Project
Inflammatory response time course, HUVEC (Wada, 2009)
Navigation
Downloads
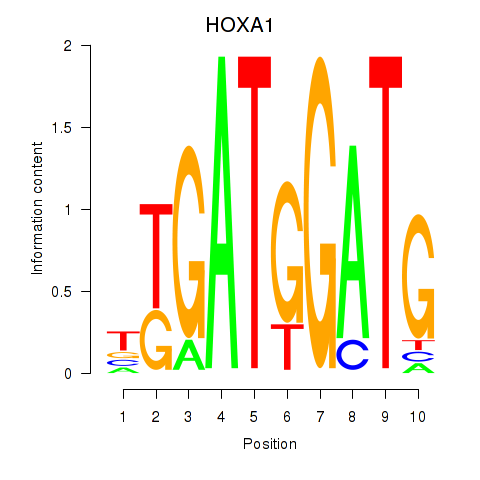
Results for HOXA1
Z-value: 0.71
Transcription factors associated with HOXA1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HOXA1
|
ENSG00000105991.10 | HOXA1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HOXA1 | hg38_v1_chr7_-_27095972_27096039 | 0.26 | 2.1e-01 | Click! |
Activity profile of HOXA1 motif
Sorted Z-values of HOXA1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of HOXA1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chrX_-_63785510 | 1.51 |
ENST00000437457.6
ENST00000374878.5 ENST00000623517.3 |
ARHGEF9
|
Cdc42 guanine nucleotide exchange factor 9 |
| chr16_+_83968244 | 1.35 |
ENST00000305202.9
|
NECAB2
|
N-terminal EF-hand calcium binding protein 2 |
| chr7_+_80135694 | 1.22 |
ENST00000457358.7
|
GNAI1
|
G protein subunit alpha i1 |
| chr2_-_75560893 | 1.16 |
ENST00000410113.5
ENST00000393913.8 |
EVA1A
|
eva-1 homolog A, regulator of programmed cell death |
| chr6_-_13487593 | 1.16 |
ENST00000379287.4
ENST00000603223.1 |
GFOD1
|
glucose-fructose oxidoreductase domain containing 1 |
| chr1_-_91886144 | 1.08 |
ENST00000212355.9
|
TGFBR3
|
transforming growth factor beta receptor 3 |
| chr4_-_137532452 | 1.07 |
ENST00000412923.6
ENST00000511115.5 ENST00000344876.9 ENST00000507846.5 ENST00000510305.5 ENST00000611581.1 |
PCDH18
|
protocadherin 18 |
| chr7_-_101165114 | 1.03 |
ENST00000445482.2
|
VGF
|
VGF nerve growth factor inducible |
| chr11_-_69819410 | 1.01 |
ENST00000334134.4
|
FGF3
|
fibroblast growth factor 3 |
| chr2_-_189580773 | 1.01 |
ENST00000261024.7
|
SLC40A1
|
solute carrier family 40 member 1 |
| chr22_-_30470577 | 0.97 |
ENST00000435069.1
ENST00000540910.5 |
SEC14L3
|
SEC14 like lipid binding 3 |
| chr1_+_18630839 | 0.89 |
ENST00000420770.7
|
PAX7
|
paired box 7 |
| chr1_+_12063285 | 0.82 |
ENST00000263932.7
|
TNFRSF8
|
TNF receptor superfamily member 8 |
| chr3_+_196639684 | 0.79 |
ENST00000328557.5
|
NRROS
|
negative regulator of reactive oxygen species |
| chr6_+_15248855 | 0.77 |
ENST00000397311.4
|
JARID2
|
jumonji and AT-rich interaction domain containing 2 |
| chr6_+_130366281 | 0.76 |
ENST00000617887.4
|
TMEM200A
|
transmembrane protein 200A |
| chr15_-_52295792 | 0.74 |
ENST00000261839.12
|
MYO5C
|
myosin VC |
| chr7_-_101165558 | 0.72 |
ENST00000611537.1
ENST00000249330.3 |
VGF
|
VGF nerve growth factor inducible |
| chr6_+_32038382 | 0.70 |
ENST00000478281.5
ENST00000471671.4 ENST00000435122.3 ENST00000644719.2 |
CYP21A2
|
cytochrome P450 family 21 subfamily A member 2 |
| chrX_-_10677720 | 0.70 |
ENST00000453318.6
|
MID1
|
midline 1 |
| chr9_-_94640130 | 0.68 |
ENST00000414122.1
|
FBP1
|
fructose-bisphosphatase 1 |
| chr13_-_77919390 | 0.67 |
ENST00000475537.2
ENST00000646605.1 |
EDNRB
|
endothelin receptor type B |
| chr5_+_36151989 | 0.61 |
ENST00000274254.9
|
SKP2
|
S-phase kinase associated protein 2 |
| chr14_-_106374129 | 0.61 |
ENST00000390616.2
|
IGHV4-34
|
immunoglobulin heavy variable 4-34 |
| chr12_+_12725897 | 0.58 |
ENST00000326765.10
|
APOLD1
|
apolipoprotein L domain containing 1 |
| chr1_+_65264694 | 0.57 |
ENST00000263441.11
ENST00000395325.7 |
DNAJC6
|
DnaJ heat shock protein family (Hsp40) member C6 |
| chr20_-_25058115 | 0.56 |
ENST00000323482.9
|
ACSS1
|
acyl-CoA synthetase short chain family member 1 |
| chr19_-_11339573 | 0.55 |
ENST00000222120.8
|
RAB3D
|
RAB3D, member RAS oncogene family |
| chr1_+_2073986 | 0.54 |
ENST00000461106.6
|
PRKCZ
|
protein kinase C zeta |
| chr9_-_10612966 | 0.54 |
ENST00000381196.9
|
PTPRD
|
protein tyrosine phosphatase receptor type D |
| chr15_+_94355956 | 0.54 |
ENST00000557742.1
|
MCTP2
|
multiple C2 and transmembrane domain containing 2 |
| chr20_+_36154630 | 0.52 |
ENST00000338074.7
ENST00000636016.2 ENST00000373945.5 |
EPB41L1
|
erythrocyte membrane protein band 4.1 like 1 |
| chr15_+_47717344 | 0.52 |
ENST00000558816.5
ENST00000536845.7 |
SEMA6D
|
semaphorin 6D |
| chr19_-_49020523 | 0.52 |
ENST00000637680.1
|
ENSG00000268655.2
|
novel protein |
| chr17_-_48613468 | 0.52 |
ENST00000498634.2
|
HOXB8
|
homeobox B8 |
| chr17_+_41819201 | 0.51 |
ENST00000455106.1
|
FKBP10
|
FKBP prolyl isomerase 10 |
| chr5_-_137736066 | 0.50 |
ENST00000309755.9
|
KLHL3
|
kelch like family member 3 |
| chr12_-_95116967 | 0.50 |
ENST00000551521.5
|
FGD6
|
FYVE, RhoGEF and PH domain containing 6 |
| chr19_+_16324817 | 0.49 |
ENST00000248071.6
ENST00000592003.1 |
KLF2
|
Kruppel like factor 2 |
| chr5_+_36152077 | 0.49 |
ENST00000546211.6
ENST00000620197.5 ENST00000678270.1 ENST00000679015.1 ENST00000678580.1 ENST00000274255.11 ENST00000508514.5 |
SKP2
|
S-phase kinase associated protein 2 |
| chr2_+_24049705 | 0.48 |
ENST00000380986.9
ENST00000452109.1 |
FKBP1B
|
FKBP prolyl isomerase 1B |
| chr11_-_5441514 | 0.48 |
ENST00000380211.1
|
OR51I1
|
olfactory receptor family 51 subfamily I member 1 |
| chr7_-_120857124 | 0.45 |
ENST00000441017.5
ENST00000424710.5 ENST00000433758.5 |
TSPAN12
|
tetraspanin 12 |
| chr6_+_17393576 | 0.44 |
ENST00000229922.7
ENST00000611958.4 |
CAP2
|
cyclase associated actin cytoskeleton regulatory protein 2 |
| chr12_-_14885845 | 0.44 |
ENST00000539261.6
ENST00000228938.5 |
MGP
|
matrix Gla protein |
| chr8_-_27611424 | 0.43 |
ENST00000405140.7
|
CLU
|
clusterin |
| chr6_+_17393505 | 0.43 |
ENST00000616440.4
|
CAP2
|
cyclase associated actin cytoskeleton regulatory protein 2 |
| chr8_-_27611325 | 0.42 |
ENST00000523500.5
|
CLU
|
clusterin |
| chr6_+_89562308 | 0.42 |
ENST00000522441.5
|
ANKRD6
|
ankyrin repeat domain 6 |
| chr11_+_5488685 | 0.41 |
ENST00000322641.5
|
OR52D1
|
olfactory receptor family 52 subfamily D member 1 |
| chr19_+_1908258 | 0.40 |
ENST00000411971.5
ENST00000588907.2 |
SCAMP4
|
secretory carrier membrane protein 4 |
| chr5_-_20575850 | 0.40 |
ENST00000507958.5
|
CDH18
|
cadherin 18 |
| chr19_-_40226682 | 0.40 |
ENST00000430325.7
ENST00000599263.6 |
CCNP
|
cyclin P |
| chr6_-_31684040 | 0.39 |
ENST00000375863.7
|
LY6G5C
|
lymphocyte antigen 6 family member G5C |
| chr17_+_7855055 | 0.39 |
ENST00000574668.1
ENST00000301599.7 |
TMEM88
|
transmembrane protein 88 |
| chr3_+_111674654 | 0.39 |
ENST00000636933.1
ENST00000393934.7 ENST00000477665.2 |
PLCXD2
|
phosphatidylinositol specific phospholipase C X domain containing 2 |
| chr17_-_3595831 | 0.38 |
ENST00000399759.7
|
TRPV1
|
transient receptor potential cation channel subfamily V member 1 |
| chr3_-_129893551 | 0.38 |
ENST00000505616.5
ENST00000426664.6 ENST00000648771.1 ENST00000393238.8 |
TMCC1
|
transmembrane and coiled-coil domain family 1 |
| chr9_+_122519141 | 0.38 |
ENST00000340750.1
|
OR1J4
|
olfactory receptor family 1 subfamily J member 4 |
| chr10_-_63269057 | 0.37 |
ENST00000542921.5
|
JMJD1C
|
jumonji domain containing 1C |
| chr11_-_2140967 | 0.37 |
ENST00000381389.5
|
IGF2
|
insulin like growth factor 2 |
| chr19_+_840991 | 0.35 |
ENST00000234347.10
|
PRTN3
|
proteinase 3 |
| chr8_+_2045058 | 0.34 |
ENST00000523438.1
|
MYOM2
|
myomesin 2 |
| chr5_-_42825884 | 0.33 |
ENST00000506577.5
|
SELENOP
|
selenoprotein P |
| chr12_+_124993633 | 0.33 |
ENST00000341446.9
ENST00000671775.2 |
BRI3BP
|
BRI3 binding protein |
| chr5_-_36151853 | 0.33 |
ENST00000296603.5
|
LMBRD2
|
LMBR1 domain containing 2 |
| chr2_-_44361555 | 0.33 |
ENST00000409957.5
|
PREPL
|
prolyl endopeptidase like |
| chr9_-_94640248 | 0.33 |
ENST00000415431.5
|
FBP1
|
fructose-bisphosphatase 1 |
| chr1_+_202203721 | 0.33 |
ENST00000255432.11
|
LGR6
|
leucine rich repeat containing G protein-coupled receptor 6 |
| chr10_+_119104075 | 0.33 |
ENST00000472379.2
ENST00000361432.3 ENST00000648560.1 |
DENND10
|
DENN domain containing 10 |
| chr19_-_42423100 | 0.32 |
ENST00000597001.1
|
LIPE
|
lipase E, hormone sensitive type |
| chr3_+_196639735 | 0.31 |
ENST00000426755.5
|
PIGX
|
phosphatidylinositol glycan anchor biosynthesis class X |
| chr7_+_94509793 | 0.31 |
ENST00000297273.9
|
CASD1
|
CAS1 domain containing 1 |
| chr3_-_49813880 | 0.31 |
ENST00000333486.4
|
UBA7
|
ubiquitin like modifier activating enzyme 7 |
| chr14_-_20305932 | 0.30 |
ENST00000258821.8
ENST00000553828.1 |
TTC5
|
tetratricopeptide repeat domain 5 |
| chr2_+_24049673 | 0.30 |
ENST00000380991.8
|
FKBP1B
|
FKBP prolyl isomerase 1B |
| chr1_-_24143112 | 0.30 |
ENST00000270800.2
|
IL22RA1
|
interleukin 22 receptor subunit alpha 1 |
| chr7_-_117323041 | 0.30 |
ENST00000491214.1
ENST00000265441.8 |
WNT2
|
Wnt family member 2 |
| chr17_+_45161864 | 0.30 |
ENST00000589230.6
ENST00000591576.5 ENST00000591070.6 ENST00000592695.1 |
HEXIM2
|
HEXIM P-TEFb complex subunit 2 |
| chr13_+_91398613 | 0.30 |
ENST00000377067.9
|
GPC5
|
glypican 5 |
| chr2_-_69961624 | 0.29 |
ENST00000320256.6
|
ASPRV1
|
aspartic peptidase retroviral like 1 |
| chr1_-_13201409 | 0.29 |
ENST00000625019.3
|
PRAMEF13
|
PRAME family member 13 |
| chr1_-_182672232 | 0.28 |
ENST00000508450.5
|
RGS8
|
regulator of G protein signaling 8 |
| chr2_+_230864921 | 0.28 |
ENST00000326427.11
ENST00000335005.10 ENST00000326407.10 |
ITM2C
|
integral membrane protein 2C |
| chr2_+_64988469 | 0.28 |
ENST00000531327.5
|
SLC1A4
|
solute carrier family 1 member 4 |
| chr8_+_42338477 | 0.28 |
ENST00000518925.5
ENST00000265421.9 |
POLB
|
DNA polymerase beta |
| chr3_+_159839847 | 0.28 |
ENST00000445224.6
|
SCHIP1
|
schwannomin interacting protein 1 |
| chr19_+_17420029 | 0.27 |
ENST00000317040.12
ENST00000529939.5 ENST00000528515.5 ENST00000543795.5 |
MVB12A
|
multivesicular body subunit 12A |
| chr7_+_105014176 | 0.27 |
ENST00000257745.8
ENST00000311117.8 ENST00000478990.5 ENST00000495267.5 ENST00000476671.5 |
KMT2E
|
lysine methyltransferase 2E (inactive) |
| chr16_-_28471175 | 0.27 |
ENST00000435324.3
|
NPIPB7
|
nuclear pore complex interacting protein family member B7 |
| chr6_+_17393607 | 0.27 |
ENST00000489374.5
ENST00000378990.6 |
CAP2
|
cyclase associated actin cytoskeleton regulatory protein 2 |
| chr19_+_13764502 | 0.27 |
ENST00000040663.8
ENST00000319545.12 |
MRI1
|
methylthioribose-1-phosphate isomerase 1 |
| chr2_-_43676406 | 0.27 |
ENST00000475092.4
|
C1GALT1C1L
|
C1GALT1 specific chaperone 1 like |
| chr22_-_50261543 | 0.26 |
ENST00000395778.3
ENST00000215659.13 |
MAPK12
|
mitogen-activated protein kinase 12 |
| chr14_+_24232921 | 0.26 |
ENST00000557854.5
ENST00000399440.7 ENST00000559104.5 ENST00000456667.7 |
GMPR2
|
guanosine monophosphate reductase 2 |
| chr20_+_9514562 | 0.25 |
ENST00000246070.3
|
LAMP5
|
lysosomal associated membrane protein family member 5 |
| chr1_+_162632454 | 0.25 |
ENST00000367921.8
ENST00000367922.7 |
DDR2
|
discoidin domain receptor tyrosine kinase 2 |
| chr1_-_44674402 | 0.25 |
ENST00000420706.1
ENST00000372235.7 ENST00000372242.7 ENST00000372243.7 ENST00000372244.3 ENST00000372237.8 |
TMEM53
|
transmembrane protein 53 |
| chr1_+_43300971 | 0.25 |
ENST00000372476.8
ENST00000538015.1 |
TIE1
|
tyrosine kinase with immunoglobulin like and EGF like domains 1 |
| chr2_-_153478753 | 0.25 |
ENST00000325926.4
|
RPRM
|
reprimo, TP53 dependent G2 arrest mediator homolog |
| chr3_+_159852933 | 0.24 |
ENST00000482804.1
|
SCHIP1
|
schwannomin interacting protein 1 |
| chr16_-_28470905 | 0.24 |
ENST00000652214.1
|
NPIPB7
|
nuclear pore complex interacting protein family member B7 |
| chr8_-_27605271 | 0.24 |
ENST00000522098.1
|
CLU
|
clusterin |
| chr2_+_180980347 | 0.24 |
ENST00000602959.5
ENST00000602479.5 ENST00000392415.6 ENST00000602291.5 |
UBE2E3
|
ubiquitin conjugating enzyme E2 E3 |
| chr17_-_76308271 | 0.23 |
ENST00000680821.2
|
QRICH2
|
glutamine rich 2 |
| chr19_+_54630960 | 0.23 |
ENST00000396317.5
ENST00000396315.5 |
LILRB1
|
leukocyte immunoglobulin like receptor B1 |
| chr2_-_169824851 | 0.23 |
ENST00000410097.5
ENST00000308099.7 ENST00000409837.5 ENST00000260953.10 ENST00000538491.1 ENST00000409965.5 ENST00000392640.6 |
METTL5
|
methyltransferase like 5 |
| chr9_-_5304713 | 0.23 |
ENST00000381627.4
|
RLN2
|
relaxin 2 |
| chr8_-_65634202 | 0.23 |
ENST00000458464.3
|
ARMC1
|
armadillo repeat containing 1 |
| chr16_+_30378312 | 0.22 |
ENST00000528032.5
ENST00000622647.3 |
ZNF48
|
zinc finger protein 48 |
| chr2_-_169824814 | 0.22 |
ENST00000409340.5
|
METTL5
|
methyltransferase like 5 |
| chr12_+_48183602 | 0.22 |
ENST00000316554.5
|
CCDC184
|
coiled-coil domain containing 184 |
| chr15_-_93073111 | 0.22 |
ENST00000557420.1
ENST00000542321.6 |
RGMA
|
repulsive guidance molecule BMP co-receptor a |
| chrX_+_135309480 | 0.22 |
ENST00000635820.1
|
ETDC
|
embryonic testis differentiation homolog C |
| chr5_+_108747879 | 0.22 |
ENST00000281092.9
|
FER
|
FER tyrosine kinase |
| chr13_-_20161038 | 0.22 |
ENST00000241125.4
|
GJA3
|
gap junction protein alpha 3 |
| chr22_-_50261711 | 0.21 |
ENST00000622558.4
|
MAPK12
|
mitogen-activated protein kinase 12 |
| chr16_+_15502266 | 0.21 |
ENST00000452191.6
|
BMERB1
|
bMERB domain containing 1 |
| chr8_-_65634138 | 0.21 |
ENST00000518908.5
ENST00000276569.8 ENST00000519352.1 |
ARMC1
|
armadillo repeat containing 1 |
| chr5_+_140834230 | 0.21 |
ENST00000356878.5
ENST00000525929.2 |
PCDHA7
|
protocadherin alpha 7 |
| chr8_+_67952028 | 0.21 |
ENST00000288368.5
|
PREX2
|
phosphatidylinositol-3,4,5-trisphosphate dependent Rac exchange factor 2 |
| chr8_+_97775829 | 0.21 |
ENST00000517924.5
|
LAPTM4B
|
lysosomal protein transmembrane 4 beta |
| chr1_-_151347234 | 0.21 |
ENST00000290524.8
ENST00000452671.7 ENST00000437327.5 ENST00000368870.6 |
RFX5
|
regulatory factor X5 |
| chr7_+_139829242 | 0.20 |
ENST00000455353.6
ENST00000458722.6 ENST00000448866.7 ENST00000411653.6 |
TBXAS1
|
thromboxane A synthase 1 |
| chr10_-_87863533 | 0.20 |
ENST00000445946.5
|
KLLN
|
killin, p53 regulated DNA replication inhibitor |
| chr4_+_70721953 | 0.20 |
ENST00000381006.8
ENST00000226328.8 |
RUFY3
|
RUN and FYVE domain containing 3 |
| chr16_-_66550112 | 0.20 |
ENST00000544898.6
ENST00000620035.5 ENST00000545043.6 |
TK2
|
thymidine kinase 2 |
| chr1_-_217631034 | 0.20 |
ENST00000366934.3
ENST00000366935.8 |
GPATCH2
|
G-patch domain containing 2 |
| chr20_+_45408276 | 0.20 |
ENST00000372710.5
ENST00000443296.1 |
DBNDD2
|
dysbindin domain containing 2 |
| chr15_+_81299416 | 0.20 |
ENST00000558332.3
|
IL16
|
interleukin 16 |
| chr7_-_44141074 | 0.20 |
ENST00000457314.5
ENST00000447951.1 ENST00000431007.1 |
MYL7
|
myosin light chain 7 |
| chr18_-_76495191 | 0.20 |
ENST00000443185.7
|
ZNF516
|
zinc finger protein 516 |
| chr1_-_13347134 | 0.19 |
ENST00000334600.7
|
PRAMEF14
|
PRAME family member 14 |
| chr20_-_23421409 | 0.19 |
ENST00000377026.4
ENST00000398425.7 ENST00000432543.6 ENST00000617876.4 |
NAPB
|
NSF attachment protein beta |
| chr19_-_38878247 | 0.19 |
ENST00000591812.2
|
RINL
|
Ras and Rab interactor like |
| chr2_+_1484663 | 0.19 |
ENST00000446278.5
ENST00000469607.3 |
TPO
|
thyroid peroxidase |
| chr6_-_168319762 | 0.19 |
ENST00000366795.4
|
DACT2
|
dishevelled binding antagonist of beta catenin 2 |
| chr8_+_97775775 | 0.19 |
ENST00000521545.7
|
LAPTM4B
|
lysosomal protein transmembrane 4 beta |
| chr6_-_41154326 | 0.19 |
ENST00000426005.6
ENST00000437044.2 ENST00000373127.8 |
TREML1
|
triggering receptor expressed on myeloid cells like 1 |
| chr15_-_22185402 | 0.18 |
ENST00000557788.2
|
IGHV4OR15-8
|
immunoglobulin heavy variable 4/OR15-8 (non-functional) |
| chr16_+_29662923 | 0.18 |
ENST00000395389.2
|
SPN
|
sialophorin |
| chr20_+_2692736 | 0.18 |
ENST00000380648.9
ENST00000497450.5 |
EBF4
|
EBF family member 4 |
| chr17_-_44023106 | 0.18 |
ENST00000585950.5
ENST00000592127.5 ENST00000589334.5 |
TMEM101
|
transmembrane protein 101 |
| chr13_+_35476740 | 0.18 |
ENST00000537702.5
|
NBEA
|
neurobeachin |
| chr6_-_31592992 | 0.18 |
ENST00000340027.10
|
NCR3
|
natural cytotoxicity triggering receptor 3 |
| chr4_-_151325488 | 0.17 |
ENST00000604030.7
|
SH3D19
|
SH3 domain containing 19 |
| chr2_-_152098810 | 0.17 |
ENST00000636442.1
ENST00000638005.1 |
CACNB4
|
calcium voltage-gated channel auxiliary subunit beta 4 |
| chr1_+_17308194 | 0.17 |
ENST00000375453.5
ENST00000375448.4 |
PADI4
|
peptidyl arginine deiminase 4 |
| chr11_+_18266254 | 0.17 |
ENST00000532858.5
ENST00000649195.1 ENST00000356524.9 ENST00000405158.2 |
SAA1
|
serum amyloid A1 |
| chr10_-_33334382 | 0.17 |
ENST00000374823.9
ENST00000374821.9 ENST00000374816.7 |
NRP1
|
neuropilin 1 |
| chr5_+_134526176 | 0.17 |
ENST00000681820.1
ENST00000512386.6 ENST00000612830.2 |
JADE2
|
jade family PHD finger 2 |
| chr1_-_19980416 | 0.17 |
ENST00000375111.7
|
PLA2G2A
|
phospholipase A2 group IIA |
| chr12_-_110468222 | 0.16 |
ENST00000228827.8
ENST00000537466.6 ENST00000550974.1 |
GPN3
|
GPN-loop GTPase 3 |
| chr19_-_38256513 | 0.16 |
ENST00000347262.8
ENST00000591585.1 |
PPP1R14A
|
protein phosphatase 1 regulatory inhibitor subunit 14A |
| chrX_+_54808359 | 0.16 |
ENST00000375058.5
ENST00000375060.5 |
MAGED2
|
MAGE family member D2 |
| chr11_+_63938971 | 0.16 |
ENST00000539656.5
ENST00000377793.9 |
NAA40
|
N-alpha-acetyltransferase 40, NatD catalytic subunit |
| chr9_-_83956677 | 0.16 |
ENST00000376344.8
|
C9orf64
|
chromosome 9 open reading frame 64 |
| chr12_+_63779894 | 0.16 |
ENST00000261234.11
|
RXYLT1
|
ribitol xylosyltransferase 1 |
| chr16_+_29663219 | 0.16 |
ENST00000436527.5
ENST00000360121.4 ENST00000652691.1 ENST00000449759.2 |
SPN
QPRT
|
sialophorin quinolinate phosphoribosyltransferase |
| chr12_-_49094792 | 0.16 |
ENST00000649637.2
|
DHH
|
desert hedgehog signaling molecule |
| chr17_-_59151794 | 0.15 |
ENST00000584089.1
|
SKA2
|
spindle and kinetochore associated complex subunit 2 |
| chr7_+_33904911 | 0.15 |
ENST00000297161.6
|
BMPER
|
BMP binding endothelial regulator |
| chr11_+_125904467 | 0.15 |
ENST00000263576.11
ENST00000530414.5 ENST00000530129.6 |
DDX25
|
DEAD-box helicase 25 |
| chr1_+_12063357 | 0.15 |
ENST00000417814.3
|
TNFRSF8
|
TNF receptor superfamily member 8 |
| chr13_-_37105664 | 0.15 |
ENST00000379800.4
|
CSNK1A1L
|
casein kinase 1 alpha 1 like |
| chr14_-_68979436 | 0.15 |
ENST00000193403.10
|
ACTN1
|
actinin alpha 1 |
| chr6_-_168319691 | 0.15 |
ENST00000610183.1
ENST00000607983.1 |
DACT2
|
dishevelled binding antagonist of beta catenin 2 |
| chr17_-_3292600 | 0.15 |
ENST00000615105.1
|
OR3A1
|
olfactory receptor family 3 subfamily A member 1 |
| chr14_+_24130659 | 0.15 |
ENST00000267426.6
|
FITM1
|
fat storage inducing transmembrane protein 1 |
| chr2_+_237692290 | 0.15 |
ENST00000420665.5
ENST00000392000.4 |
LRRFIP1
|
LRR binding FLII interacting protein 1 |
| chr16_+_28292485 | 0.15 |
ENST00000341901.5
|
SBK1
|
SH3 domain binding kinase 1 |
| chr1_-_110519175 | 0.14 |
ENST00000369771.4
|
KCNA10
|
potassium voltage-gated channel subfamily A member 10 |
| chrX_+_135520616 | 0.14 |
ENST00000370752.4
ENST00000639893.2 |
INTS6L
|
integrator complex subunit 6 like |
| chr15_+_73443149 | 0.14 |
ENST00000560581.1
ENST00000331090.11 |
REC114
|
REC114 meiotic recombination protein |
| chr6_-_31721679 | 0.14 |
ENST00000495859.1
ENST00000375819.3 |
LY6G6C
|
lymphocyte antigen 6 family member G6C |
| chr12_-_84912705 | 0.14 |
ENST00000679933.1
ENST00000680260.1 ENST00000551010.2 ENST00000679453.1 ENST00000681281.1 |
SLC6A15
|
solute carrier family 6 member 15 |
| chr17_-_8156320 | 0.14 |
ENST00000584202.1
ENST00000354903.9 ENST00000577253.5 |
PER1
|
period circadian regulator 1 |
| chr17_+_42780592 | 0.14 |
ENST00000246914.10
|
WNK4
|
WNK lysine deficient protein kinase 4 |
| chr1_+_228208054 | 0.14 |
ENST00000284548.16
ENST00000422127.5 |
OBSCN
|
obscurin, cytoskeletal calmodulin and titin-interacting RhoGEF |
| chr12_-_54984667 | 0.14 |
ENST00000524668.5
ENST00000533607.1 ENST00000449076.6 |
TESPA1
|
thymocyte expressed, positive selection associated 1 |
| chrX_+_54808334 | 0.14 |
ENST00000218439.8
|
MAGED2
|
MAGE family member D2 |
| chr6_-_31592952 | 0.13 |
ENST00000376073.8
ENST00000376072.7 |
NCR3
|
natural cytotoxicity triggering receptor 3 |
| chr8_-_96235533 | 0.13 |
ENST00000518406.5
ENST00000287022.10 ENST00000523920.1 |
UQCRB
|
ubiquinol-cytochrome c reductase binding protein |
| chr2_+_70978380 | 0.13 |
ENST00000272421.10
ENST00000441349.5 ENST00000457410.5 |
ANKRD53
|
ankyrin repeat domain 53 |
| chrM_+_4467 | 0.13 |
ENST00000361453.3
|
MT-ND2
|
mitochondrially encoded NADH:ubiquinone oxidoreductase core subunit 2 |
| chr4_+_153257339 | 0.13 |
ENST00000676374.1
ENST00000676196.1 ENST00000674935.1 ENST00000674769.1 ENST00000674896.1 ENST00000676191.1 ENST00000675312.1 ENST00000675456.1 ENST00000674847.1 ENST00000675977.1 ENST00000676264.1 ENST00000674726.1 ENST00000676252.1 ENST00000674730.1 ENST00000675738.1 ENST00000482578.3 |
TRIM2
|
tripartite motif containing 2 |
| chr13_+_111114550 | 0.13 |
ENST00000317133.9
|
ARHGEF7
|
Rho guanine nucleotide exchange factor 7 |
| chr2_-_70835808 | 0.13 |
ENST00000410009.5
|
CD207
|
CD207 molecule |
| chrX_+_37780049 | 0.13 |
ENST00000378588.5
|
CYBB
|
cytochrome b-245 beta chain |
| chr19_-_15898057 | 0.12 |
ENST00000011989.11
ENST00000221700.11 |
CYP4F2
|
cytochrome P450 family 4 subfamily F member 2 |
| chr2_+_44168866 | 0.12 |
ENST00000282412.9
ENST00000409432.7 ENST00000378551.6 ENST00000345249.8 |
PPM1B
|
protein phosphatase, Mg2+/Mn2+ dependent 1B |
| chr11_+_26188836 | 0.12 |
ENST00000672621.1
|
ANO3
|
anoctamin 3 |
| chr14_+_24310170 | 0.12 |
ENST00000530080.1
|
LTB4R2
|
leukotriene B4 receptor 2 |
| chr19_-_54242751 | 0.12 |
ENST00000245621.6
ENST00000396365.6 |
LILRA6
|
leukocyte immunoglobulin like receptor A6 |
| chr3_+_98497681 | 0.12 |
ENST00000427338.3
|
OR5K2
|
olfactory receptor family 5 subfamily K member 2 |
| chr3_-_146544701 | 0.12 |
ENST00000487389.5
|
PLSCR1
|
phospholipid scramblase 1 |
| chr4_+_81030700 | 0.12 |
ENST00000282701.4
|
BMP3
|
bone morphogenetic protein 3 |
| chr15_+_75336078 | 0.12 |
ENST00000567377.5
ENST00000562789.5 ENST00000568301.1 |
COMMD4
|
COMM domain containing 4 |
| chr8_-_78805306 | 0.12 |
ENST00000639719.1
ENST00000263851.9 |
IL7
|
interleukin 7 |
| chr1_+_161153968 | 0.12 |
ENST00000368003.6
|
UFC1
|
ubiquitin-fold modifier conjugating enzyme 1 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.4 | GO:1904021 | negative regulation of G-protein coupled receptor internalization(GO:1904021) |
| 0.3 | 1.0 | GO:0034395 | regulation of transcription from RNA polymerase II promoter in response to iron(GO:0034395) |
| 0.3 | 1.0 | GO:0045556 | TRAIL biosynthetic process(GO:0045553) regulation of TRAIL biosynthetic process(GO:0045554) positive regulation of TRAIL biosynthetic process(GO:0045556) |
| 0.3 | 0.9 | GO:0014813 | skeletal muscle satellite cell commitment(GO:0014813) |
| 0.3 | 1.1 | GO:0007181 | transforming growth factor beta receptor complex assembly(GO:0007181) |
| 0.3 | 1.0 | GO:0005986 | sucrose biosynthetic process(GO:0005986) |
| 0.2 | 1.1 | GO:1902998 | regulation of neuronal signal transduction(GO:1902847) positive regulation of neurofibrillary tangle assembly(GO:1902998) |
| 0.2 | 0.6 | GO:0019427 | acetate biosynthetic process(GO:0019413) acetyl-CoA biosynthetic process from acetate(GO:0019427) propionate biosynthetic process(GO:0019542) |
| 0.2 | 1.1 | GO:0071460 | cellular response to cell-matrix adhesion(GO:0071460) |
| 0.1 | 0.7 | GO:0035645 | enteric smooth muscle cell differentiation(GO:0035645) |
| 0.1 | 0.5 | GO:0018125 | peptidyl-cysteine methylation(GO:0018125) |
| 0.1 | 0.5 | GO:0072156 | distal tubule morphogenesis(GO:0072156) |
| 0.1 | 0.3 | GO:0001808 | negative regulation of type IV hypersensitivity(GO:0001808) |
| 0.1 | 0.5 | GO:0071409 | cellular response to cycloheximide(GO:0071409) |
| 0.1 | 0.4 | GO:0050960 | detection of temperature stimulus involved in thermoception(GO:0050960) response to capsazepine(GO:1901594) |
| 0.1 | 0.5 | GO:2001181 | positive regulation of interleukin-10 secretion(GO:2001181) |
| 0.1 | 1.2 | GO:1904321 | response to forskolin(GO:1904321) cellular response to forskolin(GO:1904322) |
| 0.1 | 0.3 | GO:0072061 | inner medullary collecting duct development(GO:0072061) negative regulation of nodal signaling pathway(GO:1900108) |
| 0.1 | 0.8 | GO:0048680 | positive regulation of axon regeneration(GO:0048680) |
| 0.1 | 0.6 | GO:0038028 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) |
| 0.1 | 0.3 | GO:0006288 | base-excision repair, DNA ligation(GO:0006288) |
| 0.1 | 0.3 | GO:0097029 | mature conventional dendritic cell differentiation(GO:0097029) |
| 0.1 | 0.1 | GO:0086100 | endothelin receptor signaling pathway(GO:0086100) |
| 0.1 | 0.2 | GO:0035494 | SNARE complex disassembly(GO:0035494) |
| 0.1 | 0.2 | GO:0035548 | gamma-delta T cell activation involved in immune response(GO:0002290) negative regulation of interferon-beta secretion(GO:0035548) regulation of gamma-delta T cell activation involved in immune response(GO:2001191) positive regulation of gamma-delta T cell activation involved in immune response(GO:2001193) |
| 0.1 | 0.2 | GO:0032304 | negative regulation of icosanoid secretion(GO:0032304) |
| 0.1 | 0.5 | GO:0007185 | transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.1 | 0.2 | GO:1990022 | RNA polymerase II complex import to nucleus(GO:0044376) RNA polymerase III complex localization to nucleus(GO:1990022) |
| 0.1 | 1.7 | GO:0002021 | response to dietary excess(GO:0002021) |
| 0.1 | 0.3 | GO:0019075 | virus maturation(GO:0019075) |
| 0.1 | 0.3 | GO:0046340 | diacylglycerol catabolic process(GO:0046340) |
| 0.1 | 0.3 | GO:0060370 | susceptibility to T cell mediated cytotoxicity(GO:0060370) |
| 0.0 | 0.3 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 0.0 | 0.2 | GO:1990523 | bone regeneration(GO:1990523) |
| 0.0 | 0.6 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 0.0 | 0.7 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.0 | 0.8 | GO:0051574 | positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.0 | 0.3 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.0 | 0.2 | GO:0018101 | protein citrullination(GO:0018101) histone citrullination(GO:0036414) |
| 0.0 | 0.3 | GO:0002074 | extraocular skeletal muscle development(GO:0002074) |
| 0.0 | 0.2 | GO:0061552 | vestibulocochlear nerve structural organization(GO:0021649) positive regulation of cytokine activity(GO:0060301) ganglion morphogenesis(GO:0061552) VEGF-activated neuropilin signaling pathway involved in axon guidance(GO:1902378) dorsal root ganglion morphogenesis(GO:1904835) otic placode development(GO:1905040) |
| 0.0 | 0.1 | GO:1904845 | response to L-glutamine(GO:1904844) cellular response to L-glutamine(GO:1904845) |
| 0.0 | 0.3 | GO:0015808 | L-alanine transport(GO:0015808) |
| 0.0 | 0.7 | GO:0008212 | mineralocorticoid biosynthetic process(GO:0006705) mineralocorticoid metabolic process(GO:0008212) |
| 0.0 | 0.3 | GO:0060424 | foregut regionalization(GO:0060423) lung field specification(GO:0060424) lung induction(GO:0060492) |
| 0.0 | 0.1 | GO:0030718 | germ-line stem cell population maintenance(GO:0030718) positive regulation of meiosis I(GO:0060903) |
| 0.0 | 0.1 | GO:0010387 | COP9 signalosome assembly(GO:0010387) |
| 0.0 | 0.2 | GO:0046092 | deoxycytidine metabolic process(GO:0046092) |
| 0.0 | 0.3 | GO:0001887 | selenium compound metabolic process(GO:0001887) |
| 0.0 | 0.5 | GO:0006975 | DNA damage induced protein phosphorylation(GO:0006975) |
| 0.0 | 0.4 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.0 | 0.1 | GO:0033591 | response to L-ascorbic acid(GO:0033591) |
| 0.0 | 0.1 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.0 | 0.1 | GO:0021569 | rhombomere 3 development(GO:0021569) |
| 0.0 | 0.1 | GO:0035261 | external genitalia morphogenesis(GO:0035261) |
| 0.0 | 0.1 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.0 | 0.3 | GO:0042985 | negative regulation of amyloid precursor protein biosynthetic process(GO:0042985) |
| 0.0 | 0.1 | GO:1990928 | response to amino acid starvation(GO:1990928) |
| 0.0 | 0.1 | GO:0050883 | negative regulation of sodium:proton antiporter activity(GO:0032416) musculoskeletal movement, spinal reflex action(GO:0050883) |
| 0.0 | 0.1 | GO:0097167 | circadian regulation of translation(GO:0097167) |
| 0.0 | 0.1 | GO:0031455 | glycine betaine biosynthetic process from choline(GO:0019285) glycine betaine metabolic process(GO:0031455) glycine betaine biosynthetic process(GO:0031456) |
| 0.0 | 0.1 | GO:0030185 | nitric oxide transport(GO:0030185) |
| 0.0 | 0.5 | GO:0021516 | dorsal spinal cord development(GO:0021516) |
| 0.0 | 0.1 | GO:0006532 | aspartate biosynthetic process(GO:0006532) |
| 0.0 | 0.1 | GO:0035524 | proline transmembrane transport(GO:0035524) |
| 0.0 | 0.7 | GO:0055026 | negative regulation of cardiac muscle tissue development(GO:0055026) |
| 0.0 | 0.3 | GO:0060159 | regulation of dopamine receptor signaling pathway(GO:0060159) |
| 0.0 | 0.4 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.0 | 0.1 | GO:0006499 | N-terminal protein myristoylation(GO:0006499) |
| 0.0 | 0.3 | GO:0090091 | positive regulation of extracellular matrix disassembly(GO:0090091) |
| 0.0 | 0.1 | GO:1990564 | protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.0 | 0.2 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 0.0 | 0.1 | GO:0010890 | positive regulation of sequestering of triglyceride(GO:0010890) |
| 0.0 | 0.1 | GO:1902412 | regulation of mitotic cytokinesis(GO:1902412) |
| 0.0 | 0.4 | GO:0001502 | cartilage condensation(GO:0001502) |
| 0.0 | 0.1 | GO:0000454 | snoRNA guided rRNA pseudouridine synthesis(GO:0000454) |
| 0.0 | 0.1 | GO:1990167 | protein K6-linked deubiquitination(GO:0044313) protein K27-linked deubiquitination(GO:1990167) |
| 0.0 | 0.1 | GO:0071442 | positive regulation of histone H3-K14 acetylation(GO:0071442) |
| 0.0 | 0.1 | GO:0035879 | plasma membrane lactate transport(GO:0035879) |
| 0.0 | 0.5 | GO:0014912 | negative regulation of smooth muscle cell migration(GO:0014912) |
| 0.0 | 0.6 | GO:0045601 | regulation of endothelial cell differentiation(GO:0045601) |
| 0.0 | 0.1 | GO:0002933 | lipid hydroxylation(GO:0002933) |
| 0.0 | 0.1 | GO:0061591 | calcium activated phospholipid scrambling(GO:0061588) calcium activated phosphatidylcholine scrambling(GO:0061590) calcium activated galactosylceramide scrambling(GO:0061591) |
| 0.0 | 0.3 | GO:0016254 | preassembly of GPI anchor in ER membrane(GO:0016254) |
| 0.0 | 0.5 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.0 | 0.1 | GO:0017062 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) |
| 0.0 | 0.0 | GO:0036309 | protein localization to M-band(GO:0036309) |
| 0.0 | 0.0 | GO:0090119 | vesicle-mediated cholesterol transport(GO:0090119) |
| 0.0 | 0.2 | GO:0048681 | negative regulation of axon regeneration(GO:0048681) |
| 0.0 | 0.2 | GO:0030238 | male sex determination(GO:0030238) Leydig cell differentiation(GO:0033327) |
| 0.0 | 0.1 | GO:0061143 | alveolar primary septum development(GO:0061143) paramesonephric duct development(GO:0061205) |
| 0.0 | 0.5 | GO:0007190 | activation of adenylate cyclase activity(GO:0007190) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.1 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.1 | 1.1 | GO:0097418 | neurofibrillary tangle(GO:0097418) |
| 0.1 | 0.3 | GO:1902560 | GMP reductase complex(GO:1902560) |
| 0.0 | 0.5 | GO:0043203 | axon hillock(GO:0043203) |
| 0.0 | 0.1 | GO:0010370 | perinucleolar chromocenter(GO:0010370) |
| 0.0 | 0.1 | GO:0032116 | SMC loading complex(GO:0032116) |
| 0.0 | 0.3 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.0 | 0.8 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 0.1 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.0 | 0.5 | GO:0042588 | zymogen granule(GO:0042588) |
| 0.0 | 1.2 | GO:0099738 | cell cortex region(GO:0099738) |
| 0.0 | 0.4 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 0.3 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 0.2 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.0 | 0.3 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 0.0 | 0.1 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.0 | 0.3 | GO:1990909 | Wnt signalosome(GO:1990909) |
| 0.0 | 1.1 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.3 | GO:0031254 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.0 | 0.3 | GO:0032982 | myosin filament(GO:0032982) |
| 0.0 | 0.1 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 0.0 | 0.1 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.0 | 0.8 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.4 | GO:0031687 | A2A adenosine receptor binding(GO:0031687) |
| 0.3 | 1.0 | GO:0042132 | fructose 1,6-bisphosphate 1-phosphatase activity(GO:0042132) |
| 0.2 | 0.8 | GO:0005219 | ryanodine-sensitive calcium-release channel activity(GO:0005219) |
| 0.1 | 0.6 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.1 | 1.0 | GO:0015093 | ferrous iron transmembrane transporter activity(GO:0015093) |
| 0.1 | 0.9 | GO:0004962 | endothelin receptor activity(GO:0004962) |
| 0.1 | 0.3 | GO:0033878 | hormone-sensitive lipase activity(GO:0033878) |
| 0.1 | 1.1 | GO:0005114 | type II transforming growth factor beta receptor binding(GO:0005114) |
| 0.1 | 1.2 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.1 | 0.2 | GO:0036134 | thromboxane-A synthase activity(GO:0004796) 12-hydroxyheptadecatrienoic acid synthase activity(GO:0036134) |
| 0.1 | 1.0 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.1 | 0.2 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.1 | 1.2 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.1 | 0.3 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 0.1 | 0.2 | GO:0030107 | HLA-A specific inhibitory MHC class I receptor activity(GO:0030107) |
| 0.1 | 0.4 | GO:0097603 | temperature-gated ion channel activity(GO:0097603) |
| 0.1 | 0.3 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.1 | 0.3 | GO:0003920 | GMP reductase activity(GO:0003920) oxidoreductase activity, acting on NAD(P)H, nitrogenous group as acceptor(GO:0016657) |
| 0.0 | 1.1 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 1.7 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 0.2 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.0 | 0.3 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.0 | 0.1 | GO:0097259 | tocopherol omega-hydroxylase activity(GO:0052870) alpha-tocopherol omega-hydroxylase activity(GO:0052871) 20-hydroxy-leukotriene B4 omega oxidase activity(GO:0097258) 20-aldehyde-leukotriene B4 20-monooxygenase activity(GO:0097259) |
| 0.0 | 0.3 | GO:0022858 | L-alanine transmembrane transporter activity(GO:0015180) alanine transmembrane transporter activity(GO:0022858) |
| 0.0 | 0.2 | GO:1990189 | peptide-serine-N-acetyltransferase activity(GO:1990189) |
| 0.0 | 0.2 | GO:0004447 | iodide peroxidase activity(GO:0004447) |
| 0.0 | 0.3 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.0 | 0.1 | GO:0034584 | piRNA binding(GO:0034584) |
| 0.0 | 0.5 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.1 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.0 | 0.1 | GO:1902271 | D3 vitamins binding(GO:1902271) |
| 0.0 | 0.2 | GO:0004137 | deoxycytidine kinase activity(GO:0004137) thymidine kinase activity(GO:0004797) |
| 0.0 | 0.3 | GO:0008430 | selenium binding(GO:0008430) |
| 0.0 | 0.3 | GO:0016413 | O-acetyltransferase activity(GO:0016413) |
| 0.0 | 0.3 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.0 | 0.9 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 1.1 | GO:0032452 | histone demethylase activity(GO:0032452) |
| 0.0 | 0.8 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.0 | 0.3 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.0 | 0.1 | GO:0004069 | L-aspartate:2-oxoglutarate aminotransferase activity(GO:0004069) |
| 0.0 | 0.6 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.0 | 0.1 | GO:0015193 | L-proline transmembrane transporter activity(GO:0015193) |
| 0.0 | 0.2 | GO:0004514 | nicotinate-nucleotide diphosphorylase (carboxylating) activity(GO:0004514) |
| 0.0 | 0.5 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 0.5 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 0.1 | GO:0036033 | mediator complex binding(GO:0036033) |
| 0.0 | 0.1 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.0 | 0.9 | GO:0000983 | transcription factor activity, RNA polymerase II core promoter sequence-specific(GO:0000983) |
| 0.0 | 0.1 | GO:0008121 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.0 | 0.4 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.5 | GO:0019198 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 0.1 | GO:0004096 | catalase activity(GO:0004096) |
| 0.0 | 0.2 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.0 | 0.2 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.0 | 0.2 | GO:1990459 | transferrin receptor binding(GO:1990459) |
| 0.0 | 0.3 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.0 | 0.1 | GO:0005221 | intracellular cyclic nucleotide activated cation channel activity(GO:0005221) cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.0 | 0.3 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.0 | 0.4 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 0.1 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.0 | 0.3 | GO:0003906 | DNA-(apurinic or apyrimidinic site) lyase activity(GO:0003906) |
| 0.0 | 0.7 | GO:0030546 | receptor activator activity(GO:0030546) |
| 0.0 | 0.2 | GO:0005113 | patched binding(GO:0005113) |
| 0.0 | 0.0 | GO:0008466 | glycogenin glucosyltransferase activity(GO:0008466) |
| 0.0 | 0.1 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) |
| 0.0 | 0.1 | GO:0051373 | FATZ binding(GO:0051373) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.2 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.0 | 1.3 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 0.5 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 1.1 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 1.6 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.7 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.0 | 1.4 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 0.5 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 0.5 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.2 | REACTOME ADENYLATE CYCLASE INHIBITORY PATHWAY | Genes involved in Adenylate cyclase inhibitory pathway |
| 0.0 | 1.0 | REACTOME FGFR1 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR1 ligand binding and activation |
| 0.0 | 0.6 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.0 | 1.5 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 0.8 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.0 | 0.8 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 1.0 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.0 | 0.5 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 0.5 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.0 | 1.1 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 0.2 | REACTOME SIGNALING BY SCF KIT | Genes involved in Signaling by SCF-KIT |
| 0.0 | 0.1 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.0 | 0.4 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.3 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.0 | 0.3 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 1.1 | REACTOME SCFSKP2 MEDIATED DEGRADATION OF P27 P21 | Genes involved in SCF(Skp2)-mediated degradation of p27/p21 |
| 0.0 | 0.3 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.8 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 0.3 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.0 | 0.3 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 1.5 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |