Project
Inflammatory response time course, HUVEC (Wada, 2009)
Navigation
Downloads
Results for HOXB4_LHX9
Z-value: 0.52
Transcription factors associated with HOXB4_LHX9
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
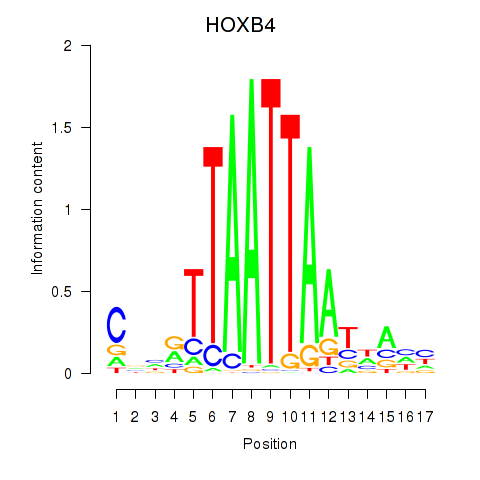
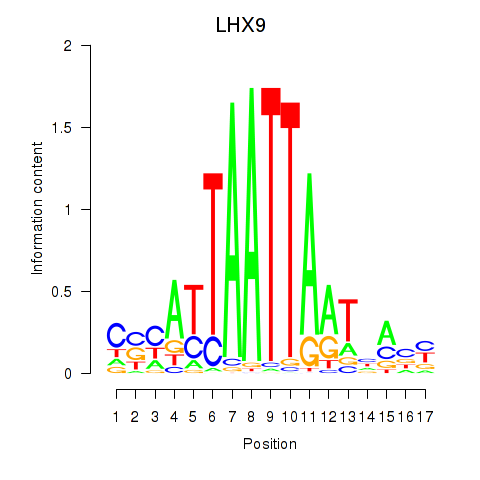
HOXB4
|
ENSG00000182742.6 | HOXB4 |
|
LHX9
|
ENSG00000143355.16 | LHX9 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| LHX9 | hg38_v1_chr1_+_197912462_197912505 | 0.24 | 2.4e-01 | Click! |
| HOXB4 | hg38_v1_chr17_-_48578341_48578356 | 0.12 | 5.8e-01 | Click! |
Activity profile of HOXB4_LHX9 motif
Sorted Z-values of HOXB4_LHX9 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of HOXB4_LHX9
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_+_155666718 | 1.35 |
ENST00000621234.4
ENST00000511108.5 |
GUCY1A1
|
guanylate cyclase 1 soluble subunit alpha 1 |
| chr12_-_9869345 | 1.11 |
ENST00000228438.3
|
CLEC2B
|
C-type lectin domain family 2 member B |
| chr18_+_44700796 | 1.10 |
ENST00000677130.1
|
SETBP1
|
SET binding protein 1 |
| chr16_-_66549839 | 1.02 |
ENST00000527800.6
ENST00000677555.1 ENST00000563369.6 |
TK2
|
thymidine kinase 2 |
| chr16_-_66550091 | 1.00 |
ENST00000564917.5
ENST00000677420.1 |
TK2
|
thymidine kinase 2 |
| chr16_-_66550005 | 0.95 |
ENST00000527284.6
|
TK2
|
thymidine kinase 2 |
| chr3_-_142029108 | 0.95 |
ENST00000497579.5
|
TFDP2
|
transcription factor Dp-2 |
| chr9_+_72577939 | 0.88 |
ENST00000645773.1
|
TMC1
|
transmembrane channel like 1 |
| chr11_+_71527267 | 0.88 |
ENST00000398536.6
|
KRTAP5-7
|
keratin associated protein 5-7 |
| chr4_-_139280179 | 0.86 |
ENST00000398955.2
|
MGARP
|
mitochondria localized glutamic acid rich protein |
| chr13_-_109786567 | 0.76 |
ENST00000375856.5
|
IRS2
|
insulin receptor substrate 2 |
| chr11_-_63608542 | 0.72 |
ENST00000540943.1
|
PLAAT3
|
phospholipase A and acyltransferase 3 |
| chr5_+_1225379 | 0.70 |
ENST00000324642.4
|
SLC6A18
|
solute carrier family 6 member 18 |
| chr2_+_90038848 | 0.64 |
ENST00000390270.2
|
IGKV3D-20
|
immunoglobulin kappa variable 3D-20 |
| chr7_+_1044542 | 0.64 |
ENST00000444847.2
|
GPR146
|
G protein-coupled receptor 146 |
| chr17_+_69502397 | 0.59 |
ENST00000613873.4
ENST00000589647.5 |
MAP2K6
|
mitogen-activated protein kinase kinase 6 |
| chr20_-_35147285 | 0.57 |
ENST00000374491.3
ENST00000374492.8 |
EDEM2
|
ER degradation enhancing alpha-mannosidase like protein 2 |
| chr15_-_19988117 | 0.56 |
ENST00000558565.2
|
IGHV3OR15-7
|
immunoglobulin heavy variable 3/OR15-7 (pseudogene) |
| chr6_+_130018565 | 0.54 |
ENST00000361794.7
ENST00000526087.5 ENST00000533560.5 |
L3MBTL3
|
L3MBTL histone methyl-lysine binding protein 3 |
| chr9_+_72577369 | 0.53 |
ENST00000651183.1
|
TMC1
|
transmembrane channel like 1 |
| chr5_+_113513674 | 0.51 |
ENST00000161863.9
ENST00000515883.5 |
YTHDC2
|
YTH domain containing 2 |
| chr12_+_38316753 | 0.51 |
ENST00000551464.1
ENST00000308742.9 |
ALG10B
|
ALG10 alpha-1,2-glucosyltransferase B |
| chr4_-_89836213 | 0.50 |
ENST00000618500.4
ENST00000508895.5 |
SNCA
|
synuclein alpha |
| chr16_-_66550142 | 0.47 |
ENST00000417693.8
ENST00000299697.12 ENST00000451102.7 |
TK2
|
thymidine kinase 2 |
| chr4_+_133149278 | 0.46 |
ENST00000264360.7
|
PCDH10
|
protocadherin 10 |
| chr11_+_7605719 | 0.45 |
ENST00000530181.5
|
PPFIBP2
|
PPFIA binding protein 2 |
| chr20_-_57690624 | 0.45 |
ENST00000414037.5
|
PMEPA1
|
prostate transmembrane protein, androgen induced 1 |
| chr7_-_150323489 | 0.42 |
ENST00000683684.1
ENST00000478393.5 |
ACTR3C
|
actin related protein 3C |
| chr7_+_150323239 | 0.41 |
ENST00000323078.7
ENST00000493307.1 ENST00000359623.9 |
LRRC61
|
leucine rich repeat containing 61 |
| chr4_-_137532452 | 0.40 |
ENST00000412923.6
ENST00000511115.5 ENST00000344876.9 ENST00000507846.5 ENST00000510305.5 ENST00000611581.1 |
PCDH18
|
protocadherin 18 |
| chr14_+_30577752 | 0.40 |
ENST00000547532.5
ENST00000555429.1 |
G2E3
|
G2/M-phase specific E3 ubiquitin protein ligase |
| chr17_+_41237998 | 0.40 |
ENST00000254072.7
|
KRTAP9-8
|
keratin associated protein 9-8 |
| chr3_-_142000353 | 0.39 |
ENST00000499676.5
|
TFDP2
|
transcription factor Dp-2 |
| chr4_+_89901979 | 0.39 |
ENST00000508372.1
|
MMRN1
|
multimerin 1 |
| chr17_+_68515399 | 0.38 |
ENST00000588188.6
|
PRKAR1A
|
protein kinase cAMP-dependent type I regulatory subunit alpha |
| chr5_-_90474765 | 0.38 |
ENST00000316610.7
|
MBLAC2
|
metallo-beta-lactamase domain containing 2 |
| chr2_+_201182873 | 0.37 |
ENST00000360132.7
|
CASP10
|
caspase 10 |
| chr7_+_102912983 | 0.37 |
ENST00000339431.9
ENST00000249377.4 |
LRRC17
|
leucine rich repeat containing 17 |
| chr6_+_47656436 | 0.36 |
ENST00000507065.5
ENST00000296862.5 |
ADGRF2
|
adhesion G protein-coupled receptor F2 |
| chr6_-_110179995 | 0.36 |
ENST00000392586.5
ENST00000419252.1 ENST00000359451.6 ENST00000392588.5 |
WASF1
|
WASP family member 1 |
| chr7_+_107583919 | 0.35 |
ENST00000491150.5
|
BCAP29
|
B cell receptor associated protein 29 |
| chr8_-_92966081 | 0.35 |
ENST00000517858.5
ENST00000378861.9 |
TRIQK
|
triple QxxK/R motif containing |
| chr8_-_30812867 | 0.34 |
ENST00000518243.5
|
PPP2CB
|
protein phosphatase 2 catalytic subunit beta |
| chr20_-_31390483 | 0.34 |
ENST00000376315.2
|
DEFB119
|
defensin beta 119 |
| chr18_-_55423757 | 0.33 |
ENST00000675707.1
|
TCF4
|
transcription factor 4 |
| chr8_-_92966129 | 0.33 |
ENST00000522925.5
ENST00000522903.5 ENST00000537541.1 ENST00000521988.6 ENST00000518748.5 ENST00000519069.5 |
TRIQK
|
triple QxxK/R motif containing |
| chr8_-_92966105 | 0.33 |
ENST00000524037.5
ENST00000520430.5 ENST00000521617.5 ENST00000523580.5 |
TRIQK
|
triple QxxK/R motif containing |
| chr2_+_66435116 | 0.33 |
ENST00000272369.14
ENST00000560281.6 |
MEIS1
|
Meis homeobox 1 |
| chr12_-_10826358 | 0.32 |
ENST00000240619.2
|
TAS2R10
|
taste 2 receptor member 10 |
| chr10_+_99732211 | 0.32 |
ENST00000370476.10
ENST00000370472.4 |
CUTC
|
cutC copper transporter |
| chr7_+_116222804 | 0.32 |
ENST00000393481.6
|
TES
|
testin LIM domain protein |
| chr5_+_154941063 | 0.32 |
ENST00000523037.6
ENST00000265229.12 ENST00000439747.7 ENST00000522038.5 |
MRPL22
|
mitochondrial ribosomal protein L22 |
| chr1_-_201171545 | 0.32 |
ENST00000367333.6
|
TMEM9
|
transmembrane protein 9 |
| chr1_+_77918128 | 0.31 |
ENST00000342754.5
|
NEXN
|
nexilin F-actin binding protein |
| chr11_-_69819410 | 0.31 |
ENST00000334134.4
|
FGF3
|
fibroblast growth factor 3 |
| chr11_-_122116215 | 0.31 |
ENST00000560104.2
|
BLID
|
BH3-like motif containing, cell death inducer |
| chr19_-_35812838 | 0.30 |
ENST00000653904.2
|
PRODH2
|
proline dehydrogenase 2 |
| chr13_+_76948500 | 0.30 |
ENST00000377462.6
|
ACOD1
|
aconitate decarboxylase 1 |
| chr19_-_51417700 | 0.30 |
ENST00000529627.1
ENST00000439889.6 |
SIGLEC10
|
sialic acid binding Ig like lectin 10 |
| chr3_+_158801926 | 0.29 |
ENST00000622669.4
ENST00000392813.8 ENST00000415822.8 ENST00000651862.1 ENST00000264266.12 |
MFSD1
|
major facilitator superfamily domain containing 1 |
| chr11_-_125111708 | 0.28 |
ENST00000531909.5
ENST00000529530.1 |
TMEM218
|
transmembrane protein 218 |
| chr6_+_130421086 | 0.28 |
ENST00000545622.5
|
TMEM200A
|
transmembrane protein 200A |
| chrX_-_18672101 | 0.28 |
ENST00000379984.4
|
RS1
|
retinoschisin 1 |
| chr16_-_66550112 | 0.28 |
ENST00000544898.6
ENST00000620035.5 ENST00000545043.6 |
TK2
|
thymidine kinase 2 |
| chr11_+_92969651 | 0.27 |
ENST00000257068.3
ENST00000528076.1 |
MTNR1B
|
melatonin receptor 1B |
| chr9_+_72577788 | 0.27 |
ENST00000645208.2
|
TMC1
|
transmembrane channel like 1 |
| chr5_-_74641419 | 0.27 |
ENST00000618628.4
ENST00000510316.5 ENST00000508331.1 |
ENC1
|
ectodermal-neural cortex 1 |
| chr9_-_74952904 | 0.26 |
ENST00000376854.6
|
C9orf40
|
chromosome 9 open reading frame 40 |
| chr10_+_96129707 | 0.26 |
ENST00000316045.9
|
ZNF518A
|
zinc finger protein 518A |
| chr12_+_26195647 | 0.25 |
ENST00000535504.1
|
SSPN
|
sarcospan |
| chr3_-_185821092 | 0.25 |
ENST00000421047.3
|
IGF2BP2
|
insulin like growth factor 2 mRNA binding protein 2 |
| chr13_-_52011337 | 0.25 |
ENST00000400366.6
ENST00000400370.8 ENST00000634844.1 ENST00000673772.1 ENST00000418097.7 ENST00000242839.10 ENST00000344297.9 ENST00000448424.7 |
ATP7B
|
ATPase copper transporting beta |
| chr8_-_100559702 | 0.25 |
ENST00000520311.5
ENST00000520552.5 ENST00000521345.1 ENST00000523000.5 ENST00000335659.7 ENST00000358990.3 ENST00000519597.5 |
ANKRD46
|
ankyrin repeat domain 46 |
| chr2_+_201183120 | 0.25 |
ENST00000272879.9
ENST00000286186.11 ENST00000374650.7 ENST00000346817.9 ENST00000313728.11 ENST00000448480.1 |
CASP10
|
caspase 10 |
| chr12_+_64404338 | 0.25 |
ENST00000332707.10
|
XPOT
|
exportin for tRNA |
| chr3_+_42979281 | 0.25 |
ENST00000488863.5
ENST00000430121.3 |
GASK1A
|
golgi associated kinase 1A |
| chr2_-_182242031 | 0.25 |
ENST00000358139.6
|
PDE1A
|
phosphodiesterase 1A |
| chr6_+_29461440 | 0.24 |
ENST00000396792.2
|
OR2H1
|
olfactory receptor family 2 subfamily H member 1 |
| chr20_-_31390580 | 0.24 |
ENST00000339144.3
ENST00000376321.4 |
DEFB119
|
defensin beta 119 |
| chr19_+_49513353 | 0.24 |
ENST00000596975.5
|
FCGRT
|
Fc fragment of IgG receptor and transporter |
| chr17_-_55722857 | 0.24 |
ENST00000424486.3
|
TMEM100
|
transmembrane protein 100 |
| chrX_+_43656289 | 0.24 |
ENST00000338702.4
|
MAOA
|
monoamine oxidase A |
| chrX_-_103255117 | 0.24 |
ENST00000372685.8
ENST00000360000.8 ENST00000451678.1 |
TCEAL8
|
transcription elongation factor A like 8 |
| chr15_+_94355956 | 0.24 |
ENST00000557742.1
|
MCTP2
|
multiple C2 and transmembrane domain containing 2 |
| chr17_-_445939 | 0.24 |
ENST00000329099.4
|
RFLNB
|
refilin B |
| chr5_+_175861628 | 0.24 |
ENST00000509837.5
|
CPLX2
|
complexin 2 |
| chr6_-_38703066 | 0.23 |
ENST00000373365.5
|
GLO1
|
glyoxalase I |
| chr1_+_42825548 | 0.23 |
ENST00000372514.7
|
ERMAP
|
erythroblast membrane associated protein (Scianna blood group) |
| chr12_+_119668109 | 0.23 |
ENST00000229328.10
ENST00000630317.1 |
PRKAB1
|
protein kinase AMP-activated non-catalytic subunit beta 1 |
| chr6_+_29111560 | 0.23 |
ENST00000377169.2
|
OR2J3
|
olfactory receptor family 2 subfamily J member 3 |
| chr1_+_152811971 | 0.23 |
ENST00000360090.4
|
LCE1B
|
late cornified envelope 1B |
| chr6_+_28124596 | 0.23 |
ENST00000340487.5
|
ZSCAN16
|
zinc finger and SCAN domain containing 16 |
| chr3_+_45886501 | 0.23 |
ENST00000395963.2
|
CCR9
|
C-C motif chemokine receptor 9 |
| chr5_-_126595237 | 0.23 |
ENST00000637206.1
ENST00000553117.5 |
ALDH7A1
|
aldehyde dehydrogenase 7 family member A1 |
| chr14_-_67412112 | 0.22 |
ENST00000216446.9
|
PLEK2
|
pleckstrin 2 |
| chr19_-_43883964 | 0.22 |
ENST00000587539.2
|
ZNF404
|
zinc finger protein 404 |
| chr2_-_88979016 | 0.22 |
ENST00000390247.2
|
IGKV3-7
|
immunoglobulin kappa variable 3-7 (non-functional) |
| chr2_+_90172802 | 0.22 |
ENST00000390277.3
|
IGKV3D-11
|
immunoglobulin kappa variable 3D-11 |
| chr6_-_110815152 | 0.22 |
ENST00000413605.6
|
CDK19
|
cyclin dependent kinase 19 |
| chr3_-_157503375 | 0.22 |
ENST00000362010.7
|
VEPH1
|
ventricular zone expressed PH domain containing 1 |
| chr16_+_3135121 | 0.22 |
ENST00000576416.5
ENST00000416391.6 |
ZNF213
|
zinc finger protein 213 |
| chr12_+_34022462 | 0.21 |
ENST00000538927.1
ENST00000266483.7 |
ALG10
|
ALG10 alpha-1,2-glucosyltransferase |
| chr9_+_93234923 | 0.21 |
ENST00000411624.5
|
WNK2
|
WNK lysine deficient protein kinase 2 |
| chr4_+_94207596 | 0.21 |
ENST00000359052.8
|
SMARCAD1
|
SWI/SNF-related, matrix-associated actin-dependent regulator of chromatin, subfamily a, containing DEAD/H box 1 |
| chr1_+_248013660 | 0.21 |
ENST00000355281.2
|
OR2L5
|
olfactory receptor family 2 subfamily L member 5 |
| chr3_-_157503339 | 0.21 |
ENST00000392833.6
|
VEPH1
|
ventricular zone expressed PH domain containing 1 |
| chrX_+_10158448 | 0.21 |
ENST00000380829.5
ENST00000421085.7 ENST00000674669.1 ENST00000454850.1 |
CLCN4
|
chloride voltage-gated channel 4 |
| chr1_-_13116854 | 0.21 |
ENST00000621994.3
|
HNRNPCL2
|
heterogeneous nuclear ribonucleoprotein C like 2 |
| chr17_+_18183052 | 0.21 |
ENST00000541285.1
|
ALKBH5
|
alkB homolog 5, RNA demethylase |
| chr4_-_149815826 | 0.21 |
ENST00000636793.2
ENST00000636414.1 |
IQCM
|
IQ motif containing M |
| chr17_+_50746534 | 0.20 |
ENST00000511974.5
|
LUC7L3
|
LUC7 like 3 pre-mRNA splicing factor |
| chrX_+_108091752 | 0.20 |
ENST00000457035.5
ENST00000372232.8 |
ATG4A
|
autophagy related 4A cysteine peptidase |
| chr14_-_106038355 | 0.20 |
ENST00000390597.3
|
IGHV2-5
|
immunoglobulin heavy variable 2-5 |
| chr1_-_212791762 | 0.20 |
ENST00000626725.1
ENST00000366977.8 ENST00000366976.3 |
NSL1
|
NSL1 component of MIS12 kinetochore complex |
| chr2_+_90234809 | 0.19 |
ENST00000443397.5
|
IGKV3D-7
|
immunoglobulin kappa variable 3D-7 |
| chr6_-_110815408 | 0.19 |
ENST00000368911.8
|
CDK19
|
cyclin dependent kinase 19 |
| chr15_+_58410543 | 0.19 |
ENST00000356113.10
ENST00000414170.7 |
LIPC
|
lipase C, hepatic type |
| chrX_+_108091665 | 0.19 |
ENST00000345734.7
|
ATG4A
|
autophagy related 4A cysteine peptidase |
| chr14_-_106211453 | 0.19 |
ENST00000390606.3
|
IGHV3-20
|
immunoglobulin heavy variable 3-20 |
| chr18_+_62539511 | 0.19 |
ENST00000586834.1
|
ZCCHC2
|
zinc finger CCHC-type containing 2 |
| chr2_-_27663594 | 0.19 |
ENST00000337768.10
ENST00000405491.5 ENST00000464789.2 ENST00000406540.5 |
SUPT7L
|
SPT7 like, STAGA complex subunit gamma |
| chr6_-_53510445 | 0.19 |
ENST00000509541.5
|
GCLC
|
glutamate-cysteine ligase catalytic subunit |
| chr6_-_52994248 | 0.19 |
ENST00000457564.1
ENST00000370960.5 |
GSTA4
|
glutathione S-transferase alpha 4 |
| chr2_-_27663817 | 0.18 |
ENST00000404798.6
|
SUPT7L
|
SPT7 like, STAGA complex subunit gamma |
| chr1_-_160954801 | 0.18 |
ENST00000368029.4
|
ITLN2
|
intelectin 2 |
| chr14_-_106470788 | 0.18 |
ENST00000434710.1
|
IGHV3-43
|
immunoglobulin heavy variable 3-43 |
| chrX_-_72239022 | 0.18 |
ENST00000373657.2
ENST00000334463.4 |
ERCC6L
|
ERCC excision repair 6 like, spindle assembly checkpoint helicase |
| chr9_-_96778053 | 0.18 |
ENST00000375231.5
ENST00000223428.9 |
ZNF510
|
zinc finger protein 510 |
| chr1_+_155308930 | 0.18 |
ENST00000465559.5
ENST00000612683.1 |
FDPS
|
farnesyl diphosphate synthase |
| chr4_+_87608529 | 0.18 |
ENST00000651931.1
|
DSPP
|
dentin sialophosphoprotein |
| chr11_-_129024157 | 0.17 |
ENST00000392657.7
|
ARHGAP32
|
Rho GTPase activating protein 32 |
| chr1_+_155308748 | 0.17 |
ENST00000611010.4
ENST00000447866.5 ENST00000368356.9 ENST00000467076.5 ENST00000491013.5 ENST00000356657.10 |
FDPS
|
farnesyl diphosphate synthase |
| chr9_+_122371014 | 0.17 |
ENST00000362012.7
|
PTGS1
|
prostaglandin-endoperoxide synthase 1 |
| chr5_+_36151989 | 0.17 |
ENST00000274254.9
|
SKP2
|
S-phase kinase associated protein 2 |
| chr7_+_117014881 | 0.17 |
ENST00000422922.5
ENST00000432298.5 |
ST7
|
suppression of tumorigenicity 7 |
| chr16_+_3134923 | 0.17 |
ENST00000574902.5
ENST00000396878.8 |
ZNF213
|
zinc finger protein 213 |
| chr12_+_101594849 | 0.17 |
ENST00000547405.5
ENST00000452455.6 ENST00000392934.7 ENST00000547509.5 ENST00000361685.6 ENST00000549145.5 ENST00000361466.7 ENST00000553190.5 ENST00000545503.6 ENST00000536007.5 ENST00000541119.5 ENST00000551300.5 ENST00000550270.1 |
MYBPC1
|
myosin binding protein C1 |
| chr6_+_29550407 | 0.16 |
ENST00000641137.1
|
OR2I1P
|
olfactory receptor family 2 subfamily I member 1 pseudogene |
| chr3_+_39383337 | 0.16 |
ENST00000650617.1
ENST00000431510.1 ENST00000645630.1 |
SLC25A38
|
solute carrier family 25 member 38 |
| chrX_+_77910656 | 0.16 |
ENST00000343533.9
ENST00000341514.11 ENST00000645454.1 ENST00000642651.1 ENST00000644362.1 |
ATP7A
PGK1
|
ATPase copper transporting alpha phosphoglycerate kinase 1 |
| chr17_+_18183803 | 0.16 |
ENST00000399138.5
|
ALKBH5
|
alkB homolog 5, RNA demethylase |
| chr5_+_140841183 | 0.16 |
ENST00000378123.4
ENST00000531613.2 |
PCDHA8
|
protocadherin alpha 8 |
| chr2_+_233729042 | 0.16 |
ENST00000482026.6
|
UGT1A3
|
UDP glucuronosyltransferase family 1 member A3 |
| chr13_-_75366973 | 0.16 |
ENST00000648194.1
|
TBC1D4
|
TBC1 domain family member 4 |
| chr14_-_94323324 | 0.16 |
ENST00000341584.4
|
SERPINA6
|
serpin family A member 6 |
| chr13_+_35476740 | 0.16 |
ENST00000537702.5
|
NBEA
|
neurobeachin |
| chr10_+_92691813 | 0.16 |
ENST00000472590.6
|
HHEX
|
hematopoietically expressed homeobox |
| chr15_-_55408245 | 0.16 |
ENST00000563171.5
ENST00000425574.7 ENST00000442196.8 ENST00000564092.1 |
CCPG1
|
cell cycle progression 1 |
| chr19_+_55836532 | 0.15 |
ENST00000301295.11
|
NLRP4
|
NLR family pyrin domain containing 4 |
| chrX_+_37780049 | 0.15 |
ENST00000378588.5
|
CYBB
|
cytochrome b-245 beta chain |
| chr5_+_36152077 | 0.15 |
ENST00000546211.6
ENST00000620197.5 ENST00000678270.1 ENST00000679015.1 ENST00000678580.1 ENST00000274255.11 ENST00000508514.5 |
SKP2
|
S-phase kinase associated protein 2 |
| chr17_-_41586887 | 0.15 |
ENST00000167586.7
|
KRT14
|
keratin 14 |
| chr2_-_89027700 | 0.15 |
ENST00000483158.1
|
IGKV3-11
|
immunoglobulin kappa variable 3-11 |
| chr6_-_109381739 | 0.15 |
ENST00000504373.2
|
CD164
|
CD164 molecule |
| chr2_+_87338511 | 0.15 |
ENST00000421835.2
|
IGKV3OR2-268
|
immunoglobulin kappa variable 3/OR2-268 (non-functional) |
| chr4_+_157076119 | 0.15 |
ENST00000541722.5
ENST00000264428.9 ENST00000512619.5 |
GLRB
|
glycine receptor beta |
| chr19_+_11374658 | 0.14 |
ENST00000674460.1
ENST00000312423.4 |
SWSAP1
|
SWIM-type zinc finger 7 associated protein 1 |
| chr2_-_160200289 | 0.14 |
ENST00000409872.1
|
ITGB6
|
integrin subunit beta 6 |
| chr8_-_30812773 | 0.14 |
ENST00000221138.9
|
PPP2CB
|
protein phosphatase 2 catalytic subunit beta |
| chr19_-_4558417 | 0.14 |
ENST00000586965.1
|
SEMA6B
|
semaphorin 6B |
| chr6_+_110180418 | 0.14 |
ENST00000368930.5
ENST00000307731.2 |
CDC40
|
cell division cycle 40 |
| chr8_-_7430348 | 0.14 |
ENST00000318124.3
|
DEFB103B
|
defensin beta 103B |
| chr12_-_23584600 | 0.14 |
ENST00000396007.6
|
SOX5
|
SRY-box transcription factor 5 |
| chr2_-_142131004 | 0.14 |
ENST00000434794.1
ENST00000389484.8 |
LRP1B
|
LDL receptor related protein 1B |
| chr14_+_56117702 | 0.13 |
ENST00000559044.5
|
PELI2
|
pellino E3 ubiquitin protein ligase family member 2 |
| chrX_+_7219431 | 0.13 |
ENST00000674499.1
ENST00000217961.5 |
STS
|
steroid sulfatase |
| chr2_+_132416795 | 0.13 |
ENST00000329321.4
|
GPR39
|
G protein-coupled receptor 39 |
| chr10_-_30629741 | 0.13 |
ENST00000647634.2
ENST00000375318.4 |
LYZL2
|
lysozyme like 2 |
| chr3_-_149576203 | 0.13 |
ENST00000472417.1
|
WWTR1
|
WW domain containing transcription regulator 1 |
| chrX_+_85003863 | 0.13 |
ENST00000373173.7
|
APOOL
|
apolipoprotein O like |
| chr1_+_27934980 | 0.13 |
ENST00000373894.8
|
SMPDL3B
|
sphingomyelin phosphodiesterase acid like 3B |
| chr2_-_201739205 | 0.13 |
ENST00000681558.1
ENST00000681495.1 |
ALS2
|
alsin Rho guanine nucleotide exchange factor ALS2 |
| chr4_+_75556048 | 0.13 |
ENST00000616557.1
ENST00000435974.2 ENST00000311623.9 |
ODAPH
|
odontogenesis associated phosphoprotein |
| chr10_+_87357720 | 0.13 |
ENST00000412718.3
ENST00000381697.7 |
NUTM2D
|
NUT family member 2D |
| chr2_+_102473219 | 0.13 |
ENST00000295269.5
|
SLC9A4
|
solute carrier family 9 member A4 |
| chr17_-_47189176 | 0.13 |
ENST00000531206.5
ENST00000527547.5 ENST00000575483.5 ENST00000066544.8 |
CDC27
|
cell division cycle 27 |
| chr17_+_50746614 | 0.13 |
ENST00000513969.5
ENST00000503728.1 |
LUC7L3
|
LUC7 like 3 pre-mRNA splicing factor |
| chr2_+_27663441 | 0.13 |
ENST00000326019.10
ENST00000613058.4 |
SLC4A1AP
|
solute carrier family 4 member 1 adaptor protein |
| chr17_+_32021005 | 0.13 |
ENST00000327564.11
ENST00000584368.5 ENST00000394713.7 ENST00000341671.11 |
LRRC37B
|
leucine rich repeat containing 37B |
| chr2_-_20823048 | 0.12 |
ENST00000402479.6
ENST00000237822.8 ENST00000626491.2 ENST00000432947.1 ENST00000403006.6 ENST00000419825.2 ENST00000381090.7 ENST00000412261.5 ENST00000619656.4 ENST00000541941.5 ENST00000440866.6 ENST00000435420.6 |
LDAH
|
lipid droplet associated hydrolase |
| chr20_+_45416551 | 0.12 |
ENST00000639292.1
|
PIGT
|
phosphatidylinositol glycan anchor biosynthesis class T |
| chr14_-_74084393 | 0.12 |
ENST00000350259.8
ENST00000553458.6 |
ALDH6A1
|
aldehyde dehydrogenase 6 family member A1 |
| chr6_+_110180116 | 0.12 |
ENST00000368932.5
|
CDC40
|
cell division cycle 40 |
| chr13_+_77741160 | 0.12 |
ENST00000314070.9
ENST00000351546.7 |
SLAIN1
|
SLAIN motif family member 1 |
| chr22_+_39901075 | 0.12 |
ENST00000344138.9
|
GRAP2
|
GRB2 related adaptor protein 2 |
| chr1_-_77219399 | 0.12 |
ENST00000359130.1
ENST00000370812.8 ENST00000445065.5 |
PIGK
|
phosphatidylinositol glycan anchor biosynthesis class K |
| chr2_+_27663880 | 0.12 |
ENST00000618046.4
ENST00000613517.4 |
SLC4A1AP
|
solute carrier family 4 member 1 adaptor protein |
| chr19_+_49513154 | 0.12 |
ENST00000426395.7
ENST00000600273.5 ENST00000599988.5 |
FCGRT
|
Fc fragment of IgG receptor and transporter |
| chr4_-_99219230 | 0.11 |
ENST00000394897.5
ENST00000508558.1 ENST00000394899.6 |
ADH6
|
alcohol dehydrogenase 6 (class V) |
| chr20_+_58907981 | 0.11 |
ENST00000656419.1
|
GNAS
|
GNAS complex locus |
| chr13_-_35855758 | 0.11 |
ENST00000615680.4
|
DCLK1
|
doublecortin like kinase 1 |
| chr15_+_48191648 | 0.11 |
ENST00000646012.1
ENST00000561127.5 ENST00000647546.1 ENST00000559641.5 ENST00000417307.3 |
SLC12A1
CTXN2
|
solute carrier family 12 member 1 cortexin 2 |
| chr7_-_36985060 | 0.11 |
ENST00000396040.6
|
ELMO1
|
engulfment and cell motility 1 |
| chr2_+_233917371 | 0.11 |
ENST00000324695.9
ENST00000433712.6 |
TRPM8
|
transient receptor potential cation channel subfamily M member 8 |
| chr18_-_35290184 | 0.11 |
ENST00000589178.5
ENST00000592278.1 ENST00000333206.10 ENST00000592211.1 ENST00000420878.7 ENST00000610712.1 ENST00000586922.2 |
ZSCAN30
ENSG00000268573.1
|
zinc finger and SCAN domain containing 30 novel transcript |
| chr7_-_87220567 | 0.11 |
ENST00000433078.5
|
TMEM243
|
transmembrane protein 243 |
| chr19_+_55857437 | 0.11 |
ENST00000587891.5
|
NLRP4
|
NLR family pyrin domain containing 4 |
| chr17_+_1771688 | 0.11 |
ENST00000572048.1
ENST00000573763.1 |
SERPINF1
|
serpin family F member 1 |
| chr10_-_13001705 | 0.10 |
ENST00000378825.5
|
CCDC3
|
coiled-coil domain containing 3 |
| chr3_-_149377637 | 0.10 |
ENST00000305366.8
|
TM4SF1
|
transmembrane 4 L six family member 1 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 3.7 | GO:0046092 | deoxycytidine metabolic process(GO:0046092) |
| 0.2 | 0.6 | GO:0072709 | cellular response to sorbitol(GO:0072709) |
| 0.2 | 0.6 | GO:0036510 | trimming of terminal mannose on C branch(GO:0036510) |
| 0.2 | 0.9 | GO:0097210 | response to gonadotropin-releasing hormone(GO:0097210) cellular response to gonadotropin-releasing hormone(GO:0097211) |
| 0.2 | 1.6 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.1 | 0.4 | GO:0060003 | copper ion export(GO:0060003) |
| 0.1 | 0.8 | GO:0010748 | regulation of plasma membrane long-chain fatty acid transport(GO:0010746) negative regulation of plasma membrane long-chain fatty acid transport(GO:0010748) |
| 0.1 | 0.4 | GO:0002416 | IgG immunoglobulin transcytosis in epithelial cells mediated by FcRn immunoglobulin receptor(GO:0002416) |
| 0.1 | 0.3 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.1 | 0.4 | GO:0035513 | oxidative RNA demethylation(GO:0035513) oxidative single-stranded RNA demethylation(GO:0035553) |
| 0.1 | 0.5 | GO:1901979 | regulation of inward rectifier potassium channel activity(GO:1901979) |
| 0.1 | 0.5 | GO:0051622 | negative regulation of dopamine uptake involved in synaptic transmission(GO:0051585) norepinephrine uptake(GO:0051620) regulation of norepinephrine uptake(GO:0051621) negative regulation of norepinephrine uptake(GO:0051622) negative regulation of catecholamine uptake involved in synaptic transmission(GO:0051945) regulation of glutathione peroxidase activity(GO:1903282) positive regulation of glutathione peroxidase activity(GO:1903284) positive regulation of hydrogen peroxide catabolic process(GO:1903285) positive regulation of peroxidase activity(GO:2000470) |
| 0.1 | 0.2 | GO:0061182 | negative regulation of chondrocyte development(GO:0061182) |
| 0.1 | 0.2 | GO:0002305 | gamma-delta intraepithelial T cell differentiation(GO:0002304) CD8-positive, gamma-delta intraepithelial T cell differentiation(GO:0002305) |
| 0.1 | 0.3 | GO:0033384 | geranyl diphosphate metabolic process(GO:0033383) geranyl diphosphate biosynthetic process(GO:0033384) farnesyl diphosphate biosynthetic process(GO:0045337) |
| 0.1 | 0.4 | GO:0051697 | protein delipidation(GO:0051697) |
| 0.1 | 0.3 | GO:0072573 | propionate metabolic process(GO:0019541) cellular response to progesterone stimulus(GO:0071393) tolerance induction to lipopolysaccharide(GO:0072573) |
| 0.1 | 0.3 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.1 | 0.2 | GO:0031456 | glycine betaine biosynthetic process from choline(GO:0019285) glycine betaine metabolic process(GO:0031455) glycine betaine biosynthetic process(GO:0031456) |
| 0.1 | 0.2 | GO:0036233 | glycine import(GO:0036233) |
| 0.1 | 0.3 | GO:0071460 | cellular response to cell-matrix adhesion(GO:0071460) |
| 0.1 | 0.2 | GO:0061011 | hepatic duct development(GO:0061011) |
| 0.1 | 0.2 | GO:1904844 | response to L-glutamine(GO:1904844) cellular response to L-glutamine(GO:1904845) |
| 0.0 | 0.2 | GO:0097069 | cellular response to thyroxine stimulus(GO:0097069) cellular response to L-phenylalanine derivative(GO:1904387) |
| 0.0 | 0.5 | GO:0044829 | positive regulation by host of viral genome replication(GO:0044829) |
| 0.0 | 0.4 | GO:0048539 | bone marrow development(GO:0048539) |
| 0.0 | 0.4 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 0.0 | 0.5 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.0 | 0.1 | GO:0097112 | gamma-aminobutyric acid receptor clustering(GO:0097112) |
| 0.0 | 0.1 | GO:0046603 | negative regulation of mitotic centrosome separation(GO:0046603) |
| 0.0 | 0.1 | GO:1903595 | positive regulation of histamine secretion by mast cell(GO:1903595) |
| 0.0 | 0.1 | GO:0039008 | pronephric nephron morphogenesis(GO:0039007) pronephric nephron tubule morphogenesis(GO:0039008) pronephric duct development(GO:0039022) pronephric duct morphogenesis(GO:0039023) Kupffer's vesicle development(GO:0070121) |
| 0.0 | 0.1 | GO:0090119 | vesicle-mediated cholesterol transport(GO:0090119) |
| 0.0 | 0.1 | GO:0019859 | pyrimidine nucleobase catabolic process(GO:0006208) thymine catabolic process(GO:0006210) thymine metabolic process(GO:0019859) |
| 0.0 | 1.4 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.0 | 0.2 | GO:0009438 | methylglyoxal metabolic process(GO:0009438) |
| 0.0 | 0.4 | GO:0010991 | negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.0 | 0.1 | GO:0035022 | positive regulation of Rac protein signal transduction(GO:0035022) |
| 0.0 | 0.3 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.0 | 0.2 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.0 | 0.4 | GO:0045835 | negative regulation of meiotic nuclear division(GO:0045835) |
| 0.0 | 0.1 | GO:0045110 | intermediate filament bundle assembly(GO:0045110) |
| 0.0 | 0.1 | GO:1903676 | regulation of cap-dependent translational initiation(GO:1903674) positive regulation of cap-dependent translational initiation(GO:1903676) |
| 0.0 | 0.1 | GO:0061419 | positive regulation of transcription from RNA polymerase II promoter in response to hypoxia(GO:0061419) |
| 0.0 | 0.1 | GO:0002184 | cytoplasmic translational termination(GO:0002184) |
| 0.0 | 0.1 | GO:0050955 | thermoception(GO:0050955) |
| 0.0 | 0.2 | GO:0031339 | negative regulation of vesicle fusion(GO:0031339) |
| 0.0 | 0.1 | GO:0071279 | cellular response to cobalt ion(GO:0071279) |
| 0.0 | 0.2 | GO:0060842 | arterial endothelial cell differentiation(GO:0060842) |
| 0.0 | 0.1 | GO:0055059 | asymmetric neuroblast division(GO:0055059) |
| 0.0 | 0.3 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.0 | 0.3 | GO:0055070 | copper ion homeostasis(GO:0055070) |
| 0.0 | 0.1 | GO:0036378 | calcitriol biosynthetic process from calciol(GO:0036378) |
| 0.0 | 0.2 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 0.0 | 0.2 | GO:2001106 | regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.0 | 0.3 | GO:0060044 | negative regulation of cardiac muscle cell proliferation(GO:0060044) |
| 0.0 | 0.1 | GO:0014005 | microglia differentiation(GO:0014004) microglia development(GO:0014005) |
| 0.0 | 0.2 | GO:0042420 | dopamine catabolic process(GO:0042420) |
| 0.0 | 0.3 | GO:0009642 | response to light intensity(GO:0009642) |
| 0.0 | 0.3 | GO:0055026 | negative regulation of cardiac muscle tissue development(GO:0055026) |
| 0.0 | 1.0 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.0 | 0.1 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.0 | 0.2 | GO:0052697 | flavonoid glucuronidation(GO:0052696) xenobiotic glucuronidation(GO:0052697) |
| 0.0 | 0.1 | GO:0032072 | plasmacytoid dendritic cell activation(GO:0002270) regulation of restriction endodeoxyribonuclease activity(GO:0032072) |
| 0.0 | 0.2 | GO:0070933 | histone H4 deacetylation(GO:0070933) |
| 0.0 | 0.0 | GO:0010387 | COP9 signalosome assembly(GO:0010387) |
| 0.0 | 0.1 | GO:0042796 | snRNA transcription from RNA polymerase III promoter(GO:0042796) |
| 0.0 | 0.1 | GO:0051001 | negative regulation of nitric-oxide synthase activity(GO:0051001) |
| 0.0 | 0.4 | GO:0051457 | maintenance of protein location in nucleus(GO:0051457) |
| 0.0 | 0.2 | GO:0001542 | ovulation from ovarian follicle(GO:0001542) |
| 0.0 | 0.2 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.0 | 0.1 | GO:1990253 | cellular response to leucine starvation(GO:1990253) |
| 0.0 | 1.1 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.0 | 0.1 | GO:0070370 | heat acclimation(GO:0010286) cellular heat acclimation(GO:0070370) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.7 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.0 | 0.2 | GO:0017109 | glutamate-cysteine ligase complex(GO:0017109) |
| 0.0 | 0.2 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.0 | 0.1 | GO:0097196 | Shu complex(GO:0097196) |
| 0.0 | 0.2 | GO:0070554 | synaptobrevin 2-SNAP-25-syntaxin-3-complexin complex(GO:0070554) |
| 0.0 | 0.2 | GO:0000444 | MIS12/MIND type complex(GO:0000444) |
| 0.0 | 0.6 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) |
| 0.0 | 0.6 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.8 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 0.4 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.4 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.0 | 0.9 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 0.1 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.0 | 0.1 | GO:0018444 | translation release factor complex(GO:0018444) |
| 0.0 | 0.4 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 1.5 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 0.6 | GO:0044322 | endoplasmic reticulum quality control compartment(GO:0044322) |
| 0.0 | 0.0 | GO:0070931 | Golgi-associated vesicle lumen(GO:0070931) |
| 0.0 | 0.1 | GO:1990131 | Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.0 | 0.1 | GO:0035363 | histone locus body(GO:0035363) |
| 0.0 | 0.3 | GO:0071004 | U2-type prespliceosome(GO:0071004) |
| 0.0 | 0.5 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 0.5 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.0 | GO:1990667 | PCSK9-AnxA2 complex(GO:1990667) |
| 0.0 | 0.1 | GO:0005927 | muscle tendon junction(GO:0005927) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 3.7 | GO:0004137 | deoxycytidine kinase activity(GO:0004137) thymidine kinase activity(GO:0004797) |
| 0.2 | 0.7 | GO:0004583 | dolichyl-phosphate-glucose-glycolipid alpha-glucosyltransferase activity(GO:0004583) |
| 0.1 | 0.4 | GO:0004008 | copper-exporting ATPase activity(GO:0004008) copper-transporting ATPase activity(GO:0043682) |
| 0.1 | 1.7 | GO:0022833 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.1 | 0.6 | GO:0097199 | cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097199) |
| 0.1 | 0.4 | GO:0035515 | oxidative RNA demethylase activity(GO:0035515) |
| 0.1 | 0.3 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.1 | 0.4 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.1 | 0.5 | GO:0060961 | phospholipase D inhibitor activity(GO:0060961) |
| 0.1 | 0.3 | GO:0004337 | dimethylallyltranstransferase activity(GO:0004161) geranyltranstransferase activity(GO:0004337) |
| 0.1 | 0.6 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.0 | 0.2 | GO:0004357 | glutamate-cysteine ligase activity(GO:0004357) |
| 0.0 | 0.2 | GO:0004666 | prostaglandin-endoperoxide synthase activity(GO:0004666) |
| 0.0 | 0.2 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.0 | 0.2 | GO:0003923 | GPI-anchor transamidase activity(GO:0003923) |
| 0.0 | 0.5 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.0 | 0.1 | GO:0008511 | sodium:potassium:chloride symporter activity(GO:0008511) |
| 0.0 | 0.1 | GO:0004773 | steryl-sulfatase activity(GO:0004773) |
| 0.0 | 0.2 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.0 | 0.2 | GO:0047273 | galactosylgalactosylglucosylceramide beta-D-acetylgalactosaminyltransferase activity(GO:0047273) |
| 0.0 | 0.1 | GO:0016933 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.0 | 0.1 | GO:0047374 | methylumbelliferyl-acetate deacetylase activity(GO:0047374) |
| 0.0 | 0.1 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.0 | 0.2 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.0 | 0.7 | GO:0005328 | neurotransmitter:sodium symporter activity(GO:0005328) |
| 0.0 | 0.2 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.0 | 0.1 | GO:1990460 | leptin receptor binding(GO:1990460) |
| 0.0 | 0.1 | GO:0030343 | vitamin D3 25-hydroxylase activity(GO:0030343) vitamin D 25-hydroxylase activity(GO:0070643) |
| 0.0 | 1.1 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 0.4 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.0 | 0.7 | GO:0043548 | phosphatidylinositol 3-kinase binding(GO:0043548) |
| 0.0 | 0.1 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.0 | 0.3 | GO:0070492 | phosphatidylinositol-5-phosphate binding(GO:0010314) oligosaccharide binding(GO:0070492) |
| 0.0 | 0.3 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.0 | 0.3 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.0 | 0.6 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.0 | 0.1 | GO:0033188 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.0 | 0.2 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.0 | 0.1 | GO:0008281 | sulfonylurea receptor activity(GO:0008281) |
| 0.0 | 0.1 | GO:0004024 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) |
| 0.0 | 0.0 | GO:0008798 | beta-aspartyl-peptidase activity(GO:0008798) |
| 0.0 | 0.7 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.1 | GO:0019958 | C-X-C chemokine binding(GO:0019958) |
| 0.0 | 0.1 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 0.1 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 0.1 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 0.6 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 1.8 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 0.7 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 0.8 | SIG IL4RECEPTOR IN B LYPHOCYTES | Genes related to IL4 rceptor signaling in B lymphocytes |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.7 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.0 | 0.6 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 0.8 | REACTOME SOS MEDIATED SIGNALLING | Genes involved in SOS-mediated signalling |
| 0.0 | 0.5 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.0 | 0.6 | REACTOME NFKB ACTIVATION THROUGH FADD RIP1 PATHWAY MEDIATED BY CASPASE 8 AND10 | Genes involved in NF-kB activation through FADD/RIP-1 pathway mediated by caspase-8 and -10 |
| 0.0 | 0.4 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.0 | 0.1 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.0 | 0.7 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.0 | 0.7 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 0.7 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.0 | 0.5 | REACTOME ACTIVATED TAK1 MEDIATES P38 MAPK ACTIVATION | Genes involved in activated TAK1 mediates p38 MAPK activation |
| 0.0 | 0.3 | REACTOME FGFR1 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR1 ligand binding and activation |
| 0.0 | 0.2 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.0 | 0.2 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 0.5 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 0.6 | REACTOME CA DEPENDENT EVENTS | Genes involved in Ca-dependent events |