Project
Inflammatory response time course, HUVEC (Wada, 2009)
Navigation
Downloads
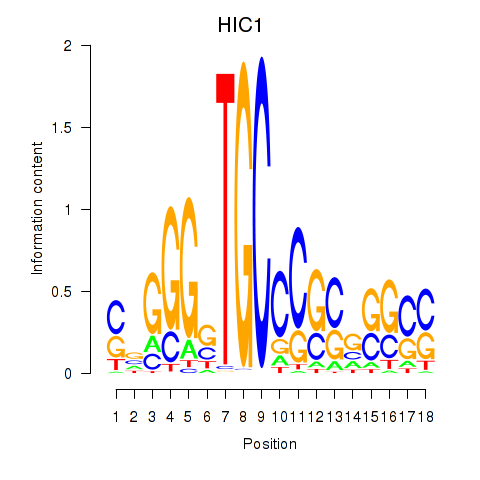
Results for HIC1
Z-value: 1.13
Transcription factors associated with HIC1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HIC1
|
ENSG00000177374.13 | HIC1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HIC1 | hg38_v1_chr17_+_2055094_2055110 | 0.30 | 1.5e-01 | Click! |
Activity profile of HIC1 motif
Sorted Z-values of HIC1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of HIC1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_+_39809602 | 6.63 |
ENST00000302541.11
ENST00000396217.7 |
MYRIP
|
myosin VIIA and Rab interacting protein |
| chr1_+_25616780 | 5.76 |
ENST00000374332.9
|
MAN1C1
|
mannosidase alpha class 1C member 1 |
| chr9_-_109013483 | 5.55 |
ENST00000325551.9
ENST00000374593.4 ENST00000374595.8 |
CTNNAL1
|
catenin alpha like 1 |
| chr4_+_41360759 | 5.28 |
ENST00000508501.5
ENST00000512946.5 ENST00000313860.11 ENST00000512632.5 ENST00000512820.5 |
LIMCH1
|
LIM and calponin homology domains 1 |
| chr3_+_32106612 | 4.88 |
ENST00000282541.10
ENST00000425459.5 ENST00000431009.1 |
GPD1L
|
glycerol-3-phosphate dehydrogenase 1 like |
| chr2_+_6917404 | 4.23 |
ENST00000320892.11
|
RNF144A
|
ring finger protein 144A |
| chr12_+_53046969 | 3.80 |
ENST00000379902.7
|
TNS2
|
tensin 2 |
| chr14_-_91060113 | 3.40 |
ENST00000536315.6
|
RPS6KA5
|
ribosomal protein S6 kinase A5 |
| chr12_+_3077355 | 3.38 |
ENST00000537971.5
ENST00000011898.10 ENST00000649909.1 |
TSPAN9
|
tetraspanin 9 |
| chr1_+_64745089 | 3.31 |
ENST00000294428.7
ENST00000371072.8 |
RAVER2
|
ribonucleoprotein, PTB binding 2 |
| chr20_+_64255728 | 3.24 |
ENST00000369758.8
ENST00000308824.11 ENST00000609372.1 ENST00000610196.1 ENST00000609764.1 |
PCMTD2
|
protein-L-isoaspartate (D-aspartate) O-methyltransferase domain containing 2 |
| chr1_+_61082398 | 2.90 |
ENST00000664149.1
|
NFIA
|
nuclear factor I A |
| chr8_+_27633863 | 2.89 |
ENST00000337221.8
|
SCARA3
|
scavenger receptor class A member 3 |
| chr9_+_136712531 | 2.86 |
ENST00000371692.9
|
DIPK1B
|
divergent protein kinase domain 1B |
| chr11_-_33869816 | 2.85 |
ENST00000395833.7
|
LMO2
|
LIM domain only 2 |
| chr15_+_57376497 | 2.83 |
ENST00000281282.6
|
CGNL1
|
cingulin like 1 |
| chr1_+_61082702 | 2.83 |
ENST00000485903.6
ENST00000371185.6 ENST00000371184.6 |
NFIA
|
nuclear factor I A |
| chr9_-_75088140 | 2.70 |
ENST00000361092.9
ENST00000376811.5 |
NMRK1
|
nicotinamide riboside kinase 1 |
| chr9_-_75088198 | 2.68 |
ENST00000376808.8
|
NMRK1
|
nicotinamide riboside kinase 1 |
| chr2_-_156342348 | 2.66 |
ENST00000409572.5
|
NR4A2
|
nuclear receptor subfamily 4 group A member 2 |
| chr2_+_241687059 | 2.49 |
ENST00000636051.1
|
ING5
|
inhibitor of growth family member 5 |
| chr11_+_95089804 | 2.48 |
ENST00000278505.5
|
ENDOD1
|
endonuclease domain containing 1 |
| chr10_+_35127023 | 2.46 |
ENST00000429130.7
ENST00000469949.6 ENST00000460270.5 |
CREM
|
cAMP responsive element modulator |
| chr12_-_29783798 | 2.32 |
ENST00000552618.5
ENST00000551659.5 ENST00000539277.6 |
TMTC1
|
transmembrane O-mannosyltransferase targeting cadherins 1 |
| chr6_+_124962420 | 2.28 |
ENST00000521654.7
ENST00000560949.5 |
RNF217
|
ring finger protein 217 |
| chr10_+_8054808 | 2.23 |
ENST00000346208.4
|
GATA3
|
GATA binding protein 3 |
| chr3_-_64268161 | 2.23 |
ENST00000564377.6
|
PRICKLE2
|
prickle planar cell polarity protein 2 |
| chr7_-_11832190 | 2.22 |
ENST00000423059.9
ENST00000617773.1 |
THSD7A
|
thrombospondin type 1 domain containing 7A |
| chr3_+_37861926 | 2.18 |
ENST00000443503.6
|
CTDSPL
|
CTD small phosphatase like |
| chr22_-_19941798 | 2.17 |
ENST00000400519.6
ENST00000334363.14 ENST00000400521.7 ENST00000400525.6 |
TXNRD2
|
thioredoxin reductase 2 |
| chr1_+_61082553 | 2.16 |
ENST00000403491.8
ENST00000371187.7 |
NFIA
|
nuclear factor I A |
| chr3_+_52246204 | 2.13 |
ENST00000409502.7
|
PPM1M
|
protein phosphatase, Mg2+/Mn2+ dependent 1M |
| chr22_+_23180365 | 2.06 |
ENST00000359540.7
ENST00000305877.13 |
BCR
|
BCR activator of RhoGEF and GTPase |
| chr9_+_76459152 | 2.06 |
ENST00000444201.6
ENST00000376730.5 |
GCNT1
|
glucosaminyl (N-acetyl) transferase 1 |
| chr12_-_94650506 | 2.06 |
ENST00000261226.9
|
TMCC3
|
transmembrane and coiled-coil domain family 3 |
| chr13_+_20703677 | 2.05 |
ENST00000682841.1
|
IL17D
|
interleukin 17D |
| chr10_+_8054668 | 2.03 |
ENST00000379328.9
|
GATA3
|
GATA binding protein 3 |
| chr3_+_37861849 | 1.98 |
ENST00000273179.10
|
CTDSPL
|
CTD small phosphatase like |
| chr4_-_148444674 | 1.98 |
ENST00000344721.8
|
NR3C2
|
nuclear receptor subfamily 3 group C member 2 |
| chr13_-_76886397 | 1.97 |
ENST00000377474.4
|
KCTD12
|
potassium channel tetramerization domain containing 12 |
| chr18_+_79863668 | 1.94 |
ENST00000316249.3
|
KCNG2
|
potassium voltage-gated channel modifier subfamily G member 2 |
| chr9_-_13279407 | 1.93 |
ENST00000546205.5
|
MPDZ
|
multiple PDZ domain crumbs cell polarity complex component |
| chr1_+_64470120 | 1.86 |
ENST00000651257.2
|
CACHD1
|
cache domain containing 1 |
| chr11_-_2139382 | 1.84 |
ENST00000416167.7
|
IGF2
|
insulin like growth factor 2 |
| chr13_-_106535653 | 1.81 |
ENST00000646441.1
|
EFNB2
|
ephrin B2 |
| chr9_-_13279642 | 1.80 |
ENST00000319217.12
|
MPDZ
|
multiple PDZ domain crumbs cell polarity complex component |
| chr9_-_13279564 | 1.77 |
ENST00000541718.5
ENST00000447879.6 |
MPDZ
|
multiple PDZ domain crumbs cell polarity complex component |
| chr2_-_157876301 | 1.76 |
ENST00000434821.7
|
ACVR1
|
activin A receptor type 1 |
| chr8_-_9150648 | 1.73 |
ENST00000310455.4
|
PPP1R3B
|
protein phosphatase 1 regulatory subunit 3B |
| chr3_+_52245721 | 1.73 |
ENST00000323588.9
|
PPM1M
|
protein phosphatase, Mg2+/Mn2+ dependent 1M |
| chr14_-_38256074 | 1.73 |
ENST00000342213.3
|
CLEC14A
|
C-type lectin domain containing 14A |
| chr17_+_63477052 | 1.71 |
ENST00000290866.10
ENST00000428043.5 |
ACE
|
angiotensin I converting enzyme |
| chr7_-_87059639 | 1.71 |
ENST00000450689.7
|
ELAPOR2
|
endosome-lysosome associated apoptosis and autophagy regulator family member 2 |
| chr8_+_27633884 | 1.68 |
ENST00000301904.4
|
SCARA3
|
scavenger receptor class A member 3 |
| chr7_-_87059515 | 1.68 |
ENST00000423294.5
|
ELAPOR2
|
endosome-lysosome associated apoptosis and autophagy regulator family member 2 |
| chr5_+_122312229 | 1.66 |
ENST00000261368.13
|
SNCAIP
|
synuclein alpha interacting protein |
| chr7_+_107044689 | 1.65 |
ENST00000265717.5
|
PRKAR2B
|
protein kinase cAMP-dependent type II regulatory subunit beta |
| chr13_-_48533165 | 1.65 |
ENST00000430805.6
ENST00000544492.5 ENST00000544904.3 |
RCBTB2
|
RCC1 and BTB domain containing protein 2 |
| chr20_+_11890785 | 1.64 |
ENST00000399006.6
ENST00000405977.5 |
BTBD3
|
BTB domain containing 3 |
| chr12_-_42589737 | 1.61 |
ENST00000345127.9
ENST00000639958.1 |
PRICKLE1
|
prickle planar cell polarity protein 1 |
| chr3_+_20040437 | 1.61 |
ENST00000263754.5
|
KAT2B
|
lysine acetyltransferase 2B |
| chr1_+_84078043 | 1.58 |
ENST00000370689.6
ENST00000370688.7 |
PRKACB
|
protein kinase cAMP-activated catalytic subunit beta |
| chr2_+_12716893 | 1.57 |
ENST00000381465.2
ENST00000155926.9 |
TRIB2
|
tribbles pseudokinase 2 |
| chr19_-_4581755 | 1.57 |
ENST00000676793.1
|
SEMA6B
|
semaphorin 6B |
| chr17_+_67825494 | 1.56 |
ENST00000306378.11
ENST00000544778.6 |
BPTF
|
bromodomain PHD finger transcription factor |
| chr7_-_87059624 | 1.54 |
ENST00000444627.5
|
ELAPOR2
|
endosome-lysosome associated apoptosis and autophagy regulator family member 2 |
| chr3_+_52246158 | 1.54 |
ENST00000296487.8
|
PPM1M
|
protein phosphatase, Mg2+/Mn2+ dependent 1M |
| chr10_+_35126791 | 1.53 |
ENST00000474362.5
ENST00000374721.7 |
CREM
|
cAMP responsive element modulator |
| chr9_-_10612966 | 1.53 |
ENST00000381196.9
|
PTPRD
|
protein tyrosine phosphatase receptor type D |
| chr13_-_48533069 | 1.52 |
ENST00000344532.8
|
RCBTB2
|
RCC1 and BTB domain containing protein 2 |
| chr10_+_35127162 | 1.51 |
ENST00000354759.7
|
CREM
|
cAMP responsive element modulator |
| chr9_-_122228845 | 1.50 |
ENST00000394319.8
ENST00000340587.7 |
LHX6
|
LIM homeobox 6 |
| chr10_-_124744280 | 1.50 |
ENST00000337318.8
|
FAM53B
|
family with sequence similarity 53 member B |
| chr4_-_98658582 | 1.50 |
ENST00000305798.8
|
TSPAN5
|
tetraspanin 5 |
| chr6_-_83709382 | 1.49 |
ENST00000520302.5
ENST00000520213.5 ENST00000439399.6 |
SNAP91
|
synaptosome associated protein 91 |
| chr12_+_93571664 | 1.47 |
ENST00000622746.4
ENST00000548537.1 |
SOCS2
|
suppressor of cytokine signaling 2 |
| chr10_+_115093404 | 1.47 |
ENST00000527407.5
|
ATRNL1
|
attractin like 1 |
| chr7_-_28958321 | 1.46 |
ENST00000539664.3
|
TRIL
|
TLR4 interactor with leucine rich repeats |
| chr12_+_52948840 | 1.46 |
ENST00000388837.6
ENST00000550600.5 |
KRT18
|
keratin 18 |
| chr8_-_28386073 | 1.45 |
ENST00000523095.5
ENST00000522795.1 |
ZNF395
|
zinc finger protein 395 |
| chr11_-_47186399 | 1.45 |
ENST00000531226.5
ENST00000524509.1 ENST00000298838.11 ENST00000528201.1 ENST00000530513.5 |
PACSIN3
|
protein kinase C and casein kinase substrate in neurons 3 |
| chr6_-_83709141 | 1.43 |
ENST00000521743.5
|
SNAP91
|
synaptosome associated protein 91 |
| chr6_+_107490103 | 1.43 |
ENST00000317357.10
|
SOBP
|
sine oculis binding protein homolog |
| chr19_-_3029013 | 1.43 |
ENST00000590536.5
ENST00000587137.5 ENST00000455444.6 |
TLE2
|
TLE family member 2, transcriptional corepressor |
| chr6_-_83709019 | 1.42 |
ENST00000519779.5
ENST00000369694.6 ENST00000195649.10 |
SNAP91
|
synaptosome associated protein 91 |
| chr14_+_63852951 | 1.41 |
ENST00000357395.7
ENST00000358025.7 ENST00000344113.8 ENST00000341472.9 ENST00000555002.6 |
SYNE2
|
spectrin repeat containing nuclear envelope protein 2 |
| chr12_+_96194501 | 1.40 |
ENST00000552142.5
|
ELK3
|
ETS transcription factor ELK3 |
| chr2_+_70258088 | 1.39 |
ENST00000433351.7
ENST00000264441.9 |
PCYOX1
|
prenylcysteine oxidase 1 |
| chr4_-_25862979 | 1.38 |
ENST00000399878.8
|
SEL1L3
|
SEL1L family member 3 |
| chr20_+_43945677 | 1.38 |
ENST00000358131.5
|
TOX2
|
TOX high mobility group box family member 2 |
| chr20_+_36092698 | 1.37 |
ENST00000430276.5
ENST00000373950.6 ENST00000373946.7 ENST00000441639.5 ENST00000628415.2 ENST00000452261.5 |
EPB41L1
|
erythrocyte membrane protein band 4.1 like 1 |
| chr11_+_33376077 | 1.37 |
ENST00000658780.2
|
KIAA1549L
|
KIAA1549 like |
| chr9_-_16870662 | 1.36 |
ENST00000380672.9
|
BNC2
|
basonuclin 2 |
| chr9_+_137077467 | 1.36 |
ENST00000409858.8
|
UAP1L1
|
UDP-N-acetylglucosamine pyrophosphorylase 1 like 1 |
| chr9_-_16870702 | 1.35 |
ENST00000380667.6
ENST00000545497.5 ENST00000486514.5 |
BNC2
|
basonuclin 2 |
| chr3_-_9553796 | 1.35 |
ENST00000287585.8
|
LHFPL4
|
LHFPL tetraspan subfamily member 4 |
| chrX_+_150363306 | 1.34 |
ENST00000370401.7
ENST00000432680.7 |
MAMLD1
|
mastermind like domain containing 1 |
| chr12_+_93571832 | 1.33 |
ENST00000549887.1
|
SOCS2
|
suppressor of cytokine signaling 2 |
| chrX_+_150363258 | 1.32 |
ENST00000683696.1
|
MAMLD1
|
mastermind like domain containing 1 |
| chr2_+_176188658 | 1.32 |
ENST00000331462.6
|
HOXD1
|
homeobox D1 |
| chr14_+_65412949 | 1.31 |
ENST00000674118.1
ENST00000553924.5 ENST00000358307.6 ENST00000557338.5 ENST00000554610.1 |
FUT8
|
fucosyltransferase 8 |
| chr6_-_42048648 | 1.30 |
ENST00000502771.1
ENST00000508143.5 ENST00000514588.1 ENST00000510503.5 |
CCND3
|
cyclin D3 |
| chr22_+_19714450 | 1.28 |
ENST00000455784.7
ENST00000406395.5 |
SEPTIN5
|
septin 5 |
| chr3_+_48990219 | 1.28 |
ENST00000383729.9
|
P4HTM
|
prolyl 4-hydroxylase, transmembrane |
| chr17_+_67825664 | 1.27 |
ENST00000321892.8
|
BPTF
|
bromodomain PHD finger transcription factor |
| chr14_-_91060578 | 1.26 |
ENST00000418736.6
ENST00000614987.5 |
RPS6KA5
|
ribosomal protein S6 kinase A5 |
| chr3_+_32238667 | 1.22 |
ENST00000458535.6
ENST00000307526.4 |
CMTM8
|
CKLF like MARVEL transmembrane domain containing 8 |
| chrX_+_10015226 | 1.22 |
ENST00000380861.9
|
WWC3
|
WWC family member 3 |
| chr19_-_3029269 | 1.21 |
ENST00000262953.11
|
TLE2
|
TLE family member 2, transcriptional corepressor |
| chr6_+_18387326 | 1.20 |
ENST00000259939.4
|
RNF144B
|
ring finger protein 144B |
| chr11_-_66347560 | 1.20 |
ENST00000311181.5
|
B4GAT1
|
beta-1,4-glucuronyltransferase 1 |
| chr8_+_6708626 | 1.19 |
ENST00000285518.11
|
AGPAT5
|
1-acylglycerol-3-phosphate O-acyltransferase 5 |
| chr2_-_98731063 | 1.19 |
ENST00000393487.6
|
MGAT4A
|
alpha-1,3-mannosyl-glycoprotein 4-beta-N-acetylglucosaminyltransferase A |
| chr3_+_50155305 | 1.18 |
ENST00000002829.8
ENST00000426511.5 |
SEMA3F
|
semaphorin 3F |
| chr14_-_100306430 | 1.17 |
ENST00000392908.7
ENST00000359232.8 |
SLC25A29
|
solute carrier family 25 member 29 |
| chr11_+_119149029 | 1.17 |
ENST00000619701.5
|
ABCG4
|
ATP binding cassette subfamily G member 4 |
| chr11_-_68121370 | 1.17 |
ENST00000265689.9
ENST00000356135.9 |
CHKA
|
choline kinase alpha |
| chr6_-_56954747 | 1.16 |
ENST00000680361.1
|
DST
|
dystonin |
| chr5_+_95731300 | 1.16 |
ENST00000379982.8
|
RHOBTB3
|
Rho related BTB domain containing 3 |
| chr4_-_25863537 | 1.16 |
ENST00000502949.5
ENST00000264868.9 ENST00000513691.1 ENST00000514872.1 |
SEL1L3
|
SEL1L family member 3 |
| chr11_+_111977300 | 1.15 |
ENST00000615255.1
|
DIXDC1
|
DIX domain containing 1 |
| chr6_+_16129077 | 1.15 |
ENST00000356840.8
ENST00000349606.4 |
MYLIP
|
myosin regulatory light chain interacting protein |
| chr6_+_7726089 | 1.15 |
ENST00000283147.7
|
BMP6
|
bone morphogenetic protein 6 |
| chr10_+_35127295 | 1.14 |
ENST00000489321.5
ENST00000427847.6 ENST00000374728.7 ENST00000345491.7 ENST00000487132.5 |
CREM
|
cAMP responsive element modulator |
| chr1_+_32539418 | 1.14 |
ENST00000373510.9
ENST00000316459.4 |
ZBTB8A
|
zinc finger and BTB domain containing 8A |
| chr19_-_13102848 | 1.13 |
ENST00000264824.5
|
LYL1
|
LYL1 basic helix-loop-helix family member |
| chr19_+_51693327 | 1.13 |
ENST00000637797.2
|
SPACA6
|
sperm acrosome associated 6 |
| chr9_+_36036899 | 1.13 |
ENST00000377966.4
|
RECK
|
reversion inducing cysteine rich protein with kazal motifs |
| chr12_-_95790755 | 1.12 |
ENST00000343702.9
ENST00000344911.8 |
NTN4
|
netrin 4 |
| chr9_+_68705230 | 1.12 |
ENST00000265382.8
|
PIP5K1B
|
phosphatidylinositol-4-phosphate 5-kinase type 1 beta |
| chr11_+_65111845 | 1.11 |
ENST00000526809.5
ENST00000524986.5 ENST00000534371.5 ENST00000279263.14 ENST00000525385.5 ENST00000345348.9 ENST00000531321.5 ENST00000529414.5 ENST00000526085.5 ENST00000530750.5 |
TM7SF2
|
transmembrane 7 superfamily member 2 |
| chr9_+_127612222 | 1.11 |
ENST00000637953.1
ENST00000636962.2 |
STXBP1
|
syntaxin binding protein 1 |
| chr13_+_26557659 | 1.11 |
ENST00000335327.6
ENST00000361042.8 ENST00000671038.1 |
WASF3
|
WASP family member 3 |
| chr1_-_236065079 | 1.11 |
ENST00000264187.7
ENST00000366595.7 |
NID1
|
nidogen 1 |
| chrX_+_46573757 | 1.11 |
ENST00000276055.4
|
CHST7
|
carbohydrate sulfotransferase 7 |
| chr3_+_50155024 | 1.10 |
ENST00000414301.5
ENST00000450338.5 ENST00000413852.5 |
SEMA3F
|
semaphorin 3F |
| chr9_+_131096476 | 1.09 |
ENST00000372309.7
ENST00000247291.8 ENST00000372302.5 ENST00000372300.5 ENST00000372298.1 |
AIF1L
|
allograft inflammatory factor 1 like |
| chr13_-_36920227 | 1.09 |
ENST00000379826.5
ENST00000350148.10 |
SMAD9
|
SMAD family member 9 |
| chr9_+_126326809 | 1.09 |
ENST00000361171.8
ENST00000489637.3 |
MVB12B
|
multivesicular body subunit 12B |
| chr11_-_64803152 | 1.09 |
ENST00000439069.5
ENST00000294066.7 ENST00000377350.7 |
MAP4K2
|
mitogen-activated protein kinase kinase kinase kinase 2 |
| chr19_-_14206168 | 1.09 |
ENST00000361434.7
ENST00000340736.10 |
ADGRL1
|
adhesion G protein-coupled receptor L1 |
| chr1_+_32741779 | 1.08 |
ENST00000401073.7
|
KIAA1522
|
KIAA1522 |
| chr17_+_6071075 | 1.08 |
ENST00000574232.5
ENST00000539421.1 |
WSCD1
|
WSC domain containing 1 |
| chr18_-_75209126 | 1.07 |
ENST00000322342.4
|
ZADH2
|
zinc binding alcohol dehydrogenase domain containing 2 |
| chr3_+_48989876 | 1.06 |
ENST00000343546.8
|
P4HTM
|
prolyl 4-hydroxylase, transmembrane |
| chr12_-_123271089 | 1.06 |
ENST00000535979.5
|
CDK2AP1
|
cyclin dependent kinase 2 associated protein 1 |
| chr22_-_26590082 | 1.05 |
ENST00000442495.5
ENST00000440953.5 ENST00000450022.1 ENST00000338754.9 |
TPST2
|
tyrosylprotein sulfotransferase 2 |
| chr5_+_69415065 | 1.05 |
ENST00000647531.1
ENST00000645446.1 ENST00000325631.10 ENST00000454295.6 |
MARVELD2
|
MARVEL domain containing 2 |
| chr2_-_75560893 | 1.03 |
ENST00000410113.5
ENST00000393913.8 |
EVA1A
|
eva-1 homolog A, regulator of programmed cell death |
| chr17_+_83079595 | 1.03 |
ENST00000320095.12
|
METRNL
|
meteorin like, glial cell differentiation regulator |
| chr22_+_19941813 | 1.02 |
ENST00000678769.1
|
COMT
|
catechol-O-methyltransferase |
| chr1_+_14924100 | 1.02 |
ENST00000361144.9
|
KAZN
|
kazrin, periplakin interacting protein |
| chr8_-_102655707 | 1.02 |
ENST00000285407.11
|
KLF10
|
Kruppel like factor 10 |
| chr12_-_42589592 | 1.01 |
ENST00000552108.6
ENST00000547113.1 ENST00000640646.1 |
PRICKLE1
|
prickle planar cell polarity protein 1 |
| chr8_-_88327475 | 1.00 |
ENST00000286614.11
|
MMP16
|
matrix metallopeptidase 16 |
| chr9_+_67859398 | 1.00 |
ENST00000620792.1
|
ANKRD20A1
|
ankyrin repeat domain 20 family member A1 |
| chr9_+_68705414 | 1.00 |
ENST00000541509.5
|
PIP5K1B
|
phosphatidylinositol-4-phosphate 5-kinase type 1 beta |
| chr13_-_45418337 | 0.99 |
ENST00000519676.6
ENST00000519547.5 |
SLC25A30
|
solute carrier family 25 member 30 |
| chr9_+_69325168 | 0.99 |
ENST00000303068.14
|
FAM189A2
|
family with sequence similarity 189 member A2 |
| chr3_+_43286512 | 0.99 |
ENST00000454177.5
ENST00000429705.6 ENST00000296088.12 ENST00000437827.1 |
SNRK
|
SNF related kinase |
| chr14_+_96039328 | 0.99 |
ENST00000553764.1
ENST00000555004.3 ENST00000556728.1 ENST00000553782.1 |
C14orf132
|
chromosome 14 open reading frame 132 |
| chr18_+_9334757 | 0.99 |
ENST00000262120.10
ENST00000581641.1 |
TWSG1
|
twisted gastrulation BMP signaling modulator 1 |
| chr7_-_149773548 | 0.99 |
ENST00000302017.4
|
ZNF467
|
zinc finger protein 467 |
| chr3_-_124887353 | 0.98 |
ENST00000296181.9
|
ITGB5
|
integrin subunit beta 5 |
| chr2_+_46297397 | 0.98 |
ENST00000263734.5
|
EPAS1
|
endothelial PAS domain protein 1 |
| chr1_-_97920986 | 0.98 |
ENST00000370192.8
|
DPYD
|
dihydropyrimidine dehydrogenase |
| chr17_+_6070361 | 0.97 |
ENST00000317744.10
|
WSCD1
|
WSC domain containing 1 |
| chr10_+_92848461 | 0.97 |
ENST00000443748.6
ENST00000371543.5 ENST00000260762.10 |
EXOC6
|
exocyst complex component 6 |
| chr10_+_35336486 | 0.97 |
ENST00000374704.8
|
CCNY
|
cyclin Y |
| chr21_+_46286623 | 0.95 |
ENST00000397691.1
|
YBEY
|
ybeY metalloendoribonuclease |
| chr16_+_81444799 | 0.95 |
ENST00000537098.8
|
CMIP
|
c-Maf inducing protein |
| chr22_+_19723525 | 0.95 |
ENST00000366425.4
|
GP1BB
|
glycoprotein Ib platelet subunit beta |
| chr15_-_34583592 | 0.94 |
ENST00000683415.1
|
GOLGA8B
|
golgin A8 family member B |
| chr14_+_92513766 | 0.94 |
ENST00000216487.12
ENST00000620541.4 ENST00000557762.1 |
RIN3
|
Ras and Rab interactor 3 |
| chr2_-_69643615 | 0.94 |
ENST00000409068.5
|
AAK1
|
AP2 associated kinase 1 |
| chr19_-_41353904 | 0.94 |
ENST00000221930.6
|
TGFB1
|
transforming growth factor beta 1 |
| chr21_-_38660656 | 0.94 |
ENST00000398919.6
|
ERG
|
ETS transcription factor ERG |
| chr11_-_119340544 | 0.94 |
ENST00000530681.2
|
C1QTNF5
|
C1q and TNF related 5 |
| chr4_-_113761724 | 0.93 |
ENST00000511664.6
|
CAMK2D
|
calcium/calmodulin dependent protein kinase II delta |
| chr2_-_164621461 | 0.93 |
ENST00000446413.6
ENST00000263915.8 |
GRB14
|
growth factor receptor bound protein 14 |
| chr6_+_117482634 | 0.93 |
ENST00000296955.12
ENST00000338728.10 |
DCBLD1
|
discoidin, CUB and LCCL domain containing 1 |
| chr9_+_127612257 | 0.93 |
ENST00000637173.2
ENST00000630492.2 ENST00000627871.2 ENST00000373302.8 ENST00000373299.5 ENST00000650920.1 ENST00000476182.3 |
STXBP1
|
syntaxin binding protein 1 |
| chr22_-_29581074 | 0.93 |
ENST00000415100.5
ENST00000216121.12 |
NIPSNAP1
|
nipsnap homolog 1 |
| chr22_+_19941733 | 0.92 |
ENST00000403184.5
ENST00000361682.11 ENST00000403710.5 ENST00000407537.5 ENST00000678868.1 |
COMT
|
catechol-O-methyltransferase |
| chr5_+_122312164 | 0.92 |
ENST00000514497.6
ENST00000261367.11 |
SNCAIP
|
synuclein alpha interacting protein |
| chr11_+_925840 | 0.92 |
ENST00000448903.7
ENST00000525796.5 ENST00000534328.5 |
AP2A2
|
adaptor related protein complex 2 subunit alpha 2 |
| chr7_-_149773207 | 0.92 |
ENST00000484747.5
|
ZNF467
|
zinc finger protein 467 |
| chr13_-_100674813 | 0.91 |
ENST00000376234.7
ENST00000423847.1 |
TMTC4
|
transmembrane O-mannosyltransferase targeting cadherins 4 |
| chr9_-_78031775 | 0.91 |
ENST00000286548.9
|
GNAQ
|
G protein subunit alpha q |
| chr8_-_73746830 | 0.91 |
ENST00000524300.6
ENST00000523558.5 ENST00000521210.5 ENST00000355780.9 ENST00000524104.5 ENST00000521736.5 ENST00000521447.5 ENST00000517542.5 ENST00000521451.5 ENST00000521419.5 ENST00000518502.5 |
STAU2
|
staufen double-stranded RNA binding protein 2 |
| chr5_+_122311740 | 0.91 |
ENST00000506272.5
ENST00000508681.5 ENST00000509154.6 |
SNCAIP
|
synuclein alpha interacting protein |
| chr2_-_69643152 | 0.91 |
ENST00000606389.7
|
AAK1
|
AP2 associated kinase 1 |
| chr17_+_30970984 | 0.91 |
ENST00000443677.6
ENST00000324689.8 ENST00000328381.10 ENST00000535306.6 ENST00000580444.2 |
RNF135
|
ring finger protein 135 |
| chr1_-_109397888 | 0.90 |
ENST00000256637.8
|
SORT1
|
sortilin 1 |
| chr10_-_125158704 | 0.90 |
ENST00000531469.5
|
CTBP2
|
C-terminal binding protein 2 |
| chr17_-_50866347 | 0.90 |
ENST00000499247.3
|
TOB1
|
transducer of ERBB2, 1 |
| chr2_+_241558716 | 0.89 |
ENST00000318407.5
|
BOK
|
BCL2 family apoptosis regulator BOK |
| chr7_-_120857124 | 0.89 |
ENST00000441017.5
ENST00000424710.5 ENST00000433758.5 |
TSPAN12
|
tetraspanin 12 |
| chr12_+_131894615 | 0.89 |
ENST00000321867.6
|
ULK1
|
unc-51 like autophagy activating kinase 1 |
| chr16_-_4614876 | 0.88 |
ENST00000591401.5
ENST00000283474.12 ENST00000591897.5 |
UBALD1
|
UBA like domain containing 1 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 4.9 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 1.5 | 4.6 | GO:0072204 | cell-cell signaling involved in kidney development(GO:0060995) Wnt signaling pathway involved in kidney development(GO:0061289) canonical Wnt signaling pathway involved in metanephric kidney development(GO:0061290) cell-cell signaling involved in metanephros development(GO:0072204) |
| 1.4 | 5.8 | GO:1904381 | Golgi apparatus mannose trimming(GO:1904381) |
| 0.9 | 4.7 | GO:0043988 | histone H3-S28 phosphorylation(GO:0043988) |
| 0.9 | 2.7 | GO:0021986 | epithalamus development(GO:0021538) habenula development(GO:0021986) |
| 0.9 | 0.9 | GO:0061441 | renal artery morphogenesis(GO:0061441) |
| 0.6 | 1.8 | GO:0030327 | prenylated protein catabolic process(GO:0030327) |
| 0.6 | 1.8 | GO:0061445 | endocardial cell fate commitment(GO:0060957) endocardial cushion cell fate commitment(GO:0061445) |
| 0.5 | 2.6 | GO:2000691 | negative regulation of cardioblast differentiation(GO:0051892) regulation of cardiac muscle cell myoblast differentiation(GO:2000690) negative regulation of cardiac muscle cell myoblast differentiation(GO:2000691) |
| 0.5 | 2.1 | GO:0043314 | negative regulation of neutrophil degranulation(GO:0043314) negative regulation of respiratory burst(GO:0060268) |
| 0.5 | 2.0 | GO:1903296 | regulation of glutamate secretion, neurotransmission(GO:1903294) positive regulation of glutamate secretion, neurotransmission(GO:1903296) |
| 0.5 | 1.5 | GO:0043000 | Golgi to plasma membrane CFTR protein transport(GO:0043000) |
| 0.5 | 6.7 | GO:0030050 | vesicle transport along actin filament(GO:0030050) |
| 0.5 | 3.2 | GO:0097338 | response to clozapine(GO:0097338) |
| 0.4 | 2.7 | GO:0050668 | cellular response to phosphate starvation(GO:0016036) positive regulation of sulfur amino acid metabolic process(GO:0031337) positive regulation of homocysteine metabolic process(GO:0050668) |
| 0.4 | 1.3 | GO:1990535 | neuron projection maintenance(GO:1990535) |
| 0.4 | 1.3 | GO:0036071 | N-glycan fucosylation(GO:0036071) |
| 0.4 | 1.2 | GO:1904772 | hepatocyte homeostasis(GO:0036333) response to tetrachloromethane(GO:1904772) |
| 0.4 | 1.6 | GO:0042091 | interleukin-10 biosynthetic process(GO:0042091) regulation of interleukin-10 biosynthetic process(GO:0045074) |
| 0.4 | 1.2 | GO:1902024 | histidine transport(GO:0015817) L-histidine transmembrane transport(GO:0089709) L-histidine transport(GO:1902024) |
| 0.4 | 1.1 | GO:0090031 | positive regulation of steroid hormone biosynthetic process(GO:0090031) |
| 0.4 | 1.9 | GO:0018076 | N-terminal peptidyl-lysine acetylation(GO:0018076) |
| 0.4 | 1.1 | GO:0071344 | diphosphate metabolic process(GO:0071344) |
| 0.4 | 1.1 | GO:0006478 | peptidyl-tyrosine sulfation(GO:0006478) |
| 0.3 | 1.7 | GO:0002019 | regulation of renal output by angiotensin(GO:0002019) |
| 0.3 | 1.0 | GO:0042412 | taurine biosynthetic process(GO:0042412) |
| 0.3 | 1.0 | GO:2000562 | negative regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000562) |
| 0.3 | 1.0 | GO:1902462 | regulation of mesenchymal stem cell proliferation(GO:1902460) positive regulation of mesenchymal stem cell proliferation(GO:1902462) |
| 0.3 | 2.3 | GO:0035290 | trunk segmentation(GO:0035290) trunk neural crest cell migration(GO:0036484) ventral trunk neural crest cell migration(GO:0036486) |
| 0.3 | 1.2 | GO:0044210 | 'de novo' CTP biosynthetic process(GO:0044210) |
| 0.3 | 0.9 | GO:0019483 | uracil catabolic process(GO:0006212) beta-alanine biosynthetic process(GO:0019483) |
| 0.3 | 0.9 | GO:0072720 | response to dithiothreitol(GO:0072720) |
| 0.3 | 1.1 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
| 0.3 | 1.1 | GO:0048627 | myoblast development(GO:0048627) |
| 0.3 | 1.6 | GO:0003199 | endocardial cushion to mesenchymal transition involved in heart valve formation(GO:0003199) positive regulation of blood vessel remodeling(GO:2000504) |
| 0.3 | 1.5 | GO:0002513 | tolerance induction to self antigen(GO:0002513) |
| 0.3 | 7.7 | GO:0072189 | ureter development(GO:0072189) |
| 0.2 | 0.7 | GO:0031064 | negative regulation of histone deacetylation(GO:0031064) |
| 0.2 | 0.7 | GO:2001153 | regulation of renal water transport(GO:2001151) positive regulation of renal water transport(GO:2001153) |
| 0.2 | 1.0 | GO:0044313 | protein K6-linked deubiquitination(GO:0044313) |
| 0.2 | 1.8 | GO:0038028 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) |
| 0.2 | 1.2 | GO:0032484 | Ral protein signal transduction(GO:0032484) regulation of Ral protein signal transduction(GO:0032485) |
| 0.2 | 0.7 | GO:1901301 | regulation of cargo loading into COPII-coated vesicle(GO:1901301) |
| 0.2 | 0.7 | GO:1902683 | regulation of receptor localization to synapse(GO:1902683) |
| 0.2 | 0.9 | GO:0009405 | pathogenesis(GO:0009405) |
| 0.2 | 1.1 | GO:0019075 | virus maturation(GO:0019075) |
| 0.2 | 1.1 | GO:2001295 | malonyl-CoA biosynthetic process(GO:2001295) |
| 0.2 | 0.8 | GO:0002270 | plasmacytoid dendritic cell activation(GO:0002270) regulation of restriction endodeoxyribonuclease activity(GO:0032072) T-helper 1 cell activation(GO:0035711) |
| 0.2 | 2.9 | GO:0021853 | cerebral cortex GABAergic interneuron migration(GO:0021853) interneuron migration(GO:1904936) |
| 0.2 | 0.8 | GO:0046379 | extracellular polysaccharide biosynthetic process(GO:0045226) extracellular polysaccharide metabolic process(GO:0046379) |
| 0.2 | 0.6 | GO:0002316 | follicular B cell differentiation(GO:0002316) |
| 0.2 | 0.6 | GO:0003192 | mitral valve formation(GO:0003192) |
| 0.2 | 0.6 | GO:0097187 | dentinogenesis(GO:0097187) |
| 0.2 | 1.5 | GO:0007185 | transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.2 | 0.8 | GO:0015772 | disaccharide transport(GO:0015766) sucrose transport(GO:0015770) oligosaccharide transport(GO:0015772) |
| 0.2 | 1.1 | GO:0030311 | poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.2 | 1.7 | GO:0060179 | male mating behavior(GO:0060179) |
| 0.2 | 1.3 | GO:0075071 | autophagy of host cells involved in interaction with symbiont(GO:0075044) autophagy involved in symbiotic interaction(GO:0075071) |
| 0.2 | 1.1 | GO:0033088 | immature T cell proliferation in thymus(GO:0033080) regulation of immature T cell proliferation in thymus(GO:0033084) negative regulation of immature T cell proliferation in thymus(GO:0033088) |
| 0.2 | 0.7 | GO:1901382 | chorionic trophoblast cell proliferation(GO:0097360) regulation of chorionic trophoblast cell proliferation(GO:1901382) |
| 0.2 | 2.2 | GO:0001955 | blood vessel maturation(GO:0001955) |
| 0.2 | 1.1 | GO:0033615 | mitochondrial proton-transporting ATP synthase complex assembly(GO:0033615) |
| 0.2 | 1.4 | GO:0099515 | nuclear migration along microfilament(GO:0031022) actin filament-based transport(GO:0099515) |
| 0.2 | 0.7 | GO:0097112 | gamma-aminobutyric acid receptor clustering(GO:0097112) |
| 0.2 | 2.1 | GO:0060352 | cell adhesion molecule production(GO:0060352) |
| 0.2 | 0.5 | GO:0043396 | corticotropin-releasing hormone secretion(GO:0043396) regulation of corticotropin-releasing hormone secretion(GO:0043397) |
| 0.2 | 0.7 | GO:0009956 | radial pattern formation(GO:0009956) |
| 0.2 | 0.8 | GO:1902498 | regulation of protein autoubiquitination(GO:1902498) |
| 0.2 | 0.2 | GO:0006447 | regulation of translational initiation by iron(GO:0006447) |
| 0.2 | 0.8 | GO:0019626 | short-chain fatty acid catabolic process(GO:0019626) |
| 0.2 | 1.3 | GO:0002329 | pre-B cell differentiation(GO:0002329) |
| 0.2 | 0.6 | GO:0021965 | spinal cord ventral commissure morphogenesis(GO:0021965) |
| 0.2 | 1.6 | GO:0021869 | forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.2 | 0.5 | GO:0048073 | regulation of eye pigmentation(GO:0048073) |
| 0.2 | 5.5 | GO:2000369 | regulation of clathrin-mediated endocytosis(GO:2000369) |
| 0.2 | 0.8 | GO:0060160 | negative regulation of dopamine receptor signaling pathway(GO:0060160) |
| 0.2 | 1.4 | GO:0032803 | regulation of low-density lipoprotein particle receptor catabolic process(GO:0032803) |
| 0.2 | 0.3 | GO:0070318 | positive regulation of G0 to G1 transition(GO:0070318) |
| 0.2 | 0.6 | GO:0018242 | protein O-linked glycosylation via serine(GO:0018242) |
| 0.2 | 0.6 | GO:0002384 | hepatic immune response(GO:0002384) |
| 0.2 | 3.9 | GO:0009650 | UV protection(GO:0009650) |
| 0.2 | 0.6 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.2 | 0.5 | GO:0002086 | diaphragm contraction(GO:0002086) |
| 0.1 | 0.9 | GO:0033490 | cholesterol biosynthetic process via desmosterol(GO:0033489) cholesterol biosynthetic process via lathosterol(GO:0033490) |
| 0.1 | 1.3 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.1 | 1.3 | GO:2001106 | regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.1 | 1.9 | GO:0072307 | metanephric nephron tubule epithelial cell differentiation(GO:0072257) regulation of metanephric nephron tubule epithelial cell differentiation(GO:0072307) |
| 0.1 | 0.9 | GO:0006574 | valine catabolic process(GO:0006574) |
| 0.1 | 1.1 | GO:0035021 | negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.1 | 1.5 | GO:0072178 | nephric duct morphogenesis(GO:0072178) |
| 0.1 | 1.5 | GO:0006657 | CDP-choline pathway(GO:0006657) |
| 0.1 | 4.7 | GO:0014850 | response to muscle activity(GO:0014850) |
| 0.1 | 1.0 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.1 | 0.4 | GO:0021593 | rhombomere morphogenesis(GO:0021593) rhombomere 3 morphogenesis(GO:0021658) |
| 0.1 | 3.0 | GO:0042789 | mRNA transcription from RNA polymerase II promoter(GO:0042789) |
| 0.1 | 0.4 | GO:0060931 | sinoatrial node cell development(GO:0060931) |
| 0.1 | 1.7 | GO:0001766 | membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) |
| 0.1 | 0.8 | GO:0016129 | ergosterol biosynthetic process(GO:0006696) ergosterol metabolic process(GO:0008204) phytosteroid metabolic process(GO:0016128) phytosteroid biosynthetic process(GO:0016129) |
| 0.1 | 0.5 | GO:0048319 | axial mesoderm morphogenesis(GO:0048319) |
| 0.1 | 0.5 | GO:0001927 | exocyst assembly(GO:0001927) |
| 0.1 | 5.2 | GO:0009435 | NAD biosynthetic process(GO:0009435) |
| 0.1 | 0.9 | GO:0032377 | regulation of intracellular lipid transport(GO:0032377) regulation of intracellular sterol transport(GO:0032380) regulation of intracellular cholesterol transport(GO:0032383) |
| 0.1 | 0.9 | GO:0030174 | regulation of DNA-dependent DNA replication initiation(GO:0030174) |
| 0.1 | 0.7 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.1 | 1.1 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.1 | 0.4 | GO:0021718 | pons maturation(GO:0021586) superior olivary nucleus development(GO:0021718) superior olivary nucleus maturation(GO:0021722) |
| 0.1 | 1.6 | GO:0098909 | regulation of cardiac muscle cell action potential involved in regulation of contraction(GO:0098909) |
| 0.1 | 0.3 | GO:0018002 | N-terminal peptidyl-serine acetylation(GO:0017198) N-terminal peptidyl-glutamic acid acetylation(GO:0018002) peptidyl-serine acetylation(GO:0030920) |
| 0.1 | 0.9 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.1 | 0.3 | GO:0036363 | transforming growth factor beta activation(GO:0036363) regulation of transforming growth factor beta activation(GO:1901388) negative regulation of transforming growth factor beta activation(GO:1901389) |
| 0.1 | 7.0 | GO:0048384 | retinoic acid receptor signaling pathway(GO:0048384) |
| 0.1 | 0.3 | GO:1902512 | positive regulation of apoptotic DNA fragmentation(GO:1902512) positive regulation of DNA catabolic process(GO:1903626) |
| 0.1 | 1.0 | GO:0032482 | Rab protein signal transduction(GO:0032482) |
| 0.1 | 0.4 | GO:0036518 | chemorepulsion of dopaminergic neuron axon(GO:0036518) |
| 0.1 | 0.3 | GO:0022018 | subpallium cell proliferation in forebrain(GO:0022012) lateral ganglionic eminence cell proliferation(GO:0022018) lambdoid suture morphogenesis(GO:0060366) sagittal suture morphogenesis(GO:0060367) anterior semicircular canal development(GO:0060873) lateral semicircular canal development(GO:0060875) |
| 0.1 | 2.8 | GO:0003416 | endochondral bone growth(GO:0003416) |
| 0.1 | 2.4 | GO:0040015 | negative regulation of multicellular organism growth(GO:0040015) |
| 0.1 | 1.3 | GO:0043129 | surfactant homeostasis(GO:0043129) |
| 0.1 | 0.4 | GO:0097089 | methyl-branched fatty acid metabolic process(GO:0097089) |
| 0.1 | 0.7 | GO:1905123 | regulation of glucosylceramidase activity(GO:1905123) |
| 0.1 | 3.7 | GO:0090083 | regulation of inclusion body assembly(GO:0090083) |
| 0.1 | 0.7 | GO:0015798 | myo-inositol transport(GO:0015798) |
| 0.1 | 0.2 | GO:0034427 | nuclear-transcribed mRNA catabolic process, exonucleolytic, 3'-5'(GO:0034427) |
| 0.1 | 0.4 | GO:0010430 | fatty acid omega-oxidation(GO:0010430) |
| 0.1 | 0.3 | GO:1903028 | asymmetric Golgi ribbon formation(GO:0090164) positive regulation of opsonization(GO:1903028) |
| 0.1 | 0.8 | GO:0042536 | negative regulation of tumor necrosis factor biosynthetic process(GO:0042536) lung vasculature development(GO:0060426) |
| 0.1 | 0.6 | GO:0034499 | late endosome to Golgi transport(GO:0034499) |
| 0.1 | 0.3 | GO:0061580 | colon epithelial cell migration(GO:0061580) |
| 0.1 | 0.4 | GO:0030200 | heparan sulfate proteoglycan catabolic process(GO:0030200) |
| 0.1 | 1.9 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.1 | 1.2 | GO:0090168 | Golgi reassembly(GO:0090168) |
| 0.1 | 1.5 | GO:0097094 | craniofacial suture morphogenesis(GO:0097094) |
| 0.1 | 1.4 | GO:0090136 | epithelial cell-cell adhesion(GO:0090136) |
| 0.1 | 0.5 | GO:1901907 | diadenosine polyphosphate catabolic process(GO:0015961) diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 0.1 | 0.5 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.1 | 0.5 | GO:0032487 | regulation of Rap protein signal transduction(GO:0032487) |
| 0.1 | 0.3 | GO:0061011 | negative regulation of transcription by transcription factor localization(GO:0010621) hepatic duct development(GO:0061011) hepatoblast differentiation(GO:0061017) |
| 0.1 | 0.4 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 0.1 | 0.3 | GO:0000710 | meiotic mismatch repair(GO:0000710) |
| 0.1 | 0.4 | GO:1901350 | cell-cell signaling involved in cell-cell junction organization(GO:1901350) |
| 0.1 | 1.2 | GO:0006975 | DNA damage induced protein phosphorylation(GO:0006975) |
| 0.1 | 0.2 | GO:0051228 | mitotic spindle disassembly(GO:0051228) spindle disassembly(GO:0051230) |
| 0.1 | 1.0 | GO:0048227 | plasma membrane to endosome transport(GO:0048227) |
| 0.1 | 1.4 | GO:0006450 | regulation of translational fidelity(GO:0006450) |
| 0.1 | 1.5 | GO:0032802 | low-density lipoprotein particle receptor catabolic process(GO:0032802) |
| 0.1 | 0.5 | GO:0090116 | C-5 methylation of cytosine(GO:0090116) |
| 0.1 | 0.2 | GO:0035494 | SNARE complex disassembly(GO:0035494) |
| 0.1 | 0.5 | GO:0033274 | response to vitamin B2(GO:0033274) heterochromatin maintenance(GO:0070829) |
| 0.1 | 0.6 | GO:0070345 | negative regulation of fat cell proliferation(GO:0070345) |
| 0.1 | 0.2 | GO:0031508 | pericentric heterochromatin assembly(GO:0031508) |
| 0.1 | 0.6 | GO:0010792 | DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) |
| 0.1 | 0.2 | GO:0003064 | regulation of heart rate by hormone(GO:0003064) negative regulation of potassium ion export(GO:1902303) |
| 0.1 | 0.3 | GO:1903288 | positive regulation of potassium ion import(GO:1903288) |
| 0.1 | 1.3 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.1 | 1.1 | GO:0032836 | glomerular basement membrane development(GO:0032836) |
| 0.1 | 0.5 | GO:0090235 | regulation of metaphase plate congression(GO:0090235) |
| 0.1 | 0.4 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.1 | 0.3 | GO:2000312 | regulation of kainate selective glutamate receptor activity(GO:2000312) |
| 0.1 | 0.4 | GO:0043385 | mycotoxin metabolic process(GO:0043385) aflatoxin metabolic process(GO:0046222) organic heteropentacyclic compound metabolic process(GO:1901376) |
| 0.1 | 0.3 | GO:0031133 | regulation of axon diameter(GO:0031133) |
| 0.1 | 0.6 | GO:0031339 | negative regulation of vesicle fusion(GO:0031339) |
| 0.1 | 0.1 | GO:0060279 | positive regulation of ovulation(GO:0060279) |
| 0.1 | 0.6 | GO:2000601 | positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.1 | 0.1 | GO:0061300 | cerebellum vasculature development(GO:0061300) |
| 0.1 | 0.7 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.1 | 0.3 | GO:0002545 | chronic inflammatory response to non-antigenic stimulus(GO:0002545) positive regulation of chronic inflammatory response(GO:0002678) regulation of chronic inflammatory response to non-antigenic stimulus(GO:0002880) |
| 0.1 | 0.1 | GO:0019065 | receptor-mediated endocytosis of virus by host cell(GO:0019065) endocytosis involved in viral entry into host cell(GO:0075509) |
| 0.1 | 0.5 | GO:0008215 | spermine metabolic process(GO:0008215) |
| 0.1 | 0.4 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.1 | 0.8 | GO:1901898 | negative regulation of relaxation of muscle(GO:1901078) negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.1 | 0.6 | GO:0032049 | cardiolipin biosynthetic process(GO:0032049) |
| 0.1 | 0.3 | GO:0018094 | protein polyglycylation(GO:0018094) |
| 0.1 | 0.2 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.1 | 0.5 | GO:1904177 | osteoblast fate commitment(GO:0002051) regulation of adipose tissue development(GO:1904177) positive regulation of adipose tissue development(GO:1904179) |
| 0.1 | 0.6 | GO:0045654 | positive regulation of megakaryocyte differentiation(GO:0045654) |
| 0.1 | 0.3 | GO:0060754 | positive regulation of mast cell chemotaxis(GO:0060754) |
| 0.1 | 0.3 | GO:0014886 | transition between slow and fast fiber(GO:0014886) |
| 0.1 | 0.3 | GO:0038030 | non-canonical Wnt signaling pathway via MAPK cascade(GO:0038030) non-canonical Wnt signaling pathway via JNK cascade(GO:0038031) |
| 0.1 | 0.2 | GO:0071139 | resolution of recombination intermediates(GO:0071139) resolution of mitotic recombination intermediates(GO:0071140) |
| 0.1 | 0.7 | GO:0061179 | negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 0.1 | 1.1 | GO:0016024 | CDP-diacylglycerol biosynthetic process(GO:0016024) |
| 0.1 | 0.4 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.1 | 0.3 | GO:0014004 | microglia differentiation(GO:0014004) microglia development(GO:0014005) |
| 0.1 | 0.5 | GO:0046959 | habituation(GO:0046959) |
| 0.1 | 0.3 | GO:0061343 | cell adhesion involved in heart morphogenesis(GO:0061343) |
| 0.1 | 0.1 | GO:0036091 | positive regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0036091) |
| 0.1 | 0.4 | GO:0033132 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.1 | 0.2 | GO:0060382 | release from viral latency(GO:0019046) regulation of DNA strand elongation(GO:0060382) |
| 0.1 | 0.3 | GO:0045080 | positive regulation of chemokine biosynthetic process(GO:0045080) |
| 0.1 | 0.8 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.1 | 0.2 | GO:0070914 | UV-damage excision repair(GO:0070914) |
| 0.0 | 0.2 | GO:0046391 | 5-phosphoribose 1-diphosphate biosynthetic process(GO:0006015) 5-phosphoribose 1-diphosphate metabolic process(GO:0046391) |
| 0.0 | 0.0 | GO:0030309 | poly-N-acetyllactosamine metabolic process(GO:0030309) |
| 0.0 | 0.2 | GO:0014049 | positive regulation of glutamate secretion(GO:0014049) |
| 0.0 | 0.0 | GO:0046010 | positive regulation of circadian sleep/wake cycle, non-REM sleep(GO:0046010) |
| 0.0 | 0.6 | GO:0010668 | ectodermal cell differentiation(GO:0010668) |
| 0.0 | 0.2 | GO:0036414 | protein citrullination(GO:0018101) histone citrullination(GO:0036414) |
| 0.0 | 0.2 | GO:0060729 | intestinal epithelial structure maintenance(GO:0060729) |
| 0.0 | 0.7 | GO:0014051 | gamma-aminobutyric acid secretion(GO:0014051) |
| 0.0 | 0.3 | GO:0000973 | posttranscriptional tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000973) nuclear retention of pre-mRNA at the site of transcription(GO:0071033) |
| 0.0 | 0.7 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 0.6 | GO:0086036 | regulation of cardiac muscle cell membrane potential(GO:0086036) |
| 0.0 | 0.5 | GO:0070307 | lens fiber cell development(GO:0070307) |
| 0.0 | 1.3 | GO:0035728 | response to hepatocyte growth factor(GO:0035728) |
| 0.0 | 0.7 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.0 | 0.5 | GO:0098535 | de novo centriole assembly(GO:0098535) |
| 0.0 | 0.2 | GO:0032290 | peripheral nervous system myelin formation(GO:0032290) |
| 0.0 | 0.5 | GO:0071880 | adenylate cyclase-activating adrenergic receptor signaling pathway(GO:0071880) |
| 0.0 | 0.8 | GO:0016226 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.0 | 0.2 | GO:1990502 | dense core granule maturation(GO:1990502) |
| 0.0 | 0.1 | GO:1901526 | positive regulation of macromitophagy(GO:1901526) positive regulation of mitophagy in response to mitochondrial depolarization(GO:1904925) |
| 0.0 | 0.8 | GO:0016446 | somatic hypermutation of immunoglobulin genes(GO:0016446) |
| 0.0 | 0.2 | GO:0031860 | telomeric 3' overhang formation(GO:0031860) |
| 0.0 | 5.2 | GO:0032436 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) |
| 0.0 | 0.2 | GO:0010807 | regulation of synaptic vesicle priming(GO:0010807) |
| 0.0 | 0.5 | GO:2000042 | negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 0.0 | 1.4 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.0 | 0.8 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.0 | 0.3 | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.0 | 0.3 | GO:0051415 | interphase microtubule nucleation by interphase microtubule organizing center(GO:0051415) microtubule nucleation by microtubule organizing center(GO:0051418) |
| 0.0 | 0.3 | GO:0006729 | tetrahydrobiopterin biosynthetic process(GO:0006729) tetrahydrobiopterin metabolic process(GO:0046146) |
| 0.0 | 0.4 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.0 | 0.8 | GO:1902373 | negative regulation of mRNA catabolic process(GO:1902373) |
| 0.0 | 0.4 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.0 | 0.2 | GO:0001923 | B-1 B cell differentiation(GO:0001923) |
| 0.0 | 0.2 | GO:0046929 | negative regulation of neurotransmitter secretion(GO:0046929) |
| 0.0 | 0.6 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.0 | 0.2 | GO:0023016 | signal transduction by trans-phosphorylation(GO:0023016) |
| 0.0 | 0.2 | GO:1903758 | regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903756) negative regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903758) |
| 0.0 | 0.2 | GO:0045048 | protein insertion into ER membrane(GO:0045048) |
| 0.0 | 0.3 | GO:0002043 | blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:0002043) |
| 0.0 | 0.3 | GO:0030259 | lipid glycosylation(GO:0030259) |
| 0.0 | 0.3 | GO:0021707 | cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.0 | 0.1 | GO:1904116 | response to vasopressin(GO:1904116) cellular response to vasopressin(GO:1904117) |
| 0.0 | 0.1 | GO:0090135 | actin filament branching(GO:0090135) |
| 0.0 | 0.7 | GO:0007130 | synaptonemal complex assembly(GO:0007130) |
| 0.0 | 0.1 | GO:0051643 | endoplasmic reticulum localization(GO:0051643) |
| 0.0 | 0.7 | GO:0006590 | thyroid hormone generation(GO:0006590) |
| 0.0 | 0.1 | GO:0099553 | trans-synaptic signaling by lipid, modulating synaptic transmission(GO:0099552) trans-synaptic signaling by endocannabinoid, modulating synaptic transmission(GO:0099553) |
| 0.0 | 0.3 | GO:0032000 | positive regulation of fatty acid beta-oxidation(GO:0032000) |
| 0.0 | 0.3 | GO:0009086 | methionine biosynthetic process(GO:0009086) |
| 0.0 | 0.6 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.0 | 0.2 | GO:0086047 | membrane depolarization during Purkinje myocyte cell action potential(GO:0086047) |
| 0.0 | 0.5 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 1.1 | GO:0000301 | retrograde transport, vesicle recycling within Golgi(GO:0000301) |
| 0.0 | 0.3 | GO:0097026 | dendritic cell dendrite assembly(GO:0097026) |
| 0.0 | 0.3 | GO:1900122 | positive regulation of receptor binding(GO:1900122) |
| 0.0 | 0.3 | GO:0072513 | positive regulation of secondary heart field cardioblast proliferation(GO:0072513) |
| 0.0 | 0.2 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.0 | 1.6 | GO:1990090 | cellular response to nerve growth factor stimulus(GO:1990090) |
| 0.0 | 0.5 | GO:0090161 | Golgi ribbon formation(GO:0090161) |
| 0.0 | 0.1 | GO:0018199 | peptidyl-glutamine modification(GO:0018199) |
| 0.0 | 0.5 | GO:0043306 | positive regulation of mast cell activation involved in immune response(GO:0033008) positive regulation of mast cell degranulation(GO:0043306) |
| 0.0 | 0.3 | GO:1902231 | positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902231) |
| 0.0 | 0.2 | GO:0048194 | Golgi vesicle budding(GO:0048194) |
| 0.0 | 0.1 | GO:0070904 | L-ascorbic acid transport(GO:0015882) molecular hydrogen transport(GO:0015993) transepithelial L-ascorbic acid transport(GO:0070904) |
| 0.0 | 1.7 | GO:0006692 | prostanoid metabolic process(GO:0006692) prostaglandin metabolic process(GO:0006693) |
| 0.0 | 0.2 | GO:0006196 | AMP catabolic process(GO:0006196) |
| 0.0 | 0.1 | GO:0031296 | positive regulation of germinal center formation(GO:0002636) B cell costimulation(GO:0031296) |
| 0.0 | 0.1 | GO:0003057 | regulation of the force of heart contraction by chemical signal(GO:0003057) |
| 0.0 | 1.3 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.0 | 0.4 | GO:0007144 | female meiosis I(GO:0007144) |
| 0.0 | 0.5 | GO:0048642 | negative regulation of skeletal muscle tissue development(GO:0048642) |
| 0.0 | 0.7 | GO:1901522 | positive regulation of transcription from RNA polymerase II promoter involved in cellular response to chemical stimulus(GO:1901522) |
| 0.0 | 0.1 | GO:0032455 | nerve growth factor processing(GO:0032455) |
| 0.0 | 0.1 | GO:0009439 | cyanate metabolic process(GO:0009439) cyanate catabolic process(GO:0009440) |
| 0.0 | 0.1 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.0 | 0.8 | GO:0007342 | fusion of sperm to egg plasma membrane(GO:0007342) |
| 0.0 | 0.4 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 0.0 | 2.1 | GO:0016574 | histone ubiquitination(GO:0016574) |
| 0.0 | 0.7 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.0 | 0.6 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.0 | 1.0 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.0 | 1.1 | GO:0034142 | toll-like receptor 4 signaling pathway(GO:0034142) |
| 0.0 | 1.2 | GO:0090503 | RNA phosphodiester bond hydrolysis, exonucleolytic(GO:0090503) |
| 0.0 | 0.5 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.0 | 0.1 | GO:2000393 | negative regulation of lamellipodium morphogenesis(GO:2000393) |
| 0.0 | 0.0 | GO:0003256 | regulation of transcription from RNA polymerase II promoter involved in myocardial precursor cell differentiation(GO:0003256) |
| 0.0 | 0.3 | GO:0061732 | mitochondrial acetyl-CoA biosynthetic process from pyruvate(GO:0061732) |
| 0.0 | 0.1 | GO:0034472 | snRNA 3'-end processing(GO:0034472) |
| 0.0 | 0.3 | GO:0006750 | glutathione biosynthetic process(GO:0006750) |
| 0.0 | 0.2 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 0.0 | 0.2 | GO:1903025 | regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903025) |
| 0.0 | 0.7 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.0 | 2.7 | GO:0043966 | histone H3 acetylation(GO:0043966) |
| 0.0 | 0.1 | GO:0003290 | atrial septum secundum morphogenesis(GO:0003290) |
| 0.0 | 0.2 | GO:0045040 | protein import into mitochondrial outer membrane(GO:0045040) |
| 0.0 | 0.2 | GO:0036089 | cleavage furrow formation(GO:0036089) |
| 0.0 | 0.9 | GO:0032878 | regulation of establishment or maintenance of cell polarity(GO:0032878) |
| 0.0 | 0.8 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 0.6 | GO:0006044 | N-acetylglucosamine metabolic process(GO:0006044) |
| 0.0 | 0.1 | GO:0010796 | regulation of multivesicular body size(GO:0010796) |
| 0.0 | 0.1 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.0 | 0.3 | GO:0017182 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.0 | 0.3 | GO:0000733 | DNA strand renaturation(GO:0000733) |
| 0.0 | 0.1 | GO:0044240 | multicellular organism lipid catabolic process(GO:0044240) |
| 0.0 | 0.3 | GO:0010042 | response to manganese ion(GO:0010042) |
| 0.0 | 1.3 | GO:0045646 | regulation of erythrocyte differentiation(GO:0045646) |
| 0.0 | 0.2 | GO:0060295 | regulation of cilium movement involved in cell motility(GO:0060295) regulation of cilium beat frequency involved in ciliary motility(GO:0060296) regulation of cilium-dependent cell motility(GO:1902019) |
| 0.0 | 0.2 | GO:0070164 | negative regulation of adiponectin secretion(GO:0070164) |
| 0.0 | 0.2 | GO:0015705 | iodide transport(GO:0015705) |
| 0.0 | 0.5 | GO:0006071 | glycerol metabolic process(GO:0006071) |
| 0.0 | 0.1 | GO:0072229 | proximal convoluted tubule development(GO:0072019) metanephric proximal convoluted tubule development(GO:0072229) |
| 0.0 | 1.4 | GO:0046513 | ceramide biosynthetic process(GO:0046513) |
| 0.0 | 0.2 | GO:2000346 | negative regulation of hepatocyte proliferation(GO:2000346) |
| 0.0 | 0.1 | GO:0030576 | Cajal body organization(GO:0030576) |
| 0.0 | 0.2 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.0 | 0.1 | GO:0001189 | RNA polymerase I transcriptional preinitiation complex assembly(GO:0001188) RNA polymerase I transcriptional preinitiation complex assembly at the promoter for the nuclear large rRNA transcript(GO:0001189) |
| 0.0 | 1.0 | GO:0090102 | cochlea development(GO:0090102) |
| 0.0 | 0.8 | GO:0090129 | positive regulation of synapse maturation(GO:0090129) |
| 0.0 | 0.9 | GO:0033344 | cholesterol efflux(GO:0033344) |
| 0.0 | 0.3 | GO:0010867 | positive regulation of triglyceride biosynthetic process(GO:0010867) |
| 0.0 | 0.2 | GO:2001135 | regulation of endocytic recycling(GO:2001135) |
| 0.0 | 0.1 | GO:1905098 | regulation of guanyl-nucleotide exchange factor activity(GO:1905097) negative regulation of guanyl-nucleotide exchange factor activity(GO:1905098) |
| 0.0 | 0.4 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.0 | 0.3 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 0.0 | 0.2 | GO:0044597 | polyketide metabolic process(GO:0030638) aminoglycoside antibiotic metabolic process(GO:0030647) daunorubicin metabolic process(GO:0044597) doxorubicin metabolic process(GO:0044598) |
| 0.0 | 0.1 | GO:0015853 | adenine transport(GO:0015853) |
| 0.0 | 0.1 | GO:0035720 | intraciliary anterograde transport(GO:0035720) |
| 0.0 | 0.1 | GO:0071502 | cellular response to temperature stimulus(GO:0071502) |
| 0.0 | 1.1 | GO:0045540 | regulation of cholesterol biosynthetic process(GO:0045540) |
| 0.0 | 1.3 | GO:0046626 | regulation of insulin receptor signaling pathway(GO:0046626) |
| 0.0 | 0.6 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 0.0 | 1.0 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.0 | 2.6 | GO:0070268 | cornification(GO:0070268) |
| 0.0 | 0.4 | GO:0030497 | fatty acid elongation(GO:0030497) |
| 0.0 | 0.3 | GO:0043568 | positive regulation of insulin-like growth factor receptor signaling pathway(GO:0043568) |
| 0.0 | 0.2 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.0 | 0.4 | GO:2000651 | positive regulation of sodium ion transmembrane transporter activity(GO:2000651) |
| 0.0 | 0.6 | GO:0001893 | maternal placenta development(GO:0001893) |
| 0.0 | 0.4 | GO:0097320 | membrane tubulation(GO:0097320) |
| 0.0 | 0.5 | GO:0060612 | adipose tissue development(GO:0060612) |
| 0.0 | 0.1 | GO:0071286 | cellular response to magnesium ion(GO:0071286) |
| 0.0 | 0.2 | GO:0046133 | pyrimidine ribonucleoside catabolic process(GO:0046133) |
| 0.0 | 0.1 | GO:0021943 | formation of radial glial scaffolds(GO:0021943) |
| 0.0 | 0.1 | GO:1903416 | response to glycoside(GO:1903416) |
| 0.0 | 0.1 | GO:0042421 | norepinephrine biosynthetic process(GO:0042421) |
| 0.0 | 0.1 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.0 | 0.2 | GO:0035563 | positive regulation of chromatin binding(GO:0035563) |
| 0.0 | 0.4 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.0 | 0.1 | GO:0071799 | response to prostaglandin D(GO:0071798) cellular response to prostaglandin D stimulus(GO:0071799) |
| 0.0 | 0.6 | GO:1901570 | icosanoid biosynthetic process(GO:0046456) fatty acid derivative biosynthetic process(GO:1901570) |
| 0.0 | 1.0 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.0 | 0.0 | GO:2000584 | regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000583) negative regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000584) |
| 0.0 | 0.1 | GO:0045872 | positive regulation of rhodopsin gene expression(GO:0045872) |
| 0.0 | 0.1 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.0 | 0.4 | GO:2000505 | regulation of energy homeostasis(GO:2000505) |
| 0.0 | 0.1 | GO:0000379 | tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 0.0 | 0.2 | GO:0050961 | detection of temperature stimulus involved in sensory perception(GO:0050961) detection of temperature stimulus involved in sensory perception of pain(GO:0050965) |
| 0.0 | 0.1 | GO:0098838 | reduced folate transmembrane transport(GO:0098838) |
| 0.0 | 0.3 | GO:0032516 | positive regulation of phosphoprotein phosphatase activity(GO:0032516) |
| 0.0 | 1.1 | GO:0070206 | protein trimerization(GO:0070206) |
| 0.0 | 0.2 | GO:0072520 | seminiferous tubule development(GO:0072520) |
| 0.0 | 0.1 | GO:0051541 | elastin metabolic process(GO:0051541) |
| 0.0 | 0.1 | GO:0019064 | fusion of virus membrane with host plasma membrane(GO:0019064) membrane fusion involved in viral entry into host cell(GO:0039663) multi-organism membrane fusion(GO:0044800) |
| 0.0 | 0.1 | GO:0097498 | endothelial tube lumen extension(GO:0097498) |
| 0.0 | 0.3 | GO:0046051 | UTP metabolic process(GO:0046051) |
| 0.0 | 0.1 | GO:0015744 | succinate transport(GO:0015744) sulfide oxidation(GO:0019418) sulfide oxidation, using sulfide:quinone oxidoreductase(GO:0070221) succinate transmembrane transport(GO:0071422) |
| 0.0 | 0.3 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.0 | 0.3 | GO:0005980 | glycogen catabolic process(GO:0005980) |
| 0.0 | 0.4 | GO:0044803 | multi-organism membrane organization(GO:0044803) |
| 0.0 | 0.5 | GO:0022400 | regulation of rhodopsin mediated signaling pathway(GO:0022400) |
| 0.0 | 0.1 | GO:0002115 | store-operated calcium entry(GO:0002115) |
| 0.0 | 0.3 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.0 | 0.2 | GO:2000781 | positive regulation of double-strand break repair(GO:2000781) |
| 0.0 | 0.3 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.2 | GO:0035810 | positive regulation of urine volume(GO:0035810) |
| 0.0 | 0.0 | GO:0006987 | activation of signaling protein activity involved in unfolded protein response(GO:0006987) |
| 0.0 | 0.0 | GO:0070493 | thrombin receptor signaling pathway(GO:0070493) |
| 0.0 | 0.3 | GO:1902884 | positive regulation of response to oxidative stress(GO:1902884) |
| 0.0 | 0.3 | GO:0045724 | positive regulation of cilium assembly(GO:0045724) |
| 0.0 | 0.1 | GO:0006621 | protein retention in ER lumen(GO:0006621) maintenance of protein localization in endoplasmic reticulum(GO:0035437) |
| 0.0 | 0.0 | GO:1903333 | regulation of protein refolding(GO:0061083) negative regulation of protein refolding(GO:0061084) regulation of protein folding(GO:1903332) negative regulation of protein folding(GO:1903333) |
| 0.0 | 0.4 | GO:0014003 | oligodendrocyte development(GO:0014003) |
| 0.0 | 0.1 | GO:1902410 | mitotic cytokinetic process(GO:1902410) |
| 0.0 | 0.0 | GO:1903121 | regulation of TRAIL-activated apoptotic signaling pathway(GO:1903121) positive regulation of TRAIL-activated apoptotic signaling pathway(GO:1903984) |
| 0.0 | 0.3 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 0.4 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.0 | 0.1 | GO:0080182 | histone H3-K4 trimethylation(GO:0080182) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 4.9 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.4 | 8.3 | GO:0043220 | Schmidt-Lanterman incisure(GO:0043220) |
| 0.4 | 2.3 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.4 | 1.8 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.3 | 1.2 | GO:0097196 | Shu complex(GO:0097196) |
| 0.3 | 7.2 | GO:0031045 | dense core granule(GO:0031045) |
| 0.3 | 3.0 | GO:0016589 | NURF complex(GO:0016589) |
| 0.3 | 0.8 | GO:0036117 | hyaluranon cable(GO:0036117) |
| 0.3 | 0.8 | GO:0071148 | TEAD-1-YAP complex(GO:0071148) |
| 0.3 | 3.0 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.2 | 1.4 | GO:0031673 | H zone(GO:0031673) |
| 0.2 | 1.6 | GO:0000308 | cytoplasmic cyclin-dependent protein kinase holoenzyme complex(GO:0000308) |
| 0.2 | 0.9 | GO:0097629 | extrinsic component of omegasome membrane(GO:0097629) |
| 0.2 | 1.9 | GO:0000125 | PCAF complex(GO:0000125) |
| 0.2 | 2.6 | GO:0070775 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.2 | 0.7 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.2 | 1.5 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.1 | 0.9 | GO:0071458 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) |
| 0.1 | 3.2 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.1 | 0.7 | GO:0034666 | integrin alpha2-beta1 complex(GO:0034666) |
| 0.1 | 0.6 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.1 | 0.4 | GO:0034657 | GID complex(GO:0034657) |
| 0.1 | 7.2 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.1 | 1.3 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.1 | 0.3 | GO:0031251 | PAN complex(GO:0031251) |
| 0.1 | 0.3 | GO:1990917 | sperm head plasma membrane(GO:1990913) ooplasm(GO:1990917) |
| 0.1 | 0.8 | GO:0043196 | varicosity(GO:0043196) |
| 0.1 | 1.2 | GO:0008274 | gamma-tubulin large complex(GO:0000931) gamma-tubulin ring complex(GO:0008274) |
| 0.1 | 2.0 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.1 | 0.6 | GO:0020016 | ciliary pocket(GO:0020016) ciliary pocket membrane(GO:0020018) |
| 0.1 | 0.3 | GO:1905103 | integral component of lysosomal membrane(GO:1905103) |
| 0.1 | 2.1 | GO:0031254 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.1 | 1.4 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.1 | 0.3 | GO:0032449 | CBM complex(GO:0032449) |
| 0.1 | 0.2 | GO:0030312 | external encapsulating structure(GO:0030312) |
| 0.1 | 0.7 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.1 | 1.2 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.1 | 0.2 | GO:1902937 | inward rectifier potassium channel complex(GO:1902937) |
| 0.1 | 0.4 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.1 | 1.2 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.1 | 0.2 | GO:0000939 | condensed chromosome inner kinetochore(GO:0000939) |
| 0.1 | 0.4 | GO:0098837 | postsynaptic recycling endosome(GO:0098837) |
| 0.1 | 1.5 | GO:0036020 | endolysosome membrane(GO:0036020) |
| 0.1 | 0.4 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.1 | 0.3 | GO:0005873 | plus-end kinesin complex(GO:0005873) |
| 0.1 | 0.3 | GO:0071595 | Nem1-Spo7 phosphatase complex(GO:0071595) |
| 0.1 | 2.2 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.1 | 1.3 | GO:0044232 | organelle membrane contact site(GO:0044232) |
| 0.1 | 0.3 | GO:0032301 | MutSalpha complex(GO:0032301) |
| 0.1 | 0.3 | GO:0032311 | angiogenin-PRI complex(GO:0032311) |
| 0.0 | 0.5 | GO:0043190 | ATP-binding cassette (ABC) transporter complex(GO:0043190) |
| 0.0 | 0.8 | GO:0097470 | ribbon synapse(GO:0097470) |
| 0.0 | 0.4 | GO:0005683 | U7 snRNP(GO:0005683) |
| 0.0 | 0.8 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.0 | 0.8 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.0 | 0.2 | GO:0019034 | viral replication complex(GO:0019034) |
| 0.0 | 0.9 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.0 | 1.2 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 0.2 | GO:0097059 | CNTFR-CLCF1 complex(GO:0097059) |
| 0.0 | 1.1 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.0 | 0.3 | GO:0031415 | NatA complex(GO:0031415) |
| 0.0 | 4.6 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.0 | 0.8 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 0.7 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.1 | GO:0055087 | Ski complex(GO:0055087) |
| 0.0 | 5.1 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.0 | 1.4 | GO:0034361 | very-low-density lipoprotein particle(GO:0034361) triglyceride-rich lipoprotein particle(GO:0034385) |
| 0.0 | 0.3 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.0 | 0.6 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.5 | GO:0033276 | transcription factor TFTC complex(GO:0033276) |
| 0.0 | 1.2 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.4 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.0 | 0.2 | GO:0061673 | mitotic spindle astral microtubule(GO:0061673) |
| 0.0 | 0.5 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.0 | 0.2 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.0 | 0.1 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.0 | 0.9 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.0 | 0.4 | GO:0030478 | actin cap(GO:0030478) |
| 0.0 | 0.2 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 0.0 | 0.8 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.0 | 3.9 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 3.2 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.1 | GO:0000814 | ESCRT II complex(GO:0000814) |
| 0.0 | 1.2 | GO:0008305 | integrin complex(GO:0008305) |
| 0.0 | 0.1 | GO:0005784 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 0.0 | 0.4 | GO:0001673 | male germ cell nucleus(GO:0001673) |
| 0.0 | 0.6 | GO:0036038 | MKS complex(GO:0036038) |
| 0.0 | 1.5 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.0 | 0.8 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 0.4 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.0 | 0.2 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.0 | 2.2 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.0 | 0.1 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 0.0 | 0.8 | GO:0005763 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 1.6 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 2.4 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.0 | 0.3 | GO:0000177 | cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.0 | 0.2 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.0 | 0.3 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 0.4 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.0 | 2.4 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 0.3 | GO:0001520 | outer dense fiber(GO:0001520) |
| 0.0 | 0.2 | GO:0098799 | outer mitochondrial membrane protein complex(GO:0098799) |
| 0.0 | 1.3 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 5.8 | GO:0000151 | ubiquitin ligase complex(GO:0000151) |
| 0.0 | 0.3 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.0 | 0.2 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.0 | 2.4 | GO:0031985 | Golgi cisterna(GO:0031985) |
| 0.0 | 0.3 | GO:0008091 | spectrin(GO:0008091) |
| 0.0 | 0.5 | GO:0097381 | photoreceptor disc membrane(GO:0097381) |
| 0.0 | 0.1 | GO:0030130 | clathrin coat of trans-Golgi network vesicle(GO:0030130) |
| 0.0 | 0.4 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.0 | 0.4 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.0 | 0.2 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.0 | 0.5 | GO:0033202 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 0.0 | 0.4 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.0 | 0.4 | GO:0030137 | COPI-coated vesicle(GO:0030137) |
| 0.0 | 0.3 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 0.9 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 1.1 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.0 | 0.0 | GO:0035189 | Rb-E2F complex(GO:0035189) |
| 0.0 | 0.2 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 0.1 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.0 | 0.2 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 0.2 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 0.3 | GO:0010369 | chromocenter(GO:0010369) |
| 0.0 | 1.9 | GO:0101002 | ficolin-1-rich granule(GO:0101002) ficolin-1-rich granule lumen(GO:1904813) |
| 0.0 | 0.2 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 0.4 | GO:0005921 | gap junction(GO:0005921) |
| 0.0 | 0.3 | GO:0044298 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.0 | 0.8 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.0 | 0.1 | GO:0031094 | platelet dense tubular network(GO:0031094) |
| 0.0 | 0.1 | GO:1990745 | EARP complex(GO:1990745) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 5.4 | GO:0050262 | ribosylnicotinamide kinase activity(GO:0050262) ribosylnicotinate kinase activity(GO:0061769) |
| 0.7 | 2.1 | GO:0052810 | 1-phosphatidylinositol-5-kinase activity(GO:0052810) |
| 0.7 | 2.1 | GO:0003829 | beta-1,3-galactosyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity(GO:0003829) |
| 0.6 | 3.2 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.6 | 5.5 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.6 | 4.3 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.5 | 0.5 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.5 | 2.2 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.5 | 1.5 | GO:0004782 | sulfinoalanine decarboxylase activity(GO:0004782) |
| 0.5 | 4.9 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.4 | 1.3 | GO:0008424 | glycoprotein 6-alpha-L-fucosyltransferase activity(GO:0008424) alpha-(1->6)-fucosyltransferase activity(GO:0046921) |
| 0.4 | 3.2 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.4 | 1.9 | GO:0070573 | metallodipeptidase activity(GO:0070573) |
| 0.4 | 6.4 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.4 | 1.1 | GO:0008903 | hydroxypyruvate isomerase activity(GO:0008903) |
| 0.4 | 1.1 | GO:0050613 | delta14-sterol reductase activity(GO:0050613) |
| 0.4 | 1.1 | GO:0008476 | protein-tyrosine sulfotransferase activity(GO:0008476) |
| 0.4 | 2.8 | GO:0008269 | JAK pathway signal transduction adaptor activity(GO:0008269) |
| 0.3 | 1.7 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.3 | 1.6 | GO:1903135 | cupric ion binding(GO:1903135) |
| 0.3 | 1.2 | GO:0003883 | CTP synthase activity(GO:0003883) |
| 0.3 | 2.7 | GO:0016206 | catechol O-methyltransferase activity(GO:0016206) |
| 0.3 | 1.2 | GO:0034041 | sterol-transporting ATPase activity(GO:0034041) |
| 0.3 | 1.2 | GO:0033265 | choline binding(GO:0033265) |
| 0.3 | 1.4 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.3 | 1.1 | GO:0016524 | latrotoxin receptor activity(GO:0016524) |
| 0.3 | 1.1 | GO:0047493 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.3 | 0.5 | GO:0015924 | mannosyl-oligosaccharide mannosidase activity(GO:0015924) |
| 0.3 | 1.8 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.2 | 2.2 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.2 | 1.0 | GO:0048763 | calcium-induced calcium release activity(GO:0048763) |
| 0.2 | 0.9 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.2 | 0.7 | GO:0030158 | protein xylosyltransferase activity(GO:0030158) |
| 0.2 | 1.8 | GO:0036132 | 13-prostaglandin reductase activity(GO:0036132) 15-oxoprostaglandin 13-oxidase activity(GO:0047522) |
| 0.2 | 0.9 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 0.2 | 1.1 | GO:0003989 | acetyl-CoA carboxylase activity(GO:0003989) |
| 0.2 | 0.6 | GO:0051717 | inositol-1,3,4,5-tetrakisphosphate 3-phosphatase activity(GO:0051717) phosphatidylinositol-3,4-bisphosphate 3-phosphatase activity(GO:0051800) |
| 0.2 | 0.6 | GO:0070119 | ciliary neurotrophic factor binding(GO:0070119) |
| 0.2 | 1.6 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.2 | 0.8 | GO:0015154 | sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.2 | 1.1 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.2 | 0.7 | GO:0035033 | histone deacetylase regulator activity(GO:0035033) |
| 0.2 | 1.6 | GO:0016670 | oxidoreductase activity, acting on a sulfur group of donors, oxygen as acceptor(GO:0016670) |
| 0.2 | 1.1 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.2 | 0.9 | GO:0047273 | galactosylgalactosylglucosylceramide beta-D-acetylgalactosaminyltransferase activity(GO:0047273) |
| 0.2 | 0.5 | GO:0080023 | 3R-hydroxyacyl-CoA dehydratase activity(GO:0080023) |
| 0.2 | 0.5 | GO:0000248 | C-5 sterol desaturase activity(GO:0000248) sterol desaturase activity(GO:0070704) |
| 0.2 | 0.5 | GO:0005055 | laminin receptor activity(GO:0005055) |
| 0.2 | 0.8 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.2 | 1.0 | GO:0004301 | epoxide hydrolase activity(GO:0004301) |
| 0.2 | 0.5 | GO:0002094 | polyprenyltransferase activity(GO:0002094) |
| 0.2 | 0.8 | GO:0031750 | D3 dopamine receptor binding(GO:0031750) |
| 0.2 | 0.5 | GO:0010698 | acetyltransferase activator activity(GO:0010698) |
| 0.2 | 1.1 | GO:0030267 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 0.2 | 0.9 | GO:0010465 | nerve growth factor receptor activity(GO:0010465) |
| 0.1 | 1.2 | GO:0010858 | calcium-dependent protein kinase regulator activity(GO:0010858) |
| 0.1 | 6.6 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.1 | 0.7 | GO:0016933 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.1 | 0.6 | GO:0032450 | maltose alpha-glucosidase activity(GO:0032450) |
| 0.1 | 2.9 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.1 | 1.0 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.1 | 1.4 | GO:0070569 | uridylyltransferase activity(GO:0070569) |
| 0.1 | 1.4 | GO:0002161 | aminoacyl-tRNA editing activity(GO:0002161) |
| 0.1 | 2.0 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.1 | 0.5 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.1 | 0.8 | GO:0051996 | farnesyl-diphosphate farnesyltransferase activity(GO:0004310) squalene synthase activity(GO:0051996) |
| 0.1 | 0.9 | GO:1904288 | BAT3 complex binding(GO:1904288) |
| 0.1 | 0.4 | GO:0071209 | U7 snRNA binding(GO:0071209) |
| 0.1 | 0.4 | GO:0004566 | beta-glucuronidase activity(GO:0004566) heparanase activity(GO:0030305) |
| 0.1 | 0.6 | GO:0043208 | glycosphingolipid binding(GO:0043208) |
| 0.1 | 2.3 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.1 | 0.5 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.1 | 0.8 | GO:0004075 | biotin carboxylase activity(GO:0004075) |
| 0.1 | 0.4 | GO:0016534 | cyclin-dependent protein kinase 5 activator activity(GO:0016534) |
| 0.1 | 0.3 | GO:1990190 | peptide-glutamate-N-acetyltransferase activity(GO:1990190) |
| 0.1 | 0.3 | GO:0002135 | CTP binding(GO:0002135) |
| 0.1 | 1.8 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.1 | 3.9 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.1 | 0.3 | GO:0004505 | phenylalanine 4-monooxygenase activity(GO:0004505) |
| 0.1 | 1.1 | GO:0034481 | chondroitin sulfotransferase activity(GO:0034481) |
| 0.1 | 0.3 | GO:0008466 | glycogenin glucosyltransferase activity(GO:0008466) |
| 0.1 | 0.4 | GO:0047391 | alkylglycerophosphoethanolamine phosphodiesterase activity(GO:0047391) |
| 0.1 | 1.5 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.1 | 1.6 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.1 | 2.0 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 0.1 | 1.5 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.1 | 1.0 | GO:0015651 | quaternary ammonium group transmembrane transporter activity(GO:0015651) |
| 0.1 | 0.4 | GO:0022865 | transmembrane electron transfer carrier(GO:0022865) |
| 0.1 | 0.3 | GO:0051990 | (R)-2-hydroxyglutarate dehydrogenase activity(GO:0051990) |
| 0.1 | 0.1 | GO:0031755 | endothelial differentiation G-protein coupled receptor binding(GO:0031753) Edg-2 lysophosphatidic acid receptor binding(GO:0031755) |
| 0.1 | 0.5 | GO:0046624 | sphingolipid transporter activity(GO:0046624) |
| 0.1 | 0.6 | GO:0004522 | ribonuclease A activity(GO:0004522) |
| 0.1 | 1.3 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.1 | 0.5 | GO:0052844 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) inositol diphosphate tetrakisphosphate diphosphatase activity(GO:0052840) inositol bisdiphosphate tetrakisphosphate diphosphatase activity(GO:0052841) inositol diphosphate pentakisphosphate diphosphatase activity(GO:0052842) inositol-1-diphosphate-2,3,4,5,6-pentakisphosphate diphosphatase activity(GO:0052843) inositol-3-diphosphate-1,2,4,5,6-pentakisphosphate diphosphatase activity(GO:0052844) inositol-5-diphosphate-1,2,3,4,6-pentakisphosphate diphosphatase activity(GO:0052845) inositol-1,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 1-diphosphatase activity(GO:0052846) inositol-1,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 5-diphosphatase activity(GO:0052847) inositol-3,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 5-diphosphatase activity(GO:0052848) |
| 0.1 | 0.7 | GO:0034713 | type I transforming growth factor beta receptor binding(GO:0034713) |
| 0.1 | 0.5 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.1 | 0.8 | GO:0043426 | MRF binding(GO:0043426) |
| 0.1 | 0.5 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.1 | 1.4 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.1 | 0.3 | GO:0004142 | diacylglycerol cholinephosphotransferase activity(GO:0004142) |
| 0.1 | 0.3 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.1 | 1.3 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.1 | 1.7 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.1 | 1.7 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.1 | 0.4 | GO:0097108 | hedgehog family protein binding(GO:0097108) |
| 0.1 | 0.7 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 0.1 | 1.0 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.1 | 0.4 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.1 | 1.3 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.1 | 0.2 | GO:0005010 | insulin-like growth factor-activated receptor activity(GO:0005010) |
| 0.1 | 0.3 | GO:0044378 | non-sequence-specific DNA binding, bending(GO:0044378) |
| 0.1 | 1.9 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.1 | 0.3 | GO:0003839 | gamma-glutamylcyclotransferase activity(GO:0003839) |
| 0.1 | 0.5 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.1 | 0.4 | GO:0016520 | growth hormone-releasing hormone receptor activity(GO:0016520) |
| 0.1 | 0.2 | GO:0004174 | electron-transferring-flavoprotein dehydrogenase activity(GO:0004174) oxidoreductase activity, acting on the CH-NH group of donors, quinone or similar compound as acceptor(GO:0016649) |
| 0.1 | 2.8 | GO:0031418 | L-ascorbic acid binding(GO:0031418) |
| 0.1 | 0.5 | GO:0004489 | methylenetetrahydrofolate reductase (NAD(P)H) activity(GO:0004489) |
| 0.1 | 0.8 | GO:0004844 | uracil DNA N-glycosylase activity(GO:0004844) deaminated base DNA N-glycosylase activity(GO:0097506) |
| 0.1 | 0.3 | GO:0004487 | methylenetetrahydrofolate dehydrogenase (NAD+) activity(GO:0004487) |
| 0.1 | 0.6 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.1 | 1.0 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.1 | 0.8 | GO:0004118 | cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) |
| 0.1 | 0.6 | GO:0019788 | NEDD8 transferase activity(GO:0019788) |
| 0.1 | 0.3 | GO:0004739 | pyruvate dehydrogenase (acetyl-transferring) activity(GO:0004739) |
| 0.1 | 0.8 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.1 | 0.3 | GO:0003943 | N-acetylgalactosamine-4-sulfatase activity(GO:0003943) |
| 0.1 | 0.3 | GO:0047192 | 1-alkylglycerophosphocholine O-acetyltransferase activity(GO:0047192) |
| 0.1 | 0.5 | GO:0004873 | asialoglycoprotein receptor activity(GO:0004873) |
| 0.1 | 1.8 | GO:0031432 | titin binding(GO:0031432) |
| 0.1 | 0.2 | GO:0055100 | adiponectin binding(GO:0055100) |
| 0.1 | 1.7 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.1 | 0.3 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.1 | 4.6 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.1 | 0.4 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.1 | 0.9 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.1 | 0.2 | GO:0002060 | purine nucleobase binding(GO:0002060) glycogen phosphorylase activity(GO:0008184) |
| 0.1 | 0.9 | GO:0032183 | SUMO binding(GO:0032183) |
| 0.1 | 0.7 | GO:0031386 | protein tag(GO:0031386) |
| 0.1 | 2.7 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.1 | 4.3 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.1 | 0.6 | GO:0001162 | RNA polymerase II intronic transcription regulatory region sequence-specific DNA binding(GO:0001162) |
| 0.1 | 0.9 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.1 | 0.3 | GO:0070735 | protein-glycine ligase activity(GO:0070735) |
| 0.1 | 1.6 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.1 | 1.5 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.1 | 1.6 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.1 | 1.8 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.1 | 0.4 | GO:1904929 | coreceptor activity involved in Wnt signaling pathway, planar cell polarity pathway(GO:1904929) |
| 0.1 | 1.6 | GO:0016628 | oxidoreductase activity, acting on the CH-CH group of donors, NAD or NADP as acceptor(GO:0016628) |
| 0.1 | 1.7 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.1 | 0.7 | GO:0015166 | polyol transmembrane transporter activity(GO:0015166) |
| 0.1 | 0.6 | GO:0038132 | neuregulin binding(GO:0038132) |
| 0.1 | 1.1 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.1 | 0.2 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.1 | 0.2 | GO:0102345 | 3-hydroxy-behenoyl-CoA dehydratase activity(GO:0102344) 3-hydroxy-lignoceroyl-CoA dehydratase activity(GO:0102345) |
| 0.1 | 0.4 | GO:0070728 | leucine binding(GO:0070728) |
| 0.1 | 0.3 | GO:0032143 | single thymine insertion binding(GO:0032143) |
| 0.1 | 2.0 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.1 | 0.7 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 0.6 | GO:0003996 | acyl-CoA ligase activity(GO:0003996) |
| 0.0 | 0.1 | GO:0052739 | phosphatidylserine 1-acylhydrolase activity(GO:0052739) |
| 0.0 | 0.6 | GO:0016755 | transferase activity, transferring amino-acyl groups(GO:0016755) |
| 0.0 | 0.3 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) |
| 0.0 | 1.5 | GO:0001530 | lipopolysaccharide binding(GO:0001530) |
| 0.0 | 0.2 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.0 | 0.5 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.0 | 0.6 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.0 | 0.4 | GO:0004996 | thyroid-stimulating hormone receptor activity(GO:0004996) |
| 0.0 | 1.2 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 1.4 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.2 | GO:0016402 | pristanoyl-CoA oxidase activity(GO:0016402) |
| 0.0 | 3.4 | GO:0051287 | NAD binding(GO:0051287) |
| 0.0 | 0.4 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.0 | 0.2 | GO:0015111 | iodide transmembrane transporter activity(GO:0015111) |
| 0.0 | 0.3 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.0 | 0.2 | GO:0016434 | rRNA (cytosine) methyltransferase activity(GO:0016434) |
| 0.0 | 1.3 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.0 | 0.1 | GO:0001537 | N-acetylgalactosamine 4-O-sulfotransferase activity(GO:0001537) |
| 0.0 | 0.1 | GO:0016784 | 3-mercaptopyruvate sulfurtransferase activity(GO:0016784) |
| 0.0 | 1.2 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.0 | 0.9 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 0.5 | GO:0050072 | m7G(5')pppN diphosphatase activity(GO:0050072) |
| 0.0 | 0.6 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.0 | 0.2 | GO:0086062 | voltage-gated sodium channel activity involved in Purkinje myocyte action potential(GO:0086062) |
| 0.0 | 0.3 | GO:0010385 | double-stranded methylated DNA binding(GO:0010385) |
| 0.0 | 0.7 | GO:0008391 | arachidonic acid monooxygenase activity(GO:0008391) arachidonic acid epoxygenase activity(GO:0008392) |
| 0.0 | 0.1 | GO:0070890 | L-ascorbate:sodium symporter activity(GO:0008520) sodium-dependent multivitamin transmembrane transporter activity(GO:0008523) L-ascorbic acid transporter activity(GO:0015229) sodium-dependent L-ascorbate transmembrane transporter activity(GO:0070890) |
| 0.0 | 0.3 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 0.0 | 0.7 | GO:0005542 | folic acid binding(GO:0005542) |
| 0.0 | 0.1 | GO:0016715 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced ascorbate as one donor, and incorporation of one atom of oxygen(GO:0016715) |
| 0.0 | 0.6 | GO:0055103 | ligase regulator activity(GO:0055103) ubiquitin-protein transferase regulator activity(GO:0055106) |
| 0.0 | 0.7 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.0 | 0.7 | GO:0050750 | low-density lipoprotein particle receptor binding(GO:0050750) |
| 0.0 | 0.1 | GO:0005471 | ATP:ADP antiporter activity(GO:0005471) adenine transmembrane transporter activity(GO:0015207) |
| 0.0 | 0.2 | GO:0030169 | low-density lipoprotein particle binding(GO:0030169) |
| 0.0 | 0.6 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.0 | 0.4 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.0 | 0.4 | GO:0003678 | DNA helicase activity(GO:0003678) |
| 0.0 | 0.6 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.0 | 1.4 | GO:0043425 | bHLH transcription factor binding(GO:0043425) |
| 0.0 | 0.3 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.0 | 0.5 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.0 | 0.3 | GO:0036310 | annealing helicase activity(GO:0036310) |
| 0.0 | 0.7 | GO:0000175 | 3'-5'-exoribonuclease activity(GO:0000175) |
| 0.0 | 0.9 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.0 | 0.2 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.0 | 0.1 | GO:1904455 | ubiquitin-specific protease activity involved in negative regulation of ERAD pathway(GO:1904455) |
| 0.0 | 0.2 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.0 | 2.9 | GO:0008094 | DNA-dependent ATPase activity(GO:0008094) |
| 0.0 | 0.1 | GO:0005046 | KDEL sequence binding(GO:0005046) |
| 0.0 | 0.2 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.0 | 0.3 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.0 | 0.1 | GO:0003985 | acetyl-CoA C-acetyltransferase activity(GO:0003985) |
| 0.0 | 0.1 | GO:0015275 | stretch-activated, cation-selective, calcium channel activity(GO:0015275) |
| 0.0 | 0.4 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.0 | 0.0 | GO:0004915 | interleukin-6 receptor activity(GO:0004915) interleukin-6 binding(GO:0019981) |
| 0.0 | 2.5 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.0 | 0.2 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.0 | 0.2 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.9 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.2 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.0 | 5.2 | GO:0004721 | phosphoprotein phosphatase activity(GO:0004721) |
| 0.0 | 0.1 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.0 | 0.2 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.1 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.0 | 0.7 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.0 | 0.5 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 0.1 | GO:0015140 | malate transmembrane transporter activity(GO:0015140) |
| 0.0 | 5.4 | GO:0008022 | protein C-terminus binding(GO:0008022) |
| 0.0 | 0.2 | GO:0015926 | glucosidase activity(GO:0015926) |
| 0.0 | 0.1 | GO:0044736 | acid-sensing ion channel activity(GO:0044736) |
| 0.0 | 0.2 | GO:0072542 | protein phosphatase activator activity(GO:0072542) |
| 0.0 | 0.1 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.0 | 0.1 | GO:0071987 | WD40-repeat domain binding(GO:0071987) |
| 0.0 | 0.1 | GO:0004983 | neuropeptide Y receptor activity(GO:0004983) |
| 0.0 | 0.4 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.7 | GO:0042165 | neurotransmitter binding(GO:0042165) |
| 0.0 | 0.6 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.0 | 0.3 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.0 | 0.1 | GO:0015440 | peptide-transporting ATPase activity(GO:0015440) |
| 0.0 | 3.1 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 0.3 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 0.8 | GO:0008146 | sulfotransferase activity(GO:0008146) |
| 0.0 | 0.4 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 0.2 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 0.9 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 0.5 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.0 | 0.0 | GO:0004051 | arachidonate 5-lipoxygenase activity(GO:0004051) |
| 0.0 | 0.4 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.0 | 0.3 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 0.1 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.0 | 1.0 | GO:0019212 | phosphatase inhibitor activity(GO:0019212) |
| 0.0 | 0.1 | GO:0047066 | phospholipid-hydroperoxide glutathione peroxidase activity(GO:0047066) |
| 0.0 | 0.3 | GO:0004177 | aminopeptidase activity(GO:0004177) |
| 0.0 | 0.1 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.0 | 0.2 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.0 | 1.3 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 0.1 | GO:0008453 | alanine-glyoxylate transaminase activity(GO:0008453) |
| 0.0 | 0.0 | GO:0031177 | phosphopantetheine binding(GO:0031177) |
| 0.0 | 0.3 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.2 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.0 | 0.1 | GO:0004532 | exoribonuclease activity(GO:0004532) |
| 0.0 | 3.3 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 0.4 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.0 | 5.5 | GO:0001077 | transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) |
| 0.0 | 0.1 | GO:0022821 | calcium, potassium:sodium antiporter activity(GO:0008273) potassium ion antiporter activity(GO:0022821) |
| 0.0 | 0.2 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.0 | 0.3 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.0 | 0.4 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.1 | GO:0034618 | arginine binding(GO:0034618) |
| 0.0 | 0.7 | GO:0003899 | DNA-directed RNA polymerase activity(GO:0003899) |
| 0.0 | 0.1 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.0 | 0.4 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 3.8 | GO:0031625 | ubiquitin protein ligase binding(GO:0031625) |
| 0.0 | 0.1 | GO:0098639 | C-X3-C chemokine binding(GO:0019960) collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.0 | 1.5 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 0.4 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 4.5 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.2 | 1.0 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.1 | 0.3 | ST T CELL SIGNAL TRANSDUCTION | T Cell Signal Transduction |
| 0.1 | 1.4 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.1 | 4.0 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.1 | 1.9 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.1 | 8.4 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.1 | 7.3 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.1 | 3.1 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.1 | 0.9 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.1 | 3.7 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.1 | 0.4 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.1 | 4.0 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.1 | 0.8 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.0 | 0.6 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 0.3 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.0 | 0.2 | PID AVB3 INTEGRIN PATHWAY | Integrins in angiogenesis |
| 0.0 | 1.7 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.0 | 0.7 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 0.7 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.0 | 8.1 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.4 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 0.3 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 1.2 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
| 0.0 | 1.8 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.0 | 0.3 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 0.6 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.0 | 0.4 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.0 | 0.8 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 0.9 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 2.0 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 2.7 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 1.0 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 0.9 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.0 | 0.1 | PID THROMBIN PAR1 PATHWAY | PAR1-mediated thrombin signaling events |
| 0.0 | 1.9 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 0.1 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 0.6 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 0.8 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.3 | PID VEGFR1 PATHWAY | VEGFR1 specific signals |
| 0.0 | 0.2 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.0 | 0.3 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 0.4 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.5 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.1 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.0 | 0.8 | PID INTEGRIN1 PATHWAY | Beta1 integrin cell surface interactions |
| 0.0 | 0.4 | SIG BCR SIGNALING PATHWAY | Members of the BCR signaling pathway |
| 0.0 | 0.1 | PID TXA2PATHWAY | Thromboxane A2 receptor signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 4.6 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.2 | 6.3 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.1 | 3.0 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.1 | 7.3 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.1 | 2.5 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.1 | 0.8 | REACTOME SHC MEDIATED SIGNALLING | Genes involved in SHC-mediated signalling |
| 0.1 | 5.6 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.1 | 3.1 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.1 | 4.3 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.1 | 1.2 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.1 | 2.6 | REACTOME GROWTH HORMONE RECEPTOR SIGNALING | Genes involved in Growth hormone receptor signaling |
| 0.1 | 1.8 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.1 | 1.6 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.1 | 1.2 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.1 | 0.9 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.1 | 1.7 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.1 | 1.0 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.1 | 2.8 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.1 | 2.5 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.1 | 1.4 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.1 | 0.4 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.1 | 0.6 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.1 | 1.9 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.1 | 0.1 | REACTOME SIGNALING BY GPCR | Genes involved in Signaling by GPCR |
| 0.1 | 0.2 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.1 | 1.1 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.1 | 1.8 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 1.5 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF CAMKII | Genes involved in CREB phosphorylation through the activation of CaMKII |
| 0.0 | 0.8 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 1.4 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 1.2 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.0 | 1.5 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 0.3 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.0 | 4.0 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 1.3 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 1.0 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.6 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 0.6 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.0 | 0.6 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.0 | 0.5 | REACTOME NFKB ACTIVATION THROUGH FADD RIP1 PATHWAY MEDIATED BY CASPASE 8 AND10 | Genes involved in NF-kB activation through FADD/RIP-1 pathway mediated by caspase-8 and -10 |
| 0.0 | 1.3 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.0 | 1.0 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 0.8 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 3.6 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 0.9 | REACTOME ADP SIGNALLING THROUGH P2RY1 | Genes involved in ADP signalling through P2Y purinoceptor 1 |
| 0.0 | 2.9 | REACTOME PHASE II CONJUGATION | Genes involved in Phase II conjugation |
| 0.0 | 0.6 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 0.3 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.0 | 0.6 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 1.0 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.0 | 0.6 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 0.9 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 1.3 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.1 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.0 | 0.5 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 1.1 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.4 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 1.0 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 0.3 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 0.3 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.0 | 0.6 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 0.5 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 0.3 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.2 | REACTOME E2F ENABLED INHIBITION OF PRE REPLICATION COMPLEX FORMATION | Genes involved in E2F-enabled inhibition of pre-replication complex formation |
| 0.0 | 0.9 | REACTOME ION CHANNEL TRANSPORT | Genes involved in Ion channel transport |
| 0.0 | 0.4 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.0 | 0.6 | REACTOME ACTIVATION OF GENES BY ATF4 | Genes involved in Activation of Genes by ATF4 |
| 0.0 | 1.6 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 0.4 | REACTOME NUCLEOTIDE LIKE PURINERGIC RECEPTORS | Genes involved in Nucleotide-like (purinergic) receptors |
| 0.0 | 0.4 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 0.6 | REACTOME REGULATION OF INSULIN SECRETION BY GLUCAGON LIKE PEPTIDE1 | Genes involved in Regulation of Insulin Secretion by Glucagon-like Peptide-1 |
| 0.0 | 0.3 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 0.0 | 0.4 | REACTOME SIGNALING BY FGFR MUTANTS | Genes involved in Signaling by FGFR mutants |
| 0.0 | 0.1 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 0.3 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.0 | 0.2 | REACTOME REGULATION OF WATER BALANCE BY RENAL AQUAPORINS | Genes involved in Regulation of Water Balance by Renal Aquaporins |
| 0.0 | 0.7 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.0 | 0.2 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.7 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.6 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |