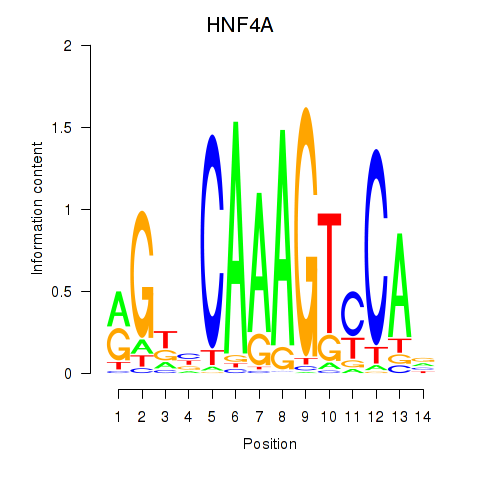
|
chr1_+_78045956
Show fit
|
4.08 |
ENST00000370759.4
|
GIPC2
|
GIPC PDZ domain containing family member 2
|
|
chr1_+_61082398
Show fit
|
3.81 |
ENST00000664149.1
|
NFIA
|
nuclear factor I A
|
|
chr12_-_94616061
Show fit
|
3.48 |
ENST00000551457.1
|
TMCC3
|
transmembrane and coiled-coil domain family 3
|
|
chr1_+_61082553
Show fit
|
3.29 |
ENST00000403491.8
ENST00000371187.7
|
NFIA
|
nuclear factor I A
|
|
chr1_+_61082702
Show fit
|
3.12 |
ENST00000485903.6
ENST00000371185.6
ENST00000371184.6
|
NFIA
|
nuclear factor I A
|
|
chr2_-_221572272
Show fit
|
2.12 |
ENST00000409854.5
ENST00000443796.5
ENST00000281821.7
|
EPHA4
|
EPH receptor A4
|
|
chr14_+_38207893
Show fit
|
1.84 |
ENST00000267377.3
|
SSTR1
|
somatostatin receptor 1
|
|
chrX_+_118974608
Show fit
|
1.55 |
ENST00000304778.11
ENST00000371628.8
|
LONRF3
|
LON peptidase N-terminal domain and ring finger 3
|
|
chr17_+_63477052
Show fit
|
1.55 |
ENST00000290866.10
ENST00000428043.5
|
ACE
|
angiotensin I converting enzyme
|
|
chr17_-_7179544
Show fit
|
1.55 |
ENST00000619926.4
|
ASGR1
|
asialoglycoprotein receptor 1
|
|
chr10_-_17129786
Show fit
|
1.53 |
ENST00000377833.10
|
CUBN
|
cubilin
|
|
chr20_+_64063481
Show fit
|
1.50 |
ENST00000415602.5
|
TCEA2
|
transcription elongation factor A2
|
|
chr18_+_32190015
Show fit
|
1.50 |
ENST00000581447.1
|
MEP1B
|
meprin A subunit beta
|
|
chr2_+_190927649
Show fit
|
1.49 |
ENST00000409428.5
ENST00000409215.5
|
GLS
|
glutaminase
|
|
chr10_-_114684612
Show fit
|
1.48 |
ENST00000533213.6
ENST00000369252.8
|
ABLIM1
|
actin binding LIM protein 1
|
|
chr18_+_32190033
Show fit
|
1.48 |
ENST00000269202.11
|
MEP1B
|
meprin A subunit beta
|
|
chr8_-_17002327
Show fit
|
1.38 |
ENST00000180166.6
|
FGF20
|
fibroblast growth factor 20
|
|
chr19_-_48363914
Show fit
|
1.36 |
ENST00000377431.6
ENST00000293261.8
|
TMEM143
|
transmembrane protein 143
|
|
chr6_+_30914205
Show fit
|
1.35 |
ENST00000672801.1
ENST00000321897.9
ENST00000625423.2
ENST00000676266.1
ENST00000428017.5
|
VARS2
|
valyl-tRNA synthetase 2, mitochondrial
|
|
chr17_-_7179348
Show fit
|
1.29 |
ENST00000573083.1
ENST00000574388.5
ENST00000269299.8
|
ASGR1
|
asialoglycoprotein receptor 1
|
|
chr11_-_63229652
Show fit
|
1.25 |
ENST00000306494.10
|
SLC22A25
|
solute carrier family 22 member 25
|
|
chr6_+_30914329
Show fit
|
1.24 |
ENST00000541562.6
|
VARS2
|
valyl-tRNA synthetase 2, mitochondrial
|
|
chr19_-_48364034
Show fit
|
1.21 |
ENST00000435956.7
|
TMEM143
|
transmembrane protein 143
|
|
chr13_-_52011337
Show fit
|
1.15 |
ENST00000400366.6
ENST00000400370.8
ENST00000634844.1
ENST00000673772.1
ENST00000418097.7
ENST00000242839.10
ENST00000344297.9
ENST00000448424.7
|
ATP7B
|
ATPase copper transporting beta
|
|
chr1_+_204073104
Show fit
|
1.15 |
ENST00000367204.6
|
SOX13
|
SRY-box transcription factor 13
|
|
chr6_+_143677935
Show fit
|
1.09 |
ENST00000440869.6
ENST00000367582.7
ENST00000451827.6
|
PHACTR2
|
phosphatase and actin regulator 2
|
|
chr3_+_50269140
Show fit
|
1.07 |
ENST00000616701.5
ENST00000433753.4
ENST00000611067.4
|
SEMA3B
|
semaphorin 3B
|
|
chr14_+_64214136
Show fit
|
1.06 |
ENST00000557084.1
ENST00000458046.6
|
SYNE2
|
spectrin repeat containing nuclear envelope protein 2
|
|
chr6_-_31897200
Show fit
|
1.00 |
ENST00000395728.7
ENST00000375528.8
|
EHMT2
|
euchromatic histone lysine methyltransferase 2
|
|
chr16_+_8674605
Show fit
|
0.97 |
ENST00000268251.13
|
ABAT
|
4-aminobutyrate aminotransferase
|
|
chr8_+_95024977
Show fit
|
0.95 |
ENST00000396124.9
|
NDUFAF6
|
NADH:ubiquinone oxidoreductase complex assembly factor 6
|
|
chr5_-_42811884
Show fit
|
0.93 |
ENST00000514985.6
ENST00000511224.5
ENST00000507920.5
ENST00000510965.1
|
SELENOP
|
selenoprotein P
|
|
chr3_-_126056786
Show fit
|
0.90 |
ENST00000383598.6
|
SLC41A3
|
solute carrier family 41 member 3
|
|
chr2_+_158456939
Show fit
|
0.88 |
ENST00000389759.8
ENST00000628904.2
ENST00000389757.7
|
PKP4
|
plakophilin 4
|
|
chr1_+_94418375
Show fit
|
0.88 |
ENST00000370214.9
|
ABCD3
|
ATP binding cassette subfamily D member 3
|
|
chr1_+_28369705
Show fit
|
0.83 |
ENST00000373839.8
|
PHACTR4
|
phosphatase and actin regulator 4
|
|
chr12_-_6556034
Show fit
|
0.83 |
ENST00000619571.5
ENST00000336604.8
ENST00000396840.6
ENST00000356896.8
|
IFFO1
|
intermediate filament family orphan 1
|
|
chr1_+_94418435
Show fit
|
0.81 |
ENST00000647998.2
|
ABCD3
|
ATP binding cassette subfamily D member 3
|
|
chr1_+_94418467
Show fit
|
0.79 |
ENST00000315713.5
|
ABCD3
|
ATP binding cassette subfamily D member 3
|
|
chr20_+_64063105
Show fit
|
0.78 |
ENST00000395053.7
ENST00000343484.10
ENST00000339217.8
|
TCEA2
|
transcription elongation factor A2
|
|
chr20_-_37527723
Show fit
|
0.77 |
ENST00000397135.1
ENST00000397137.5
|
BLCAP
|
BLCAP apoptosis inducing factor
|
|
chr15_-_56245074
Show fit
|
0.74 |
ENST00000674082.1
|
RFX7
|
regulatory factor X7
|
|
chr1_+_20633450
Show fit
|
0.74 |
ENST00000321556.5
|
PINK1
|
PTEN induced kinase 1
|
|
chr13_+_113122791
Show fit
|
0.71 |
ENST00000375559.8
ENST00000409306.5
ENST00000375551.7
|
F10
|
coagulation factor X
|
|
chr11_+_118998126
Show fit
|
0.71 |
ENST00000334418.6
|
CENATAC
|
centrosomal AT-AC splicing factor
|
|
chr19_-_14062028
Show fit
|
0.68 |
ENST00000669674.2
|
PALM3
|
paralemmin 3
|
|
chr19_-_14090963
Show fit
|
0.67 |
ENST00000269724.5
|
SAMD1
|
sterile alpha motif domain containing 1
|
|
chr19_-_15125095
Show fit
|
0.67 |
ENST00000600984.5
|
ILVBL
|
ilvB acetolactate synthase like
|
|
chr22_+_19962537
Show fit
|
0.66 |
ENST00000449653.5
|
COMT
|
catechol-O-methyltransferase
|
|
chr2_+_218270392
Show fit
|
0.65 |
ENST00000248451.7
ENST00000273077.9
|
PNKD
|
PNKD metallo-beta-lactamase domain containing
|
|
chr8_+_22579100
Show fit
|
0.65 |
ENST00000452226.5
ENST00000397760.8
|
PDLIM2
|
PDZ and LIM domain 2
|
|
chr13_+_113105782
Show fit
|
0.65 |
ENST00000541084.5
ENST00000346342.8
ENST00000375581.3
|
F7
|
coagulation factor VII
|
|
chr3_-_49813880
Show fit
|
0.65 |
ENST00000333486.4
|
UBA7
|
ubiquitin like modifier activating enzyme 7
|
|
chr12_-_53727476
Show fit
|
0.64 |
ENST00000549784.5
ENST00000262059.8
|
CALCOCO1
|
calcium binding and coiled-coil domain 1
|
|
chr9_-_113303271
Show fit
|
0.63 |
ENST00000297894.5
ENST00000489339.2
|
RNF183
|
ring finger protein 183
|
|
chr8_+_22579139
Show fit
|
0.63 |
ENST00000397761.6
|
PDLIM2
|
PDZ and LIM domain 2
|
|
chr7_-_15561986
Show fit
|
0.63 |
ENST00000342526.8
|
AGMO
|
alkylglycerol monooxygenase
|
|
chr10_-_102120246
Show fit
|
0.62 |
ENST00000425280.2
|
LDB1
|
LIM domain binding 1
|
|
chr12_-_108826161
Show fit
|
0.62 |
ENST00000546697.1
|
SSH1
|
slingshot protein phosphatase 1
|
|
chr6_-_30113086
Show fit
|
0.62 |
ENST00000376734.4
|
TRIM31
|
tripartite motif containing 31
|
|
chr12_-_53727428
Show fit
|
0.61 |
ENST00000548263.5
ENST00000430117.6
ENST00000549173.5
ENST00000550804.6
ENST00000551900.5
ENST00000546619.5
ENST00000548177.5
ENST00000549349.5
|
CALCOCO1
|
calcium binding and coiled-coil domain 1
|
|
chr20_-_37527862
Show fit
|
0.60 |
ENST00000373537.7
ENST00000445723.5
ENST00000414080.1
|
BLCAP
|
BLCAP apoptosis inducing factor
|
|
chr17_+_28744034
Show fit
|
0.59 |
ENST00000444415.7
ENST00000262396.10
|
TRAF4
|
TNF receptor associated factor 4
|
|
chr8_+_11808417
Show fit
|
0.52 |
ENST00000525954.5
|
FDFT1
|
farnesyl-diphosphate farnesyltransferase 1
|
|
chr2_-_62506136
Show fit
|
0.52 |
ENST00000335390.6
|
TMEM17
|
transmembrane protein 17
|
|
chr2_+_172860038
Show fit
|
0.51 |
ENST00000538974.5
ENST00000540783.5
|
RAPGEF4
|
Rap guanine nucleotide exchange factor 4
|
|
chr10_+_100347225
Show fit
|
0.51 |
ENST00000370355.3
|
SCD
|
stearoyl-CoA desaturase
|
|
chr22_+_29767351
Show fit
|
0.51 |
ENST00000330029.6
ENST00000401406.3
|
UQCR10
|
ubiquinol-cytochrome c reductase, complex III subunit X
|
|
chr1_-_50023875
Show fit
|
0.51 |
ENST00000371836.1
ENST00000371838.5
|
AGBL4
|
ATP/GTP binding protein like 4
|
|
chr10_+_102644462
Show fit
|
0.50 |
ENST00000643721.2
ENST00000302424.12
|
TRIM8
|
tripartite motif containing 8
|
|
chr17_+_75610690
Show fit
|
0.50 |
ENST00000642007.1
|
MYO15B
|
myosin XVB
|
|
chr17_+_29043124
Show fit
|
0.48 |
ENST00000323372.9
|
PIPOX
|
pipecolic acid and sarcosine oxidase
|
|
chr8_+_69492793
Show fit
|
0.48 |
ENST00000616868.1
ENST00000419716.7
ENST00000402687.9
|
SULF1
|
sulfatase 1
|
|
chr9_+_27109200
Show fit
|
0.47 |
ENST00000380036.10
|
TEK
|
TEK receptor tyrosine kinase
|
|
chr17_-_29176752
Show fit
|
0.47 |
ENST00000533112.5
|
MYO18A
|
myosin XVIIIA
|
|
chrX_-_154412083
Show fit
|
0.46 |
ENST00000451865.5
ENST00000432135.1
ENST00000369809.5
ENST00000393638.5
ENST00000424626.5
ENST00000309585.9
|
DNASE1L1
|
deoxyribonuclease 1 like 1
|
|
chr20_-_38033424
Show fit
|
0.45 |
ENST00000373447.8
ENST00000373448.6
|
TTI1
|
TELO2 interacting protein 1
|
|
chr3_+_124094663
Show fit
|
0.45 |
ENST00000460856.5
ENST00000240874.7
|
KALRN
|
kalirin RhoGEF kinase
|
|
chr9_+_27109135
Show fit
|
0.45 |
ENST00000519097.5
ENST00000615002.4
|
TEK
|
TEK receptor tyrosine kinase
|
|
chr19_+_18386150
Show fit
|
0.45 |
ENST00000252809.3
|
GDF15
|
growth differentiation factor 15
|
|
chr1_-_167090370
Show fit
|
0.45 |
ENST00000367868.4
|
GPA33
|
glycoprotein A33
|
|
chr1_+_241532121
Show fit
|
0.45 |
ENST00000366558.7
|
KMO
|
kynurenine 3-monooxygenase
|
|
chr3_-_53844617
Show fit
|
0.44 |
ENST00000481668.5
ENST00000467802.1
|
CHDH
|
choline dehydrogenase
|
|
chr2_-_218568291
Show fit
|
0.43 |
ENST00000418019.5
ENST00000454775.5
ENST00000338465.5
ENST00000415516.5
ENST00000258399.8
|
USP37
|
ubiquitin specific peptidase 37
|
|
chr16_+_66566393
Show fit
|
0.43 |
ENST00000332695.11
ENST00000336328.10
ENST00000457188.6
ENST00000528324.5
ENST00000529506.5
ENST00000531885.5
ENST00000533666.5
ENST00000533953.5
ENST00000379500.7
ENST00000328020.10
|
CMTM1
|
CKLF like MARVEL transmembrane domain containing 1
|
|
chr6_+_31723344
Show fit
|
0.43 |
ENST00000480039.5
ENST00000375810.8
ENST00000375805.6
ENST00000375809.7
ENST00000649779.1
ENST00000375804.6
ENST00000375814.7
|
MPIG6B
|
megakaryocyte and platelet inhibitory receptor G6b
|
|
chr5_+_80654644
Show fit
|
0.42 |
ENST00000265081.7
|
MSH3
|
mutS homolog 3
|
|
chr2_+_27890716
Show fit
|
0.42 |
ENST00000344773.6
ENST00000379624.6
ENST00000342045.6
ENST00000379632.6
ENST00000361704.6
|
BABAM2
|
BRISC and BRCA1 A complex member 2
|
|
chr13_+_75760362
Show fit
|
0.42 |
ENST00000534657.5
|
LMO7
|
LIM domain 7
|
|
chr1_+_200027702
Show fit
|
0.41 |
ENST00000367362.8
|
NR5A2
|
nuclear receptor subfamily 5 group A member 2
|
|
chr3_+_46877705
Show fit
|
0.41 |
ENST00000449590.6
|
PTH1R
|
parathyroid hormone 1 receptor
|
|
chr11_-_116823293
Show fit
|
0.40 |
ENST00000357780.5
|
APOA4
|
apolipoprotein A4
|
|
chr7_+_101020073
Show fit
|
0.39 |
ENST00000306151.9
|
MUC17
|
mucin 17, cell surface associated
|
|
chr9_+_136945234
Show fit
|
0.39 |
ENST00000371634.7
|
C8G
|
complement C8 gamma chain
|
|
chr12_-_118372883
Show fit
|
0.39 |
ENST00000542532.5
ENST00000392533.8
|
TAOK3
|
TAO kinase 3
|
|
chr1_+_200027605
Show fit
|
0.39 |
ENST00000236914.7
|
NR5A2
|
nuclear receptor subfamily 5 group A member 2
|
|
chr9_-_101442403
Show fit
|
0.37 |
ENST00000648758.1
|
ALDOB
|
aldolase, fructose-bisphosphate B
|
|
chr3_+_124094696
Show fit
|
0.37 |
ENST00000360013.7
ENST00000684186.1
ENST00000684276.1
|
KALRN
|
kalirin RhoGEF kinase
|
|
chr5_+_154858482
Show fit
|
0.37 |
ENST00000519211.5
ENST00000522458.5
ENST00000519903.5
ENST00000521450.5
ENST00000403027.6
|
CNOT8
|
CCR4-NOT transcription complex subunit 8
|
|
chr1_-_177969907
Show fit
|
0.37 |
ENST00000308284.10
|
SEC16B
|
SEC16 homolog B, endoplasmic reticulum export factor
|
|
chr19_-_40750302
Show fit
|
0.36 |
ENST00000598485.6
ENST00000378313.7
ENST00000470681.5
|
C19orf54
|
chromosome 19 open reading frame 54
|
|
chr5_+_154858218
Show fit
|
0.36 |
ENST00000523698.5
ENST00000517876.5
ENST00000520472.5
|
CNOT8
|
CCR4-NOT transcription complex subunit 8
|
|
chr6_+_31982057
Show fit
|
0.36 |
ENST00000428956.7
ENST00000498271.1
|
C4A
|
complement C4A (Rodgers blood group)
|
|
chr19_-_35812838
Show fit
|
0.35 |
ENST00000653904.2
|
PRODH2
|
proline dehydrogenase 2
|
|
chr5_+_154858594
Show fit
|
0.34 |
ENST00000519430.5
ENST00000520671.5
ENST00000521583.5
ENST00000285896.11
ENST00000518028.5
ENST00000519404.5
ENST00000519394.5
ENST00000518775.5
|
CNOT8
|
CCR4-NOT transcription complex subunit 8
|
|
chr4_+_94455245
Show fit
|
0.34 |
ENST00000508216.5
ENST00000514743.5
|
PDLIM5
|
PDZ and LIM domain 5
|
|
chr17_+_50548041
Show fit
|
0.33 |
ENST00000634597.1
|
SPATA20
|
spermatogenesis associated 20
|
|
chr3_+_63911929
Show fit
|
0.33 |
ENST00000487717.5
|
ATXN7
|
ataxin 7
|
|
chr1_+_241532370
Show fit
|
0.33 |
ENST00000366559.9
ENST00000366557.8
|
KMO
|
kynurenine 3-monooxygenase
|
|
chr12_-_14696571
Show fit
|
0.32 |
ENST00000261170.5
|
GUCY2C
|
guanylate cyclase 2C
|
|
chr2_+_44275473
Show fit
|
0.32 |
ENST00000260649.11
|
SLC3A1
|
solute carrier family 3 member 1
|
|
chr2_+_48568981
Show fit
|
0.32 |
ENST00000394754.5
|
STON1-GTF2A1L
|
STON1-GTF2A1L readthrough
|
|
chr11_-_10294194
Show fit
|
0.31 |
ENST00000676387.1
ENST00000256190.13
ENST00000675281.1
|
SBF2
|
SET binding factor 2
|
|
chr2_-_180007254
Show fit
|
0.31 |
ENST00000410053.8
|
CWC22
|
CWC22 spliceosome associated protein homolog
|
|
chr4_+_112231748
Show fit
|
0.30 |
ENST00000274000.10
ENST00000309703.10
|
AP1AR
|
adaptor related protein complex 1 associated regulatory protein
|
|
chr11_+_1834804
Show fit
|
0.30 |
ENST00000341958.3
|
SYT8
|
synaptotagmin 8
|
|
chr13_+_75760659
Show fit
|
0.29 |
ENST00000526202.5
ENST00000465261.6
|
LMO7
|
LIM domain 7
|
|
chr19_-_38812936
Show fit
|
0.29 |
ENST00000307751.9
ENST00000594209.1
|
LGALS4
|
galectin 4
|
|
chr1_-_23368301
Show fit
|
0.29 |
ENST00000374608.3
|
ZNF436
|
zinc finger protein 436
|
|
chr1_-_27366917
Show fit
|
0.28 |
ENST00000357582.3
|
MAP3K6
|
mitogen-activated protein kinase kinase kinase 6
|
|
chrX_-_77786198
Show fit
|
0.28 |
ENST00000624032.3
ENST00000624668.3
ENST00000373344.11
ENST00000395603.7
ENST00000624166.3
ENST00000623321.3
ENST00000622960.1
|
ATRX
|
ATRX chromatin remodeler
|
|
chr13_+_75760431
Show fit
|
0.28 |
ENST00000321797.12
|
LMO7
|
LIM domain 7
|
|
chrX_-_53281524
Show fit
|
0.28 |
ENST00000498281.2
|
IQSEC2
|
IQ motif and Sec7 domain ArfGEF 2
|
|
chr1_-_155301423
Show fit
|
0.27 |
ENST00000342741.6
|
PKLR
|
pyruvate kinase L/R
|
|
chr14_+_23185316
Show fit
|
0.26 |
ENST00000399910.5
ENST00000492621.5
|
RNF212B
|
ring finger protein 212B
|
|
chr4_+_108620584
Show fit
|
0.26 |
ENST00000394665.5
|
RPL34
|
ribosomal protein L34
|
|
chr11_+_119185469
Show fit
|
0.26 |
ENST00000525131.5
ENST00000355547.10
ENST00000531114.5
ENST00000322712.4
|
PDZD3
|
PDZ domain containing 3
|
|
chr4_+_108620567
Show fit
|
0.26 |
ENST00000502534.5
ENST00000394667.8
|
RPL34
|
ribosomal protein L34
|
|
chr14_+_39267055
Show fit
|
0.26 |
ENST00000396158.6
ENST00000280083.7
ENST00000341502.9
|
MIA2
|
MIA SH3 domain ER export factor 2
|
|
chr9_-_68540879
Show fit
|
0.25 |
ENST00000377311.4
|
TMEM252
|
transmembrane protein 252
|
|
chr6_+_32014795
Show fit
|
0.25 |
ENST00000435363.7
ENST00000425700.3
|
C4B
|
complement C4B (Chido blood group)
|
|
chr6_-_29375291
Show fit
|
0.25 |
ENST00000396806.3
|
OR12D3
|
olfactory receptor family 12 subfamily D member 3
|
|
chr16_-_16223467
Show fit
|
0.25 |
ENST00000575728.1
ENST00000574094.6
ENST00000205557.12
|
ABCC6
|
ATP binding cassette subfamily C member 6
|
|
chr3_+_63912588
Show fit
|
0.24 |
ENST00000522345.2
|
ATXN7
|
ataxin 7
|
|
chr5_+_154858537
Show fit
|
0.24 |
ENST00000517568.5
ENST00000524105.5
|
CNOT8
|
CCR4-NOT transcription complex subunit 8
|
|
chr22_-_30667795
Show fit
|
0.24 |
ENST00000404885.5
ENST00000403268.1
ENST00000407308.1
ENST00000334679.4
ENST00000342474.4
|
DUSP18
|
dual specificity phosphatase 18
|
|
chrX_+_154429092
Show fit
|
0.23 |
ENST00000619046.5
|
ATP6AP1
|
ATPase H+ transporting accessory protein 1
|
|
chr11_+_1834415
Show fit
|
0.23 |
ENST00000381968.7
ENST00000381978.7
|
SYT8
|
synaptotagmin 8
|
|
chr20_-_45857196
Show fit
|
0.23 |
ENST00000457981.5
ENST00000426915.1
ENST00000217455.9
|
ACOT8
|
acyl-CoA thioesterase 8
|
|
chr19_-_35135180
Show fit
|
0.23 |
ENST00000392225.7
|
LGI4
|
leucine rich repeat LGI family member 4
|
|
chr1_-_161223408
Show fit
|
0.23 |
ENST00000491350.1
|
APOA2
|
apolipoprotein A2
|
|
chr2_-_42792558
Show fit
|
0.23 |
ENST00000431905.1
ENST00000294973.11
|
HAAO
|
3-hydroxyanthranilate 3,4-dioxygenase
|
|
chr5_+_80319997
Show fit
|
0.22 |
ENST00000296739.6
|
SPZ1
|
spermatogenic leucine zipper 1
|
|
chr17_+_1742836
Show fit
|
0.22 |
ENST00000324015.7
ENST00000450523.6
ENST00000453723.5
ENST00000453066.6
ENST00000382061.5
|
SERPINF2
|
serpin family F member 2
|
|
chr2_-_27890348
Show fit
|
0.21 |
ENST00000302188.8
|
RBKS
|
ribokinase
|
|
chr20_-_3663399
Show fit
|
0.21 |
ENST00000290417.7
ENST00000319242.8
|
GFRA4
|
GDNF family receptor alpha 4
|
|
chr2_-_208129824
Show fit
|
0.21 |
ENST00000282141.4
|
CRYGC
|
crystallin gamma C
|
|
chr1_+_180632001
Show fit
|
0.21 |
ENST00000367590.9
ENST00000367589.3
|
XPR1
|
xenotropic and polytropic retrovirus receptor 1
|
|
chr19_-_35134861
Show fit
|
0.21 |
ENST00000591633.2
|
LGI4
|
leucine rich repeat LGI family member 4
|
|
chr10_-_102056116
Show fit
|
0.21 |
ENST00000370033.9
ENST00000311122.5
|
ARMH3
|
armadillo like helical domain containing 3
|
|
chr4_+_108620616
Show fit
|
0.21 |
ENST00000506397.5
ENST00000394668.2
|
RPL34
|
ribosomal protein L34
|
|
chr9_-_29739
Show fit
|
0.20 |
ENST00000442898.5
|
WASHC1
|
WASH complex subunit 1
|
|
chrX_+_154411519
Show fit
|
0.20 |
ENST00000612460.5
ENST00000601016.6
ENST00000613002.4
ENST00000475699.6
ENST00000612012.5
ENST00000369776.8
ENST00000439735.2
ENST00000616020.5
ENST00000652358.1
ENST00000652390.1
|
TAZ
|
tafazzin
|
|
chr2_+_61162185
Show fit
|
0.20 |
ENST00000432605.3
ENST00000488469.4
|
C2orf74
|
chromosome 2 open reading frame 74
|
|
chr19_+_7595830
Show fit
|
0.20 |
ENST00000160298.9
ENST00000446248.4
|
CAMSAP3
|
calmodulin regulated spectrin associated protein family member 3
|
|
chr19_+_15641280
Show fit
|
0.20 |
ENST00000585846.1
|
CYP4F3
|
cytochrome P450 family 4 subfamily F member 3
|
|
chr19_+_39391323
Show fit
|
0.19 |
ENST00000615911.4
ENST00000315588.11
ENST00000594368.5
ENST00000596297.1
|
MED29
|
mediator complex subunit 29
|
|
chr15_-_63156774
Show fit
|
0.19 |
ENST00000462430.5
|
RPS27L
|
ribosomal protein S27 like
|
|
chr7_-_78771265
Show fit
|
0.18 |
ENST00000630991.2
ENST00000629359.2
|
MAGI2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2
|
|
chr3_-_194351290
Show fit
|
0.18 |
ENST00000429275.1
ENST00000323830.4
|
CPN2
|
carboxypeptidase N subunit 2
|
|
chr14_+_100019375
Show fit
|
0.17 |
ENST00000544450.6
|
EVL
|
Enah/Vasp-like
|
|
chr5_+_177384430
Show fit
|
0.17 |
ENST00000512593.5
ENST00000324417.6
|
SLC34A1
|
solute carrier family 34 member 1
|
|
chr15_+_51341648
Show fit
|
0.17 |
ENST00000335449.11
ENST00000560215.5
|
GLDN
|
gliomedin
|
|
chr6_+_31959130
Show fit
|
0.17 |
ENST00000375394.7
ENST00000628157.1
|
SKIV2L
|
Ski2 like RNA helicase
|
|
chrX_-_43882411
Show fit
|
0.16 |
ENST00000378069.5
|
MAOB
|
monoamine oxidase B
|
|
chr18_+_56651385
Show fit
|
0.16 |
ENST00000615645.4
|
WDR7
|
WD repeat domain 7
|
|
chr11_+_63369779
Show fit
|
0.16 |
ENST00000279178.4
|
SLC22A9
|
solute carrier family 22 member 9
|
|
chr19_-_35135011
Show fit
|
0.16 |
ENST00000310123.8
|
LGI4
|
leucine rich repeat LGI family member 4
|
|
chr18_+_56651335
Show fit
|
0.16 |
ENST00000589935.1
ENST00000254442.8
ENST00000357574.7
|
WDR7
|
WD repeat domain 7
|
|
chr17_+_50346099
Show fit
|
0.16 |
ENST00000017003.7
ENST00000509778.1
ENST00000507602.5
|
XYLT2
|
xylosyltransferase 2
|
|
chr7_-_87475839
Show fit
|
0.15 |
ENST00000359206.8
|
ABCB4
|
ATP binding cassette subfamily B member 4
|
|
chr19_+_15615211
Show fit
|
0.15 |
ENST00000617645.4
ENST00000612078.5
|
CYP4F8
|
cytochrome P450 family 4 subfamily F member 8
|
|
chr15_+_50424377
Show fit
|
0.14 |
ENST00000560297.5
ENST00000396444.7
ENST00000425032.7
ENST00000307179.9
ENST00000625664.2
|
USP8
|
ubiquitin specific peptidase 8
|
|
chr22_+_24607602
Show fit
|
0.14 |
ENST00000447416.5
|
GGT1
|
gamma-glutamyltransferase 1
|
|
chr18_+_59225492
Show fit
|
0.14 |
ENST00000456142.3
ENST00000530323.1
|
GRP
|
gastrin releasing peptide
|
|
chr19_-_35852495
Show fit
|
0.14 |
ENST00000378910.10
|
NPHS1
|
NPHS1 adhesion molecule, nephrin
|
|
chr1_-_40665435
Show fit
|
0.14 |
ENST00000372683.1
|
RIMS3
|
regulating synaptic membrane exocytosis 3
|
|
chr11_+_63888515
Show fit
|
0.14 |
ENST00000509502.6
ENST00000512060.1
|
MARK2
|
microtubule affinity regulating kinase 2
|
|
chr12_-_53200443
Show fit
|
0.14 |
ENST00000550743.6
|
ITGB7
|
integrin subunit beta 7
|
|
chr6_-_32178080
Show fit
|
0.13 |
ENST00000336984.6
|
AGPAT1
|
1-acylglycerol-3-phosphate O-acyltransferase 1
|
|
chr11_-_707063
Show fit
|
0.13 |
ENST00000683307.1
|
DEAF1
|
DEAF1 transcription factor
|
|
chr1_+_48222685
Show fit
|
0.13 |
ENST00000533824.5
ENST00000236495.9
ENST00000438567.7
|
SLC5A9
|
solute carrier family 5 member 9
|
|
chr12_+_120978537
Show fit
|
0.12 |
ENST00000257555.11
ENST00000400024.6
|
HNF1A
|
HNF1 homeobox A
|
|
chr15_-_73633310
Show fit
|
0.11 |
ENST00000345330.9
|
NPTN
|
neuroplastin
|
|
chr5_+_90899183
Show fit
|
0.11 |
ENST00000640815.1
|
ADGRV1
|
adhesion G protein-coupled receptor V1
|
|
chr6_+_30626842
Show fit
|
0.11 |
ENST00000318999.11
ENST00000376485.8
ENST00000330083.6
ENST00000319027.9
ENST00000376483.8
ENST00000329992.12
|
ATAT1
|
alpha tubulin acetyltransferase 1
|
|
chr2_-_158456702
Show fit
|
0.10 |
ENST00000409889.1
ENST00000283233.10
|
CCDC148
|
coiled-coil domain containing 148
|
|
chr7_-_143647646
Show fit
|
0.10 |
ENST00000636941.1
|
TCAF2C
|
TRPM8 channel associated factor 2C
|
|
chr6_-_149484965
Show fit
|
0.10 |
ENST00000409806.8
|
ZC3H12D
|
zinc finger CCCH-type containing 12D
|
|
chr5_+_1801387
Show fit
|
0.10 |
ENST00000274137.10
ENST00000469176.1
|
NDUFS6
|
NADH:ubiquinone oxidoreductase subunit S6
|
|
chr6_+_43298326
Show fit
|
0.10 |
ENST00000372574.7
|
SLC22A7
|
solute carrier family 22 member 7
|
|
chr12_+_108880085
Show fit
|
0.10 |
ENST00000228476.8
ENST00000547768.5
|
DAO
|
D-amino acid oxidase
|
|
chr16_-_88686453
Show fit
|
0.09 |
ENST00000332281.6
|
SNAI3
|
snail family transcriptional repressor 3
|
|
chr14_+_77181780
Show fit
|
0.09 |
ENST00000298351.5
ENST00000554346.5
|
TMEM63C
|
transmembrane protein 63C
|
|
chr19_-_7632971
Show fit
|
0.09 |
ENST00000598935.5
|
PCP2
|
Purkinje cell protein 2
|
|
chr5_-_16508812
Show fit
|
0.09 |
ENST00000683414.1
|
RETREG1
|
reticulophagy regulator 1
|
|
chr19_-_48835823
Show fit
|
0.08 |
ENST00000595764.1
|
HSD17B14
|
hydroxysteroid 17-beta dehydrogenase 14
|
|
chr2_+_74421721
Show fit
|
0.08 |
ENST00000409791.5
ENST00000348227.4
|
WDR54
|
WD repeat domain 54
|
|
chr18_+_3262098
Show fit
|
0.08 |
ENST00000237500.10
|
MYL12B
|
myosin light chain 12B
|
|
chr3_-_9878765
Show fit
|
0.07 |
ENST00000430427.6
ENST00000383817.5
ENST00000679265.1
|
CIDEC
|
cell death inducing DFFA like effector c
|