|
chr2_+_12716893
Show fit
|
0.83 |
ENST00000381465.2
ENST00000155926.9
|
TRIB2
|
tribbles pseudokinase 2
|
|
chr4_-_89836213
Show fit
|
0.62 |
ENST00000618500.4
ENST00000508895.5
|
SNCA
|
synuclein alpha
|
|
chr4_-_89835617
Show fit
|
0.61 |
ENST00000611107.1
ENST00000345009.8
ENST00000505199.5
ENST00000502987.5
|
SNCA
|
synuclein alpha
|
|
chr17_+_7855055
Show fit
|
0.58 |
ENST00000574668.1
ENST00000301599.7
|
TMEM88
|
transmembrane protein 88
|
|
chr1_-_91886144
Show fit
|
0.46 |
ENST00000212355.9
|
TGFBR3
|
transforming growth factor beta receptor 3
|
|
chr18_+_59220149
Show fit
|
0.43 |
ENST00000256857.7
ENST00000529320.2
ENST00000420468.6
|
GRP
|
gastrin releasing peptide
|
|
chr8_+_76683779
Show fit
|
0.40 |
ENST00000523885.2
|
ZFHX4
|
zinc finger homeobox 4
|
|
chr20_+_43945677
Show fit
|
0.35 |
ENST00000358131.5
|
TOX2
|
TOX high mobility group box family member 2
|
|
chr12_+_101594849
Show fit
|
0.32 |
ENST00000547405.5
ENST00000452455.6
ENST00000392934.7
ENST00000547509.5
ENST00000361685.6
ENST00000549145.5
ENST00000361466.7
ENST00000553190.5
ENST00000545503.6
ENST00000536007.5
ENST00000541119.5
ENST00000551300.5
ENST00000550270.1
|
MYBPC1
|
myosin binding protein C1
|
|
chr15_-_93073111
Show fit
|
0.30 |
ENST00000557420.1
ENST00000542321.6
|
RGMA
|
repulsive guidance molecule BMP co-receptor a
|
|
chrX_-_10677720
Show fit
|
0.29 |
ENST00000453318.6
|
MID1
|
midline 1
|
|
chr7_+_80135694
Show fit
|
0.28 |
ENST00000457358.7
|
GNAI1
|
G protein subunit alpha i1
|
|
chr20_+_36154630
Show fit
|
0.27 |
ENST00000338074.7
ENST00000636016.2
ENST00000373945.5
|
EPB41L1
|
erythrocyte membrane protein band 4.1 like 1
|
|
chr20_+_36214373
Show fit
|
0.25 |
ENST00000432603.1
|
EPB41L1
|
erythrocyte membrane protein band 4.1 like 1
|
|
chr7_-_120857124
Show fit
|
0.25 |
ENST00000441017.5
ENST00000424710.5
ENST00000433758.5
|
TSPAN12
|
tetraspanin 12
|
|
chr7_+_86643902
Show fit
|
0.22 |
ENST00000361669.7
|
GRM3
|
glutamate metabotropic receptor 3
|
|
chr5_-_88785493
Show fit
|
0.22 |
ENST00000503554.4
|
MEF2C
|
myocyte enhancer factor 2C
|
|
chr17_-_44199206
Show fit
|
0.20 |
ENST00000589805.1
|
ATXN7L3
|
ataxin 7 like 3
|
|
chr2_-_44361555
Show fit
|
0.20 |
ENST00000409957.5
|
PREPL
|
prolyl endopeptidase like
|
|
chr15_+_47717344
Show fit
|
0.19 |
ENST00000558816.5
ENST00000536845.7
|
SEMA6D
|
semaphorin 6D
|
|
chr7_-_86965872
Show fit
|
0.18 |
ENST00000398276.6
ENST00000416314.5
ENST00000425689.1
|
ELAPOR2
|
endosome-lysosome associated apoptosis and autophagy regulator family member 2
|
|
chr12_-_14885845
Show fit
|
0.17 |
ENST00000539261.6
ENST00000228938.5
|
MGP
|
matrix Gla protein
|
|
chr2_-_153478753
Show fit
|
0.17 |
ENST00000325926.4
|
RPRM
|
reprimo, TP53 dependent G2 arrest mediator homolog
|
|
chr4_-_137532452
Show fit
|
0.17 |
ENST00000412923.6
ENST00000511115.5
ENST00000344876.9
ENST00000507846.5
ENST00000510305.5
ENST00000611581.1
|
PCDH18
|
protocadherin 18
|
|
chr1_+_51617079
Show fit
|
0.16 |
ENST00000447887.5
ENST00000428468.6
ENST00000453295.5
|
OSBPL9
|
oxysterol binding protein like 9
|
|
chr13_-_77919390
Show fit
|
0.16 |
ENST00000475537.2
ENST00000646605.1
|
EDNRB
|
endothelin receptor type B
|
|
chr18_-_55403682
Show fit
|
0.16 |
ENST00000564228.5
ENST00000630828.2
|
TCF4
|
transcription factor 4
|
|
chr16_-_49856105
Show fit
|
0.16 |
ENST00000563137.7
|
ZNF423
|
zinc finger protein 423
|
|
chr16_+_15502266
Show fit
|
0.15 |
ENST00000452191.6
|
BMERB1
|
bMERB domain containing 1
|
|
chr1_+_64470120
Show fit
|
0.14 |
ENST00000651257.2
|
CACHD1
|
cache domain containing 1
|
|
chr5_-_126595237
Show fit
|
0.13 |
ENST00000637206.1
ENST00000553117.5
|
ALDH7A1
|
aldehyde dehydrogenase 7 family member A1
|
|
chr5_-_179617581
Show fit
|
0.12 |
ENST00000523449.5
|
HNRNPH1
|
heterogeneous nuclear ribonucleoprotein H1
|
|
chr7_-_144410227
Show fit
|
0.12 |
ENST00000467773.1
ENST00000483238.5
|
NOBOX
|
NOBOX oogenesis homeobox
|
|
chr10_+_35175586
Show fit
|
0.12 |
ENST00000494479.5
ENST00000463314.5
ENST00000342105.7
ENST00000495301.1
|
CREM
|
cAMP responsive element modulator
|
|
chr10_+_122560639
Show fit
|
0.11 |
ENST00000344338.7
ENST00000330163.8
ENST00000652446.2
ENST00000666315.1
ENST00000368955.7
ENST00000368909.7
ENST00000368956.6
ENST00000619379.1
|
DMBT1
|
deleted in malignant brain tumors 1
|
|
chr10_+_122560679
Show fit
|
0.11 |
ENST00000657942.1
|
DMBT1
|
deleted in malignant brain tumors 1
|
|
chr5_-_126595185
Show fit
|
0.11 |
ENST00000510111.6
ENST00000635851.1
ENST00000637964.1
ENST00000413020.6
ENST00000637782.1
ENST00000409134.8
ENST00000637272.1
ENST00000636879.1
ENST00000636743.1
ENST00000636886.1
ENST00000509270.2
|
ALDH7A1
|
aldehyde dehydrogenase 7 family member A1
|
|
chr10_+_122560751
Show fit
|
0.11 |
ENST00000338354.10
ENST00000664692.1
ENST00000653442.1
ENST00000664974.1
|
DMBT1
|
deleted in malignant brain tumors 1
|
|
chr6_-_112254485
Show fit
|
0.10 |
ENST00000521398.5
ENST00000424408.6
ENST00000243219.7
|
LAMA4
|
laminin subunit alpha 4
|
|
chr3_+_189789672
Show fit
|
0.10 |
ENST00000434928.5
|
TP63
|
tumor protein p63
|
|
chr2_+_26401909
Show fit
|
0.10 |
ENST00000288710.7
|
DRC1
|
dynein regulatory complex subunit 1
|
|
chr10_-_63269057
Show fit
|
0.09 |
ENST00000542921.5
|
JMJD1C
|
jumonji domain containing 1C
|
|
chr10_-_77637633
Show fit
|
0.08 |
ENST00000638223.1
ENST00000639544.1
ENST00000640807.1
ENST00000434208.6
ENST00000626620.3
ENST00000638575.1
ENST00000638759.1
|
KCNMA1
|
potassium calcium-activated channel subfamily M alpha 1
|
|
chr3_-_51779187
Show fit
|
0.07 |
ENST00000398780.5
ENST00000668964.1
ENST00000667863.2
ENST00000647442.1
|
IQCF6
|
IQ motif containing F6
|
|
chr17_+_60677822
Show fit
|
0.07 |
ENST00000407086.8
ENST00000589222.5
ENST00000626960.2
ENST00000390652.9
|
BCAS3
|
BCAS3 microtubule associated cell migration factor
|
|
chr1_-_107965009
Show fit
|
0.07 |
ENST00000527011.5
ENST00000370056.9
|
VAV3
|
vav guanine nucleotide exchange factor 3
|
|
chr6_+_32038382
Show fit
|
0.07 |
ENST00000478281.5
ENST00000471671.4
ENST00000435122.3
ENST00000644719.2
|
CYP21A2
|
cytochrome P450 family 21 subfamily A member 2
|
|
chr12_+_57089094
Show fit
|
0.07 |
ENST00000342556.6
ENST00000300131.8
|
NAB2
|
NGFI-A binding protein 2
|
|
chr9_+_26947039
Show fit
|
0.07 |
ENST00000519968.5
ENST00000433700.5
|
IFT74
|
intraflagellar transport 74
|
|
chr7_-_27102669
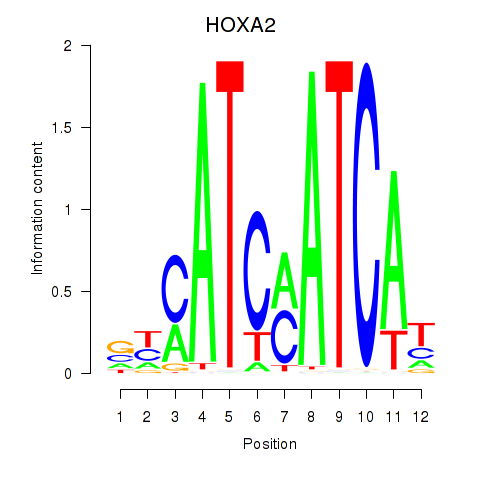
Show fit
|
0.07 |
ENST00000222718.7
|
HOXA2
|
homeobox A2
|
|
chr6_-_112254647
Show fit
|
0.07 |
ENST00000455073.1
ENST00000522006.5
ENST00000519932.5
|
LAMA4
|
laminin subunit alpha 4
|
|
chr6_+_122996227
Show fit
|
0.07 |
ENST00000275162.10
|
CLVS2
|
clavesin 2
|
|
chr6_+_151239951
Show fit
|
0.07 |
ENST00000402676.7
|
AKAP12
|
A-kinase anchoring protein 12
|
|
chr19_-_51009586
Show fit
|
0.07 |
ENST00000594211.2
|
KLK9
|
kallikrein related peptidase 9
|
|
chr8_-_66962563
Show fit
|
0.06 |
ENST00000563496.2
|
TCF24
|
transcription factor 24
|
|
chr20_-_32584795
Show fit
|
0.06 |
ENST00000201961.6
ENST00000621426.6
|
NOL4L
|
nucleolar protein 4 like
|
|
chr6_-_32128191
Show fit
|
0.06 |
ENST00000453203.2
ENST00000375203.8
ENST00000375201.8
|
ATF6B
|
activating transcription factor 6 beta
|
|
chr5_+_148202771
Show fit
|
0.06 |
ENST00000514389.5
ENST00000621437.4
|
SPINK6
|
serine peptidase inhibitor Kazal type 6
|
|
chr10_-_121596117
Show fit
|
0.06 |
ENST00000351936.11
|
FGFR2
|
fibroblast growth factor receptor 2
|
|
chr11_-_2140967
Show fit
|
0.06 |
ENST00000381389.5
|
IGF2
|
insulin like growth factor 2
|
|
chr13_-_35476682
Show fit
|
0.05 |
ENST00000379919.6
|
MAB21L1
|
mab-21 like 1
|
|
chr2_+_134254065
Show fit
|
0.05 |
ENST00000281923.4
|
MGAT5
|
alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase
|
|
chr3_+_148827800
Show fit
|
0.05 |
ENST00000282957.9
ENST00000468341.1
|
CPB1
|
carboxypeptidase B1
|
|
chr10_+_101131284
Show fit
|
0.05 |
ENST00000370196.11
ENST00000467928.2
|
TLX1
|
T cell leukemia homeobox 1
|
|
chr6_-_112254555
Show fit
|
0.05 |
ENST00000230538.12
ENST00000389463.9
ENST00000368638.5
ENST00000431543.6
ENST00000453937.2
|
LAMA4
|
laminin subunit alpha 4
|
|
chr1_+_243256034
Show fit
|
0.05 |
ENST00000366541.8
|
SDCCAG8
|
SHH signaling and ciliogenesis regulator SDCCAG8
|
|
chr3_+_150546765
Show fit
|
0.05 |
ENST00000406576.7
ENST00000460851.6
ENST00000482093.5
ENST00000273435.9
|
EIF2A
|
eukaryotic translation initiation factor 2A
|
|
chr11_+_26188836
Show fit
|
0.05 |
ENST00000672621.1
|
ANO3
|
anoctamin 3
|
|
chr8_-_78805306
Show fit
|
0.04 |
ENST00000639719.1
ENST00000263851.9
|
IL7
|
interleukin 7
|
|
chr10_+_100745570
Show fit
|
0.04 |
ENST00000427256.6
ENST00000355243.8
|
PAX2
|
paired box 2
|
|
chr8_-_66613229
Show fit
|
0.04 |
ENST00000517885.5
|
MYBL1
|
MYB proto-oncogene like 1
|
|
chr3_+_150546671
Show fit
|
0.04 |
ENST00000487799.5
|
EIF2A
|
eukaryotic translation initiation factor 2A
|
|
chr12_-_27971970
Show fit
|
0.04 |
ENST00000395872.5
ENST00000201015.8
|
PTHLH
|
parathyroid hormone like hormone
|
|
chr13_+_35476740
Show fit
|
0.04 |
ENST00000537702.5
|
NBEA
|
neurobeachin
|
|
chr15_-_74208969
Show fit
|
0.04 |
ENST00000423167.6
ENST00000432245.6
|
STRA6
|
signaling receptor and transporter of retinol STRA6
|
|
chrX_+_100644183
Show fit
|
0.04 |
ENST00000640889.1
ENST00000373004.5
|
SRPX2
|
sushi repeat containing protein X-linked 2
|
|
chr17_-_39197652
Show fit
|
0.04 |
ENST00000394303.8
|
CACNB1
|
calcium voltage-gated channel auxiliary subunit beta 1
|
|
chr12_+_7130341
Show fit
|
0.04 |
ENST00000266546.11
|
CLSTN3
|
calsyntenin 3
|
|
chr16_+_85611401
Show fit
|
0.04 |
ENST00000405402.6
|
GSE1
|
Gse1 coiled-coil protein
|
|
chr10_+_49988318
Show fit
|
0.04 |
ENST00000374056.10
ENST00000679810.1
|
AGAP6
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 6
|
|
chr19_+_37506931
Show fit
|
0.04 |
ENST00000627814.3
ENST00000587143.5
|
ZNF793
|
zinc finger protein 793
|
|
chr10_+_100745711
Show fit
|
0.03 |
ENST00000370296.6
ENST00000428433.5
|
PAX2
|
paired box 2
|
|
chr14_+_22516273
Show fit
|
0.03 |
ENST00000390510.1
|
TRAJ27
|
T cell receptor alpha joining 27
|
|
chrX_+_126819720
Show fit
|
0.03 |
ENST00000371125.4
|
PRR32
|
proline rich 32
|
|
chr19_-_49441508
Show fit
|
0.03 |
ENST00000221485.8
|
SLC17A7
|
solute carrier family 17 member 7
|
|
chr8_-_78805515
Show fit
|
0.03 |
ENST00000379113.6
ENST00000541183.2
|
IL7
|
interleukin 7
|
|
chr5_+_148203024
Show fit
|
0.03 |
ENST00000325630.3
|
SPINK6
|
serine peptidase inhibitor Kazal type 6
|
|
chr17_+_41819201
Show fit
|
0.03 |
ENST00000455106.1
|
FKBP10
|
FKBP prolyl isomerase 10
|
|
chr7_-_101237827
Show fit
|
0.03 |
ENST00000611078.4
|
CLDN15
|
claudin 15
|
|
chr4_+_76435216
Show fit
|
0.03 |
ENST00000296043.7
|
SHROOM3
|
shroom family member 3
|
|
chr17_+_81528370
Show fit
|
0.03 |
ENST00000417245.7
ENST00000334850.7
|
FSCN2
|
fascin actin-bundling protein 2, retinal
|
|
chr11_-_128842467
Show fit
|
0.03 |
ENST00000392664.2
|
KCNJ1
|
potassium inwardly rectifying channel subfamily J member 1
|
|
chr12_-_57742120
Show fit
|
0.03 |
ENST00000257897.7
|
AGAP2
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 2
|
|
chrX_-_66033664
Show fit
|
0.03 |
ENST00000427538.5
|
VSIG4
|
V-set and immunoglobulin domain containing 4
|
|
chr7_-_144259722
Show fit
|
0.03 |
ENST00000493325.1
|
OR2A7
|
olfactory receptor family 2 subfamily A member 7
|
|
chr8_+_142449430
Show fit
|
0.03 |
ENST00000643448.1
ENST00000517894.5
|
ADGRB1
|
adhesion G protein-coupled receptor B1
|
|
chr15_+_81299416
Show fit
|
0.02 |
ENST00000558332.3
|
IL16
|
interleukin 16
|
|
chr17_+_45161864
Show fit
|
0.02 |
ENST00000589230.6
ENST00000591576.5
ENST00000591070.6
ENST00000592695.1
|
HEXIM2
|
HEXIM P-TEFb complex subunit 2
|
|
chr1_-_109041986
Show fit
|
0.02 |
ENST00000400794.7
ENST00000528747.1
ENST00000361054.7
|
WDR47
|
WD repeat domain 47
|
|
chr7_-_13989891
Show fit
|
0.02 |
ENST00000405218.6
|
ETV1
|
ETS variant transcription factor 1
|
|
chr9_+_110668854
Show fit
|
0.02 |
ENST00000189978.10
ENST00000374440.7
|
MUSK
|
muscle associated receptor tyrosine kinase
|
|
chr7_-_13989658
Show fit
|
0.02 |
ENST00000430479.6
ENST00000433547.1
ENST00000405192.6
|
ETV1
|
ETS variant transcription factor 1
|
|
chrX_-_111412162
Show fit
|
0.02 |
ENST00000637570.1
ENST00000356220.8
ENST00000636035.2
ENST00000637453.1
ENST00000635795.1
|
DCX
|
doublecortin
|
|
chr9_+_110668779
Show fit
|
0.02 |
ENST00000416899.7
ENST00000374448.9
|
MUSK
|
muscle associated receptor tyrosine kinase
|
|
chr3_+_189789734
Show fit
|
0.02 |
ENST00000437221.5
ENST00000392463.6
ENST00000392461.7
ENST00000449992.5
ENST00000456148.1
|
TP63
|
tumor protein p63
|
|
chr11_-_123059413
Show fit
|
0.02 |
ENST00000524552.5
|
HSPA8
|
heat shock protein family A (Hsp70) member 8
|
|
chr11_+_57597563
Show fit
|
0.01 |
ENST00000619430.2
ENST00000457869.1
ENST00000340687.10
ENST00000278407.9
ENST00000378323.8
ENST00000378324.6
ENST00000403558.1
|
SERPING1
|
serpin family G member 1
|
|
chr10_-_77637721
Show fit
|
0.01 |
ENST00000638848.1
ENST00000639406.1
ENST00000618048.2
ENST00000639120.1
ENST00000640834.1
ENST00000639601.1
ENST00000638514.1
ENST00000457953.6
ENST00000639090.1
ENST00000639489.1
ENST00000372440.6
ENST00000404771.8
ENST00000638203.1
ENST00000638306.1
ENST00000638351.1
ENST00000638606.1
ENST00000639591.1
ENST00000640182.1
ENST00000640605.1
ENST00000640141.1
|
KCNMA1
|
potassium calcium-activated channel subfamily M alpha 1
|
|
chr21_+_18244828
Show fit
|
0.01 |
ENST00000299295.7
|
CHODL
|
chondrolectin
|
|
chr10_+_94089034
Show fit
|
0.01 |
ENST00000676102.1
ENST00000371385.8
|
PLCE1
|
phospholipase C epsilon 1
|
|
chr7_-_151167692
Show fit
|
0.01 |
ENST00000297537.5
|
GBX1
|
gastrulation brain homeobox 1
|
|
chr10_-_77637789
Show fit
|
0.01 |
ENST00000481070.1
ENST00000640969.1
ENST00000286628.14
ENST00000638991.1
ENST00000639913.1
ENST00000480683.2
|
KCNMA1
|
potassium calcium-activated channel subfamily M alpha 1
|
|
chr13_-_77919459
Show fit
|
0.01 |
ENST00000643890.1
|
EDNRB
|
endothelin receptor type B
|
|
chr18_+_21363593
Show fit
|
0.01 |
ENST00000580732.6
|
GREB1L
|
GREB1 like retinoic acid receptor coactivator
|
|
chr11_+_68460712
Show fit
|
0.01 |
ENST00000528635.5
ENST00000533127.5
ENST00000529907.5
ENST00000529344.5
ENST00000534534.5
ENST00000524845.5
ENST00000393800.7
ENST00000265637.8
ENST00000524904.5
ENST00000393801.7
ENST00000265636.9
ENST00000529710.5
|
PPP6R3
|
protein phosphatase 6 regulatory subunit 3
|
|
chr8_-_66613208
Show fit
|
0.01 |
ENST00000522677.8
|
MYBL1
|
MYB proto-oncogene like 1
|
|
chr17_-_39197696
Show fit
|
0.01 |
ENST00000394310.7
ENST00000622445.4
ENST00000344140.5
|
CACNB1
|
calcium voltage-gated channel auxiliary subunit beta 1
|
|
chr5_-_179618032
Show fit
|
0.01 |
ENST00000519033.5
|
HNRNPH1
|
heterogeneous nuclear ribonucleoprotein H1
|
|
chr1_+_66924895
Show fit
|
0.01 |
ENST00000355977.10
ENST00000371018.7
ENST00000357692.6
ENST00000371016.5
ENST00000371014.5
ENST00000371012.6
ENST00000401041.6
|
MIER1
|
MIER1 transcriptional regulator
|
|
chr10_+_94089067
Show fit
|
0.01 |
ENST00000371375.1
ENST00000675218.1
|
PLCE1
|
phospholipase C epsilon 1
|
|
chr17_-_1684807
Show fit
|
0.01 |
ENST00000577001.1
ENST00000572621.5
ENST00000304992.11
|
PRPF8
|
pre-mRNA processing factor 8
|
|
chr15_-_74209019
Show fit
|
0.01 |
ENST00000323940.9
|
STRA6
|
signaling receptor and transporter of retinol STRA6
|
|
chr1_-_44788168
Show fit
|
0.01 |
ENST00000372207.4
|
BEST4
|
bestrophin 4
|
|
chr1_-_66924791
Show fit
|
0.01 |
ENST00000371023.7
ENST00000371022.3
ENST00000371026.8
|
DNAI4
|
dynein axonemal intermediate chain 4
|
|
chr11_-_123058991
Show fit
|
0.01 |
ENST00000526686.1
|
HSPA8
|
heat shock protein family A (Hsp70) member 8
|
|
chr7_-_126533850
Show fit
|
0.01 |
ENST00000444921.3
|
GRM8
|
glutamate metabotropic receptor 8
|
|
chr12_+_56021005
Show fit
|
0.00 |
ENST00000547167.6
|
IKZF4
|
IKAROS family zinc finger 4
|
|
chr2_-_152098810
Show fit
|
0.00 |
ENST00000636442.1
ENST00000638005.1
|
CACNB4
|
calcium voltage-gated channel auxiliary subunit beta 4
|
|
chr12_-_114406133
Show fit
|
0.00 |
ENST00000405440.7
|
TBX5
|
T-box transcription factor 5
|
|
chrX_+_70062457
Show fit
|
0.00 |
ENST00000338352.3
|
OTUD6A
|
OTU deubiquitinase 6A
|
|
chr20_+_21303303
Show fit
|
0.00 |
ENST00000377191.5
|
XRN2
|
5'-3' exoribonuclease 2
|
|
chr8_-_142917843
Show fit
|
0.00 |
ENST00000323110.2
|
CYP11B2
|
cytochrome P450 family 11 subfamily B member 2
|
|
chr10_-_45847473
Show fit
|
0.00 |
ENST00000448048.7
|
AGAP4
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 4
|
|
chr8_-_142879820
Show fit
|
0.00 |
ENST00000377675.3
ENST00000517471.5
ENST00000292427.10
|
CYP11B1
|
cytochrome P450 family 11 subfamily B member 1
|
|
chr12_-_27972725
Show fit
|
0.00 |
ENST00000545234.6
|
PTHLH
|
parathyroid hormone like hormone
|
|
chr1_-_217076889
Show fit
|
0.00 |
ENST00000493748.5
ENST00000463665.5
|
ESRRG
|
estrogen related receptor gamma
|
|
chr10_-_99235783
Show fit
|
0.00 |
ENST00000370546.5
ENST00000614306.1
|
HPSE2
|
heparanase 2 (inactive)
|