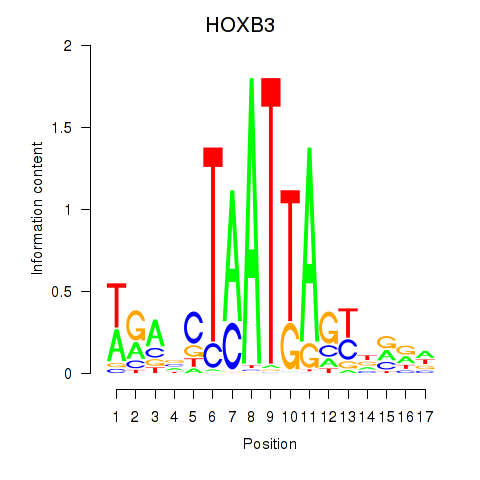
|
chr1_+_34792990
Show fit
|
0.78 |
ENST00000450137.1
ENST00000342280.5
|
GJA4
|
gap junction protein alpha 4
|
|
chr2_+_8682046
Show fit
|
0.69 |
ENST00000331129.3
ENST00000396290.2
|
ID2
|
inhibitor of DNA binding 2
|
|
chr11_-_118264282
Show fit
|
0.68 |
ENST00000278937.7
|
MPZL2
|
myelin protein zero like 2
|
|
chr12_-_122716790
Show fit
|
0.66 |
ENST00000528880.3
|
HCAR3
|
hydroxycarboxylic acid receptor 3
|
|
chr5_+_62578810
Show fit
|
0.60 |
ENST00000334994.6
ENST00000409534.1
|
LRRC70
IPO11
|
leucine rich repeat containing 70
importin 11
|
|
chr2_-_119525301
Show fit
|
0.50 |
ENST00000627145.1
|
SCTR
|
secretin receptor
|
|
chr9_+_5231413
Show fit
|
0.48 |
ENST00000239316.4
|
INSL4
|
insulin like 4
|
|
chr7_+_5879827
Show fit
|
0.48 |
ENST00000416608.5
|
OCM
|
oncomodulin
|
|
chr17_+_32021005
Show fit
|
0.47 |
ENST00000327564.11
ENST00000584368.5
ENST00000394713.7
ENST00000341671.11
|
LRRC37B
|
leucine rich repeat containing 37B
|
|
chr9_+_128149447
Show fit
|
0.45 |
ENST00000277480.7
ENST00000372998.1
|
LCN2
|
lipocalin 2
|
|
chr6_-_53510445
Show fit
|
0.40 |
ENST00000509541.5
|
GCLC
|
glutamate-cysteine ligase catalytic subunit
|
|
chr7_+_2519763
Show fit
|
0.39 |
ENST00000222725.10
ENST00000359574.7
|
LFNG
|
LFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase
|
|
chr9_-_110337808
Show fit
|
0.38 |
ENST00000374510.8
ENST00000374507.4
ENST00000423740.7
ENST00000374511.7
|
TXNDC8
|
thioredoxin domain containing 8
|
|
chr3_-_161105070
Show fit
|
0.36 |
ENST00000651430.1
ENST00000650695.1
ENST00000651689.1
ENST00000651916.1
ENST00000488170.5
ENST00000652377.1
ENST00000652669.1
|
B3GALNT1
|
beta-1,3-N-acetylgalactosaminyltransferase 1 (globoside blood group)
|
|
chr13_-_109786567
Show fit
|
0.36 |
ENST00000375856.5
|
IRS2
|
insulin receptor substrate 2
|
|
chr3_-_105868964
Show fit
|
0.35 |
ENST00000394030.8
|
CBLB
|
Cbl proto-oncogene B
|
|
chr6_+_110180418
Show fit
|
0.35 |
ENST00000368930.5
ENST00000307731.2
|
CDC40
|
cell division cycle 40
|
|
chr2_+_233636502
Show fit
|
0.34 |
ENST00000373445.1
|
UGT1A10
|
UDP glucuronosyltransferase family 1 member A10
|
|
chr12_-_10826358
Show fit
|
0.34 |
ENST00000240619.2
|
TAS2R10
|
taste 2 receptor member 10
|
|
chr11_+_121576760
Show fit
|
0.33 |
ENST00000532694.5
ENST00000534286.5
|
SORL1
|
sortilin related receptor 1
|
|
chr3_-_161105399
Show fit
|
0.33 |
ENST00000652593.1
|
B3GALNT1
|
beta-1,3-N-acetylgalactosaminyltransferase 1 (globoside blood group)
|
|
chr11_+_95790459
Show fit
|
0.33 |
ENST00000325486.9
ENST00000325542.10
ENST00000544522.5
ENST00000541365.5
|
CEP57
|
centrosomal protein 57
|
|
chr3_-_151316795
Show fit
|
0.33 |
ENST00000260843.5
|
GPR87
|
G protein-coupled receptor 87
|
|
chr13_+_77741160
Show fit
|
0.33 |
ENST00000314070.9
ENST00000351546.7
|
SLAIN1
|
SLAIN motif family member 1
|
|
chr3_-_161105224
Show fit
|
0.32 |
ENST00000651254.1
ENST00000651178.1
ENST00000476999.6
ENST00000652596.1
ENST00000651305.1
ENST00000652111.1
ENST00000651292.1
ENST00000651282.1
ENST00000651380.1
ENST00000494173.7
ENST00000484127.5
ENST00000650733.1
ENST00000494818.6
ENST00000492353.5
ENST00000652143.1
ENST00000473142.5
ENST00000651147.1
ENST00000468268.5
ENST00000460353.2
ENST00000651953.1
ENST00000651972.1
ENST00000652730.1
ENST00000651460.1
ENST00000652059.1
ENST00000651509.1
ENST00000651801.1
ENST00000651686.1
ENST00000320474.10
ENST00000392781.7
ENST00000392779.6
ENST00000651791.1
ENST00000651117.1
ENST00000652032.1
|
B3GALNT1
|
beta-1,3-N-acetylgalactosaminyltransferase 1 (globoside blood group)
|
|
chr6_+_110180116
Show fit
|
0.32 |
ENST00000368932.5
|
CDC40
|
cell division cycle 40
|
|
chr9_-_122828539
Show fit
|
0.32 |
ENST00000259467.9
|
PDCL
|
phosducin like
|
|
chr8_-_56211257
Show fit
|
0.31 |
ENST00000316981.8
ENST00000423799.6
ENST00000429357.2
|
PLAG1
|
PLAG1 zinc finger
|
|
chr1_+_47023659
Show fit
|
0.31 |
ENST00000371901.4
|
CYP4X1
|
cytochrome P450 family 4 subfamily X member 1
|
|
chr9_-_76906041
Show fit
|
0.31 |
ENST00000443509.6
ENST00000428286.5
ENST00000376713.3
|
PRUNE2
|
prune homolog 2 with BCH domain
|
|
chr12_-_7936177
Show fit
|
0.31 |
ENST00000544291.1
ENST00000075120.12
|
SLC2A3
|
solute carrier family 2 member 3
|
|
chr12_+_15546344
Show fit
|
0.31 |
ENST00000674388.1
ENST00000542557.5
ENST00000445537.6
ENST00000544244.5
ENST00000442921.7
|
PTPRO
|
protein tyrosine phosphatase receptor type O
|
|
chr6_-_111483700
Show fit
|
0.30 |
ENST00000435970.5
ENST00000358835.7
|
REV3L
|
REV3 like, DNA directed polymerase zeta catalytic subunit
|
|
chr4_-_137532452
Show fit
|
0.30 |
ENST00000412923.6
ENST00000511115.5
ENST00000344876.9
ENST00000507846.5
ENST00000510305.5
ENST00000611581.1
|
PCDH18
|
protocadherin 18
|
|
chr11_+_95789965
Show fit
|
0.30 |
ENST00000537677.5
|
CEP57
|
centrosomal protein 57
|
|
chr12_-_76423256
Show fit
|
0.29 |
ENST00000546946.5
|
OSBPL8
|
oxysterol binding protein like 8
|
|
chr14_+_61187544
Show fit
|
0.28 |
ENST00000555185.5
ENST00000557294.5
ENST00000556778.5
|
PRKCH
|
protein kinase C eta
|
|
chr21_+_30396030
Show fit
|
0.28 |
ENST00000355459.4
|
KRTAP13-1
|
keratin associated protein 13-1
|
|
chr2_+_137964446
Show fit
|
0.28 |
ENST00000280096.5
ENST00000280097.5
|
HNMT
|
histamine N-methyltransferase
|
|
chr11_-_63608542
Show fit
|
0.27 |
ENST00000540943.1
|
PLAAT3
|
phospholipase A and acyltransferase 3
|
|
chr14_-_89619118
Show fit
|
0.27 |
ENST00000345097.8
ENST00000555855.5
ENST00000555353.5
|
FOXN3
|
forkhead box N3
|
|
chr11_-_125111708
Show fit
|
0.27 |
ENST00000531909.5
ENST00000529530.1
|
TMEM218
|
transmembrane protein 218
|
|
chr2_-_119524483
Show fit
|
0.27 |
ENST00000019103.8
|
SCTR
|
secretin receptor
|
|
chr7_-_100167222
Show fit
|
0.26 |
ENST00000411994.1
|
GAL3ST4
|
galactose-3-O-sulfotransferase 4
|
|
chr12_-_122703346
Show fit
|
0.26 |
ENST00000328880.6
|
HCAR2
|
hydroxycarboxylic acid receptor 2
|
|
chr9_+_128275343
Show fit
|
0.25 |
ENST00000495313.5
ENST00000372898.6
|
SWI5
|
SWI5 homologous recombination repair protein
|
|
chr3_+_42979281
Show fit
|
0.25 |
ENST00000488863.5
ENST00000430121.3
|
GASK1A
|
golgi associated kinase 1A
|
|
chr1_+_174874434
Show fit
|
0.25 |
ENST00000478442.5
ENST00000465412.5
|
RABGAP1L
|
RAB GTPase activating protein 1 like
|
|
chr5_+_140848360
Show fit
|
0.24 |
ENST00000532602.2
|
PCDHA9
|
protocadherin alpha 9
|
|
chr4_+_40196907
Show fit
|
0.24 |
ENST00000622175.4
ENST00000619474.4
ENST00000615083.4
ENST00000610353.4
ENST00000614836.1
|
RHOH
|
ras homolog family member H
|
|
chrX_-_77634229
Show fit
|
0.24 |
ENST00000675732.1
|
ATRX
|
ATRX chromatin remodeler
|
|
chr3_-_151329539
Show fit
|
0.24 |
ENST00000325602.6
|
P2RY13
|
purinergic receptor P2Y13
|
|
chr1_+_159008978
Show fit
|
0.23 |
ENST00000447473.6
|
IFI16
|
interferon gamma inducible protein 16
|
|
chr6_-_110815152
Show fit
|
0.23 |
ENST00000413605.6
|
CDK19
|
cyclin dependent kinase 19
|
|
chr6_+_130018565
Show fit
|
0.22 |
ENST00000361794.7
ENST00000526087.5
ENST00000533560.5
|
L3MBTL3
|
L3MBTL histone methyl-lysine binding protein 3
|
|
chr5_-_22853320
Show fit
|
0.22 |
ENST00000504376.6
ENST00000382254.6
|
CDH12
|
cadherin 12
|
|
chr2_+_11542662
Show fit
|
0.22 |
ENST00000389825.7
ENST00000381483.6
|
GREB1
|
growth regulating estrogen receptor binding 1
|
|
chr11_-_118264445
Show fit
|
0.22 |
ENST00000438295.2
|
MPZL2
|
myelin protein zero like 2
|
|
chr4_-_163613505
Show fit
|
0.22 |
ENST00000339875.9
|
MARCHF1
|
membrane associated ring-CH-type finger 1
|
|
chr8_-_17722217
Show fit
|
0.22 |
ENST00000381861.7
|
MTUS1
|
microtubule associated scaffold protein 1
|
|
chr6_-_52840843
Show fit
|
0.22 |
ENST00000370989.6
|
GSTA5
|
glutathione S-transferase alpha 5
|
|
chr4_+_40197023
Show fit
|
0.21 |
ENST00000381799.10
|
RHOH
|
ras homolog family member H
|
|
chr16_+_53099100
Show fit
|
0.21 |
ENST00000565832.5
|
CHD9
|
chromodomain helicase DNA binding protein 9
|
|
chr13_+_39655627
Show fit
|
0.21 |
ENST00000416691.5
ENST00000455146.8
ENST00000630730.1
|
COG6
|
component of oligomeric golgi complex 6
|
|
chr17_+_46295099
Show fit
|
0.21 |
ENST00000393465.7
ENST00000320254.5
|
LRRC37A
|
leucine rich repeat containing 37A
|
|
chr2_+_28395511
Show fit
|
0.21 |
ENST00000436647.1
|
FOSL2
|
FOS like 2, AP-1 transcription factor subunit
|
|
chr4_+_143433491
Show fit
|
0.21 |
ENST00000512843.1
|
GAB1
|
GRB2 associated binding protein 1
|
|
chr4_+_173168800
Show fit
|
0.20 |
ENST00000512285.5
ENST00000265000.9
|
GALNT7
|
polypeptide N-acetylgalactosaminyltransferase 7
|
|
chr16_+_590056
Show fit
|
0.20 |
ENST00000248139.8
ENST00000568586.5
ENST00000538492.5
|
RAB40C
|
RAB40C, member RAS oncogene family
|
|
chr14_+_101809795
Show fit
|
0.20 |
ENST00000350249.7
ENST00000557621.5
ENST00000556946.1
|
PPP2R5C
|
protein phosphatase 2 regulatory subunit B'gamma
|
|
chr9_+_470291
Show fit
|
0.20 |
ENST00000382303.5
|
KANK1
|
KN motif and ankyrin repeat domains 1
|
|
chr6_-_110815408
Show fit
|
0.20 |
ENST00000368911.8
|
CDK19
|
cyclin dependent kinase 19
|
|
chr4_-_142846275
Show fit
|
0.20 |
ENST00000513000.5
ENST00000509777.5
ENST00000503927.5
|
INPP4B
|
inositol polyphosphate-4-phosphatase type II B
|
|
chr12_-_118359639
Show fit
|
0.20 |
ENST00000541786.5
ENST00000419821.6
ENST00000541878.5
|
TAOK3
|
TAO kinase 3
|
|
chr12_+_64404338
Show fit
|
0.20 |
ENST00000332707.10
|
XPOT
|
exportin for tRNA
|
|
chr20_+_9514562
Show fit
|
0.20 |
ENST00000246070.3
|
LAMP5
|
lysosomal associated membrane protein family member 5
|
|
chr20_-_56525925
Show fit
|
0.19 |
ENST00000243913.8
|
GCNT7
|
glucosaminyl (N-acetyl) transferase family member 7
|
|
chr1_+_78649818
Show fit
|
0.19 |
ENST00000370747.9
ENST00000438486.1
|
IFI44
|
interferon induced protein 44
|
|
chr1_-_248277976
Show fit
|
0.19 |
ENST00000641220.1
|
OR2T33
|
olfactory receptor family 2 subfamily T member 33
|
|
chr3_-_185821092
Show fit
|
0.19 |
ENST00000421047.3
|
IGF2BP2
|
insulin like growth factor 2 mRNA binding protein 2
|
|
chr19_+_17527250
Show fit
|
0.19 |
ENST00000599164.6
ENST00000449408.6
ENST00000600871.5
ENST00000599124.1
|
NIBAN3
|
niban apoptosis regulator 3
|
|
chr12_+_54854505
Show fit
|
0.19 |
ENST00000308796.11
ENST00000619042.1
|
MUCL1
|
mucin like 1
|
|
chr15_+_67548992
Show fit
|
0.18 |
ENST00000354498.9
|
MAP2K5
|
mitogen-activated protein kinase kinase 5
|
|
chr15_+_40765620
Show fit
|
0.18 |
ENST00000558467.1
|
GCHFR
|
GTP cyclohydrolase I feedback regulator
|
|
chrX_+_136648214
Show fit
|
0.18 |
ENST00000370628.2
|
CD40LG
|
CD40 ligand
|
|
chr6_-_110179995
Show fit
|
0.18 |
ENST00000392586.5
ENST00000419252.1
ENST00000359451.6
ENST00000392588.5
|
WASF1
|
WASP family member 1
|
|
chr4_-_25863537
Show fit
|
0.18 |
ENST00000502949.5
ENST00000264868.9
ENST00000513691.1
ENST00000514872.1
|
SEL1L3
|
SEL1L family member 3
|
|
chr7_+_26291850
Show fit
|
0.18 |
ENST00000338523.9
ENST00000446848.6
|
SNX10
|
sorting nexin 10
|
|
chr2_+_27275429
Show fit
|
0.18 |
ENST00000420191.5
ENST00000296097.8
|
DNAJC5G
|
DnaJ heat shock protein family (Hsp40) member C5 gamma
|
|
chr10_-_27240505
Show fit
|
0.17 |
ENST00000375888.5
ENST00000676732.1
|
ACBD5
|
acyl-CoA binding domain containing 5
|
|
chr5_+_149357999
Show fit
|
0.17 |
ENST00000274569.9
|
PCYOX1L
|
prenylcysteine oxidase 1 like
|
|
chr8_-_85341659
Show fit
|
0.17 |
ENST00000522389.5
|
CA1
|
carbonic anhydrase 1
|
|
chr1_-_201171545
Show fit
|
0.17 |
ENST00000367333.6
|
TMEM9
|
transmembrane protein 9
|
|
chr8_+_97869040
Show fit
|
0.17 |
ENST00000254898.7
ENST00000524308.5
ENST00000522025.6
|
MATN2
|
matrilin 2
|
|
chr16_-_21857418
Show fit
|
0.17 |
ENST00000415645.6
|
NPIPB4
|
nuclear pore complex interacting protein family member B4
|
|
chr8_-_13276491
Show fit
|
0.17 |
ENST00000512044.6
|
DLC1
|
DLC1 Rho GTPase activating protein
|
|
chr5_+_154941063
Show fit
|
0.17 |
ENST00000523037.6
ENST00000265229.12
ENST00000439747.7
ENST00000522038.5
|
MRPL22
|
mitochondrial ribosomal protein L22
|
|
chr12_+_28257195
Show fit
|
0.17 |
ENST00000381259.5
|
CCDC91
|
coiled-coil domain containing 91
|
|
chr16_+_31713231
Show fit
|
0.17 |
ENST00000539915.5
ENST00000316491.13
ENST00000398696.3
ENST00000534369.1
ENST00000530881.5
ENST00000529515.1
|
ZNF720
|
zinc finger protein 720
|
|
chrX_+_77910656
Show fit
|
0.17 |
ENST00000343533.9
ENST00000341514.11
ENST00000645454.1
ENST00000642651.1
ENST00000644362.1
|
ATP7A
PGK1
|
ATPase copper transporting alpha
phosphoglycerate kinase 1
|
|
chr1_+_161721563
Show fit
|
0.17 |
ENST00000367948.6
|
FCRLB
|
Fc receptor like B
|
|
chr16_-_29404029
Show fit
|
0.17 |
ENST00000524087.5
|
NPIPB11
|
nuclear pore complex interacting protein family member B11
|
|
chr16_+_22513523
Show fit
|
0.17 |
ENST00000538606.5
ENST00000451409.5
ENST00000424340.5
ENST00000517539.5
ENST00000528249.5
|
NPIPB5
|
nuclear pore complex interacting protein family member B5
|
|
chr3_+_158801926
Show fit
|
0.17 |
ENST00000622669.4
ENST00000392813.8
ENST00000415822.8
ENST00000651862.1
ENST00000264266.12
|
MFSD1
|
major facilitator superfamily domain containing 1
|
|
chr3_-_44477639
Show fit
|
0.16 |
ENST00000396077.8
|
ZNF445
|
zinc finger protein 445
|
|
chr9_-_5339874
Show fit
|
0.16 |
ENST00000223862.2
|
RLN1
|
relaxin 1
|
|
chr16_+_28638065
Show fit
|
0.16 |
ENST00000683297.1
|
NPIPB8
|
nuclear pore complex interacting protein family member B8
|
|
chr14_-_54441325
Show fit
|
0.16 |
ENST00000556113.1
ENST00000553660.5
ENST00000216416.9
ENST00000395573.8
ENST00000557690.5
|
CNIH1
|
cornichon family AMPA receptor auxiliary protein 1
|
|
chr7_+_105963253
Show fit
|
0.16 |
ENST00000478080.5
ENST00000317716.14
|
CDHR3
|
cadherin related family member 3
|
|
chr16_-_21857657
Show fit
|
0.16 |
ENST00000341400.11
ENST00000518761.8
ENST00000682606.1
|
NPIPB4
|
nuclear pore complex interacting protein family member B4
|
|
chr1_+_15764419
Show fit
|
0.16 |
ENST00000441801.6
|
FBLIM1
|
filamin binding LIM protein 1
|
|
chr20_-_31390483
Show fit
|
0.16 |
ENST00000376315.2
|
DEFB119
|
defensin beta 119
|
|
chr2_+_64988469
Show fit
|
0.15 |
ENST00000531327.5
|
SLC1A4
|
solute carrier family 1 member 4
|
|
chr11_-_96343170
Show fit
|
0.15 |
ENST00000524717.6
|
MAML2
|
mastermind like transcriptional coactivator 2
|
|
chrX_+_139530730
Show fit
|
0.15 |
ENST00000218099.7
|
F9
|
coagulation factor IX
|
|
chr11_-_95789744
Show fit
|
0.15 |
ENST00000358780.10
ENST00000542135.5
|
FAM76B
|
family with sequence similarity 76 member B
|
|
chr11_+_59436469
Show fit
|
0.15 |
ENST00000641045.1
|
OR5A1
|
olfactory receptor family 5 subfamily A member 1
|
|
chr22_-_28712136
Show fit
|
0.15 |
ENST00000464581.6
|
CHEK2
|
checkpoint kinase 2
|
|
chr12_+_119668109
Show fit
|
0.15 |
ENST00000229328.10
ENST00000630317.1
|
PRKAB1
|
protein kinase AMP-activated non-catalytic subunit beta 1
|
|
chr7_+_100119607
Show fit
|
0.14 |
ENST00000262932.5
|
CNPY4
|
canopy FGF signaling regulator 4
|
|
chr8_+_12104389
Show fit
|
0.14 |
ENST00000400085.7
|
ZNF705D
|
zinc finger protein 705D
|
|
chr10_+_4963406
Show fit
|
0.14 |
ENST00000380872.9
ENST00000442997.5
|
AKR1C1
|
aldo-keto reductase family 1 member C1
|
|
chr12_-_95551417
Show fit
|
0.14 |
ENST00000258499.8
|
USP44
|
ubiquitin specific peptidase 44
|
|
chr20_-_290717
Show fit
|
0.14 |
ENST00000360321.7
ENST00000400269.4
|
C20orf96
|
chromosome 20 open reading frame 96
|
|
chr4_+_76435216
Show fit
|
0.14 |
ENST00000296043.7
|
SHROOM3
|
shroom family member 3
|
|
chr9_-_92404559
Show fit
|
0.14 |
ENST00000262551.8
ENST00000375561.10
|
OGN
|
osteoglycin
|
|
chr10_-_30999469
Show fit
|
0.13 |
ENST00000538351.6
|
ZNF438
|
zinc finger protein 438
|
|
chr3_+_43690880
Show fit
|
0.13 |
ENST00000458276.7
|
ABHD5
|
abhydrolase domain containing 5, lysophosphatidic acid acyltransferase
|
|
chr10_-_50251500
Show fit
|
0.13 |
ENST00000682911.1
|
ASAH2
|
N-acylsphingosine amidohydrolase 2
|
|
chrX_+_136648138
Show fit
|
0.13 |
ENST00000370629.7
|
CD40LG
|
CD40 ligand
|
|
chr12_+_56128217
Show fit
|
0.13 |
ENST00000267113.4
ENST00000394048.10
|
ESYT1
|
extended synaptotagmin 1
|
|
chr12_+_16347665
Show fit
|
0.13 |
ENST00000535309.5
ENST00000540056.5
ENST00000396209.5
ENST00000540126.5
|
MGST1
|
microsomal glutathione S-transferase 1
|
|
chr5_-_34043205
Show fit
|
0.13 |
ENST00000382065.8
ENST00000231338.7
|
C1QTNF3
|
C1q and TNF related 3
|
|
chr4_+_94207596
Show fit
|
0.13 |
ENST00000359052.8
|
SMARCAD1
|
SWI/SNF-related, matrix-associated actin-dependent regulator of chromatin, subfamily a, containing DEAD/H box 1
|
|
chr12_+_22625182
Show fit
|
0.13 |
ENST00000538218.2
|
ETNK1
|
ethanolamine kinase 1
|
|
chr7_-_100167284
Show fit
|
0.12 |
ENST00000413800.5
|
GAL3ST4
|
galactose-3-O-sulfotransferase 4
|
|
chr21_+_46286623
Show fit
|
0.12 |
ENST00000397691.1
|
YBEY
|
ybeY metalloendoribonuclease
|
|
chr14_+_32329341
Show fit
|
0.12 |
ENST00000557354.5
ENST00000557102.1
ENST00000557272.1
|
AKAP6
|
A-kinase anchoring protein 6
|
|
chr1_+_155033824
Show fit
|
0.12 |
ENST00000295542.6
ENST00000423025.6
ENST00000368419.2
|
DCST1
|
DC-STAMP domain containing 1
|
|
chr11_+_112176364
Show fit
|
0.12 |
ENST00000526088.5
ENST00000532593.5
ENST00000531169.5
|
BCO2
|
beta-carotene oxygenase 2
|
|
chr1_-_68232539
Show fit
|
0.12 |
ENST00000370976.7
ENST00000354777.6
|
WLS
|
Wnt ligand secretion mediator
|
|
chr3_+_148791058
Show fit
|
0.12 |
ENST00000491148.5
|
CPB1
|
carboxypeptidase B1
|
|
chr4_-_149815826
Show fit
|
0.12 |
ENST00000636793.2
ENST00000636414.1
|
IQCM
|
IQ motif containing M
|
|
chr6_+_54018910
Show fit
|
0.11 |
ENST00000514921.5
ENST00000274897.9
ENST00000370877.6
|
MLIP
|
muscular LMNA interacting protein
|
|
chr22_-_28711931
Show fit
|
0.11 |
ENST00000434810.5
ENST00000456369.5
|
CHEK2
|
checkpoint kinase 2
|
|
chrX_+_43656289
Show fit
|
0.11 |
ENST00000338702.4
|
MAOA
|
monoamine oxidase A
|
|
chr16_-_28363508
Show fit
|
0.11 |
ENST00000532254.1
|
NPIPB6
|
nuclear pore complex interacting protein family member B6
|
|
chr2_+_186694007
Show fit
|
0.11 |
ENST00000304698.10
|
FAM171B
|
family with sequence similarity 171 member B
|
|
chr5_-_97142579
Show fit
|
0.11 |
ENST00000274382.9
|
LIX1
|
limb and CNS expressed 1
|
|
chr1_-_68232514
Show fit
|
0.11 |
ENST00000262348.9
ENST00000370973.2
ENST00000370971.1
|
WLS
|
Wnt ligand secretion mediator
|
|
chr12_-_10807286
Show fit
|
0.11 |
ENST00000240615.3
|
TAS2R8
|
taste 2 receptor member 8
|
|
chr19_+_1077394
Show fit
|
0.11 |
ENST00000590577.2
|
ARHGAP45
|
Rho GTPase activating protein 45
|
|
chr16_+_48244264
Show fit
|
0.11 |
ENST00000285737.9
|
LONP2
|
lon peptidase 2, peroxisomal
|
|
chr12_-_50249883
Show fit
|
0.10 |
ENST00000550592.1
|
LIMA1
|
LIM domain and actin binding 1
|
|
chr3_+_195720867
Show fit
|
0.10 |
ENST00000436408.6
|
MUC20
|
mucin 20, cell surface associated
|
|
chr17_-_59151794
Show fit
|
0.10 |
ENST00000584089.1
|
SKA2
|
spindle and kinetochore associated complex subunit 2
|
|
chr3_-_143848442
Show fit
|
0.10 |
ENST00000474151.1
ENST00000316549.11
|
SLC9A9
|
solute carrier family 9 member A9
|
|
chr2_-_20051610
Show fit
|
0.10 |
ENST00000175091.5
|
LAPTM4A
|
lysosomal protein transmembrane 4 alpha
|
|
chr2_-_113052867
Show fit
|
0.10 |
ENST00000259213.9
ENST00000327407.2
|
IL36B
|
interleukin 36 beta
|
|
chr17_+_45148468
Show fit
|
0.10 |
ENST00000332499.4
|
HEXIM1
|
HEXIM P-TEFb complex subunit 1
|
|
chr4_-_174283614
Show fit
|
0.10 |
ENST00000513696.1
ENST00000393674.7
ENST00000503293.5
|
FBXO8
|
F-box protein 8
|
|
chr2_+_200889411
Show fit
|
0.10 |
ENST00000409357.5
ENST00000409129.2
|
NIF3L1
|
NGG1 interacting factor 3 like 1
|
|
chr2_+_200889327
Show fit
|
0.10 |
ENST00000426253.5
ENST00000454952.1
ENST00000651500.1
ENST00000409020.6
ENST00000359683.8
|
NIF3L1
|
NGG1 interacting factor 3 like 1
|
|
chr19_-_45584810
Show fit
|
0.10 |
ENST00000323060.3
|
OPA3
|
outer mitochondrial membrane lipid metabolism regulator OPA3
|
|
chr13_+_51861963
Show fit
|
0.10 |
ENST00000242819.7
|
CCDC70
|
coiled-coil domain containing 70
|
|
chr1_+_207053229
Show fit
|
0.09 |
ENST00000367080.8
ENST00000367079.3
|
PFKFB2
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 2
|
|
chr20_-_41618362
Show fit
|
0.09 |
ENST00000373222.3
ENST00000373233.8
|
CHD6
|
chromodomain helicase DNA binding protein 6
|
|
chr9_+_72577369
Show fit
|
0.09 |
ENST00000651183.1
|
TMC1
|
transmembrane channel like 1
|
|
chr2_-_200889136
Show fit
|
0.09 |
ENST00000409361.5
|
PPIL3
|
peptidylprolyl isomerase like 3
|
|
chr2_+_137964279
Show fit
|
0.09 |
ENST00000329366.8
|
HNMT
|
histamine N-methyltransferase
|
|
chr15_+_78438199
Show fit
|
0.09 |
ENST00000258886.13
|
IREB2
|
iron responsive element binding protein 2
|
|
chr14_-_106038355
Show fit
|
0.09 |
ENST00000390597.3
|
IGHV2-5
|
immunoglobulin heavy variable 2-5
|
|
chr1_-_92792396
Show fit
|
0.09 |
ENST00000370331.5
ENST00000540033.2
|
EVI5
|
ecotropic viral integration site 5
|
|
chrX_-_103502853
Show fit
|
0.09 |
ENST00000372633.1
|
RAB40A
|
RAB40A, member RAS oncogene family
|
|
chr12_+_16347637
Show fit
|
0.09 |
ENST00000543076.5
ENST00000396210.8
|
MGST1
|
microsomal glutathione S-transferase 1
|
|
chr6_+_26204552
Show fit
|
0.09 |
ENST00000615164.2
|
H4C5
|
H4 clustered histone 5
|
|
chr8_-_48921419
Show fit
|
0.09 |
ENST00000020945.4
|
SNAI2
|
snail family transcriptional repressor 2
|
|
chr4_+_112860912
Show fit
|
0.09 |
ENST00000671951.1
|
ANK2
|
ankyrin 2
|
|
chr1_+_207325629
Show fit
|
0.09 |
ENST00000618707.2
|
CD55
|
CD55 molecule (Cromer blood group)
|
|
chr16_-_81220370
Show fit
|
0.08 |
ENST00000337114.8
|
PKD1L2
|
polycystin 1 like 2 (gene/pseudogene)
|
|
chr6_+_29550407
Show fit
|
0.08 |
ENST00000641137.1
|
OR2I1P
|
olfactory receptor family 2 subfamily I member 1 pseudogene
|
|
chr3_-_131026726
Show fit
|
0.08 |
ENST00000514044.5
ENST00000264992.8
|
ASTE1
|
asteroid homolog 1
|
|
chr14_-_94476240
Show fit
|
0.08 |
ENST00000424550.6
ENST00000337425.10
ENST00000674164.1
ENST00000546329.2
|
SERPINA9
|
serpin family A member 9
|
|
chr4_+_150078426
Show fit
|
0.08 |
ENST00000296550.12
|
DCLK2
|
doublecortin like kinase 2
|
|
chr2_-_200888993
Show fit
|
0.08 |
ENST00000409264.6
ENST00000392283.9
|
PPIL3
|
peptidylprolyl isomerase like 3
|
|
chr16_-_21425278
Show fit
|
0.08 |
ENST00000504841.6
ENST00000419180.6
|
NPIPB3
|
nuclear pore complex interacting protein family member B3
|
|
chr19_-_17255448
Show fit
|
0.08 |
ENST00000594059.1
|
ENSG00000269095.1
|
novel protein
|
|
chr2_+_233636445
Show fit
|
0.08 |
ENST00000344644.9
|
UGT1A10
|
UDP glucuronosyltransferase family 1 member A10
|
|
chr7_+_6615576
Show fit
|
0.08 |
ENST00000457543.4
|
ZNF853
|
zinc finger protein 853
|
|
chr19_-_14848922
Show fit
|
0.08 |
ENST00000641129.1
|
OR7A10
|
olfactory receptor family 7 subfamily A member 10
|
|
chr7_-_100641507
Show fit
|
0.08 |
ENST00000431692.5
ENST00000223051.8
|
TFR2
|
transferrin receptor 2
|
|
chr2_+_29113989
Show fit
|
0.08 |
ENST00000404424.5
|
CLIP4
|
CAP-Gly domain containing linker protein family member 4
|
|
chr16_-_29505820
Show fit
|
0.08 |
ENST00000550665.5
|
NPIPB12
|
nuclear pore complex interacting protein family member B12
|
|
chr1_+_15617415
Show fit
|
0.08 |
ENST00000480945.6
|
DDI2
|
DNA damage inducible 1 homolog 2
|
|
chr4_-_65670478
Show fit
|
0.08 |
ENST00000613740.5
ENST00000622150.4
ENST00000511294.1
|
EPHA5
|
EPH receptor A5
|
|
chr17_+_7407838
Show fit
|
0.08 |
ENST00000302926.7
|
NLGN2
|
neuroligin 2
|
|
chr4_-_65670339
Show fit
|
0.07 |
ENST00000273854.7
|
EPHA5
|
EPH receptor A5
|
|
chr4_+_112861053
Show fit
|
0.07 |
ENST00000672221.1
|
ANK2
|
ankyrin 2
|
|
chr4_+_112860981
Show fit
|
0.07 |
ENST00000671704.1
|
ANK2
|
ankyrin 2
|
|
chr12_+_22625075
Show fit
|
0.07 |
ENST00000671733.1
ENST00000335148.8
ENST00000672951.1
ENST00000266517.9
|
ETNK1
|
ethanolamine kinase 1
|