Project
Inflammatory response time course, HUVEC (Wada, 2009)
Navigation
Downloads
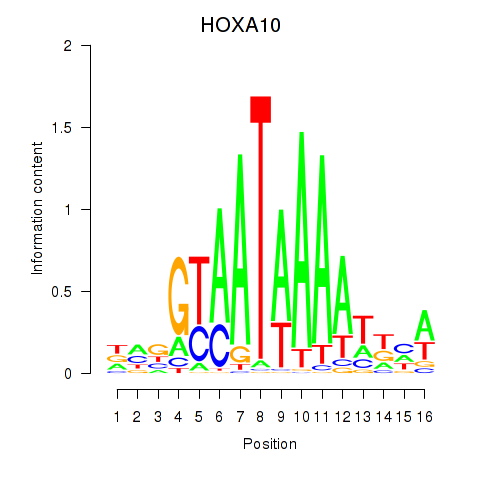
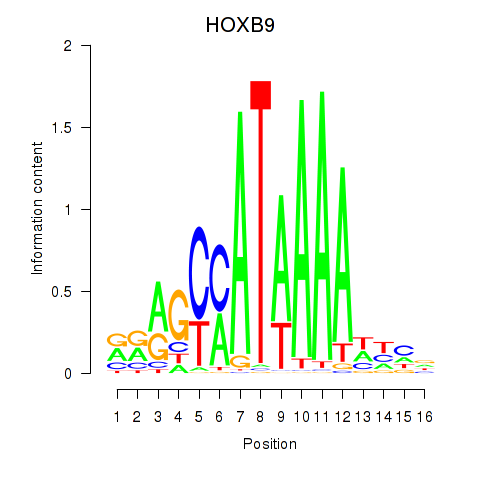
Results for HOXA10_HOXB9
Z-value: 0.45
Transcription factors associated with HOXA10_HOXB9
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HOXA10
|
ENSG00000253293.5 | HOXA10 |
|
HOXB9
|
ENSG00000170689.10 | HOXB9 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HOXA10 | hg38_v1_chr7_-_27174274_27174335, hg38_v1_chr7_-_27174253_27174267 | -0.45 | 2.3e-02 | Click! |
| HOXB9 | hg38_v1_chr17_-_48626325_48626379 | 0.06 | 7.6e-01 | Click! |
Activity profile of HOXA10_HOXB9 motif
Sorted Z-values of HOXA10_HOXB9 motif
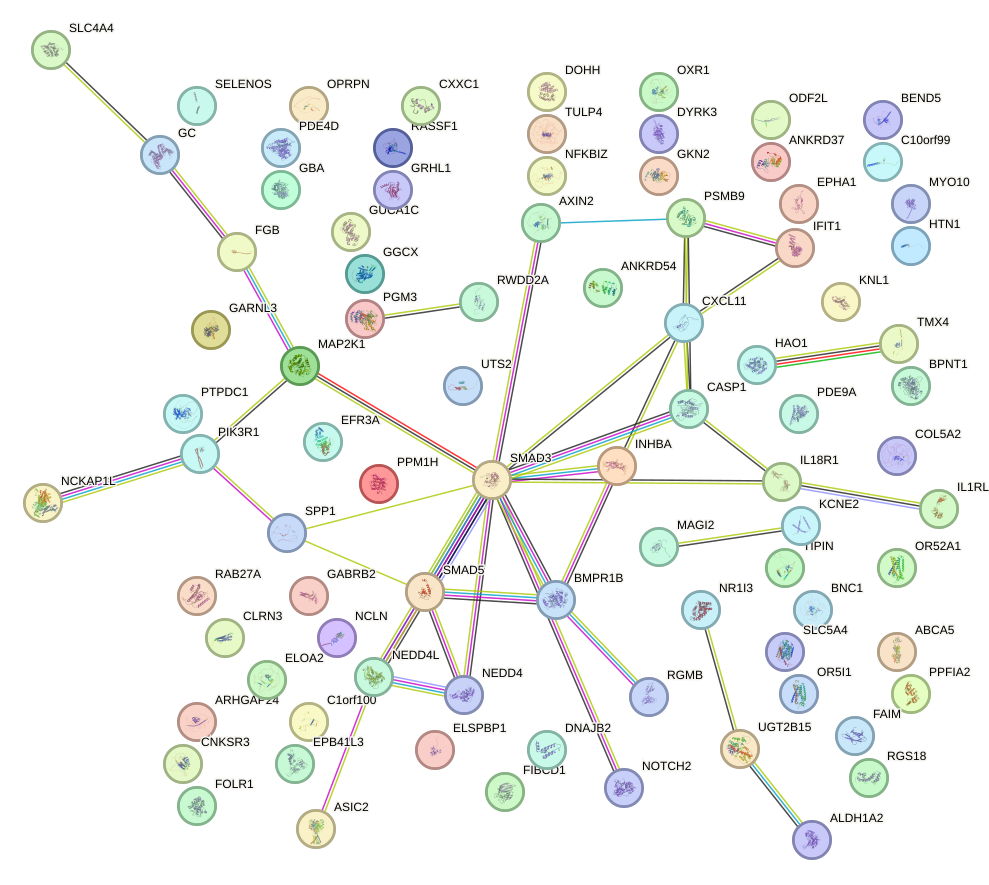
Network of associatons between targets according to the STRING database.
First level regulatory network of HOXA10_HOXB9
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr15_+_67125707 | 1.33 |
ENST00000540846.6
|
SMAD3
|
SMAD family member 3 |
| chr6_+_32844789 | 1.29 |
ENST00000414474.5
|
PSMB9
|
proteasome 20S subunit beta 9 |
| chr15_-_55249029 | 1.08 |
ENST00000566877.5
|
RAB27A
|
RAB27A, member RAS oncogene family |
| chr18_-_5396265 | 0.97 |
ENST00000579951.2
|
EPB41L3
|
erythrocyte membrane protein band 4.1 like 3 |
| chr4_-_76036060 | 0.91 |
ENST00000306621.8
|
CXCL11
|
C-X-C motif chemokine ligand 11 |
| chr1_-_120054225 | 0.90 |
ENST00000602566.6
|
NOTCH2
|
notch receptor 2 |
| chr4_+_70050431 | 0.72 |
ENST00000511674.5
ENST00000246896.8 |
HTN1
|
histatin 1 |
| chr1_+_244352627 | 0.71 |
ENST00000366537.5
ENST00000308105.5 |
C1orf100
|
chromosome 1 open reading frame 100 |
| chr5_+_98773651 | 0.56 |
ENST00000513185.3
|
RGMB
|
repulsive guidance molecule BMP co-receptor b |
| chr1_+_206635521 | 0.55 |
ENST00000367109.8
|
DYRK3
|
dual specificity tyrosine phosphorylation regulated kinase 3 |
| chr7_-_41703062 | 0.52 |
ENST00000242208.5
|
INHBA
|
inhibin subunit beta A |
| chr18_+_58196736 | 0.51 |
ENST00000675221.1
|
NEDD4L
|
NEDD4 like E3 ubiquitin protein ligase |
| chr11_+_111245725 | 0.49 |
ENST00000280325.7
|
C11orf53
|
chromosome 11 open reading frame 53 |
| chr10_+_89392546 | 0.47 |
ENST00000546318.2
ENST00000371804.4 |
IFIT1
|
interferon induced protein with tetratricopeptide repeats 1 |
| chr11_+_55635113 | 0.46 |
ENST00000641760.1
|
OR4P4
|
olfactory receptor family 4 subfamily P member 4 |
| chr4_+_95051671 | 0.45 |
ENST00000440890.7
|
BMPR1B
|
bone morphogenetic protein receptor type 1B |
| chr5_-_58999885 | 0.44 |
ENST00000317118.12
|
PDE4D
|
phosphodiesterase 4D |
| chr6_-_154430495 | 0.38 |
ENST00000424998.3
|
CNKSR3
|
CNKSR family member 3 |
| chr6_+_29301701 | 0.37 |
ENST00000641895.1
|
OR14J1
|
olfactory receptor family 14 subfamily J member 1 |
| chr11_-_55936400 | 0.34 |
ENST00000301532.3
|
OR5I1
|
olfactory receptor family 5 subfamily I member 1 |
| chr2_+_102311502 | 0.33 |
ENST00000404917.6
ENST00000410040.5 |
IL1RL1
IL18R1
|
interleukin 1 receptor like 1 interleukin 18 receptor 1 |
| chr2_-_216013582 | 0.30 |
ENST00000620139.4
|
MREG
|
melanoregulin |
| chr2_-_189179754 | 0.29 |
ENST00000374866.9
ENST00000618828.1 |
COL5A2
|
collagen type V alpha 2 chain |
| chr19_+_47994625 | 0.29 |
ENST00000339841.7
|
ELSPBP1
|
epididymal sperm binding protein 1 |
| chr10_-_127892930 | 0.29 |
ENST00000368671.4
|
CLRN3
|
clarin 3 |
| chr19_+_47994696 | 0.28 |
ENST00000596043.5
ENST00000597519.5 |
ELSPBP1
|
epididymal sperm binding protein 1 |
| chr11_-_105035113 | 0.26 |
ENST00000526568.5
ENST00000531166.5 ENST00000534497.5 ENST00000527979.5 ENST00000533400.6 ENST00000528974.1 ENST00000525825.5 ENST00000353247.9 ENST00000446369.5 ENST00000436863.7 |
CASP1
|
caspase 1 |
| chr12_+_69825221 | 0.26 |
ENST00000552032.7
|
MYRFL
|
myelin regulatory factor like |
| chr11_-_5154757 | 0.26 |
ENST00000380367.3
|
OR52A1
|
olfactory receptor family 52 subfamily A member 1 |
| chr17_-_65561137 | 0.25 |
ENST00000580513.1
|
AXIN2
|
axin 2 |
| chr22_-_37844308 | 0.25 |
ENST00000411961.6
ENST00000434930.1 ENST00000215941.9 |
ANKRD54
|
ankyrin repeat domain 54 |
| chr15_+_66386902 | 0.23 |
ENST00000307102.10
|
MAP2K1
|
mitogen-activated protein kinase kinase 1 |
| chr5_+_68290637 | 0.23 |
ENST00000336483.9
|
PIK3R1
|
phosphoinositide-3-kinase regulatory subunit 1 |
| chr3_+_101827982 | 0.23 |
ENST00000461724.5
ENST00000483180.5 ENST00000394054.6 |
NFKBIZ
|
NFKB inhibitor zeta |
| chr15_+_40594001 | 0.22 |
ENST00000346991.9
ENST00000528975.5 |
KNL1
|
kinetochore scaffold 1 |
| chr10_-_72088533 | 0.22 |
ENST00000373109.7
|
SPOCK2
|
SPARC (osteonectin), cwcv and kazal like domains proteoglycan 2 |
| chr4_+_71062642 | 0.21 |
ENST00000649996.1
|
SLC4A4
|
solute carrier family 4 member 4 |
| chr12_+_69825273 | 0.21 |
ENST00000547771.6
|
MYRFL
|
myelin regulatory factor like |
| chr3_+_138621225 | 0.21 |
ENST00000479848.1
|
FAIM
|
Fas apoptotic inhibitory molecule |
| chr4_+_94974984 | 0.21 |
ENST00000672698.1
|
BMPR1B
|
bone morphogenetic protein receptor type 1B |
| chr6_-_83193882 | 0.20 |
ENST00000506587.5
ENST00000507554.1 |
PGM3
|
phosphoglucomutase 3 |
| chr17_-_10549652 | 0.20 |
ENST00000245503.10
|
MYH2
|
myosin heavy chain 2 |
| chr1_-_48776811 | 0.19 |
ENST00000371833.4
|
BEND5
|
BEN domain containing 5 |
| chr9_-_130939205 | 0.19 |
ENST00000372338.9
|
FIBCD1
|
fibrinogen C domain containing 1 |
| chr6_+_158312459 | 0.19 |
ENST00000367097.8
|
TULP4
|
TUB like protein 4 |
| chr12_+_54498766 | 0.19 |
ENST00000545638.2
|
NCKAP1L
|
NCK associated protein 1 like |
| chr3_-_108953870 | 0.19 |
ENST00000261047.8
|
GUCA1C
|
guanylate cyclase activator 1C |
| chr2_+_161136901 | 0.19 |
ENST00000259075.6
ENST00000432002.5 |
TANK
|
TRAF family member associated NFKB activator |
| chr12_-_21775581 | 0.19 |
ENST00000537950.1
ENST00000665145.1 |
KCNJ8
|
potassium inwardly rectifying channel subfamily J member 8 |
| chr22_-_32255344 | 0.19 |
ENST00000266086.6
|
SLC5A4
|
solute carrier family 5 member 4 |
| chr8_-_103501890 | 0.19 |
ENST00000649416.1
|
ENSG00000285982.1
|
novel protein |
| chr4_-_71784046 | 0.19 |
ENST00000513476.5
ENST00000273951.13 |
GC
|
GC vitamin D binding protein |
| chr5_+_136160986 | 0.18 |
ENST00000507637.1
|
SMAD5
|
SMAD family member 5 |
| chr4_-_68670648 | 0.18 |
ENST00000338206.6
|
UGT2B15
|
UDP glucuronosyltransferase family 2 member B15 |
| chr4_+_70397931 | 0.18 |
ENST00000399575.7
|
OPRPN
|
opiorphin prepropeptide |
| chr5_-_161546671 | 0.18 |
ENST00000517547.5
|
GABRB2
|
gamma-aminobutyric acid type A receptor subunit beta2 |
| chr15_-_101277287 | 0.18 |
ENST00000528346.1
ENST00000531964.5 ENST00000398226.7 ENST00000526049.6 |
SELENOS
|
selenoprotein S |
| chr5_-_161546970 | 0.17 |
ENST00000675303.1
|
GABRB2
|
gamma-aminobutyric acid type A receptor subunit beta2 |
| chr10_+_84173793 | 0.17 |
ENST00000372126.4
|
C10orf99
|
chromosome 10 open reading frame 99 |
| chr21_+_34364003 | 0.17 |
ENST00000290310.4
|
KCNE2
|
potassium voltage-gated channel subfamily E regulatory subunit 2 |
| chr17_-_10549612 | 0.17 |
ENST00000532183.6
ENST00000397183.6 ENST00000420805.1 |
MYH2
|
myosin heavy chain 2 |
| chr18_+_58341038 | 0.17 |
ENST00000679791.1
|
NEDD4L
|
NEDD4 like E3 ubiquitin protein ligase |
| chr7_-_143408848 | 0.17 |
ENST00000275815.4
|
EPHA1
|
EPH receptor A1 |
| chr21_+_42653585 | 0.17 |
ENST00000291539.11
|
PDE9A
|
phosphodiesterase 9A |
| chr2_+_219279330 | 0.17 |
ENST00000425450.5
ENST00000392086.8 ENST00000421532.5 ENST00000336576.10 |
DNAJB2
|
DnaJ heat shock protein family (Hsp40) member B2 |
| chr3_-_50338226 | 0.16 |
ENST00000395126.7
|
RASSF1
|
Ras association domain family member 1 |
| chr4_+_70383123 | 0.16 |
ENST00000304915.8
|
SMR3B
|
submaxillary gland androgen regulated protein 3B |
| chr15_-_58279245 | 0.16 |
ENST00000558231.5
|
ALDH1A2
|
aldehyde dehydrogenase 1 family member A2 |
| chr19_+_3185911 | 0.16 |
ENST00000246117.9
ENST00000588428.5 |
NCLN
|
nicalin |
| chr1_-_161238085 | 0.15 |
ENST00000512372.5
ENST00000437437.6 ENST00000412844.6 ENST00000428574.6 ENST00000442691.6 ENST00000505005.5 ENST00000508740.5 ENST00000367981.7 ENST00000502985.5 ENST00000504010.5 ENST00000508387.5 ENST00000511676.5 ENST00000511748.5 ENST00000511944.5 ENST00000515621.5 ENST00000367984.8 ENST00000367985.7 |
NR1I3
|
nuclear receptor subfamily 1 group I member 3 |
| chr12_-_62935117 | 0.15 |
ENST00000228705.7
|
PPM1H
|
protein phosphatase, Mg2+/Mn2+ dependent 1H |
| chr8_+_131939865 | 0.15 |
ENST00000520362.5
ENST00000519656.1 |
EFR3A
|
EFR3 homolog A |
| chr9_+_127264740 | 0.14 |
ENST00000373387.9
|
GARNL3
|
GTPase activating Rap/RanGAP domain like 3 |
| chr9_+_94084458 | 0.14 |
ENST00000620992.5
ENST00000288976.3 |
PTPDC1
|
protein tyrosine phosphatase domain containing 1 |
| chr5_-_16738341 | 0.14 |
ENST00000515803.5
|
MYO10
|
myosin X |
| chr15_-_55917080 | 0.14 |
ENST00000506154.1
|
NEDD4
|
NEDD4 E3 ubiquitin protein ligase |
| chr2_-_164773733 | 0.13 |
ENST00000480873.1
|
COBLL1
|
cordon-bleu WH2 repeat protein like 1 |
| chr3_-_108953762 | 0.13 |
ENST00000393963.7
ENST00000471108.1 |
GUCA1C
|
guanylate cyclase activator 1C |
| chr20_-_7940444 | 0.13 |
ENST00000378789.4
|
HAO1
|
hydroxyacid oxidase 1 |
| chr17_-_10549694 | 0.13 |
ENST00000622564.4
|
MYH2
|
myosin heavy chain 2 |
| chr11_+_72189528 | 0.13 |
ENST00000312293.9
|
FOLR1
|
folate receptor alpha |
| chr15_+_40594241 | 0.13 |
ENST00000532056.5
ENST00000527044.5 ENST00000399668.7 |
KNL1
|
kinetochore scaffold 1 |
| chr15_-_83283449 | 0.13 |
ENST00000569704.2
|
BNC1
|
basonuclin 1 |
| chr18_-_47035621 | 0.13 |
ENST00000332567.6
|
ELOA2
|
elongin A2 |
| chr14_+_22168387 | 0.13 |
ENST00000557168.1
|
TRAV30
|
T cell receptor alpha variable 30 |
| chr2_-_68952880 | 0.13 |
ENST00000481498.1
ENST00000328895.9 |
GKN2
|
gastrokine 2 |
| chr11_+_72189659 | 0.13 |
ENST00000393681.6
|
FOLR1
|
folate receptor alpha |
| chr1_+_192158448 | 0.12 |
ENST00000367460.4
|
RGS18
|
regulator of G protein signaling 18 |
| chr20_-_8019744 | 0.12 |
ENST00000246024.7
|
TMX4
|
thioredoxin related transmembrane protein 4 |
| chr1_-_161238163 | 0.12 |
ENST00000367982.8
|
NR1I3
|
nuclear receptor subfamily 1 group I member 3 |
| chr1_-_86383078 | 0.12 |
ENST00000460698.6
|
ODF2L
|
outer dense fiber of sperm tails 2 like |
| chr1_-_161238196 | 0.12 |
ENST00000367983.9
ENST00000506209.5 ENST00000367980.6 ENST00000628566.2 |
NR1I3
|
nuclear receptor subfamily 1 group I member 3 |
| chr4_+_154563003 | 0.12 |
ENST00000302068.9
ENST00000509493.1 |
FGB
|
fibrinogen beta chain |
| chr1_-_220089818 | 0.12 |
ENST00000498791.6
ENST00000480959.6 |
BPNT1
|
3'(2'), 5'-bisphosphate nucleotidase 1 |
| chr7_-_78771265 | 0.12 |
ENST00000630991.2
ENST00000629359.2 |
MAGI2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2 |
| chr12_-_81598332 | 0.12 |
ENST00000443686.7
|
PPFIA2
|
PTPRF interacting protein alpha 2 |
| chr1_+_22052627 | 0.12 |
ENST00000400259.5
ENST00000344548.7 ENST00000315554.13 ENST00000656825.1 ENST00000651171.1 ENST00000652582.1 ENST00000667384.1 |
CDC42
|
cell division cycle 42 |
| chr9_-_101435760 | 0.11 |
ENST00000647789.2
ENST00000616752.1 |
ALDOB
|
aldolase, fructose-bisphosphate B |
| chr2_+_9961165 | 0.11 |
ENST00000405379.6
|
GRHL1
|
grainyhead like transcription factor 1 |
| chr1_-_7853054 | 0.11 |
ENST00000361696.10
|
UTS2
|
urotensin 2 |
| chr17_-_69268812 | 0.11 |
ENST00000586811.1
|
ABCA5
|
ATP binding cassette subfamily A member 5 |
| chr6_+_43770707 | 0.11 |
ENST00000324450.11
ENST00000417285.7 ENST00000413642.8 ENST00000372055.9 ENST00000482630.7 ENST00000425836.7 ENST00000372064.9 ENST00000372077.8 ENST00000519767.5 |
VEGFA
|
vascular endothelial growth factor A |
| chr6_+_43770202 | 0.11 |
ENST00000372067.8
ENST00000672860.2 |
VEGFA
|
vascular endothelial growth factor A |
| chr4_+_87975667 | 0.11 |
ENST00000237623.11
ENST00000682655.1 ENST00000508233.6 ENST00000360804.4 ENST00000395080.8 |
SPP1
|
secreted phosphoprotein 1 |
| chr15_-_66386668 | 0.11 |
ENST00000568216.5
ENST00000562124.5 ENST00000570251.1 |
TIPIN
|
TIMELESS interacting protein |
| chr19_-_3500664 | 0.11 |
ENST00000427575.6
|
DOHH
|
deoxyhypusine hydroxylase |
| chr6_+_83193331 | 0.11 |
ENST00000369724.5
|
RWDD2A
|
RWD domain containing 2A |
| chr4_+_85604146 | 0.10 |
ENST00000512201.5
|
ARHGAP24
|
Rho GTPase activating protein 24 |
| chr15_-_55917129 | 0.10 |
ENST00000338963.6
ENST00000508342.5 |
NEDD4
|
NEDD4 E3 ubiquitin protein ligase |
| chr4_+_185395979 | 0.10 |
ENST00000507753.1
|
ANKRD37
|
ankyrin repeat domain 37 |
| chr5_-_161546708 | 0.10 |
ENST00000393959.6
|
GABRB2
|
gamma-aminobutyric acid type A receptor subunit beta2 |
| chr14_-_56805648 | 0.10 |
ENST00000554788.5
ENST00000554845.1 ENST00000408990.8 |
OTX2
|
orthodenticle homeobox 2 |
| chr8_+_106726012 | 0.10 |
ENST00000449762.6
ENST00000297447.10 |
OXR1
|
oxidation resistance 1 |
| chr9_+_92974476 | 0.10 |
ENST00000337352.10
|
FGD3
|
FYVE, RhoGEF and PH domain containing 3 |
| chr2_+_225399684 | 0.10 |
ENST00000636099.1
|
NYAP2
|
neuronal tyrosine-phosphorylated phosphoinositide-3-kinase adaptor 2 |
| chr5_+_72848161 | 0.10 |
ENST00000506351.6
|
TNPO1
|
transportin 1 |
| chr19_+_21082140 | 0.10 |
ENST00000616183.1
ENST00000596053.5 |
ZNF714
|
zinc finger protein 714 |
| chr13_+_23570370 | 0.10 |
ENST00000403372.6
ENST00000248484.9 |
TNFRSF19
|
TNF receptor superfamily member 19 |
| chr6_+_26156323 | 0.09 |
ENST00000304218.6
|
H1-4
|
H1.4 linker histone, cluster member |
| chr10_-_95069489 | 0.09 |
ENST00000371270.6
ENST00000535898.5 ENST00000623108.3 |
CYP2C8
|
cytochrome P450 family 2 subfamily C member 8 |
| chr14_+_56117702 | 0.09 |
ENST00000559044.5
|
PELI2
|
pellino E3 ubiquitin protein ligase family member 2 |
| chr12_-_49187369 | 0.09 |
ENST00000547939.6
|
TUBA1A
|
tubulin alpha 1a |
| chr15_+_63122561 | 0.09 |
ENST00000557972.1
|
LACTB
|
lactamase beta |
| chr9_+_60914407 | 0.09 |
ENST00000437823.5
|
SPATA31A5
|
SPATA31 subfamily A member 5 |
| chr6_+_12717660 | 0.09 |
ENST00000674637.1
|
PHACTR1
|
phosphatase and actin regulator 1 |
| chr13_+_24270681 | 0.09 |
ENST00000343003.10
ENST00000399949.6 |
SPATA13
|
spermatogenesis associated 13 |
| chr19_-_3500625 | 0.09 |
ENST00000672935.1
|
DOHH
|
deoxyhypusine hydroxylase |
| chr12_-_84892120 | 0.09 |
ENST00000680379.1
|
SLC6A15
|
solute carrier family 6 member 15 |
| chr3_-_9880250 | 0.09 |
ENST00000423850.5
ENST00000336832.7 ENST00000675828.1 ENST00000618572.4 ENST00000455015.6 |
CIDEC
|
cell death inducing DFFA like effector c |
| chr9_+_102995308 | 0.09 |
ENST00000612124.4
ENST00000374798.8 ENST00000487798.5 |
CYLC2
|
cylicin 2 |
| chr17_-_31297231 | 0.09 |
ENST00000247271.5
|
OMG
|
oligodendrocyte myelin glycoprotein |
| chr4_-_56656448 | 0.08 |
ENST00000553379.6
|
HOPX
|
HOP homeobox |
| chr17_-_35142280 | 0.08 |
ENST00000586869.5
ENST00000442241.9 ENST00000360831.9 |
NLE1
|
notchless homolog 1 |
| chr4_-_70666492 | 0.08 |
ENST00000254801.9
ENST00000391614.7 |
JCHAIN
|
joining chain of multimeric IgA and IgM |
| chr7_-_25228485 | 0.08 |
ENST00000222674.2
|
NPVF
|
neuropeptide VF precursor |
| chr1_+_117420597 | 0.08 |
ENST00000449370.6
|
MAN1A2
|
mannosidase alpha class 1A member 2 |
| chr19_+_48393657 | 0.08 |
ENST00000263269.4
|
GRIN2D
|
glutamate ionotropic receptor NMDA type subunit 2D |
| chr12_-_91111460 | 0.08 |
ENST00000266718.5
|
LUM
|
lumican |
| chr14_-_25010604 | 0.08 |
ENST00000550887.5
|
STXBP6
|
syntaxin binding protein 6 |
| chr5_+_122129533 | 0.08 |
ENST00000296600.5
ENST00000504912.1 ENST00000505843.1 |
ZNF474
|
zinc finger protein 474 |
| chr19_+_29942205 | 0.08 |
ENST00000392271.6
|
URI1
|
URI1 prefoldin like chaperone |
| chr11_-_31811231 | 0.08 |
ENST00000438681.6
|
PAX6
|
paired box 6 |
| chr4_-_56656507 | 0.08 |
ENST00000381255.7
ENST00000317745.11 ENST00000555760.6 ENST00000556614.6 |
HOPX
|
HOP homeobox |
| chr11_-_13496018 | 0.08 |
ENST00000529816.1
|
PTH
|
parathyroid hormone |
| chr16_-_21652598 | 0.08 |
ENST00000569602.1
ENST00000268389.6 |
IGSF6
|
immunoglobulin superfamily member 6 |
| chr1_-_205121986 | 0.08 |
ENST00000367164.1
|
RBBP5
|
RB binding protein 5, histone lysine methyltransferase complex subunit |
| chr4_-_56681588 | 0.08 |
ENST00000554144.5
ENST00000381260.7 |
HOPX
|
HOP homeobox |
| chr12_-_81598360 | 0.08 |
ENST00000333447.11
ENST00000407050.8 |
PPFIA2
|
PTPRF interacting protein alpha 2 |
| chr11_-_31811034 | 0.08 |
ENST00000638250.1
|
PAX6
|
paired box 6 |
| chr8_+_76681208 | 0.07 |
ENST00000651372.2
|
ZFHX4
|
zinc finger homeobox 4 |
| chr7_+_16661182 | 0.07 |
ENST00000446596.5
ENST00000452975.6 ENST00000438834.5 |
BZW2
|
basic leucine zipper and W2 domains 2 |
| chr1_-_205121964 | 0.07 |
ENST00000264515.11
|
RBBP5
|
RB binding protein 5, histone lysine methyltransferase complex subunit |
| chr4_+_87832917 | 0.07 |
ENST00000395102.8
ENST00000497649.6 ENST00000540395.1 ENST00000560249.5 ENST00000511670.5 ENST00000361056.3 |
MEPE
|
matrix extracellular phosphoglycoprotein |
| chr12_-_8612850 | 0.07 |
ENST00000229335.11
ENST00000537228.5 |
AICDA
|
activation induced cytidine deaminase |
| chr13_+_76948500 | 0.07 |
ENST00000377462.6
|
ACOD1
|
aconitate decarboxylase 1 |
| chr1_+_23019415 | 0.07 |
ENST00000465864.2
ENST00000356634.7 ENST00000400181.9 |
KDM1A
|
lysine demethylase 1A |
| chr5_+_141966820 | 0.07 |
ENST00000513019.5
ENST00000394519.5 ENST00000356143.5 |
RNF14
|
ring finger protein 14 |
| chrX_-_54043927 | 0.07 |
ENST00000415025.5
ENST00000338946.10 |
PHF8
|
PHD finger protein 8 |
| chr13_+_45464901 | 0.07 |
ENST00000349995.10
|
COG3
|
component of oligomeric golgi complex 3 |
| chr3_-_197184131 | 0.07 |
ENST00000452595.5
|
DLG1
|
discs large MAGUK scaffold protein 1 |
| chr4_-_52020332 | 0.07 |
ENST00000682860.1
|
LRRC66
|
leucine rich repeat containing 66 |
| chr5_+_72848115 | 0.07 |
ENST00000679378.1
|
TNPO1
|
transportin 1 |
| chr11_-_13495984 | 0.07 |
ENST00000282091.6
|
PTH
|
parathyroid hormone |
| chr8_-_94208548 | 0.06 |
ENST00000027335.8
ENST00000441892.6 ENST00000521491.1 |
CDH17
|
cadherin 17 |
| chr1_-_247458105 | 0.06 |
ENST00000641149.1
ENST00000641527.1 |
OR2B11
|
olfactory receptor family 2 subfamily B member 11 |
| chr19_-_51082883 | 0.06 |
ENST00000650543.2
|
KLK14
|
kallikrein related peptidase 14 |
| chr14_+_79279403 | 0.06 |
ENST00000281127.11
|
NRXN3
|
neurexin 3 |
| chr8_+_106726115 | 0.06 |
ENST00000521592.5
|
OXR1
|
oxidation resistance 1 |
| chr12_+_55549602 | 0.06 |
ENST00000641569.1
ENST00000641851.1 |
OR6C4
|
olfactory receptor family 6 subfamily C member 4 |
| chr6_+_131637296 | 0.06 |
ENST00000358229.6
ENST00000357639.8 |
ENPP3
|
ectonucleotide pyrophosphatase/phosphodiesterase 3 |
| chr14_-_106130061 | 0.06 |
ENST00000390602.3
|
IGHV3-13
|
immunoglobulin heavy variable 3-13 |
| chr9_+_128787331 | 0.06 |
ENST00000223865.8
|
TBC1D13
|
TBC1 domain family member 13 |
| chr3_+_138621207 | 0.06 |
ENST00000464668.5
|
FAIM
|
Fas apoptotic inhibitory molecule |
| chr11_-_40294089 | 0.06 |
ENST00000278198.2
|
LRRC4C
|
leucine rich repeat containing 4C |
| chr16_-_48247533 | 0.06 |
ENST00000356608.7
ENST00000569991.1 |
ABCC11
|
ATP binding cassette subfamily C member 11 |
| chr6_+_30328889 | 0.06 |
ENST00000396547.5
ENST00000623385.3 |
TRIM39
TRIM39-RPP21
|
tripartite motif containing 39 TRIM39-RPP21 readthrough |
| chr13_-_79405784 | 0.06 |
ENST00000267229.11
|
RBM26
|
RNA binding motif protein 26 |
| chr21_+_33025927 | 0.06 |
ENST00000430860.1
ENST00000382357.4 ENST00000333337.3 |
OLIG2
|
oligodendrocyte transcription factor 2 |
| chrX_-_15600953 | 0.06 |
ENST00000679212.1
ENST00000679278.1 ENST00000678046.1 ENST00000252519.8 |
ACE2
|
angiotensin I converting enzyme 2 |
| chr11_-_31810991 | 0.06 |
ENST00000640684.1
|
PAX6
|
paired box 6 |
| chr14_-_67533720 | 0.05 |
ENST00000554278.6
|
TMEM229B
|
transmembrane protein 229B |
| chr13_+_101452569 | 0.05 |
ENST00000618057.4
|
ITGBL1
|
integrin subunit beta like 1 |
| chr2_+_196713117 | 0.05 |
ENST00000409270.5
|
CCDC150
|
coiled-coil domain containing 150 |
| chr11_-_31811112 | 0.05 |
ENST00000640242.1
ENST00000640613.1 ENST00000606377.7 |
PAX6
|
paired box 6 |
| chr12_-_18090141 | 0.05 |
ENST00000536890.1
|
RERGL
|
RERG like |
| chr13_-_103066411 | 0.05 |
ENST00000245312.5
|
SLC10A2
|
solute carrier family 10 member 2 |
| chr6_+_111087495 | 0.05 |
ENST00000612036.4
ENST00000368851.10 |
SLC16A10
|
solute carrier family 16 member 10 |
| chr1_+_209704836 | 0.05 |
ENST00000367027.5
|
HSD11B1
|
hydroxysteroid 11-beta dehydrogenase 1 |
| chr5_-_36001006 | 0.05 |
ENST00000625798.2
|
UGT3A1
|
UDP glycosyltransferase family 3 member A1 |
| chr11_-_31811140 | 0.05 |
ENST00000639916.1
|
PAX6
|
paired box 6 |
| chr16_-_3018170 | 0.05 |
ENST00000572154.1
ENST00000328796.5 |
CLDN6
|
claudin 6 |
| chr10_-_125816510 | 0.05 |
ENST00000650587.1
|
UROS
|
uroporphyrinogen III synthase |
| chr4_-_122456725 | 0.05 |
ENST00000226730.5
|
IL2
|
interleukin 2 |
| chr2_-_240820945 | 0.05 |
ENST00000428768.2
ENST00000650053.1 ENST00000650130.1 |
KIF1A
|
kinesin family member 1A |
| chr15_-_43879835 | 0.05 |
ENST00000636859.1
|
FRMD5
|
FERM domain containing 5 |
| chr9_+_122371036 | 0.05 |
ENST00000619306.5
ENST00000426608.6 ENST00000223423.8 |
PTGS1
|
prostaglandin-endoperoxide synthase 1 |
| chrX_+_11293411 | 0.05 |
ENST00000348912.4
ENST00000380714.7 ENST00000380712.7 |
AMELX
|
amelogenin X-linked |
| chr12_-_71157872 | 0.05 |
ENST00000546561.2
|
TSPAN8
|
tetraspanin 8 |
| chr1_+_154429315 | 0.05 |
ENST00000476006.5
|
IL6R
|
interleukin 6 receptor |
| chr17_-_41489907 | 0.05 |
ENST00000328119.11
|
KRT36
|
keratin 36 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.3 | GO:0061767 | negative regulation of lung blood pressure(GO:0061767) |
| 0.2 | 1.1 | GO:1903435 | positive regulation of constitutive secretory pathway(GO:1903435) |
| 0.2 | 0.5 | GO:0060279 | positive regulation of ovulation(GO:0060279) |
| 0.2 | 0.5 | GO:0051097 | negative regulation of helicase activity(GO:0051097) |
| 0.1 | 1.0 | GO:0002175 | protein localization to paranode region of axon(GO:0002175) |
| 0.1 | 0.3 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.1 | 0.7 | GO:1902731 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) negative regulation of chondrocyte proliferation(GO:1902731) |
| 0.1 | 0.5 | GO:0035617 | stress granule disassembly(GO:0035617) |
| 0.1 | 0.3 | GO:0050717 | positive regulation of interleukin-1 alpha secretion(GO:0050717) |
| 0.1 | 0.3 | GO:0060974 | neural crest cell migration involved in heart formation(GO:0003147) cell migration involved in heart formation(GO:0060974) anterior neural tube closure(GO:0061713) |
| 0.1 | 0.9 | GO:0043950 | positive regulation of cAMP-mediated signaling(GO:0043950) |
| 0.1 | 0.2 | GO:0051821 | negative regulation of transcription from RNA polymerase II promoter in response to UV-induced DNA damage(GO:0010768) transmission of virus(GO:0019089) dissemination or transmission of symbiont from host(GO:0044007) dissemination or transmission of organism from other organism involved in symbiotic interaction(GO:0051821) |
| 0.1 | 0.2 | GO:1903572 | regulation of protein kinase D signaling(GO:1903570) positive regulation of protein kinase D signaling(GO:1903572) |
| 0.1 | 0.2 | GO:0043376 | regulation of CD8-positive, alpha-beta T cell differentiation(GO:0043376) |
| 0.1 | 1.0 | GO:0002315 | marginal zone B cell differentiation(GO:0002315) |
| 0.1 | 0.7 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.1 | 0.2 | GO:1903644 | regulation of chaperone-mediated protein folding(GO:1903644) |
| 0.0 | 0.3 | GO:0038172 | interleukin-33-mediated signaling pathway(GO:0038172) |
| 0.0 | 0.4 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.0 | 0.2 | GO:1902159 | regulation of cyclic nucleotide-gated ion channel activity(GO:1902159) |
| 0.0 | 0.1 | GO:0071335 | submandibular salivary gland formation(GO:0060661) hair follicle cell proliferation(GO:0071335) regulation of hair follicle cell proliferation(GO:0071336) positive regulation of hair follicle cell proliferation(GO:0071338) |
| 0.0 | 0.1 | GO:0046005 | positive regulation of circadian sleep/wake cycle, REM sleep(GO:0046005) |
| 0.0 | 0.1 | GO:0090290 | positive regulation of osteoclast proliferation(GO:0090290) |
| 0.0 | 0.2 | GO:0006041 | glucosamine metabolic process(GO:0006041) glucose 1-phosphate metabolic process(GO:0019255) |
| 0.0 | 0.2 | GO:0090170 | regulation of Golgi inheritance(GO:0090170) |
| 0.0 | 0.2 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 0.0 | 0.2 | GO:0001880 | Mullerian duct regression(GO:0001880) |
| 0.0 | 0.5 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.0 | 0.2 | GO:0051771 | negative regulation of nitric-oxide synthase biosynthetic process(GO:0051771) |
| 0.0 | 0.1 | GO:2000866 | positive regulation of estrogen secretion(GO:2000863) positive regulation of estradiol secretion(GO:2000866) |
| 0.0 | 0.1 | GO:0090310 | negative regulation of methylation-dependent chromatin silencing(GO:0090310) |
| 0.0 | 0.1 | GO:0035574 | histone H4-K20 demethylation(GO:0035574) |
| 0.0 | 0.1 | GO:0030505 | inorganic diphosphate transport(GO:0030505) |
| 0.0 | 0.1 | GO:1904381 | Golgi apparatus mannose trimming(GO:1904381) |
| 0.0 | 0.3 | GO:0003322 | pancreatic A cell development(GO:0003322) |
| 0.0 | 0.6 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.0 | 0.4 | GO:0034501 | protein localization to kinetochore(GO:0034501) |
| 0.0 | 0.3 | GO:0031284 | positive regulation of guanylate cyclase activity(GO:0031284) |
| 0.0 | 0.1 | GO:0021529 | spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) |
| 0.0 | 0.2 | GO:0008612 | peptidyl-lysine modification to peptidyl-hypusine(GO:0008612) |
| 0.0 | 0.2 | GO:0042905 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.0 | 0.9 | GO:0050832 | defense response to fungus(GO:0050832) |
| 0.0 | 0.1 | GO:0034628 | nicotinamide nucleotide biosynthetic process from aspartate(GO:0019355) 'de novo' NAD biosynthetic process from aspartate(GO:0034628) |
| 0.0 | 0.5 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.0 | 0.1 | GO:0009441 | glycolate metabolic process(GO:0009441) |
| 0.0 | 0.1 | GO:0002933 | lipid hydroxylation(GO:0002933) |
| 0.0 | 0.1 | GO:0061181 | regulation of chondrocyte development(GO:0061181) |
| 0.0 | 0.1 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.0 | 0.1 | GO:0072573 | propionate metabolic process(GO:0019541) cellular response to progesterone stimulus(GO:0071393) tolerance induction to lipopolysaccharide(GO:0072573) |
| 0.0 | 0.1 | GO:0033184 | positive regulation of histone ubiquitination(GO:0033184) |
| 0.0 | 0.1 | GO:2001268 | inner cell mass cell differentiation(GO:0001826) negative regulation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:2001268) |
| 0.0 | 0.0 | GO:0010760 | negative regulation of macrophage chemotaxis(GO:0010760) negative regulation of macrophage colony-stimulating factor signaling pathway(GO:1902227) negative regulation of response to macrophage colony-stimulating factor(GO:1903970) negative regulation of cellular response to macrophage colony-stimulating factor stimulus(GO:1903973) |
| 0.0 | 0.1 | GO:0071418 | cellular response to amine stimulus(GO:0071418) |
| 0.0 | 0.2 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.0 | 0.2 | GO:1990573 | potassium ion import across plasma membrane(GO:1990573) |
| 0.0 | 0.4 | GO:2000651 | positive regulation of sodium ion transmembrane transporter activity(GO:2000651) |
| 0.0 | 0.0 | GO:0003099 | positive regulation of the force of heart contraction by chemical signal(GO:0003099) |
| 0.0 | 0.0 | GO:0002384 | hepatic immune response(GO:0002384) |
| 0.0 | 0.2 | GO:0051531 | NFAT protein import into nucleus(GO:0051531) |
| 0.0 | 0.1 | GO:1900245 | positive regulation of MDA-5 signaling pathway(GO:1900245) |
| 0.0 | 0.1 | GO:0006001 | fructose catabolic process(GO:0006001) fructose catabolic process to hydroxyacetone phosphate and glyceraldehyde-3-phosphate(GO:0061624) |
| 0.0 | 0.1 | GO:0032277 | negative regulation of gonadotropin secretion(GO:0032277) |
| 0.0 | 0.2 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.0 | 1.1 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0043511 | inhibin complex(GO:0043511) inhibin A complex(GO:0043512) |
| 0.2 | 1.3 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.1 | 1.3 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.1 | 0.3 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.1 | 1.1 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.1 | 0.5 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.1 | 0.3 | GO:0097179 | protease inhibitor complex(GO:0097179) |
| 0.1 | 0.2 | GO:1990578 | perinuclear endoplasmic reticulum membrane(GO:1990578) |
| 0.0 | 0.7 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.0 | 1.0 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.0 | 0.2 | GO:0036502 | Derlin-1-VIMP complex(GO:0036502) |
| 0.0 | 0.1 | GO:0033150 | cytoskeletal calyx(GO:0033150) |
| 0.0 | 0.1 | GO:0071756 | dimeric IgA immunoglobulin complex(GO:0071750) secretory dimeric IgA immunoglobulin complex(GO:0071752) IgM immunoglobulin complex(GO:0071753) IgM immunoglobulin complex, circulating(GO:0071754) pentameric IgM immunoglobulin complex(GO:0071756) |
| 0.0 | 0.2 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.0 | 0.8 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 0.2 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.2 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.0 | 0.2 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 0.2 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 0.5 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.0 | 0.1 | GO:0070449 | elongin complex(GO:0070449) |
| 0.0 | 0.3 | GO:0097381 | photoreceptor disc membrane(GO:0097381) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.3 | GO:0031962 | mineralocorticoid receptor binding(GO:0031962) |
| 0.2 | 0.9 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.1 | 0.3 | GO:0042008 | interleukin-33 binding(GO:0002113) interleukin-18 receptor activity(GO:0042008) |
| 0.1 | 0.9 | GO:0038049 | transcription factor activity, ligand-activated RNA polymerase II transcription factor binding(GO:0038049) |
| 0.1 | 0.6 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.1 | 0.4 | GO:0004882 | androgen receptor activity(GO:0004882) |
| 0.1 | 0.3 | GO:0061714 | folic acid receptor activity(GO:0061714) |
| 0.1 | 0.2 | GO:1902271 | D3 vitamins binding(GO:1902271) |
| 0.1 | 0.7 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 0.5 | GO:0005237 | inhibitory extracellular ligand-gated ion channel activity(GO:0005237) |
| 0.0 | 0.1 | GO:0052852 | (S)-2-hydroxy-acid oxidase activity(GO:0003973) very-long-chain-(S)-2-hydroxy-acid oxidase activity(GO:0052852) long-chain-(S)-2-hydroxy-long-chain-acid oxidase activity(GO:0052853) medium-chain-(S)-2-hydroxy-acid oxidase activity(GO:0052854) |
| 0.0 | 1.3 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.2 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.0 | 0.3 | GO:0008048 | calcium sensitive guanylate cyclase activator activity(GO:0008048) |
| 0.0 | 0.9 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.0 | 0.1 | GO:0032427 | GBD domain binding(GO:0032427) |
| 0.0 | 1.1 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 0.2 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.0 | 0.2 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.0 | 0.1 | GO:0061752 | telomeric repeat-containing RNA binding(GO:0061752) |
| 0.0 | 0.1 | GO:0034875 | oxidoreductase activity, acting on CH or CH2 groups, quinone or similar compound as acceptor(GO:0033695) caffeine oxidase activity(GO:0034875) |
| 0.0 | 0.8 | GO:0005549 | odorant binding(GO:0005549) |
| 0.0 | 0.1 | GO:0061609 | fructose-1-phosphate aldolase activity(GO:0061609) |
| 0.0 | 0.1 | GO:0035575 | histone demethylase activity (H4-K20 specific)(GO:0035575) |
| 0.0 | 0.2 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.2 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.0 | 0.4 | GO:1902282 | voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization(GO:1902282) |
| 0.0 | 0.2 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.0 | 0.1 | GO:0015173 | aromatic amino acid transmembrane transporter activity(GO:0015173) |
| 0.0 | 0.1 | GO:0003845 | 11-beta-hydroxysteroid dehydrogenase [NAD(P)] activity(GO:0003845) |
| 0.0 | 0.1 | GO:0000309 | nicotinamide-nucleotide adenylyltransferase activity(GO:0000309) |
| 0.0 | 0.1 | GO:0019862 | IgA binding(GO:0019862) |
| 0.0 | 0.4 | GO:0031698 | beta-2 adrenergic receptor binding(GO:0031698) |
| 0.0 | 0.0 | GO:0046848 | hydroxyapatite binding(GO:0046848) |
| 0.0 | 0.0 | GO:0070119 | ciliary neurotrophic factor binding(GO:0070119) |
| 0.0 | 0.1 | GO:0060002 | plus-end directed microfilament motor activity(GO:0060002) |
| 0.0 | 0.1 | GO:0005298 | proline:sodium symporter activity(GO:0005298) |
| 0.0 | 0.1 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.0 | 0.2 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 0.2 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.0 | 0.1 | GO:0035673 | oligopeptide transmembrane transporter activity(GO:0035673) |
| 0.0 | 0.5 | GO:0000146 | microfilament motor activity(GO:0000146) |
| 0.0 | 0.2 | GO:0008061 | chitin binding(GO:0008061) |
| 0.0 | 0.1 | GO:0004852 | uroporphyrinogen-III synthase activity(GO:0004852) |
| 0.0 | 0.1 | GO:0035529 | NADH pyrophosphatase activity(GO:0035529) |
| 0.0 | 0.0 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.0 | 0.1 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.3 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 1.2 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 0.5 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 0.5 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 0.6 | PID IL23 PATHWAY | IL23-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.0 | 0.5 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.0 | 1.3 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 1.1 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 1.1 | REACTOME CDK MEDIATED PHOSPHORYLATION AND REMOVAL OF CDC6 | Genes involved in CDK-mediated phosphorylation and removal of Cdc6 |
| 0.0 | 0.6 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.2 | REACTOME RAF MAP KINASE CASCADE | Genes involved in RAF/MAP kinase cascade |
| 0.0 | 0.9 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 0.5 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 0.8 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 0.2 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |