Project
Inflammatory response time course, HUVEC (Wada, 2009)
Navigation
Downloads
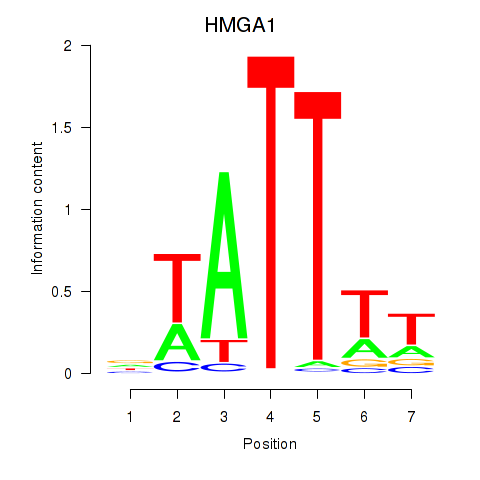
Results for HMGA1
Z-value: 1.58
Transcription factors associated with HMGA1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HMGA1
|
ENSG00000137309.20 | HMGA1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HMGA1 | hg38_v1_chr6_+_34236865_34236887 | -0.52 | 7.9e-03 | Click! |
Activity profile of HMGA1 motif
Sorted Z-values of HMGA1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of HMGA1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr9_-_41908681 | 16.74 |
ENST00000476961.5
|
CNTNAP3B
|
contactin associated protein family member 3B |
| chr4_+_41538143 | 12.38 |
ENST00000503057.6
ENST00000511496.5 |
LIMCH1
|
LIM and calponin homology domains 1 |
| chr3_-_18438767 | 8.92 |
ENST00000454909.6
|
SATB1
|
SATB homeobox 1 |
| chr4_-_89835617 | 7.65 |
ENST00000611107.1
ENST00000345009.8 ENST00000505199.5 ENST00000502987.5 |
SNCA
|
synuclein alpha |
| chr9_-_39288138 | 6.61 |
ENST00000297668.10
|
CNTNAP3
|
contactin associated protein family member 3 |
| chr4_-_185775271 | 6.07 |
ENST00000430503.5
ENST00000319454.10 ENST00000450341.5 |
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr13_+_31945826 | 5.88 |
ENST00000647500.1
|
FRY
|
FRY microtubule binding protein |
| chr2_-_187448244 | 5.73 |
ENST00000392370.8
ENST00000410068.5 ENST00000447403.5 ENST00000410102.5 |
CALCRL
|
calcitonin receptor like receptor |
| chr13_+_102656933 | 5.53 |
ENST00000650757.1
|
TPP2
|
tripeptidyl peptidase 2 |
| chr3_+_194136138 | 5.14 |
ENST00000232424.4
|
HES1
|
hes family bHLH transcription factor 1 |
| chr9_+_2159672 | 5.12 |
ENST00000634343.1
|
SMARCA2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr10_-_114684612 | 4.74 |
ENST00000533213.6
ENST00000369252.8 |
ABLIM1
|
actin binding LIM protein 1 |
| chr4_-_137532452 | 4.53 |
ENST00000412923.6
ENST00000511115.5 ENST00000344876.9 ENST00000507846.5 ENST00000510305.5 ENST00000611581.1 |
PCDH18
|
protocadherin 18 |
| chr1_+_84164962 | 4.46 |
ENST00000614872.4
ENST00000394839.6 |
PRKACB
|
protein kinase cAMP-activated catalytic subunit beta |
| chr3_-_18424533 | 4.38 |
ENST00000417717.6
|
SATB1
|
SATB homeobox 1 |
| chr18_+_75210755 | 4.37 |
ENST00000322038.5
|
TSHZ1
|
teashirt zinc finger homeobox 1 |
| chr2_+_238138661 | 4.33 |
ENST00000409223.2
|
KLHL30
|
kelch like family member 30 |
| chr1_+_84181630 | 4.30 |
ENST00000610457.1
|
PRKACB
|
protein kinase cAMP-activated catalytic subunit beta |
| chr5_+_62578810 | 4.28 |
ENST00000334994.6
ENST00000409534.1 |
LRRC70
IPO11
|
leucine rich repeat containing 70 importin 11 |
| chr10_-_114684457 | 4.14 |
ENST00000392955.7
|
ABLIM1
|
actin binding LIM protein 1 |
| chrX_-_19965142 | 4.11 |
ENST00000340625.3
|
BCLAF3
|
BCLAF1 and THRAP3 family member 3 |
| chr10_+_92834594 | 4.10 |
ENST00000371552.8
|
EXOC6
|
exocyst complex component 6 |
| chr4_-_185775890 | 3.98 |
ENST00000437304.6
|
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr4_-_185956652 | 3.92 |
ENST00000355634.9
|
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr7_-_120858066 | 3.92 |
ENST00000222747.8
|
TSPAN12
|
tetraspanin 12 |
| chr10_+_92831153 | 3.85 |
ENST00000672817.1
|
EXOC6
|
exocyst complex component 6 |
| chr1_+_60865259 | 3.65 |
ENST00000371191.5
|
NFIA
|
nuclear factor I A |
| chr8_-_3409528 | 3.60 |
ENST00000335551.11
|
CSMD1
|
CUB and Sushi multiple domains 1 |
| chr2_+_178320228 | 3.54 |
ENST00000315022.2
|
OSBPL6
|
oxysterol binding protein like 6 |
| chr18_+_75210789 | 3.37 |
ENST00000580243.3
|
TSHZ1
|
teashirt zinc finger homeobox 1 |
| chr10_+_35167516 | 3.35 |
ENST00000361599.8
|
CREM
|
cAMP responsive element modulator |
| chr9_-_42129125 | 3.31 |
ENST00000617422.4
ENST00000612828.4 ENST00000341990.8 ENST00000377561.7 ENST00000276974.7 |
CNTNAP3B
|
contactin associated protein family member 3B |
| chr13_+_32031706 | 3.29 |
ENST00000542859.6
|
FRY
|
FRY microtubule binding protein |
| chr1_+_77918128 | 3.28 |
ENST00000342754.5
|
NEXN
|
nexilin F-actin binding protein |
| chr12_+_8992029 | 3.17 |
ENST00000543895.1
|
KLRG1
|
killer cell lectin like receptor G1 |
| chr17_-_69141878 | 3.13 |
ENST00000590645.1
ENST00000284425.7 |
ABCA6
|
ATP binding cassette subfamily A member 6 |
| chr9_-_14307928 | 3.11 |
ENST00000637640.1
ENST00000493697.1 ENST00000636057.1 |
NFIB
|
nuclear factor I B |
| chr4_-_158159657 | 3.08 |
ENST00000590648.5
|
GASK1B
|
golgi associated kinase 1B |
| chr6_+_36029082 | 2.97 |
ENST00000472333.1
|
MAPK14
|
mitogen-activated protein kinase 14 |
| chr5_-_88731827 | 2.94 |
ENST00000627170.2
|
MEF2C
|
myocyte enhancer factor 2C |
| chr13_-_48413105 | 2.91 |
ENST00000620633.5
|
LPAR6
|
lysophosphatidic acid receptor 6 |
| chr2_-_98663464 | 2.87 |
ENST00000414521.6
|
MGAT4A
|
alpha-1,3-mannosyl-glycoprotein 4-beta-N-acetylglucosaminyltransferase A |
| chr7_+_94394886 | 2.86 |
ENST00000297268.11
ENST00000620463.1 |
COL1A2
|
collagen type I alpha 2 chain |
| chr1_-_153094521 | 2.75 |
ENST00000368750.8
|
SPRR2E
|
small proline rich protein 2E |
| chr12_+_78864768 | 2.75 |
ENST00000261205.9
ENST00000457153.6 |
SYT1
|
synaptotagmin 1 |
| chr9_+_87497222 | 2.60 |
ENST00000358077.9
|
DAPK1
|
death associated protein kinase 1 |
| chr4_+_133149278 | 2.59 |
ENST00000264360.7
|
PCDH10
|
protocadherin 10 |
| chr8_+_27633863 | 2.59 |
ENST00000337221.8
|
SCARA3
|
scavenger receptor class A member 3 |
| chr22_-_28711931 | 2.56 |
ENST00000434810.5
ENST00000456369.5 |
CHEK2
|
checkpoint kinase 2 |
| chr1_+_78004930 | 2.55 |
ENST00000370763.6
|
DNAJB4
|
DnaJ heat shock protein family (Hsp40) member B4 |
| chr7_-_120858303 | 2.55 |
ENST00000415871.5
ENST00000430985.1 |
TSPAN12
|
tetraspanin 12 |
| chr3_-_15798184 | 2.51 |
ENST00000624145.3
|
ANKRD28
|
ankyrin repeat domain 28 |
| chr12_+_12785652 | 2.48 |
ENST00000356591.5
|
APOLD1
|
apolipoprotein L domain containing 1 |
| chr6_-_75363003 | 2.44 |
ENST00000370020.1
|
FILIP1
|
filamin A interacting protein 1 |
| chr9_-_72953047 | 2.44 |
ENST00000297785.8
ENST00000376939.5 |
ALDH1A1
|
aldehyde dehydrogenase 1 family member A1 |
| chrX_-_117973579 | 2.42 |
ENST00000371878.5
|
KLHL13
|
kelch like family member 13 |
| chr4_+_143381939 | 2.39 |
ENST00000505913.5
|
GAB1
|
GRB2 associated binding protein 1 |
| chr5_-_88785493 | 2.38 |
ENST00000503554.4
|
MEF2C
|
myocyte enhancer factor 2C |
| chr12_+_32502114 | 2.37 |
ENST00000682739.1
ENST00000427716.7 ENST00000583694.2 |
FGD4
|
FYVE, RhoGEF and PH domain containing 4 |
| chr5_+_69415065 | 2.32 |
ENST00000647531.1
ENST00000645446.1 ENST00000325631.10 ENST00000454295.6 |
MARVELD2
|
MARVEL domain containing 2 |
| chr2_+_42494547 | 2.31 |
ENST00000405592.5
|
MTA3
|
metastasis associated 1 family member 3 |
| chr12_+_19205294 | 2.30 |
ENST00000424268.5
|
PLEKHA5
|
pleckstrin homology domain containing A5 |
| chr8_-_17676484 | 2.24 |
ENST00000634613.1
ENST00000519066.5 |
MTUS1
|
microtubule associated scaffold protein 1 |
| chr7_-_122098831 | 2.22 |
ENST00000681213.1
ENST00000679419.1 |
AASS
|
aminoadipate-semialdehyde synthase |
| chr9_+_2159850 | 2.21 |
ENST00000416751.2
|
SMARCA2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr2_+_162318884 | 2.19 |
ENST00000446271.5
ENST00000429691.6 |
GCA
|
grancalcin |
| chr13_-_44437214 | 2.17 |
ENST00000622051.1
|
TSC22D1
|
TSC22 domain family member 1 |
| chr2_-_69643152 | 2.15 |
ENST00000606389.7
|
AAK1
|
AP2 associated kinase 1 |
| chr7_+_150567347 | 2.13 |
ENST00000461940.5
|
GIMAP4
|
GTPase, IMAP family member 4 |
| chr2_+_190927649 | 2.11 |
ENST00000409428.5
ENST00000409215.5 |
GLS
|
glutaminase |
| chr8_-_17697654 | 2.10 |
ENST00000297488.10
|
MTUS1
|
microtubule associated scaffold protein 1 |
| chrX_-_117973717 | 2.08 |
ENST00000262820.7
|
KLHL13
|
kelch like family member 13 |
| chr4_-_69860138 | 2.08 |
ENST00000226444.4
|
SULT1E1
|
sulfotransferase family 1E member 1 |
| chr10_+_35195124 | 2.05 |
ENST00000487763.5
ENST00000473940.5 ENST00000488328.5 ENST00000356917.9 |
CREM
|
cAMP responsive element modulator |
| chr3_-_185821092 | 2.05 |
ENST00000421047.3
|
IGF2BP2
|
insulin like growth factor 2 mRNA binding protein 2 |
| chr7_+_150567382 | 2.03 |
ENST00000255945.4
ENST00000479232.1 |
GIMAP4
|
GTPase, IMAP family member 4 |
| chr17_-_445939 | 2.02 |
ENST00000329099.4
|
RFLNB
|
refilin B |
| chrX_-_10677720 | 2.01 |
ENST00000453318.6
|
MID1
|
midline 1 |
| chr7_+_93921720 | 2.00 |
ENST00000248564.6
|
GNG11
|
G protein subunit gamma 11 |
| chr14_-_91946989 | 2.00 |
ENST00000556154.5
|
FBLN5
|
fibulin 5 |
| chr4_+_122339221 | 2.00 |
ENST00000442707.1
|
KIAA1109
|
KIAA1109 |
| chr2_+_66435116 | 1.98 |
ENST00000272369.14
ENST00000560281.6 |
MEIS1
|
Meis homeobox 1 |
| chr16_-_3372666 | 1.98 |
ENST00000399974.5
|
MTRNR2L4
|
MT-RNR2 like 4 |
| chr3_+_132597260 | 1.91 |
ENST00000249887.3
|
ACKR4
|
atypical chemokine receptor 4 |
| chr20_-_61998132 | 1.90 |
ENST00000474089.5
|
TAF4
|
TATA-box binding protein associated factor 4 |
| chr7_+_92057602 | 1.89 |
ENST00000491695.2
|
AKAP9
|
A-kinase anchoring protein 9 |
| chr9_-_123184233 | 1.89 |
ENST00000447404.6
|
STRBP
|
spermatid perinuclear RNA binding protein |
| chr15_+_57219411 | 1.88 |
ENST00000543579.5
ENST00000537840.5 ENST00000343827.7 |
TCF12
|
transcription factor 12 |
| chr8_+_73991345 | 1.88 |
ENST00000284818.7
ENST00000518893.1 |
LY96
|
lymphocyte antigen 96 |
| chr2_-_221572272 | 1.85 |
ENST00000409854.5
ENST00000443796.5 ENST00000281821.7 |
EPHA4
|
EPH receptor A4 |
| chr1_+_65264694 | 1.84 |
ENST00000263441.11
ENST00000395325.7 |
DNAJC6
|
DnaJ heat shock protein family (Hsp40) member C6 |
| chr11_-_66347560 | 1.82 |
ENST00000311181.5
|
B4GAT1
|
beta-1,4-glucuronyltransferase 1 |
| chr2_-_69643615 | 1.81 |
ENST00000409068.5
|
AAK1
|
AP2 associated kinase 1 |
| chr4_-_109801978 | 1.81 |
ENST00000510800.1
ENST00000512148.5 ENST00000394634.7 ENST00000394635.8 ENST00000645635.1 |
CFI
ENSG00000285330.1
|
complement factor I novel protein |
| chr18_-_55510753 | 1.80 |
ENST00000543082.5
|
TCF4
|
transcription factor 4 |
| chr6_-_52994248 | 1.80 |
ENST00000457564.1
ENST00000370960.5 |
GSTA4
|
glutathione S-transferase alpha 4 |
| chr2_+_108588453 | 1.79 |
ENST00000393310.5
|
LIMS1
|
LIM zinc finger domain containing 1 |
| chr8_+_95024977 | 1.77 |
ENST00000396124.9
|
NDUFAF6
|
NADH:ubiquinone oxidoreductase complex assembly factor 6 |
| chr13_-_67230377 | 1.75 |
ENST00000544246.5
ENST00000377861.4 |
PCDH9
|
protocadherin 9 |
| chr6_+_30888672 | 1.75 |
ENST00000446312.5
|
DDR1
|
discoidin domain receptor tyrosine kinase 1 |
| chr2_+_108588286 | 1.74 |
ENST00000332345.10
|
LIMS1
|
LIM zinc finger domain containing 1 |
| chr14_-_20802402 | 1.71 |
ENST00000412779.2
|
RNASE1
|
ribonuclease A family member 1, pancreatic |
| chr5_-_88877967 | 1.68 |
ENST00000508610.5
ENST00000636294.1 |
MEF2C
|
myocyte enhancer factor 2C |
| chr13_-_44474296 | 1.67 |
ENST00000611198.4
|
TSC22D1
|
TSC22 domain family member 1 |
| chr2_-_182242031 | 1.66 |
ENST00000358139.6
|
PDE1A
|
phosphodiesterase 1A |
| chr18_-_55588535 | 1.65 |
ENST00000566286.5
ENST00000566279.5 ENST00000626595.2 ENST00000564999.5 ENST00000616053.4 ENST00000356073.8 |
TCF4
|
transcription factor 4 |
| chr7_+_116222804 | 1.64 |
ENST00000393481.6
|
TES
|
testin LIM domain protein |
| chrX_-_77634229 | 1.63 |
ENST00000675732.1
|
ATRX
|
ATRX chromatin remodeler |
| chr22_-_28306645 | 1.63 |
ENST00000612946.4
|
TTC28
|
tetratricopeptide repeat domain 28 |
| chr12_+_20810698 | 1.63 |
ENST00000540853.5
ENST00000381545.8 |
SLCO1B3
|
solute carrier organic anion transporter family member 1B3 |
| chr15_-_99249523 | 1.62 |
ENST00000560235.1
ENST00000394132.7 ENST00000560860.5 ENST00000558078.5 ENST00000560772.5 |
TTC23
|
tetratricopeptide repeat domain 23 |
| chr2_+_1414382 | 1.62 |
ENST00000423320.5
ENST00000346956.7 ENST00000382198.5 |
TPO
|
thyroid peroxidase |
| chrX_-_20218941 | 1.62 |
ENST00000457145.6
|
RPS6KA3
|
ribosomal protein S6 kinase A3 |
| chr1_+_179882040 | 1.61 |
ENST00000528443.6
|
TOR1AIP1
|
torsin 1A interacting protein 1 |
| chr22_-_28712136 | 1.60 |
ENST00000464581.6
|
CHEK2
|
checkpoint kinase 2 |
| chr12_+_59596010 | 1.56 |
ENST00000547379.6
ENST00000552432.5 |
SLC16A7
|
solute carrier family 16 member 7 |
| chrX_-_10576901 | 1.55 |
ENST00000380779.5
|
MID1
|
midline 1 |
| chr1_+_196652022 | 1.54 |
ENST00000367429.9
ENST00000630130.2 ENST00000359637.2 |
CFH
|
complement factor H |
| chr5_+_141192330 | 1.53 |
ENST00000239446.6
|
PCDHB10
|
protocadherin beta 10 |
| chr5_+_141223332 | 1.52 |
ENST00000239449.7
ENST00000624896.1 ENST00000624396.1 |
PCDHB14
ENSG00000279983.1
|
protocadherin beta 14 novel protein |
| chr15_+_47717344 | 1.49 |
ENST00000558816.5
ENST00000536845.7 |
SEMA6D
|
semaphorin 6D |
| chr8_+_27633884 | 1.49 |
ENST00000301904.4
|
SCARA3
|
scavenger receptor class A member 3 |
| chr17_+_7857987 | 1.48 |
ENST00000332439.5
|
CYB5D1
|
cytochrome b5 domain containing 1 |
| chr12_+_101594849 | 1.47 |
ENST00000547405.5
ENST00000452455.6 ENST00000392934.7 ENST00000547509.5 ENST00000361685.6 ENST00000549145.5 ENST00000361466.7 ENST00000553190.5 ENST00000545503.6 ENST00000536007.5 ENST00000541119.5 ENST00000551300.5 ENST00000550270.1 |
MYBPC1
|
myosin binding protein C1 |
| chr17_-_65826445 | 1.47 |
ENST00000317442.12
|
CEP112
|
centrosomal protein 112 |
| chr4_-_88823306 | 1.46 |
ENST00000395002.6
|
FAM13A
|
family with sequence similarity 13 member A |
| chr12_-_46372763 | 1.44 |
ENST00000256689.10
|
SLC38A2
|
solute carrier family 38 member 2 |
| chr4_-_151226427 | 1.43 |
ENST00000304527.8
ENST00000409598.8 |
SH3D19
|
SH3 domain containing 19 |
| chr13_-_44436801 | 1.43 |
ENST00000261489.6
|
TSC22D1
|
TSC22 domain family member 1 |
| chr7_-_92134737 | 1.40 |
ENST00000450723.5
|
CYP51A1
|
cytochrome P450 family 51 subfamily A member 1 |
| chr18_-_36829154 | 1.40 |
ENST00000614939.4
ENST00000610723.4 |
TPGS2
|
tubulin polyglutamylase complex subunit 2 |
| chr4_+_89894846 | 1.39 |
ENST00000264790.7
|
MMRN1
|
multimerin 1 |
| chr3_+_124584625 | 1.37 |
ENST00000291478.9
ENST00000682363.1 ENST00000454902.1 |
KALRN
|
kalirin RhoGEF kinase |
| chr12_+_59689337 | 1.33 |
ENST00000261187.8
|
SLC16A7
|
solute carrier family 16 member 7 |
| chr5_-_88823763 | 1.32 |
ENST00000635898.1
ENST00000626391.2 ENST00000628656.2 |
MEF2C
|
myocyte enhancer factor 2C |
| chr2_+_188974364 | 1.31 |
ENST00000304636.9
ENST00000317840.9 |
COL3A1
|
collagen type III alpha 1 chain |
| chr6_-_132798587 | 1.31 |
ENST00000275227.9
|
SLC18B1
|
solute carrier family 18 member B1 |
| chr12_+_56267674 | 1.31 |
ENST00000546544.5
ENST00000553234.1 |
COQ10A
|
coenzyme Q10A |
| chr1_+_179882275 | 1.30 |
ENST00000606911.7
ENST00000271583.7 |
TOR1AIP1
|
torsin 1A interacting protein 1 |
| chr9_+_73151833 | 1.30 |
ENST00000456643.5
ENST00000257497.11 ENST00000415424.5 |
ANXA1
|
annexin A1 |
| chr3_-_197573323 | 1.30 |
ENST00000358186.6
ENST00000431056.5 |
BDH1
|
3-hydroxybutyrate dehydrogenase 1 |
| chr9_-_21202205 | 1.29 |
ENST00000239347.3
|
IFNA7
|
interferon alpha 7 |
| chr5_-_111758061 | 1.27 |
ENST00000509979.5
ENST00000513100.5 ENST00000508161.5 ENST00000455559.6 |
NREP
|
neuronal regeneration related protein |
| chr14_-_89619118 | 1.26 |
ENST00000345097.8
ENST00000555855.5 ENST00000555353.5 |
FOXN3
|
forkhead box N3 |
| chr18_-_55321640 | 1.25 |
ENST00000637169.2
|
TCF4
|
transcription factor 4 |
| chr5_+_140834230 | 1.25 |
ENST00000356878.5
ENST00000525929.2 |
PCDHA7
|
protocadherin alpha 7 |
| chr1_-_179143044 | 1.24 |
ENST00000504405.5
ENST00000512653.5 ENST00000344730.7 |
ABL2
|
ABL proto-oncogene 2, non-receptor tyrosine kinase |
| chr8_+_11809135 | 1.24 |
ENST00000528643.5
ENST00000525777.5 |
FDFT1
|
farnesyl-diphosphate farnesyltransferase 1 |
| chr14_-_75069478 | 1.24 |
ENST00000555463.1
|
ACYP1
|
acylphosphatase 1 |
| chr13_+_75804221 | 1.22 |
ENST00000489941.6
ENST00000525373.5 |
LMO7
|
LIM domain 7 |
| chr12_-_12338674 | 1.21 |
ENST00000545735.1
|
MANSC1
|
MANSC domain containing 1 |
| chr1_+_207770322 | 1.21 |
ENST00000462968.2
|
CD46
|
CD46 molecule |
| chr7_+_107583919 | 1.21 |
ENST00000491150.5
|
BCAP29
|
B cell receptor associated protein 29 |
| chr11_+_9575496 | 1.21 |
ENST00000681684.1
|
WEE1
|
WEE1 G2 checkpoint kinase |
| chr12_-_70754631 | 1.21 |
ENST00000440835.6
ENST00000549308.5 ENST00000550661.1 ENST00000378778.5 |
PTPRR
|
protein tyrosine phosphatase receptor type R |
| chr1_+_51617079 | 1.20 |
ENST00000447887.5
ENST00000428468.6 ENST00000453295.5 |
OSBPL9
|
oxysterol binding protein like 9 |
| chr6_-_116254063 | 1.18 |
ENST00000420283.3
|
TSPYL4
|
TSPY like 4 |
| chr2_+_172735912 | 1.18 |
ENST00000409036.5
|
RAPGEF4
|
Rap guanine nucleotide exchange factor 4 |
| chr4_+_89901979 | 1.18 |
ENST00000508372.1
|
MMRN1
|
multimerin 1 |
| chr17_-_69060906 | 1.17 |
ENST00000495634.5
ENST00000453985.6 ENST00000340001.9 ENST00000585714.1 |
ABCA9
|
ATP binding cassette subfamily A member 9 |
| chr13_-_67230313 | 1.17 |
ENST00000377865.7
|
PCDH9
|
protocadherin 9 |
| chr7_+_117014881 | 1.14 |
ENST00000422922.5
ENST00000432298.5 |
ST7
|
suppression of tumorigenicity 7 |
| chr10_-_125158704 | 1.14 |
ENST00000531469.5
|
CTBP2
|
C-terminal binding protein 2 |
| chr20_+_11917859 | 1.13 |
ENST00000618296.4
ENST00000378226.7 |
BTBD3
|
BTB domain containing 3 |
| chr14_-_21537206 | 1.13 |
ENST00000614342.1
|
SALL2
|
spalt like transcription factor 2 |
| chr7_+_116862552 | 1.13 |
ENST00000361183.8
ENST00000639546.1 ENST00000490693.5 |
CAPZA2
|
capping actin protein of muscle Z-line subunit alpha 2 |
| chr14_-_20305932 | 1.11 |
ENST00000258821.8
ENST00000553828.1 |
TTC5
|
tetratricopeptide repeat domain 5 |
| chr4_-_25863537 | 1.11 |
ENST00000502949.5
ENST00000264868.9 ENST00000513691.1 ENST00000514872.1 |
SEL1L3
|
SEL1L family member 3 |
| chr14_-_34713759 | 1.11 |
ENST00000673315.1
|
CFL2
|
cofilin 2 |
| chr6_-_69699124 | 1.11 |
ENST00000651675.1
|
LMBRD1
|
LMBR1 domain containing 1 |
| chr22_+_40177917 | 1.10 |
ENST00000454349.7
ENST00000335727.13 |
TNRC6B
|
trinucleotide repeat containing adaptor 6B |
| chr3_+_148865288 | 1.10 |
ENST00000296046.4
|
CPA3
|
carboxypeptidase A3 |
| chr1_+_145964675 | 1.10 |
ENST00000369314.2
ENST00000369313.7 |
POLR3GL
|
RNA polymerase III subunit GL |
| chr5_+_119476530 | 1.09 |
ENST00000645099.1
ENST00000513628.5 |
HSD17B4
|
hydroxysteroid 17-beta dehydrogenase 4 |
| chr6_+_152697888 | 1.09 |
ENST00000367245.5
ENST00000529453.1 |
MYCT1
|
MYC target 1 |
| chrX_+_37780049 | 1.09 |
ENST00000378588.5
|
CYBB
|
cytochrome b-245 beta chain |
| chr11_-_104898670 | 1.08 |
ENST00000422698.6
|
CASP12
|
caspase 12 (gene/pseudogene) |
| chr10_+_119819244 | 1.08 |
ENST00000637174.1
|
INPP5F
|
inositol polyphosphate-5-phosphatase F |
| chr1_+_163068775 | 1.06 |
ENST00000421743.6
|
RGS4
|
regulator of G protein signaling 4 |
| chr6_+_152697934 | 1.05 |
ENST00000532295.1
|
MYCT1
|
MYC target 1 |
| chr9_-_110337808 | 1.05 |
ENST00000374510.8
ENST00000374507.4 ENST00000423740.7 ENST00000374511.7 |
TXNDC8
|
thioredoxin domain containing 8 |
| chr8_-_81483226 | 1.05 |
ENST00000256104.5
|
FABP4
|
fatty acid binding protein 4 |
| chr17_+_3475959 | 1.04 |
ENST00000263080.3
|
ASPA
|
aspartoacylase |
| chr9_+_72628020 | 1.03 |
ENST00000646619.1
|
TMC1
|
transmembrane channel like 1 |
| chr3_-_123980727 | 1.01 |
ENST00000620893.4
|
ROPN1
|
rhophilin associated tail protein 1 |
| chr7_+_832470 | 1.01 |
ENST00000401592.6
|
SUN1
|
Sad1 and UNC84 domain containing 1 |
| chr3_+_148791058 | 1.01 |
ENST00000491148.5
|
CPB1
|
carboxypeptidase B1 |
| chr12_+_12725897 | 1.00 |
ENST00000326765.10
|
APOLD1
|
apolipoprotein L domain containing 1 |
| chr3_-_149377637 | 1.00 |
ENST00000305366.8
|
TM4SF1
|
transmembrane 4 L six family member 1 |
| chrX_-_101617921 | 0.99 |
ENST00000361910.9
ENST00000538627.5 ENST00000539247.5 |
ARMCX6
|
armadillo repeat containing X-linked 6 |
| chr12_-_102197827 | 0.99 |
ENST00000329406.5
|
PMCH
|
pro-melanin concentrating hormone |
| chr10_-_45594760 | 0.98 |
ENST00000319836.7
|
MARCHF8
|
membrane associated ring-CH-type finger 8 |
| chr2_+_209580024 | 0.98 |
ENST00000392194.5
|
MAP2
|
microtubule associated protein 2 |
| chrX_-_81201886 | 0.98 |
ENST00000451455.1
ENST00000358130.7 ENST00000436386.5 |
HMGN5
|
high mobility group nucleosome binding domain 5 |
| chr11_+_58622659 | 0.98 |
ENST00000361987.6
|
CNTF
|
ciliary neurotrophic factor |
| chrX_+_10158448 | 0.97 |
ENST00000380829.5
ENST00000421085.7 ENST00000674669.1 ENST00000454850.1 |
CLCN4
|
chloride voltage-gated channel 4 |
| chr7_-_56051544 | 0.97 |
ENST00000395471.7
|
PSPH
|
phosphoserine phosphatase |
| chr2_+_172735838 | 0.97 |
ENST00000397081.8
|
RAPGEF4
|
Rap guanine nucleotide exchange factor 4 |
| chr3_-_69080350 | 0.97 |
ENST00000630585.1
ENST00000361055.9 ENST00000415609.6 ENST00000349511.8 |
UBA3
|
ubiquitin like modifier activating enzyme 3 |
| chr2_-_201739205 | 0.96 |
ENST00000681558.1
ENST00000681495.1 |
ALS2
|
alsin Rho guanine nucleotide exchange factor ALS2 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.3 | 9.2 | GO:1904428 | negative regulation of tubulin deacetylation(GO:1904428) |
| 1.7 | 5.1 | GO:0042668 | trochlear nerve development(GO:0021558) auditory receptor cell fate determination(GO:0042668) negative regulation of auditory receptor cell differentiation(GO:0045608) regulation of timing of neuron differentiation(GO:0060164) negative regulation of pro-B cell differentiation(GO:2000974) |
| 1.4 | 4.2 | GO:1903926 | signal transduction involved in intra-S DNA damage checkpoint(GO:0072428) response to bisphenol A(GO:1903925) cellular response to bisphenol A(GO:1903926) |
| 1.3 | 7.7 | GO:0060023 | soft palate development(GO:0060023) |
| 1.3 | 7.7 | GO:1903284 | negative regulation of dopamine uptake involved in synaptic transmission(GO:0051585) norepinephrine uptake(GO:0051620) regulation of norepinephrine uptake(GO:0051621) negative regulation of norepinephrine uptake(GO:0051622) negative regulation of catecholamine uptake involved in synaptic transmission(GO:0051945) regulation of glutathione peroxidase activity(GO:1903282) positive regulation of glutathione peroxidase activity(GO:1903284) positive regulation of hydrogen peroxide catabolic process(GO:1903285) positive regulation of peroxidase activity(GO:2000470) |
| 1.3 | 8.8 | GO:0097338 | response to clozapine(GO:0097338) |
| 1.0 | 8.3 | GO:0003138 | primary heart field specification(GO:0003138) sinoatrial valve development(GO:0003172) sinoatrial valve morphogenesis(GO:0003185) |
| 1.0 | 1.0 | GO:0046668 | regulation of retinal cell programmed cell death(GO:0046668) |
| 0.9 | 2.8 | GO:0098746 | fast, calcium ion-dependent exocytosis of neurotransmitter(GO:0098746) |
| 0.8 | 13.3 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.7 | 2.2 | GO:0019878 | lysine biosynthetic process(GO:0009085) lysine biosynthetic process via aminoadipic acid(GO:0019878) |
| 0.7 | 2.1 | GO:0006711 | estrogen catabolic process(GO:0006711) |
| 0.7 | 2.0 | GO:0061182 | negative regulation of chondrocyte development(GO:0061182) |
| 0.6 | 1.9 | GO:0097156 | fasciculation of motor neuron axon(GO:0097156) |
| 0.5 | 1.6 | GO:0034183 | negative regulation of maintenance of sister chromatid cohesion(GO:0034092) negative regulation of maintenance of mitotic sister chromatid cohesion(GO:0034183) maintenance of mitotic sister chromatid cohesion, telomeric(GO:0099403) mitotic sister chromatid cohesion, telomeric(GO:0099404) regulation of maintenance of mitotic sister chromatid cohesion, telomeric(GO:1904907) negative regulation of maintenance of mitotic sister chromatid cohesion, telomeric(GO:1904908) |
| 0.5 | 3.0 | GO:0014835 | myoblast differentiation involved in skeletal muscle regeneration(GO:0014835) positive regulation of interleukin-12 secretion(GO:2001184) |
| 0.4 | 5.7 | GO:0009744 | response to sucrose(GO:0009744) response to disaccharide(GO:0034285) |
| 0.4 | 1.3 | GO:0036292 | DNA rewinding(GO:0036292) |
| 0.4 | 2.4 | GO:0061624 | fructose catabolic process(GO:0006001) fructose catabolic process to hydroxyacetone phosphate and glyceraldehyde-3-phosphate(GO:0061624) |
| 0.4 | 1.1 | GO:0009106 | lipoate metabolic process(GO:0009106) |
| 0.4 | 1.1 | GO:0036111 | very long-chain fatty-acyl-CoA metabolic process(GO:0036111) |
| 0.4 | 1.1 | GO:1904845 | response to L-glutamine(GO:1904844) cellular response to L-glutamine(GO:1904845) |
| 0.4 | 2.9 | GO:0006848 | pyruvate transport(GO:0006848) pyruvate transmembrane transport(GO:1901475) |
| 0.4 | 2.1 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 0.3 | 1.0 | GO:0006533 | aspartate catabolic process(GO:0006533) |
| 0.3 | 13.2 | GO:0061049 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.3 | 1.0 | GO:2000502 | negative regulation of natural killer cell chemotaxis(GO:2000502) |
| 0.3 | 1.8 | GO:0030311 | poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.3 | 0.9 | GO:0097274 | urea homeostasis(GO:0097274) |
| 0.3 | 2.9 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.3 | 3.1 | GO:2000795 | negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.3 | 1.1 | GO:0031161 | phosphatidylinositol catabolic process(GO:0031161) |
| 0.3 | 1.9 | GO:0032497 | detection of lipopolysaccharide(GO:0032497) |
| 0.3 | 0.8 | GO:0001812 | positive regulation of type I hypersensitivity(GO:0001812) |
| 0.3 | 0.8 | GO:0014813 | skeletal muscle satellite cell commitment(GO:0014813) |
| 0.3 | 0.8 | GO:1903676 | regulation of cap-dependent translational initiation(GO:1903674) positive regulation of cap-dependent translational initiation(GO:1903676) |
| 0.3 | 4.3 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.2 | 0.7 | GO:0090234 | regulation of kinetochore assembly(GO:0090234) |
| 0.2 | 1.0 | GO:0007113 | endomitotic cell cycle(GO:0007113) |
| 0.2 | 1.0 | GO:2000342 | negative regulation of chemokine (C-X-C motif) ligand 2 production(GO:2000342) |
| 0.2 | 3.6 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.2 | 0.7 | GO:0071206 | establishment of protein localization to juxtaparanode region of axon(GO:0071206) |
| 0.2 | 1.1 | GO:0038016 | insulin receptor internalization(GO:0038016) |
| 0.2 | 1.3 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.2 | 0.9 | GO:0031456 | glycine betaine biosynthetic process from choline(GO:0019285) glycine betaine metabolic process(GO:0031455) glycine betaine biosynthetic process(GO:0031456) |
| 0.2 | 4.1 | GO:0009650 | UV protection(GO:0009650) |
| 0.2 | 0.6 | GO:0035526 | retrograde transport, plasma membrane to Golgi(GO:0035526) |
| 0.2 | 0.8 | GO:1901842 | negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.2 | 1.2 | GO:0006696 | ergosterol biosynthetic process(GO:0006696) ergosterol metabolic process(GO:0008204) |
| 0.2 | 1.2 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.2 | 1.2 | GO:1903385 | regulation of homophilic cell adhesion(GO:1903385) |
| 0.2 | 2.0 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.2 | 0.8 | GO:0044861 | protein transport into plasma membrane raft(GO:0044861) |
| 0.2 | 0.6 | GO:0002581 | negative regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002581) regulation of antigen processing and presentation of peptide antigen via MHC class II(GO:0002586) |
| 0.2 | 1.3 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.2 | 3.0 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.2 | 2.4 | GO:0043589 | skin morphogenesis(GO:0043589) |
| 0.2 | 0.8 | GO:0061760 | antifungal innate immune response(GO:0061760) |
| 0.2 | 1.8 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 0.2 | 0.8 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.1 | 0.4 | GO:0035262 | gonad morphogenesis(GO:0035262) |
| 0.1 | 0.4 | GO:0070682 | proteasome regulatory particle assembly(GO:0070682) |
| 0.1 | 0.6 | GO:0051563 | smooth endoplasmic reticulum calcium ion homeostasis(GO:0051563) |
| 0.1 | 5.9 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.1 | 1.6 | GO:0090292 | nucleokinesis involved in cell motility in cerebral cortex radial glia guided migration(GO:0021817) nuclear migration along microtubule(GO:0030473) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.1 | 1.3 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.1 | 2.0 | GO:0051661 | maintenance of centrosome location(GO:0051661) |
| 0.1 | 1.4 | GO:0032328 | alanine transport(GO:0032328) |
| 0.1 | 0.9 | GO:0050917 | sensory perception of sweet taste(GO:0050916) sensory perception of umami taste(GO:0050917) |
| 0.1 | 0.4 | GO:0060931 | sinoatrial node cell development(GO:0060931) |
| 0.1 | 0.7 | GO:0010751 | negative regulation of nitric oxide mediated signal transduction(GO:0010751) |
| 0.1 | 0.8 | GO:0071461 | cellular response to redox state(GO:0071461) |
| 0.1 | 1.4 | GO:0060125 | negative regulation of growth hormone secretion(GO:0060125) |
| 0.1 | 0.5 | GO:2000393 | negative regulation of lamellipodium morphogenesis(GO:2000393) |
| 0.1 | 0.7 | GO:0035022 | positive regulation of Rac protein signal transduction(GO:0035022) |
| 0.1 | 1.1 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.1 | 3.7 | GO:0072189 | ureter development(GO:0072189) |
| 0.1 | 2.1 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.1 | 1.1 | GO:0060054 | positive regulation of epithelial cell proliferation involved in wound healing(GO:0060054) |
| 0.1 | 0.4 | GO:0002378 | immunoglobulin biosynthetic process(GO:0002378) |
| 0.1 | 0.6 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.1 | 0.5 | GO:0006663 | platelet activating factor biosynthetic process(GO:0006663) |
| 0.1 | 0.5 | GO:2000563 | positive regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000563) |
| 0.1 | 1.1 | GO:0043382 | positive regulation of memory T cell differentiation(GO:0043382) |
| 0.1 | 0.8 | GO:0016139 | glycoside catabolic process(GO:0016139) |
| 0.1 | 0.5 | GO:0036518 | chemorepulsion of dopaminergic neuron axon(GO:0036518) |
| 0.1 | 1.5 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.1 | 0.8 | GO:0032383 | regulation of intracellular lipid transport(GO:0032377) regulation of intracellular sterol transport(GO:0032380) regulation of intracellular cholesterol transport(GO:0032383) |
| 0.1 | 1.1 | GO:0030043 | actin filament fragmentation(GO:0030043) |
| 0.1 | 0.5 | GO:1904491 | protein localization to ciliary transition zone(GO:1904491) |
| 0.1 | 0.6 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.1 | 0.7 | GO:0051552 | flavone metabolic process(GO:0051552) |
| 0.1 | 0.6 | GO:0051697 | protein delipidation(GO:0051697) |
| 0.1 | 1.1 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.1 | 1.0 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.1 | 0.5 | GO:1900245 | positive regulation of MDA-5 signaling pathway(GO:1900245) |
| 0.1 | 7.3 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.1 | 0.5 | GO:0045356 | positive regulation of interferon-alpha biosynthetic process(GO:0045356) |
| 0.1 | 1.3 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.1 | 5.4 | GO:0048384 | retinoic acid receptor signaling pathway(GO:0048384) |
| 0.1 | 0.6 | GO:0010166 | wax biosynthetic process(GO:0010025) wax metabolic process(GO:0010166) |
| 0.1 | 4.8 | GO:0006699 | bile acid biosynthetic process(GO:0006699) |
| 0.1 | 1.1 | GO:0006662 | glycerol ether metabolic process(GO:0006662) |
| 0.1 | 1.9 | GO:0060044 | negative regulation of cardiac muscle tissue growth(GO:0055022) negative regulation of cardiac muscle cell proliferation(GO:0060044) negative regulation of heart growth(GO:0061117) |
| 0.1 | 3.1 | GO:2000369 | regulation of clathrin-mediated endocytosis(GO:2000369) |
| 0.1 | 2.7 | GO:0071447 | cellular response to hydroperoxide(GO:0071447) |
| 0.1 | 2.6 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.1 | 0.5 | GO:0033088 | negative regulation of immature T cell proliferation in thymus(GO:0033088) |
| 0.1 | 2.5 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.1 | 0.4 | GO:1902896 | terminal web assembly(GO:1902896) |
| 0.1 | 0.6 | GO:0045541 | negative regulation of cholesterol biosynthetic process(GO:0045541) negative regulation of cholesterol metabolic process(GO:0090206) |
| 0.1 | 0.1 | GO:0052047 | interaction with other organism via secreted substance involved in symbiotic interaction(GO:0052047) |
| 0.1 | 0.6 | GO:0006990 | positive regulation of transcription from RNA polymerase II promoter involved in unfolded protein response(GO:0006990) |
| 0.1 | 3.5 | GO:0051894 | positive regulation of focal adhesion assembly(GO:0051894) |
| 0.1 | 0.7 | GO:0070649 | formin-nucleated actin cable assembly(GO:0070649) |
| 0.1 | 0.7 | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.1 | 1.5 | GO:0006012 | galactose metabolic process(GO:0006012) |
| 0.1 | 1.0 | GO:0071285 | cellular response to lithium ion(GO:0071285) |
| 0.1 | 1.6 | GO:0006590 | thyroid hormone generation(GO:0006590) |
| 0.1 | 14.2 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.1 | 0.3 | GO:0021965 | spinal cord ventral commissure morphogenesis(GO:0021965) |
| 0.1 | 2.9 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.1 | 8.2 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.1 | 1.3 | GO:0031507 | heterochromatin assembly(GO:0031507) |
| 0.1 | 0.5 | GO:1902459 | positive regulation of stem cell population maintenance(GO:1902459) |
| 0.1 | 0.3 | GO:1990414 | replication-born double-strand break repair via sister chromatid exchange(GO:1990414) |
| 0.1 | 2.2 | GO:0035728 | response to hepatocyte growth factor(GO:0035728) |
| 0.1 | 1.1 | GO:0072672 | neutrophil extravasation(GO:0072672) |
| 0.1 | 0.6 | GO:0034465 | response to carbon monoxide(GO:0034465) |
| 0.1 | 1.7 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.1 | 0.2 | GO:0007538 | primary sex determination(GO:0007538) |
| 0.1 | 1.1 | GO:0034390 | smooth muscle cell apoptotic process(GO:0034390) regulation of smooth muscle cell apoptotic process(GO:0034391) |
| 0.1 | 1.4 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.1 | 0.6 | GO:0001956 | positive regulation of neurotransmitter secretion(GO:0001956) |
| 0.1 | 1.8 | GO:1901687 | glutathione derivative metabolic process(GO:1901685) glutathione derivative biosynthetic process(GO:1901687) |
| 0.1 | 1.2 | GO:0060347 | heart trabecula formation(GO:0060347) |
| 0.1 | 0.3 | GO:0006127 | glycerophosphate shuttle(GO:0006127) |
| 0.1 | 1.1 | GO:0035278 | miRNA mediated inhibition of translation(GO:0035278) negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) |
| 0.1 | 0.2 | GO:0070101 | positive regulation of chemokine-mediated signaling pathway(GO:0070101) |
| 0.1 | 1.1 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.1 | 1.0 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.1 | 1.3 | GO:0002003 | angiotensin maturation(GO:0002003) |
| 0.1 | 0.3 | GO:0090362 | positive regulation of platelet-derived growth factor production(GO:0090362) |
| 0.1 | 1.9 | GO:0002323 | natural killer cell activation involved in immune response(GO:0002323) |
| 0.1 | 0.8 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.1 | 0.4 | GO:0032071 | regulation of endodeoxyribonuclease activity(GO:0032071) |
| 0.1 | 1.6 | GO:0043555 | regulation of translation in response to stress(GO:0043555) |
| 0.1 | 0.5 | GO:0070131 | positive regulation of mitochondrial translation(GO:0070131) |
| 0.1 | 0.3 | GO:1900738 | positive regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900738) |
| 0.1 | 0.6 | GO:1901409 | positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.1 | 0.6 | GO:0042985 | negative regulation of amyloid precursor protein biosynthetic process(GO:0042985) |
| 0.0 | 0.6 | GO:0051964 | negative regulation of synapse assembly(GO:0051964) |
| 0.0 | 1.1 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 0.0 | 9.4 | GO:0007286 | spermatid development(GO:0007286) |
| 0.0 | 0.6 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.0 | 0.5 | GO:0034551 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) |
| 0.0 | 1.3 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.0 | 1.5 | GO:0014912 | negative regulation of smooth muscle cell migration(GO:0014912) |
| 0.0 | 0.2 | GO:0007198 | adenylate cyclase-inhibiting serotonin receptor signaling pathway(GO:0007198) |
| 0.0 | 0.1 | GO:0044278 | cell wall disruption in other organism(GO:0044278) |
| 0.0 | 0.9 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.0 | 1.9 | GO:0051123 | RNA polymerase II transcriptional preinitiation complex assembly(GO:0051123) |
| 0.0 | 0.3 | GO:2000270 | negative regulation of fibroblast apoptotic process(GO:2000270) |
| 0.0 | 0.7 | GO:0075522 | IRES-dependent translational initiation(GO:0002192) IRES-dependent viral translational initiation(GO:0075522) |
| 0.0 | 0.3 | GO:0032534 | regulation of microvillus assembly(GO:0032534) |
| 0.0 | 0.4 | GO:0030643 | cellular phosphate ion homeostasis(GO:0030643) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.0 | 0.5 | GO:0060287 | epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 0.0 | 1.8 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.0 | 1.9 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.0 | 1.1 | GO:0030947 | regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030947) |
| 0.0 | 1.0 | GO:0090083 | regulation of inclusion body assembly(GO:0090083) |
| 0.0 | 0.1 | GO:0001579 | medium-chain fatty acid transport(GO:0001579) |
| 0.0 | 0.1 | GO:0035691 | macrophage migration inhibitory factor signaling pathway(GO:0035691) |
| 0.0 | 0.6 | GO:0097435 | fibril organization(GO:0097435) |
| 0.0 | 2.4 | GO:0046847 | filopodium assembly(GO:0046847) |
| 0.0 | 0.6 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.0 | 1.1 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.0 | 0.8 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.0 | 0.1 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.0 | 0.5 | GO:0035721 | intraciliary retrograde transport(GO:0035721) |
| 0.0 | 0.3 | GO:0050713 | negative regulation of interleukin-1 beta secretion(GO:0050713) |
| 0.0 | 0.2 | GO:0098735 | positive regulation of the force of heart contraction(GO:0098735) |
| 0.0 | 2.1 | GO:0032781 | positive regulation of ATPase activity(GO:0032781) |
| 0.0 | 0.5 | GO:1902659 | regulation of glucose mediated signaling pathway(GO:1902659) |
| 0.0 | 0.8 | GO:0000729 | DNA double-strand break processing(GO:0000729) |
| 0.0 | 0.2 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 | 0.1 | GO:0035927 | RNA import into mitochondrion(GO:0035927) |
| 0.0 | 1.5 | GO:0071377 | cellular response to glucagon stimulus(GO:0071377) |
| 0.0 | 1.2 | GO:0051353 | positive regulation of oxidoreductase activity(GO:0051353) |
| 0.0 | 7.2 | GO:0031032 | actomyosin structure organization(GO:0031032) |
| 0.0 | 0.9 | GO:0072663 | protein targeting to peroxisome(GO:0006625) protein localization to peroxisome(GO:0072662) establishment of protein localization to peroxisome(GO:0072663) |
| 0.0 | 0.1 | GO:0018312 | peptidyl-serine ADP-ribosylation(GO:0018312) |
| 0.0 | 0.1 | GO:0006169 | adenosine salvage(GO:0006169) dATP biosynthetic process(GO:0006175) |
| 0.0 | 0.5 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.0 | 0.4 | GO:1900364 | negative regulation of mRNA polyadenylation(GO:1900364) |
| 0.0 | 0.3 | GO:1903608 | protein localization to cytoplasmic stress granule(GO:1903608) |
| 0.0 | 5.0 | GO:0065004 | protein-DNA complex assembly(GO:0065004) |
| 0.0 | 2.0 | GO:0097031 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 0.1 | GO:0070885 | negative regulation of calcineurin-NFAT signaling cascade(GO:0070885) |
| 0.0 | 0.2 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.0 | 0.5 | GO:0030207 | chondroitin sulfate catabolic process(GO:0030207) |
| 0.0 | 0.8 | GO:0034260 | negative regulation of GTPase activity(GO:0034260) |
| 0.0 | 0.1 | GO:0039513 | modulation by virus of host molecular function(GO:0039506) suppression by virus of host molecular function(GO:0039507) suppression by virus of host catalytic activity(GO:0039513) modulation by virus of host catalytic activity(GO:0039516) suppression by virus of host cysteine-type endopeptidase activity involved in apoptotic process(GO:0039650) negative regulation by symbiont of host catalytic activity(GO:0052053) negative regulation by symbiont of host molecular function(GO:0052056) modulation by symbiont of host catalytic activity(GO:0052148) |
| 0.0 | 7.2 | GO:0000209 | protein polyubiquitination(GO:0000209) |
| 0.0 | 1.0 | GO:0034219 | carbohydrate transmembrane transport(GO:0034219) |
| 0.0 | 1.8 | GO:1902036 | regulation of hematopoietic stem cell differentiation(GO:1902036) |
| 0.0 | 1.0 | GO:1903959 | regulation of anion transmembrane transport(GO:1903959) |
| 0.0 | 1.1 | GO:0006383 | transcription from RNA polymerase III promoter(GO:0006383) |
| 0.0 | 0.3 | GO:2000821 | regulation of grooming behavior(GO:2000821) |
| 0.0 | 0.8 | GO:0006333 | chromatin assembly or disassembly(GO:0006333) |
| 0.0 | 0.2 | GO:0051988 | regulation of attachment of spindle microtubules to kinetochore(GO:0051988) |
| 0.0 | 0.3 | GO:1901798 | positive regulation of signal transduction by p53 class mediator(GO:1901798) |
| 0.0 | 0.2 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) |
| 0.0 | 0.3 | GO:0033189 | response to vitamin A(GO:0033189) |
| 0.0 | 0.3 | GO:0035886 | vascular smooth muscle cell differentiation(GO:0035886) |
| 0.0 | 0.2 | GO:0097105 | presynaptic membrane assembly(GO:0097105) |
| 0.0 | 1.0 | GO:0045540 | regulation of cholesterol biosynthetic process(GO:0045540) |
| 0.0 | 0.4 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.0 | 0.1 | GO:0007144 | female meiosis I(GO:0007144) |
| 0.0 | 1.1 | GO:0061178 | regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061178) |
| 0.0 | 0.7 | GO:0097352 | autophagosome maturation(GO:0097352) |
| 0.0 | 0.1 | GO:0006682 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.0 | 0.1 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
| 0.0 | 0.2 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.0 | 1.1 | GO:0048207 | vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) |
| 0.0 | 0.0 | GO:0071395 | response to jasmonic acid(GO:0009753) cellular response to jasmonic acid stimulus(GO:0071395) |
| 0.0 | 0.1 | GO:0060252 | positive regulation of glial cell proliferation(GO:0060252) |
| 0.0 | 0.3 | GO:0007191 | adenylate cyclase-activating dopamine receptor signaling pathway(GO:0007191) |
| 0.0 | 0.1 | GO:0042994 | cytoplasmic sequestering of transcription factor(GO:0042994) |
| 0.0 | 0.2 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.0 | 1.6 | GO:0030010 | establishment of cell polarity(GO:0030010) |
| 0.0 | 0.2 | GO:0010839 | negative regulation of keratinocyte proliferation(GO:0010839) |
| 0.0 | 0.2 | GO:1904668 | positive regulation of ubiquitin protein ligase activity(GO:1904668) |
| 0.0 | 1.0 | GO:0042787 | protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0042787) |
| 0.0 | 0.5 | GO:0007194 | negative regulation of adenylate cyclase activity(GO:0007194) |
| 0.0 | 0.7 | GO:0010507 | negative regulation of autophagy(GO:0010507) |
| 0.0 | 0.2 | GO:0007253 | cytoplasmic sequestering of NF-kappaB(GO:0007253) |
| 0.0 | 0.2 | GO:0050860 | negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.0 | 0.1 | GO:0045722 | positive regulation of gluconeogenesis(GO:0045722) |
| 0.0 | 1.2 | GO:0030449 | regulation of complement activation(GO:0030449) |
| 0.0 | 0.2 | GO:0090312 | positive regulation of protein deacetylation(GO:0090312) |
| 0.0 | 0.2 | GO:0031572 | G2 DNA damage checkpoint(GO:0031572) |
| 0.0 | 0.6 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 1.9 | GO:0007605 | sensory perception of sound(GO:0007605) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 2.8 | GO:0060201 | clathrin-sculpted acetylcholine transport vesicle(GO:0060200) clathrin-sculpted acetylcholine transport vesicle membrane(GO:0060201) |
| 0.5 | 1.9 | GO:0044307 | dendritic branch(GO:0044307) |
| 0.4 | 1.6 | GO:0031933 | telomeric heterochromatin(GO:0031933) |
| 0.4 | 8.5 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.3 | 1.0 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 0.3 | 12.6 | GO:0005720 | nuclear heterochromatin(GO:0005720) |
| 0.3 | 7.5 | GO:0000145 | exocyst(GO:0000145) |
| 0.3 | 3.6 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.2 | 2.3 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.2 | 2.0 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.2 | 1.9 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.2 | 1.2 | GO:0031417 | NatC complex(GO:0031417) |
| 0.2 | 7.3 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.2 | 1.2 | GO:0002079 | inner acrosomal membrane(GO:0002079) |
| 0.2 | 1.4 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.2 | 1.1 | GO:0097169 | AIM2 inflammasome complex(GO:0097169) |
| 0.2 | 7.4 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.2 | 3.5 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.2 | 2.1 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.1 | 0.4 | GO:0071148 | TEAD-1-YAP complex(GO:0071148) |
| 0.1 | 1.1 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.1 | 0.9 | GO:0000308 | cytoplasmic cyclin-dependent protein kinase holoenzyme complex(GO:0000308) |
| 0.1 | 3.1 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.1 | 1.3 | GO:0005677 | chromatin silencing complex(GO:0005677) |
| 0.1 | 0.8 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.1 | 1.9 | GO:0033276 | transcription factor TFTC complex(GO:0033276) |
| 0.1 | 8.7 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.1 | 1.2 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.1 | 3.2 | GO:0090545 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.1 | 1.6 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.1 | 1.1 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.1 | 10.2 | GO:0032432 | actin filament bundle(GO:0032432) |
| 0.1 | 2.5 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.1 | 0.3 | GO:0071920 | cleavage body(GO:0071920) |
| 0.1 | 1.2 | GO:0097433 | dense body(GO:0097433) |
| 0.1 | 0.3 | GO:0005602 | complement component C1 complex(GO:0005602) |
| 0.1 | 1.1 | GO:0097470 | ribbon synapse(GO:0097470) |
| 0.1 | 0.3 | GO:0005944 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) |
| 0.1 | 0.8 | GO:0044354 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.1 | 0.7 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.1 | 0.6 | GO:1990761 | growth cone lamellipodium(GO:1990761) |
| 0.1 | 1.3 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.1 | 0.5 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.1 | 14.6 | GO:0030018 | Z disc(GO:0030018) |
| 0.1 | 1.0 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.1 | 0.2 | GO:0097679 | other organism cytoplasm(GO:0097679) |
| 0.1 | 12.9 | GO:0030427 | site of polarized growth(GO:0030427) |
| 0.0 | 0.8 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.0 | 0.3 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.0 | 0.5 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.0 | 4.4 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.0 | 4.2 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.5 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.0 | 0.1 | GO:0005753 | mitochondrial proton-transporting ATP synthase complex(GO:0005753) proton-transporting ATP synthase complex(GO:0045259) |
| 0.0 | 1.1 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 0.3 | GO:0035867 | alphav-beta3 integrin-IGF-1-IGF1R complex(GO:0035867) |
| 0.0 | 2.2 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 0.7 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 0.7 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.3 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.3 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.0 | 0.6 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 2.4 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 0.6 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 2.9 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.0 | 2.5 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 1.8 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 0.6 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.0 | 0.6 | GO:0043205 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.0 | 0.2 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.0 | 0.6 | GO:0036038 | MKS complex(GO:0036038) |
| 0.0 | 2.1 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 0.5 | GO:0036020 | endolysosome membrane(GO:0036020) |
| 0.0 | 0.2 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.0 | 0.4 | GO:0071439 | clathrin complex(GO:0071439) |
| 0.0 | 4.5 | GO:0032993 | protein-DNA complex(GO:0032993) |
| 0.0 | 3.8 | GO:0000781 | chromosome, telomeric region(GO:0000781) |
| 0.0 | 0.4 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.0 | 3.3 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 7.5 | GO:0005819 | spindle(GO:0005819) |
| 0.0 | 0.2 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.0 | 0.0 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.0 | 0.9 | GO:0045334 | clathrin-coated endocytic vesicle(GO:0045334) |
| 0.0 | 0.4 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 1.7 | GO:0035578 | azurophil granule lumen(GO:0035578) |
| 0.0 | 1.7 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.0 | 0.7 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 0.8 | GO:0005766 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 0.0 | 0.2 | GO:0008091 | spectrin(GO:0008091) |
| 0.0 | 0.3 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) |
| 0.0 | 0.3 | GO:0071339 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.0 | 0.2 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 1.1 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 6.1 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 1.7 | GO:0031902 | late endosome membrane(GO:0031902) |
| 0.0 | 2.6 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 0.2 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 5.7 | GO:0004948 | calcitonin receptor activity(GO:0004948) |
| 1.3 | 7.7 | GO:0060961 | phospholipase D inhibitor activity(GO:0060961) |
| 1.1 | 5.5 | GO:0008240 | tripeptidyl-peptidase activity(GO:0008240) |
| 1.0 | 2.9 | GO:0005477 | pyruvate secondary active transmembrane transporter activity(GO:0005477) |
| 0.5 | 8.8 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.5 | 1.6 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.5 | 1.5 | GO:0033858 | N-acetylgalactosamine kinase activity(GO:0033858) |
| 0.5 | 14.7 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.5 | 2.8 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.4 | 8.3 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.4 | 1.3 | GO:0003858 | 3-hydroxybutyrate dehydrogenase activity(GO:0003858) |
| 0.4 | 2.4 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 0.4 | 1.1 | GO:0016979 | lipoate-protein ligase activity(GO:0016979) |
| 0.4 | 1.1 | GO:0033989 | 3alpha,7alpha,12alpha-trihydroxy-5beta-cholest-24-enoyl-CoA hydratase activity(GO:0033989) 17-beta-hydroxysteroid dehydrogenase (NAD+) activity(GO:0044594) |
| 0.4 | 2.1 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.3 | 2.1 | GO:0047894 | flavonol 3-sulfotransferase activity(GO:0047894) |
| 0.3 | 1.6 | GO:0004447 | iodide peroxidase activity(GO:0004447) |
| 0.3 | 5.4 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.3 | 0.9 | GO:0080130 | L-phenylalanine:2-oxoglutarate aminotransferase activity(GO:0080130) |
| 0.3 | 1.8 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.3 | 1.8 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.3 | 1.7 | GO:0004522 | ribonuclease A activity(GO:0004522) |
| 0.3 | 4.3 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.3 | 1.7 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.3 | 3.0 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.3 | 2.9 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.3 | 1.3 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.3 | 0.8 | GO:0070538 | oleic acid binding(GO:0070538) |
| 0.2 | 6.7 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.2 | 1.9 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.2 | 4.7 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.2 | 1.2 | GO:0051996 | farnesyl-diphosphate farnesyltransferase activity(GO:0004310) squalene synthase activity(GO:0051996) |
| 0.2 | 1.2 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.2 | 4.3 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.2 | 1.0 | GO:0004647 | phosphoserine phosphatase activity(GO:0004647) |
| 0.2 | 2.1 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.2 | 2.4 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.2 | 0.7 | GO:0016524 | latrotoxin receptor activity(GO:0016524) |
| 0.2 | 1.1 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.2 | 1.1 | GO:0030267 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 0.1 | 5.5 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.1 | 1.0 | GO:0005127 | ciliary neurotrophic factor receptor binding(GO:0005127) |
| 0.1 | 1.1 | GO:0015235 | cobalamin transporter activity(GO:0015235) |
| 0.1 | 1.1 | GO:0008934 | inositol monophosphate 1-phosphatase activity(GO:0008934) inositol monophosphate 3-phosphatase activity(GO:0052832) inositol monophosphate 4-phosphatase activity(GO:0052833) inositol monophosphate phosphatase activity(GO:0052834) |
| 0.1 | 2.1 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.1 | 1.3 | GO:0048039 | ubiquinone binding(GO:0048039) |
| 0.1 | 1.0 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.1 | 2.2 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.1 | 0.6 | GO:0080019 | fatty-acyl-CoA reductase (alcohol-forming) activity(GO:0080019) |
| 0.1 | 1.9 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.1 | 4.7 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.1 | 3.8 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.1 | 1.0 | GO:0019788 | NEDD8 transferase activity(GO:0019788) |
| 0.1 | 0.8 | GO:0019763 | immunoglobulin receptor activity(GO:0019763) IgE binding(GO:0019863) |
| 0.1 | 7.8 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.1 | 1.1 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 0.1 | 0.7 | GO:0043295 | glutathione binding(GO:0043295) oligopeptide binding(GO:1900750) |
| 0.1 | 1.2 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.1 | 0.7 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 0.1 | 0.7 | GO:0016929 | SUMO-specific protease activity(GO:0016929) |
| 0.1 | 0.4 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) |
| 0.1 | 7.1 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.1 | 0.8 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.1 | 0.8 | GO:0008310 | single-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008310) |
| 0.1 | 0.5 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.1 | 1.5 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.1 | 1.0 | GO:0022833 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.1 | 0.3 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.1 | 0.3 | GO:0052590 | sn-glycerol-3-phosphate:ubiquinone oxidoreductase activity(GO:0052590) sn-glycerol-3-phosphate:ubiquinone-8 oxidoreductase activity(GO:0052591) |
| 0.1 | 0.2 | GO:0004796 | thromboxane-A synthase activity(GO:0004796) 12-hydroxyheptadecatrienoic acid synthase activity(GO:0036134) |
| 0.1 | 0.9 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.1 | 0.5 | GO:0001165 | RNA polymerase I upstream control element sequence-specific DNA binding(GO:0001165) |
| 0.1 | 1.9 | GO:0031005 | filamin binding(GO:0031005) |
| 0.1 | 0.5 | GO:0016681 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.1 | 1.9 | GO:0050664 | oxidoreductase activity, acting on NAD(P)H, oxygen as acceptor(GO:0050664) |
| 0.1 | 1.5 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.1 | 24.0 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.1 | 2.2 | GO:0016646 | oxidoreductase activity, acting on the CH-NH group of donors, NAD or NADP as acceptor(GO:0016646) |
| 0.1 | 2.2 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.1 | 0.6 | GO:0060072 | large conductance calcium-activated potassium channel activity(GO:0060072) |
| 0.1 | 1.6 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.1 | 1.6 | GO:0005521 | lamin binding(GO:0005521) |
| 0.1 | 0.5 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.1 | 2.0 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.1 | 0.5 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 0.1 | 1.3 | GO:0031432 | titin binding(GO:0031432) |
| 0.1 | 0.6 | GO:0051425 | PTB domain binding(GO:0051425) |
| 0.1 | 2.7 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 0.6 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 1.6 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 0.5 | GO:0008970 | phosphatidylcholine 1-acylhydrolase activity(GO:0008970) |
| 0.0 | 0.2 | GO:0001093 | TFIIB-class transcription factor binding(GO:0001093) |
| 0.0 | 0.1 | GO:0052815 | medium-chain acyl-CoA hydrolase activity(GO:0052815) long-chain acyl-CoA hydrolase activity(GO:0052816) |
| 0.0 | 2.1 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.0 | 0.9 | GO:0004029 | aldehyde dehydrogenase (NAD) activity(GO:0004029) |
| 0.0 | 0.3 | GO:0009378 | four-way junction helicase activity(GO:0009378) |
| 0.0 | 1.9 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.4 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) |
| 0.0 | 0.6 | GO:0032395 | MHC class II receptor activity(GO:0032395) |
| 0.0 | 0.2 | GO:0042328 | heparan sulfate N-acetylglucosaminyltransferase activity(GO:0042328) |
| 0.0 | 1.0 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.8 | GO:0000983 | transcription factor activity, RNA polymerase II core promoter sequence-specific(GO:0000983) |
| 0.0 | 2.7 | GO:0016303 | 1-phosphatidylinositol-3-kinase activity(GO:0016303) |
| 0.0 | 0.8 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 1.1 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 0.2 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 0.0 | 0.7 | GO:0000339 | RNA cap binding(GO:0000339) |
| 0.0 | 0.3 | GO:0089720 | caspase binding(GO:0089720) |
| 0.0 | 0.3 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.0 | 1.4 | GO:0032451 | demethylase activity(GO:0032451) |
| 0.0 | 2.0 | GO:0004407 | histone deacetylase activity(GO:0004407) |
| 0.0 | 0.5 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.2 | GO:0099580 | ion antiporter activity involved in regulation of postsynaptic membrane potential(GO:0099580) |
| 0.0 | 0.9 | GO:0042288 | MHC class I protein binding(GO:0042288) |
| 0.0 | 0.8 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.0 | 0.1 | GO:0004001 | adenosine kinase activity(GO:0004001) |
| 0.0 | 0.8 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.7 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.5 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.0 | 0.3 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.0 | 0.2 | GO:0043176 | amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 0.0 | 0.1 | GO:0015252 | hydrogen ion channel activity(GO:0015252) acid-sensing ion channel activity(GO:0044736) |
| 0.0 | 2.7 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.0 | 17.2 | GO:0003779 | actin binding(GO:0003779) |
| 0.0 | 0.7 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.0 | 0.3 | GO:0022889 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.0 | 0.6 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 2.3 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.4 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.0 | 0.7 | GO:0001848 | complement binding(GO:0001848) |
| 0.0 | 0.9 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.0 | 0.4 | GO:0019213 | deacetylase activity(GO:0019213) |
| 0.0 | 0.2 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.0 | 0.1 | GO:0015450 | P-P-bond-hydrolysis-driven protein transmembrane transporter activity(GO:0015450) |
| 0.0 | 0.4 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.0 | 16.1 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 0.7 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 0.4 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.0 | 0.3 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.1 | GO:0001595 | angiotensin receptor activity(GO:0001595) angiotensin type II receptor activity(GO:0004945) |
| 0.0 | 0.3 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 1.0 | GO:1901476 | carbohydrate transmembrane transporter activity(GO:0015144) carbohydrate transporter activity(GO:1901476) |
| 0.0 | 0.1 | GO:0047756 | chondroitin 4-sulfotransferase activity(GO:0047756) |
| 0.0 | 0.1 | GO:0035877 | death effector domain binding(GO:0035877) cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097199) |
| 0.0 | 1.4 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.0 | 0.8 | GO:0008375 | acetylglucosaminyltransferase activity(GO:0008375) |
| 0.0 | 0.8 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.0 | 0.1 | GO:0008281 | sulfonylurea receptor activity(GO:0008281) |
| 0.0 | 0.3 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.0 | 1.0 | GO:0051219 | phosphoprotein binding(GO:0051219) |
| 0.0 | 0.3 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 2.0 | GO:0005516 | calmodulin binding(GO:0005516) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 12.4 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.2 | 8.8 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.2 | 8.6 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.1 | 2.9 | PID LYMPH ANGIOGENESIS PATHWAY | VEGFR3 signaling in lymphatic endothelium |
| 0.1 | 7.1 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.1 | 14.7 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.1 | 4.2 | PID ATM PATHWAY | ATM pathway |
| 0.1 | 1.3 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.1 | 2.4 | PID EPO PATHWAY | EPO signaling pathway |
| 0.1 | 7.1 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.1 | 1.5 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.1 | 4.8 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.1 | 4.4 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.1 | 4.2 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.1 | 2.1 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.1 | 5.9 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.1 | 2.9 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.1 | 1.9 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.1 | 3.9 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.1 | 0.8 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.1 | 1.4 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.1 | 3.3 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.1 | 2.4 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.1 | 1.0 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 1.9 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.0 | 0.3 | PID IGF1 PATHWAY | IGF1 pathway |
| 0.0 | 1.1 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 0.9 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 0.4 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 1.1 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 1.5 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.1 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 1.1 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.0 | 2.0 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 0.5 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.0 | 0.6 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.0 | 0.5 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.2 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 0.2 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 0.4 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.0 | 0.4 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.0 | 0.6 | ST FAS SIGNALING PATHWAY | Fas Signaling Pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 10.6 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.3 | 11.7 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.3 | 5.1 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.3 | 12.9 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.3 | 7.9 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.2 | 6.3 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.2 | 5.1 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.2 | 2.9 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.2 | 8.9 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.2 | 2.6 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.2 | 3.2 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.1 | 2.1 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.1 | 3.5 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.1 | 2.9 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.1 | 1.9 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.1 | 10.4 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.1 | 3.5 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.1 | 6.2 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.1 | 2.0 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.1 | 4.2 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.1 | 1.9 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.1 | 1.9 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.1 | 0.9 | REACTOME NFKB ACTIVATION THROUGH FADD RIP1 PATHWAY MEDIATED BY CASPASE 8 AND10 | Genes involved in NF-kB activation through FADD/RIP-1 pathway mediated by caspase-8 and -10 |
| 0.1 | 0.5 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.1 | 2.4 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.1 | 2.1 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.1 | 4.7 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.1 | 2.2 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.1 | 1.7 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.1 | 1.1 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.1 | 2.6 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.1 | 0.8 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.1 | 1.5 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.1 | 1.8 | REACTOME G BETA GAMMA SIGNALLING THROUGH PI3KGAMMA | Genes involved in G beta:gamma signalling through PI3Kgamma |
| 0.1 | 0.9 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 1.9 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.0 | 1.9 | REACTOME RAS ACTIVATION UOPN CA2 INFUX THROUGH NMDA RECEPTOR | Genes involved in Ras activation uopn Ca2+ infux through NMDA receptor |
| 0.0 | 5.0 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 1.1 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.0 | 1.1 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 3.7 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 1.5 | REACTOME CA DEPENDENT EVENTS | Genes involved in Ca-dependent events |
| 0.0 | 1.1 | REACTOME REGULATORY RNA PATHWAYS | Genes involved in Regulatory RNA pathways |
| 0.0 | 0.5 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.0 | 1.8 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.0 | 1.0 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 0.2 | REACTOME ADENYLATE CYCLASE INHIBITORY PATHWAY | Genes involved in Adenylate cyclase inhibitory pathway |
| 0.0 | 0.6 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 2.0 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 2.3 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.6 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 0.3 | REACTOME SIGNALLING TO P38 VIA RIT AND RIN | Genes involved in Signalling to p38 via RIT and RIN |
| 0.0 | 1.1 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.0 | 0.6 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 0.4 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.4 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 0.7 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.6 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.4 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 0.9 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.3 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GLP1 | Genes involved in Synthesis, Secretion, and Inactivation of Glucagon-like Peptide-1 (GLP-1) |
| 0.0 | 3.9 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 0.1 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.0 | 0.3 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |