 | negative regulation of fat cell differentiation(GO:0045599) |
| 0.1 | 0.5 | GO:0032815 | negative regulation of natural killer cell activation(GO:0032815) |
| 0.1 | 0.5 | GO:0090073 | positive regulation of protein homodimerization activity(GO:0090073) |
| 0.1 | 0.7 | GO:1900028 | negative regulation of ruffle assembly(GO:1900028) |
| 0.1 | 0.1 | GO:0086100 | endothelin receptor signaling pathway(GO:0086100) |
| 0.1 | 0.8 | GO:2000774 | positive regulation of cellular senescence(GO:2000774) |
| 0.1 | 0.3 | GO:0034499 | late endosome to Golgi transport(GO:0034499) |
| 0.0 | 0.5 | GO:0006689 | ganglioside catabolic process(GO:0006689) |
| 0.0 | 0.1 | GO:0006447 | regulation of translational initiation by iron(GO:0006447) |
| 0.0 | 0.4 | GO:2000825 | positive regulation of androgen receptor activity(GO:2000825) |
| 0.0 | 0.6 | GO:0001561 | fatty acid alpha-oxidation(GO:0001561) |
| 0.0 | 0.5 | GO:1904779 | regulation of protein localization to centrosome(GO:1904779) |
| 0.0 | 0.2 | GO:1902559 | 3'-phosphoadenosine 5'-phosphosulfate transport(GO:0046963) 3'-phospho-5'-adenylyl sulfate transmembrane transport(GO:1902559) |
| 0.0 | 0.2 | GO:0052651 | monoacylglycerol metabolic process(GO:0046462) monoacylglycerol catabolic process(GO:0052651) |
| 0.0 | 0.2 | GO:1903445 | intermembrane transport(GO:0046909) protein transport from ciliary membrane to plasma membrane(GO:1903445) |
| 0.0 | 0.2 | GO:0030821 | negative regulation of cyclic nucleotide catabolic process(GO:0030806) negative regulation of cAMP catabolic process(GO:0030821) |
| 0.0 | 0.4 | GO:0033132 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.0 | 0.4 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.0 | 0.1 | GO:0046452 | dihydrofolate metabolic process(GO:0046452) |
| 0.0 | 0.1 | GO:0014022 | neural plate elongation(GO:0014022) convergent extension involved in neural plate elongation(GO:0022007) |
| 0.0 | 0.2 | GO:0035711 | plasmacytoid dendritic cell activation(GO:0002270) regulation of restriction endodeoxyribonuclease activity(GO:0032072) positive regulation of mismatch repair(GO:0032425) T-helper 1 cell activation(GO:0035711) |
| 0.0 | 0.1 | GO:0002071 | glandular epithelial cell maturation(GO:0002071) positive regulation of type B pancreatic cell development(GO:2000078) |
| 0.0 | 0.4 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.0 | 0.1 | GO:0019878 | lysine biosynthetic process(GO:0009085) lysine biosynthetic process via aminoadipic acid(GO:0019878) |
| 0.0 | 0.4 | GO:0006335 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 | 0.7 | GO:0042407 | cristae formation(GO:0042407) |
| 0.0 | 0.2 | GO:0030241 | skeletal muscle myosin thick filament assembly(GO:0030241) |
| 0.0 | 0.5 | GO:0031053 | primary miRNA processing(GO:0031053) |
| 0.0 | 0.2 | GO:0019086 | late viral transcription(GO:0019086) |
| 0.0 | 0.3 | GO:0000707 | meiotic DNA recombinase assembly(GO:0000707) |
| 0.0 | 0.3 | GO:0038026 | reelin-mediated signaling pathway(GO:0038026) |
| 0.0 | 0.1 | GO:0042418 | phthalate metabolic process(GO:0018963) epinephrine biosynthetic process(GO:0042418) phytoalexin metabolic process(GO:0052314) |
| 0.0 | 1.8 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.0 | 0.9 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.0 | 0.3 | GO:1903025 | regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903025) |
| 0.0 | 1.0 | GO:0030208 | dermatan sulfate biosynthetic process(GO:0030208) |
| 0.0 | 0.9 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.0 | 0.3 | GO:0008215 | spermine metabolic process(GO:0008215) |
| 0.0 | 0.5 | GO:0030949 | positive regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030949) |
| 0.0 | 0.1 | GO:0015770 | disaccharide transport(GO:0015766) sucrose transport(GO:0015770) oligosaccharide transport(GO:0015772) |
| 0.0 | 2.2 | GO:0071470 | cellular response to osmotic stress(GO:0071470) |
| 0.0 | 0.2 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.0 | 0.2 | GO:0035965 | cardiolipin acyl-chain remodeling(GO:0035965) |
| 0.0 | 0.8 | GO:0035729 | cellular response to hepatocyte growth factor stimulus(GO:0035729) |
| 0.0 | 0.8 | GO:0007216 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.0 | 0.4 | GO:0006983 | ER overload response(GO:0006983) |
| 0.0 | 0.4 | GO:1904424 | regulation of GTP binding(GO:1904424) |
| 0.0 | 0.4 | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.0 | 0.3 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 0.0 | 0.1 | GO:0019065 | receptor-mediated endocytosis of virus by host cell(GO:0019065) endocytosis involved in viral entry into host cell(GO:0075509) |
| 0.0 | 0.2 | GO:0090037 | positive regulation of protein kinase C signaling(GO:0090037) |
| 0.0 | 0.1 | GO:0098905 | regulation of bundle of His cell action potential(GO:0098905) |
| 0.0 | 0.6 | GO:0006590 | thyroid hormone generation(GO:0006590) |
| 0.0 | 0.1 | GO:0002032 | desensitization of G-protein coupled receptor protein signaling pathway by arrestin(GO:0002032) |
| 0.0 | 0.1 | GO:1990519 | pyrimidine nucleotide transport(GO:0006864) mitochondrial pyrimidine nucleotide import(GO:1990519) |
| 0.0 | 0.2 | GO:0090206 | negative regulation of cholesterol biosynthetic process(GO:0045541) negative regulation of cholesterol metabolic process(GO:0090206) |
| 0.0 | 0.2 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 | 0.1 | GO:1904761 | negative regulation of myofibroblast differentiation(GO:1904761) |
| 0.0 | 0.1 | GO:0036233 | glycine import(GO:0036233) |
| 0.0 | 0.6 | GO:0014029 | neural crest formation(GO:0014029) |
| 0.0 | 0.4 | GO:0035268 | protein mannosylation(GO:0035268) |
| 0.0 | 0.3 | GO:0031507 | heterochromatin assembly(GO:0031507) |
| 0.0 | 0.2 | GO:0060050 | positive regulation of protein glycosylation(GO:0060050) |
| 0.0 | 0.9 | GO:0071425 | hematopoietic stem cell proliferation(GO:0071425) |
| 0.0 | 0.1 | GO:0035509 | negative regulation of myosin-light-chain-phosphatase activity(GO:0035509) negative regulation of bicellular tight junction assembly(GO:1903347) |
| 0.0 | 0.3 | GO:0090336 | positive regulation of brown fat cell differentiation(GO:0090336) |
| 0.0 | 0.3 | GO:0006657 | CDP-choline pathway(GO:0006657) |
| 0.0 | 0.2 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.0 | 0.2 | GO:0035879 | plasma membrane lactate transport(GO:0035879) |
| 0.0 | 0.4 | GO:0090385 | phagosome-lysosome fusion(GO:0090385) |
| 0.0 | 1.0 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.0 | 0.1 | GO:2000182 | regulation of progesterone biosynthetic process(GO:2000182) |
| 0.0 | 0.4 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.0 | 0.4 | GO:0071447 | cellular response to hydroperoxide(GO:0071447) |
| 0.0 | 0.6 | GO:0043153 | entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.0 | 0.1 | GO:0072365 | regulation of cellular ketone metabolic process by negative regulation of transcription from RNA polymerase II promoter(GO:0072365) |
| 0.0 | 0.3 | GO:0035634 | response to stilbenoid(GO:0035634) |
| 0.0 | 0.1 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.0 | 0.1 | GO:0045082 | interleukin-10 biosynthetic process(GO:0042091) interleukin-13 biosynthetic process(GO:0042231) regulation of interleukin-10 biosynthetic process(GO:0045074) positive regulation of interleukin-10 biosynthetic process(GO:0045082) |
| 0.0 | 0.2 | GO:1902174 | positive regulation of keratinocyte apoptotic process(GO:1902174) |
| 0.0 | 0.3 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 0.4 | GO:0032957 | inositol trisphosphate metabolic process(GO:0032957) |
| 0.0 | 0.3 | GO:1904886 | beta-catenin destruction complex disassembly(GO:1904886) |
| 0.0 | 0.2 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.0 | 0.3 | GO:0036109 | alpha-linolenic acid metabolic process(GO:0036109) |
| 0.0 | 0.2 | GO:0060992 | response to fungicide(GO:0060992) |
| 0.0 | 0.3 | GO:0042983 | amyloid precursor protein biosynthetic process(GO:0042983) regulation of amyloid precursor protein biosynthetic process(GO:0042984) |
| 0.0 | 0.2 | GO:0072711 | cellular response to hydroxyurea(GO:0072711) |
| 0.0 | 1.1 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 0.2 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.0 | 0.6 | GO:0006144 | purine nucleobase metabolic process(GO:0006144) |
| 0.0 | 0.1 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.0 | 0.2 | GO:0032286 | central nervous system myelin maintenance(GO:0032286) |
| 0.0 | 0.1 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.0 | 0.1 | GO:2000535 | regulation of entry of bacterium into host cell(GO:2000535) |
| 0.0 | 0.1 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.0 | 0.3 | GO:0006450 | regulation of translational fidelity(GO:0006450) |
| 0.0 | 0.7 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.0 | 0.3 | GO:0036066 | protein O-linked fucosylation(GO:0036066) |
| 0.0 | 0.1 | GO:0043652 | engulfment of apoptotic cell(GO:0043652) |
| 0.0 | 0.4 | GO:1901663 | ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.0 | 0.1 | GO:1903758 | regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903756) negative regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903758) |
| 0.0 | 0.3 | GO:1904714 | regulation of chaperone-mediated autophagy(GO:1904714) |
| 0.0 | 0.1 | GO:0043084 | penile erection(GO:0043084) |
| 0.0 | 0.7 | GO:0000305 | response to oxygen radical(GO:0000305) |
| 0.0 | 0.6 | GO:0007638 | mechanosensory behavior(GO:0007638) |
| 0.0 | 1.0 | GO:0045652 | regulation of megakaryocyte differentiation(GO:0045652) |
| 0.0 | 0.3 | GO:0050812 | regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) regulation of acyl-CoA biosynthetic process(GO:0050812) |
| 0.0 | 0.3 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.0 | 0.3 | GO:0046033 | AMP metabolic process(GO:0046033) |
| 0.0 | 1.0 | GO:0006635 | fatty acid beta-oxidation(GO:0006635) |
| 0.0 | 0.2 | GO:0070493 | thrombin receptor signaling pathway(GO:0070493) |
| 0.0 | 0.1 | GO:1901970 | positive regulation of mitotic metaphase/anaphase transition(GO:0045842) positive regulation of mitotic sister chromatid separation(GO:1901970) positive regulation of metaphase/anaphase transition of cell cycle(GO:1902101) |
| 0.0 | 0.1 | GO:0051198 | negative regulation of glycolytic process(GO:0045820) negative regulation of cofactor metabolic process(GO:0051195) negative regulation of coenzyme metabolic process(GO:0051198) |
| 0.0 | 0.2 | GO:0015886 | heme transport(GO:0015886) |
| 0.0 | 0.3 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.0 | 0.1 | GO:0031548 | regulation of brain-derived neurotrophic factor receptor signaling pathway(GO:0031548) |
| 0.0 | 0.2 | GO:0038092 | nodal signaling pathway(GO:0038092) |
| 0.0 | 0.1 | GO:0042796 | snRNA transcription from RNA polymerase III promoter(GO:0042796) |
| 0.0 | 0.2 | GO:0060019 | radial glial cell differentiation(GO:0060019) |
| 0.0 | 0.1 | GO:0070914 | UV-damage excision repair(GO:0070914) |
| 0.0 | 0.2 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 0.2 | GO:1902603 | carnitine transmembrane transport(GO:1902603) |
| 0.0 | 0.7 | GO:0097120 | receptor localization to synapse(GO:0097120) |
| 0.0 | 0.1 | GO:0072321 | chaperone-mediated protein transport(GO:0072321) |
| 0.0 | 0.1 | GO:0097029 | mature conventional dendritic cell differentiation(GO:0097029) |
| 0.0 | 0.0 | GO:0003162 | atrioventricular node development(GO:0003162) |
| 0.0 | 0.1 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.0 | 0.0 | GO:0006679 | glucosylceramide biosynthetic process(GO:0006679) |
| 0.0 | 0.0 | GO:0070837 | dehydroascorbic acid transport(GO:0070837) |
| 0.0 | 0.2 | GO:0018206 | peptidyl-methionine modification(GO:0018206) |
| 0.0 | 0.3 | GO:1901522 | positive regulation of transcription from RNA polymerase II promoter involved in cellular response to chemical stimulus(GO:1901522) |
| 0.0 | 0.1 | GO:0061088 | regulation of sequestering of zinc ion(GO:0061088) |
| 0.0 | 0.8 | GO:1901222 | regulation of NIK/NF-kappaB signaling(GO:1901222) |
| 0.0 | 0.4 | GO:0042994 | cytoplasmic sequestering of transcription factor(GO:0042994) |
| 0.0 | 0.1 | GO:0045627 | positive regulation of T-helper 1 cell differentiation(GO:0045627) negative regulation of T-helper 2 cell differentiation(GO:0045629) |
| 0.0 | 0.1 | GO:0043983 | histone H4-K12 acetylation(GO:0043983) |
| 0.0 | 0.1 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 0.0 | 0.1 | GO:0031022 | nuclear migration along microfilament(GO:0031022) |
| 0.0 | 0.1 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
| 0.0 | 0.1 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.0 | 0.2 | GO:0007184 | SMAD protein import into nucleus(GO:0007184) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0097598 | sperm cytoplasmic droplet(GO:0097598) |
| 0.2 | 0.9 | GO:0034666 | integrin alpha2-beta1 complex(GO:0034666) |
| 0.2 | 1.6 | GO:0000125 | PCAF complex(GO:0000125) |
| 0.1 | 0.7 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.1 | 1.5 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.1 | 0.7 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.1 | 0.4 | GO:0097545 | axonemal outer doublet(GO:0097545) |
| 0.1 | 0.3 | GO:1990723 | cytoplasmic periphery of the nuclear pore complex(GO:1990723) |
| 0.1 | 0.3 | GO:0031084 | BLOC-2 complex(GO:0031084) |
| 0.1 | 0.4 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.1 | 0.8 | GO:0001940 | male pronucleus(GO:0001940) |
| 0.1 | 3.9 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.1 | 0.4 | GO:1902560 | GMP reductase complex(GO:1902560) |
| 0.1 | 0.7 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.1 | 0.6 | GO:0005851 | eukaryotic translation initiation factor 2B complex(GO:0005851) |
| 0.1 | 1.8 | GO:0036038 | MKS complex(GO:0036038) |
| 0.1 | 0.7 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.1 | 0.7 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.1 | 0.7 | GO:0005879 | axonemal microtubule(GO:0005879) |
| 0.1 | 0.2 | GO:0000799 | nuclear condensin complex(GO:0000799) |
| 0.1 | 1.7 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.1 | 0.2 | GO:0097135 | cyclin E2-CDK2 complex(GO:0097135) |
| 0.1 | 1.9 | GO:0030057 | desmosome(GO:0030057) |
| 0.1 | 0.2 | GO:0005745 | m-AAA complex(GO:0005745) |
| 0.1 | 0.8 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 3.4 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.2 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.0 | 0.3 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 0.0 | 0.8 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.0 | 0.3 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.0 | 0.5 | GO:0045179 | apical cortex(GO:0045179) |
| 0.0 | 0.8 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 0.8 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 0.4 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.0 | 0.3 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.0 | 4.1 | GO:0031903 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 0.2 | GO:0000796 | condensin complex(GO:0000796) |
| 0.0 | 0.5 | GO:0098799 | outer mitochondrial membrane protein complex(GO:0098799) |
| 0.0 | 0.8 | GO:0031045 | dense core granule(GO:0031045) |
| 0.0 | 0.7 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.0 | 0.9 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 0.5 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 3.4 | GO:0101003 | ficolin-1-rich granule membrane(GO:0101003) |
| 0.0 | 0.5 | GO:0097470 | ribbon synapse(GO:0097470) |
| 0.0 | 0.2 | GO:1903440 | calcitonin family receptor complex(GO:1903439) amylin receptor complex(GO:1903440) |
| 0.0 | 0.7 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.3 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.0 | 0.6 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.0 | 0.2 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.0 | 0.0 | GO:0000805 | X chromosome(GO:0000805) |
| 0.0 | 0.1 | GO:0000839 | Hrd1p ubiquitin ligase ERAD-L complex(GO:0000839) |
| 0.0 | 0.3 | GO:0005677 | chromatin silencing complex(GO:0005677) |
| 0.0 | 0.4 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.0 | 0.5 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.0 | 2.2 | GO:0031672 | A band(GO:0031672) |
| 0.0 | 0.7 | GO:0005763 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 0.1 | GO:0032021 | NELF complex(GO:0032021) |
| 0.0 | 0.2 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 0.4 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.0 | 0.4 | GO:1990909 | Wnt signalosome(GO:1990909) |
| 0.0 | 1.0 | GO:0030660 | Golgi-associated vesicle membrane(GO:0030660) |
| 0.0 | 0.6 | GO:0016460 | myosin II complex(GO:0016460) |
| 0.0 | 0.3 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 0.7 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 0.1 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.0 | 1.5 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.0 | 0.2 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 0.4 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 0.1 | GO:0001405 | presequence translocase-associated import motor(GO:0001405) |
| 0.0 | 0.5 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 0.1 | GO:0045298 | tubulin complex(GO:0045298) |
| 0.0 | 0.2 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 0.4 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.0 | 0.4 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.0 | 1.2 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 0.7 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 0.2 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 0.0 | GO:0044305 | calyx of Held(GO:0044305) |
| 0.0 | 0.1 | GO:1990745 | EARP complex(GO:1990745) |
| 0.0 | 0.3 | GO:0097381 | photoreceptor disc membrane(GO:0097381) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 7.2 | GO:0047291 | neolactotetraosylceramide alpha-2,3-sialyltransferase activity(GO:0004513) lactosylceramide alpha-2,3-sialyltransferase activity(GO:0047291) |
| 0.3 | 0.9 | GO:0070506 | high-density lipoprotein particle receptor activity(GO:0070506) |
| 0.3 | 1.2 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.3 | 1.4 | GO:0003896 | DNA primase activity(GO:0003896) |
| 0.2 | 0.7 | GO:0052725 | inositol tetrakisphosphate 1-kinase activity(GO:0047325) inositol-1,3,4-trisphosphate 6-kinase activity(GO:0052725) inositol-1,3,4-trisphosphate 5-kinase activity(GO:0052726) inositol-1,3,4,5,6-pentakisphosphate 1-phosphatase activity(GO:0052825) inositol-1,3,4,6-tetrakisphosphate 6-phosphatase activity(GO:0052830) inositol-1,3,4,6-tetrakisphosphate 1-phosphatase activity(GO:0052831) inositol-3,4,6-trisphosphate 1-kinase activity(GO:0052835) |
| 0.2 | 0.7 | GO:0072590 | N-acetyl-L-aspartate-L-glutamate ligase activity(GO:0072590) |
| 0.2 | 0.9 | GO:0004583 | dolichyl-phosphate-glucose-glycolipid alpha-glucosyltransferase activity(GO:0004583) |
| 0.2 | 0.7 | GO:0004782 | sulfinoalanine decarboxylase activity(GO:0004782) |
| 0.2 | 0.7 | GO:0016534 | cyclin-dependent protein kinase 5 activator activity(GO:0016534) |
| 0.2 | 1.7 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.2 | 1.5 | GO:0001025 | RNA polymerase III transcription factor binding(GO:0001025) |
| 0.2 | 0.6 | GO:0019777 | Atg12 transferase activity(GO:0019777) |
| 0.2 | 1.3 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.2 | 0.7 | GO:0032450 | maltose alpha-glucosidase activity(GO:0032450) |
| 0.2 | 1.2 | GO:0004962 | endothelin receptor activity(GO:0004962) |
| 0.2 | 0.7 | GO:0047493 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.2 | 0.9 | GO:0043208 | glycosphingolipid binding(GO:0043208) |
| 0.2 | 4.1 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.1 | 0.4 | GO:0008240 | tripeptidyl-peptidase activity(GO:0008240) |
| 0.1 | 1.6 | GO:0097100 | supercoiled DNA binding(GO:0097100) |
| 0.1 | 0.6 | GO:0070573 | metallodipeptidase activity(GO:0070573) |
| 0.1 | 0.5 | GO:0004487 | methylenetetrahydrofolate dehydrogenase (NAD+) activity(GO:0004487) |
| 0.1 | 0.9 | GO:0047522 | 13-prostaglandin reductase activity(GO:0036132) 15-oxoprostaglandin 13-oxidase activity(GO:0047522) |
| 0.1 | 0.8 | GO:0030267 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 0.1 | 0.3 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.1 | 1.0 | GO:0070569 | uridylyltransferase activity(GO:0070569) |
| 0.1 | 0.3 | GO:0000248 | C-5 sterol desaturase activity(GO:0000248) sterol desaturase activity(GO:0070704) |
| 0.1 | 0.7 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.1 | 0.5 | GO:0052795 | exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 0.1 | 0.8 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.1 | 0.6 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 0.1 | 1.0 | GO:0046703 | natural killer cell lectin-like receptor binding(GO:0046703) |
| 0.1 | 0.5 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.1 | 0.6 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.1 | 1.0 | GO:0000099 | sulfur amino acid transmembrane transporter activity(GO:0000099) |
| 0.1 | 0.8 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.1 | 0.5 | GO:0004525 | ribonuclease III activity(GO:0004525) double-stranded RNA-specific ribonuclease activity(GO:0032296) |
| 0.1 | 0.4 | GO:0016657 | GMP reductase activity(GO:0003920) oxidoreductase activity, acting on NAD(P)H, nitrogenous group as acceptor(GO:0016657) |
| 0.1 | 0.4 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.1 | 0.9 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.1 | 1.2 | GO:0003857 | 3-hydroxyacyl-CoA dehydrogenase activity(GO:0003857) |
| 0.1 | 0.7 | GO:0043426 | MRF binding(GO:0043426) |
| 0.1 | 1.3 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.1 | 0.4 | GO:0004122 | cystathionine beta-synthase activity(GO:0004122) |
| 0.1 | 0.4 | GO:0003989 | acetyl-CoA carboxylase activity(GO:0003989) |
| 0.1 | 1.0 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.1 | 0.3 | GO:0047708 | biotinidase activity(GO:0047708) |
| 0.1 | 0.3 | GO:0016418 | S-acetyltransferase activity(GO:0016418) |
| 0.1 | 0.3 | GO:0008518 | reduced folate carrier activity(GO:0008518) |
| 0.1 | 0.4 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.1 | 0.3 | GO:0004001 | adenosine kinase activity(GO:0004001) |
| 0.1 | 0.8 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.1 | 0.2 | GO:0004948 | calcitonin receptor activity(GO:0004948) calcitonin receptor binding(GO:0031716) |
| 0.1 | 0.4 | GO:0071535 | RING-like zinc finger domain binding(GO:0071535) |
| 0.1 | 0.5 | GO:0042285 | UDP-xylosyltransferase activity(GO:0035252) xylosyltransferase activity(GO:0042285) |
| 0.1 | 0.9 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.1 | 0.6 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 0.1 | 0.5 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.1 | 1.2 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.1 | 2.0 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.1 | 0.3 | GO:0038025 | reelin receptor activity(GO:0038025) |
| 0.1 | 0.6 | GO:0030976 | thiamine pyrophosphate binding(GO:0030976) |
| 0.1 | 0.3 | GO:0046899 | nucleoside triphosphate adenylate kinase activity(GO:0046899) |
| 0.0 | 0.9 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 0.2 | GO:0046964 | 3'-phosphoadenosine 5'-phosphosulfate transmembrane transporter activity(GO:0046964) |
| 0.0 | 1.4 | GO:0000030 | mannosyltransferase activity(GO:0000030) |
| 0.0 | 0.2 | GO:0050254 | rhodopsin kinase activity(GO:0050254) |
| 0.0 | 0.9 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.0 | 0.1 | GO:0004638 | phosphoribosylaminoimidazole carboxylase activity(GO:0004638) phosphoribosylaminoimidazolesuccinocarboxamide synthase activity(GO:0004639) |
| 0.0 | 0.1 | GO:0005010 | insulin-like growth factor-activated receptor activity(GO:0005010) |
| 0.0 | 1.1 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.6 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.0 | 0.3 | GO:0004305 | ethanolamine kinase activity(GO:0004305) |
| 0.0 | 0.6 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.0 | 0.5 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.0 | 0.8 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.0 | 0.4 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.0 | 0.7 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 0.0 | 1.0 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.2 | GO:0015142 | citrate transmembrane transporter activity(GO:0015137) tricarboxylic acid transmembrane transporter activity(GO:0015142) |
| 0.0 | 0.1 | GO:0008515 | sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.0 | 0.1 | GO:0004146 | dihydrofolate reductase activity(GO:0004146) |
| 0.0 | 0.2 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.0 | 0.6 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.0 | 0.4 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.2 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.0 | 0.4 | GO:0016413 | O-acetyltransferase activity(GO:0016413) |
| 0.0 | 0.5 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.0 | 0.8 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.0 | 0.3 | GO:0002161 | aminoacyl-tRNA editing activity(GO:0002161) |
| 0.0 | 0.5 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.1 | GO:0015218 | pyrimidine nucleotide transmembrane transporter activity(GO:0015218) |
| 0.0 | 0.8 | GO:0008409 | 5'-3' exonuclease activity(GO:0008409) |
| 0.0 | 0.2 | GO:0004301 | epoxide hydrolase activity(GO:0004301) |
| 0.0 | 0.8 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.8 | GO:0016628 | oxidoreductase activity, acting on the CH-CH group of donors, NAD or NADP as acceptor(GO:0016628) |
| 0.0 | 0.1 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 0.0 | 1.0 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.0 | 0.2 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.0 | 0.2 | GO:0030346 | protein phosphatase 2B binding(GO:0030346) |
| 0.0 | 0.3 | GO:0022851 | GABA-gated chloride ion channel activity(GO:0022851) |
| 0.0 | 0.1 | GO:0035650 | AP-1 adaptor complex binding(GO:0035650) |
| 0.0 | 0.4 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.0 | 0.3 | GO:0005542 | folic acid binding(GO:0005542) |
| 0.0 | 1.2 | GO:0016409 | palmitoyltransferase activity(GO:0016409) |
| 0.0 | 0.3 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.0 | 0.1 | GO:0034739 | histone deacetylase activity (H4-K16 specific)(GO:0034739) |
| 0.0 | 1.3 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.0 | 0.1 | GO:0015199 | amino-acid betaine transmembrane transporter activity(GO:0015199) carnitine transmembrane transporter activity(GO:0015226) |
| 0.0 | 0.7 | GO:0016505 | peptidase activator activity involved in apoptotic process(GO:0016505) |
| 0.0 | 0.6 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 0.3 | GO:0043225 | anion transmembrane-transporting ATPase activity(GO:0043225) |
| 0.0 | 0.1 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.0 | 0.2 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.0 | 2.5 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.2 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.0 | 0.3 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.0 | 0.2 | GO:0004673 | protein histidine kinase activity(GO:0004673) |
| 0.0 | 0.1 | GO:0034617 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced pteridine as one donor, and incorporation of one atom of oxygen(GO:0016714) tetrahydrobiopterin binding(GO:0034617) |
| 0.0 | 0.2 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.7 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.4 | GO:0001056 | RNA polymerase III activity(GO:0001056) |
| 0.0 | 0.2 | GO:0004931 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.0 | 0.3 | GO:0000400 | four-way junction DNA binding(GO:0000400) |
| 0.0 | 0.8 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 0.3 | GO:0016641 | oxidoreductase activity, acting on the CH-NH2 group of donors, oxygen as acceptor(GO:0016641) |
| 0.0 | 0.0 | GO:0047322 | [hydroxymethylglutaryl-CoA reductase (NADPH)] kinase activity(GO:0047322) [acetyl-CoA carboxylase] kinase activity(GO:0050405) |
| 0.0 | 0.5 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.6 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.0 | 0.1 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.0 | 0.1 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.0 | 0.4 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.3 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.3 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 0.2 | GO:0022820 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.0 | 0.1 | GO:0048403 | brain-derived neurotrophic factor binding(GO:0048403) |
| 0.0 | 0.1 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.0 | 0.2 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.0 | 0.7 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
| 0.0 | 0.3 | GO:0019198 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 0.1 | GO:0000182 | rDNA binding(GO:0000182) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.1 | 3.3 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.1 | 5.0 | PID IGF1 PATHWAY | IGF1 pathway |
| 0.1 | 0.9 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.1 | 1.2 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.1 | 4.8 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.1 | 2.0 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 0.6 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.0 | 1.3 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 2.6 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 0.6 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.0 | 2.2 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.0 | 0.5 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 1.3 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 1.3 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.0 | 0.9 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 0.6 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 1.3 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 1.1 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.2 | PID LYMPH ANGIOGENESIS PATHWAY | VEGFR3 signaling in lymphatic endothelium |
| 0.0 | 1.0 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.0 | 0.7 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.0 | 0.4 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 0.5 | PID ATM PATHWAY | ATM pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.6 | REACTOME SIGNALING BY ACTIVATED POINT MUTANTS OF FGFR1 | Genes involved in Signaling by activated point mutants of FGFR1 |
| 0.1 | 1.0 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.1 | 3.3 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.1 | 2.9 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.1 | 1.2 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.1 | 1.9 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.1 | 0.9 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.1 | 0.9 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.1 | 1.3 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 0.9 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 0.9 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.0 | 1.0 | REACTOME SOS MEDIATED SIGNALLING | Genes involved in SOS-mediated signalling |
| 0.0 | 0.5 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.0 | 0.4 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.0 | 1.0 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 1.5 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 0.6 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.0 | 0.3 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.0 | 1.2 | REACTOME FATTY ACYL COA BIOSYNTHESIS | Genes involved in Fatty Acyl-CoA Biosynthesis |
| 0.0 | 0.6 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 0.6 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.0 | 0.6 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 0.7 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.5 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 0.3 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 0.6 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.0 | 1.4 | REACTOME CIRCADIAN CLOCK | Genes involved in Circadian Clock |
| 0.0 | 1.1 | REACTOME ACTIVATION OF ATR IN RESPONSE TO REPLICATION STRESS | Genes involved in Activation of ATR in response to replication stress |
| 0.0 | 0.5 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 0.7 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.0 | 0.4 | REACTOME G ALPHA S SIGNALLING EVENTS | Genes involved in G alpha (s) signalling events |
| 0.0 | 1.0 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |
| 0.0 | 0.5 | REACTOME PYRUVATE METABOLISM | Genes involved in Pyruvate metabolism |
| 0.0 | 0.5 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 0.7 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.0 | 0.4 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 0.2 | REACTOME OPSINS | Genes involved in Opsins |
| 0.0 | 0.4 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.0 | 0.5 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 1.2 | REACTOME REGULATION OF INSULIN SECRETION BY GLUCAGON LIKE PEPTIDE1 | Genes involved in Regulation of Insulin Secretion by Glucagon-like Peptide-1 |
| 0.0 | 0.2 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.3 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.0 | 0.3 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.0 | 0.3 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.6 | REACTOME SIGNAL TRANSDUCTION BY L1 | Genes involved in Signal transduction by L1 |
| 0.0 | 0.8 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.0 | 0.1 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.7 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |