Project
Inflammatory response time course, HUVEC (Wada, 2009)
Navigation
Downloads
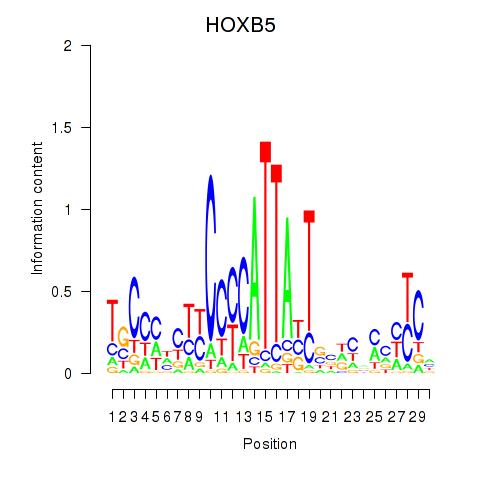
Results for HOXB5
Z-value: 0.46
Transcription factors associated with HOXB5
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HOXB5
|
ENSG00000120075.6 | HOXB5 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HOXB5 | hg38_v1_chr17_-_48593748_48593786 | -0.34 | 1.0e-01 | Click! |
Activity profile of HOXB5 motif
Sorted Z-values of HOXB5 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of HOXB5
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_-_127822455 | 2.49 |
ENST00000265052.10
|
MGLL
|
monoglyceride lipase |
| chr19_+_49363923 | 0.78 |
ENST00000597546.1
|
DKKL1
|
dickkopf like acrosomal protein 1 |
| chr11_-_6030758 | 0.71 |
ENST00000641900.1
|
OR56A1
|
olfactory receptor family 56 subfamily A member 1 |
| chr16_-_84618067 | 0.70 |
ENST00000262428.5
|
COTL1
|
coactosin like F-actin binding protein 1 |
| chr16_-_84618041 | 0.69 |
ENST00000564057.1
|
COTL1
|
coactosin like F-actin binding protein 1 |
| chr5_-_35047935 | 0.54 |
ENST00000510428.1
ENST00000231420.11 |
AGXT2
|
alanine--glyoxylate aminotransferase 2 |
| chr11_-_128522189 | 0.52 |
ENST00000526145.6
|
ETS1
|
ETS proto-oncogene 1, transcription factor |
| chr1_+_155610218 | 0.51 |
ENST00000649846.1
ENST00000245564.8 ENST00000368341.8 |
MSTO1
|
misato mitochondrial distribution and morphology regulator 1 |
| chr3_-_165196689 | 0.51 |
ENST00000241274.3
|
SLITRK3
|
SLIT and NTRK like family member 3 |
| chr11_-_57322197 | 0.48 |
ENST00000532437.1
|
TNKS1BP1
|
tankyrase 1 binding protein 1 |
| chr1_+_21981099 | 0.45 |
ENST00000400277.2
|
CELA3B
|
chymotrypsin like elastase 3B |
| chr1_+_4654732 | 0.45 |
ENST00000378190.7
|
AJAP1
|
adherens junctions associated protein 1 |
| chr10_+_112376193 | 0.43 |
ENST00000433418.6
ENST00000356116.6 ENST00000354273.5 |
ACSL5
|
acyl-CoA synthetase long chain family member 5 |
| chr19_+_49363730 | 0.42 |
ENST00000596402.1
ENST00000221498.7 |
DKKL1
|
dickkopf like acrosomal protein 1 |
| chr5_-_161546970 | 0.38 |
ENST00000675303.1
|
GABRB2
|
gamma-aminobutyric acid type A receptor subunit beta2 |
| chr14_-_23551729 | 0.37 |
ENST00000419474.5
|
ZFHX2
|
zinc finger homeobox 2 |
| chr11_-_35418966 | 0.36 |
ENST00000531628.2
|
SLC1A2
|
solute carrier family 1 member 2 |
| chr1_+_146938744 | 0.33 |
ENST00000617931.4
|
NBPF12
|
NBPF member 12 |
| chr15_-_50765656 | 0.33 |
ENST00000261854.10
|
SPPL2A
|
signal peptide peptidase like 2A |
| chr11_-_35419098 | 0.31 |
ENST00000606205.6
ENST00000645303.1 |
SLC1A2
|
solute carrier family 1 member 2 |
| chr12_-_101407727 | 0.29 |
ENST00000539055.5
ENST00000551688.1 ENST00000551671.5 ENST00000261636.13 |
ARL1
|
ADP ribosylation factor like GTPase 1 |
| chr1_+_4654601 | 0.29 |
ENST00000378191.5
|
AJAP1
|
adherens junctions associated protein 1 |
| chr3_+_51385282 | 0.28 |
ENST00000528157.7
|
MANF
|
mesencephalic astrocyte derived neurotrophic factor |
| chr7_-_102611591 | 0.28 |
ENST00000461209.5
|
RASA4
|
RAS p21 protein activator 4 |
| chr15_+_41621492 | 0.27 |
ENST00000570161.6
|
MGA
|
MAX dimerization protein MGA |
| chr1_-_247458105 | 0.27 |
ENST00000641149.1
ENST00000641527.1 |
OR2B11
|
olfactory receptor family 2 subfamily B member 11 |
| chr16_-_1771495 | 0.27 |
ENST00000564628.1
ENST00000219302.8 ENST00000563498.1 |
NME3
|
NME/NM23 nucleoside diphosphate kinase 3 |
| chr1_-_155688475 | 0.26 |
ENST00000405763.7
ENST00000368340.9 ENST00000454523.5 ENST00000368339.9 ENST00000443231.5 ENST00000347088.9 ENST00000361831.9 |
YY1AP1
|
YY1 associated protein 1 |
| chr16_-_57971121 | 0.26 |
ENST00000251102.13
|
CNGB1
|
cyclic nucleotide gated channel subunit beta 1 |
| chr9_-_127937800 | 0.23 |
ENST00000373110.4
ENST00000314392.13 |
DPM2
|
dolichyl-phosphate mannosyltransferase subunit 2, regulatory |
| chr12_+_54561442 | 0.23 |
ENST00000550620.1
|
PDE1B
|
phosphodiesterase 1B |
| chr10_-_101843777 | 0.22 |
ENST00000356640.7
|
KCNIP2
|
potassium voltage-gated channel interacting protein 2 |
| chrX_+_47218232 | 0.22 |
ENST00000457458.6
ENST00000522883.1 |
CDK16
|
cyclin dependent kinase 16 |
| chr9_+_4662281 | 0.21 |
ENST00000381883.5
|
PLPP6
|
phospholipid phosphatase 6 |
| chr17_-_44066595 | 0.21 |
ENST00000585388.2
ENST00000293406.8 |
LSM12
|
LSM12 homolog |
| chr14_-_102238439 | 0.21 |
ENST00000522534.5
|
MOK
|
MOK protein kinase |
| chr15_+_41621134 | 0.21 |
ENST00000566718.6
|
MGA
|
MAX dimerization protein MGA |
| chr12_-_52321395 | 0.20 |
ENST00000293670.3
|
KRT83
|
keratin 83 |
| chr9_-_110208156 | 0.19 |
ENST00000400613.5
|
C9orf152
|
chromosome 9 open reading frame 152 |
| chrM_+_12329 | 0.19 |
ENST00000361567.2
|
MT-ND5
|
mitochondrially encoded NADH:ubiquinone oxidoreductase core subunit 5 |
| chr7_+_130381092 | 0.19 |
ENST00000484324.1
|
CPA1
|
carboxypeptidase A1 |
| chr16_-_57971086 | 0.18 |
ENST00000564448.5
ENST00000311183.8 |
CNGB1
|
cyclic nucleotide gated channel subunit beta 1 |
| chr1_-_36385872 | 0.18 |
ENST00000373129.7
|
STK40
|
serine/threonine kinase 40 |
| chr17_+_48892761 | 0.18 |
ENST00000355938.9
ENST00000393366.7 ENST00000503641.5 ENST00000514808.5 ENST00000506855.1 |
ATP5MC1
|
ATP synthase membrane subunit c locus 1 |
| chr1_-_36385887 | 0.18 |
ENST00000373130.7
ENST00000373132.4 |
STK40
|
serine/threonine kinase 40 |
| chr3_+_130894382 | 0.17 |
ENST00000509662.5
ENST00000328560.12 ENST00000428331.6 ENST00000359644.7 ENST00000422190.6 |
ATP2C1
|
ATPase secretory pathway Ca2+ transporting 1 |
| chr18_+_34978244 | 0.17 |
ENST00000436190.6
|
MAPRE2
|
microtubule associated protein RP/EB family member 2 |
| chr1_+_154257071 | 0.16 |
ENST00000428595.1
|
UBAP2L
|
ubiquitin associated protein 2 like |
| chr7_+_5282935 | 0.15 |
ENST00000396872.8
ENST00000444741.5 ENST00000297195.8 ENST00000406453.3 |
SLC29A4
|
solute carrier family 29 member 4 |
| chr8_+_117135259 | 0.15 |
ENST00000519688.5
|
SLC30A8
|
solute carrier family 30 member 8 |
| chr1_-_155688727 | 0.15 |
ENST00000404643.5
ENST00000359205.9 ENST00000407221.5 ENST00000355499.9 |
YY1AP1
|
YY1 associated protein 1 |
| chr1_-_155688294 | 0.15 |
ENST00000311573.9
|
YY1AP1
|
YY1 associated protein 1 |
| chr1_-_145859061 | 0.15 |
ENST00000393045.7
|
PIAS3
|
protein inhibitor of activated STAT 3 |
| chr1_-_145859043 | 0.14 |
ENST00000369298.5
|
PIAS3
|
protein inhibitor of activated STAT 3 |
| chr11_+_63506046 | 0.14 |
ENST00000674247.1
|
LGALS12
|
galectin 12 |
| chr18_-_35497591 | 0.14 |
ENST00000589273.1
ENST00000586489.5 |
INO80C
|
INO80 complex subunit C |
| chr1_+_235328959 | 0.13 |
ENST00000643758.1
ENST00000497327.1 |
TBCE
GGPS1
|
tubulin folding cofactor E geranylgeranyl diphosphate synthase 1 |
| chr12_-_6689359 | 0.13 |
ENST00000683879.1
|
ZNF384
|
zinc finger protein 384 |
| chr12_-_6689244 | 0.13 |
ENST00000361959.7
ENST00000436774.6 ENST00000544482.1 |
ZNF384
|
zinc finger protein 384 |
| chr14_+_21030509 | 0.13 |
ENST00000481535.5
|
TPPP2
|
tubulin polymerization promoting protein family member 2 |
| chr3_-_195811857 | 0.13 |
ENST00000349607.8
ENST00000346145.8 |
MUC4
|
mucin 4, cell surface associated |
| chr6_-_31541937 | 0.12 |
ENST00000456662.5
ENST00000431908.5 ENST00000456976.5 ENST00000428450.5 ENST00000418897.5 ENST00000396172.6 ENST00000419020.1 ENST00000428098.5 |
DDX39B
|
DExD-box helicase 39B |
| chr15_-_74725370 | 0.11 |
ENST00000567032.5
ENST00000564596.5 ENST00000566503.1 ENST00000395049.8 ENST00000379727.8 ENST00000617691.4 ENST00000395048.6 |
CYP1A1
|
cytochrome P450 family 1 subfamily A member 1 |
| chr20_+_8789517 | 0.11 |
ENST00000437439.2
|
PLCB1
|
phospholipase C beta 1 |
| chr12_-_6689450 | 0.11 |
ENST00000355772.8
ENST00000417772.7 ENST00000319770.7 ENST00000396801.7 |
ZNF384
|
zinc finger protein 384 |
| chr3_-_195811916 | 0.11 |
ENST00000463781.8
|
MUC4
|
mucin 4, cell surface associated |
| chr19_-_12156710 | 0.11 |
ENST00000455799.1
ENST00000439556.3 |
ZNF625
|
zinc finger protein 625 |
| chr2_-_97589862 | 0.11 |
ENST00000258459.11
ENST00000438709.6 |
ANKRD36B
|
ankyrin repeat domain 36B |
| chr17_-_7207245 | 0.10 |
ENST00000649971.1
|
DLG4
|
discs large MAGUK scaffold protein 4 |
| chrX_+_14529516 | 0.09 |
ENST00000218075.9
|
GLRA2
|
glycine receptor alpha 2 |
| chr1_-_235328147 | 0.09 |
ENST00000264183.9
ENST00000418304.1 ENST00000349213.7 |
ARID4B
|
AT-rich interaction domain 4B |
| chr2_-_219308963 | 0.09 |
ENST00000423636.6
ENST00000442029.5 ENST00000412847.5 |
PTPRN
|
protein tyrosine phosphatase receptor type N |
| chr2_-_46941710 | 0.09 |
ENST00000409207.5
|
MCFD2
|
multiple coagulation factor deficiency 2, ER cargo receptor complex subunit |
| chr2_+_17541157 | 0.09 |
ENST00000406397.1
|
VSNL1
|
visinin like 1 |
| chr19_-_13953302 | 0.08 |
ENST00000585607.1
ENST00000538517.6 ENST00000587458.1 ENST00000538371.6 |
PODNL1
|
podocan like 1 |
| chr15_-_48811038 | 0.08 |
ENST00000380950.7
|
CEP152
|
centrosomal protein 152 |
| chr14_+_21030201 | 0.06 |
ENST00000321760.11
ENST00000460647.6 ENST00000530140.6 ENST00000472458.5 |
TPPP2
|
tubulin polymerization promoting protein family member 2 |
| chr11_-_125778788 | 0.06 |
ENST00000436890.2
|
PATE2
|
prostate and testis expressed 2 |
| chr11_-_125778818 | 0.06 |
ENST00000358524.8
|
PATE2
|
prostate and testis expressed 2 |
| chr17_+_6776209 | 0.06 |
ENST00000321535.5
|
FBXO39
|
F-box protein 39 |
| chr6_-_31542339 | 0.06 |
ENST00000458640.5
|
DDX39B
|
DExD-box helicase 39B |
| chr20_-_6054026 | 0.03 |
ENST00000378858.5
|
LRRN4
|
leucine rich repeat neuronal 4 |
| chr12_+_57591158 | 0.03 |
ENST00000422156.7
ENST00000354947.10 ENST00000540759.6 ENST00000551772.5 ENST00000550465.5 |
PIP4K2C
|
phosphatidylinositol-5-phosphate 4-kinase type 2 gamma |
| chr19_-_49763295 | 0.03 |
ENST00000246801.8
|
TSKS
|
testis specific serine kinase substrate |
| chr1_+_157993601 | 0.03 |
ENST00000359209.11
|
KIRREL1
|
kirre like nephrin family adhesion molecule 1 |
| chr19_-_42220109 | 0.03 |
ENST00000595337.5
|
DEDD2
|
death effector domain containing 2 |
| chrX_+_48521817 | 0.02 |
ENST00000446158.5
ENST00000414061.1 |
EBP
|
EBP cholestenol delta-isomerase |
| chr1_-_161223408 | 0.01 |
ENST00000491350.1
|
APOA2
|
apolipoprotein A2 |
| chr1_-_161223559 | 0.01 |
ENST00000469730.2
ENST00000463273.5 ENST00000464492.5 ENST00000367990.7 ENST00000470459.6 ENST00000463812.1 ENST00000468465.5 |
APOA2
|
apolipoprotein A2 |
| chr12_-_7665897 | 0.00 |
ENST00000229304.5
|
APOBEC1
|
apolipoprotein B mRNA editing enzyme catalytic subunit 1 |
| chr2_+_17540670 | 0.00 |
ENST00000451533.5
ENST00000295156.9 |
VSNL1
|
visinin like 1 |
| chr1_+_24502320 | 0.00 |
ENST00000538532.6
ENST00000618490.4 |
RCAN3
|
RCAN family member 3 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.5 | GO:2000124 | regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.1 | 0.5 | GO:0009436 | glyoxylate catabolic process(GO:0009436) |
| 0.1 | 0.7 | GO:0030860 | regulation of polarized epithelial cell differentiation(GO:0030860) |
| 0.1 | 0.7 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.0 | 0.4 | GO:0035845 | photoreceptor cell outer segment organization(GO:0035845) |
| 0.0 | 0.5 | GO:1904996 | positive regulation of leukocyte adhesion to vascular endothelial cell(GO:1904996) |
| 0.0 | 0.7 | GO:0070777 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.0 | 0.1 | GO:0033385 | geranylgeranyl diphosphate metabolic process(GO:0033385) geranylgeranyl diphosphate biosynthetic process(GO:0033386) |
| 0.0 | 0.2 | GO:0019348 | dolichol metabolic process(GO:0019348) |
| 0.0 | 0.1 | GO:0043449 | cellular alkene metabolic process(GO:0043449) |
| 0.0 | 0.3 | GO:0031584 | activation of phospholipase D activity(GO:0031584) |
| 0.0 | 0.1 | GO:1904692 | positive regulation of type B pancreatic cell proliferation(GO:1904692) |
| 0.0 | 0.2 | GO:0032468 | Golgi calcium ion homeostasis(GO:0032468) |
| 0.0 | 1.4 | GO:0050832 | defense response to fungus(GO:0050832) |
| 0.0 | 0.1 | GO:0090427 | activation of meiosis(GO:0090427) |
| 0.0 | 0.2 | GO:0045163 | clustering of voltage-gated potassium channels(GO:0045163) |
| 0.0 | 0.1 | GO:0097368 | establishment of Sertoli cell barrier(GO:0097368) |
| 0.0 | 0.5 | GO:0048311 | mitochondrion distribution(GO:0048311) |
| 0.0 | 0.2 | GO:0097011 | cellular response to granulocyte macrophage colony-stimulating factor stimulus(GO:0097011) response to granulocyte macrophage colony-stimulating factor(GO:0097012) |
| 0.0 | 0.2 | GO:2000002 | negative regulation of DNA damage checkpoint(GO:2000002) |
| 0.0 | 0.2 | GO:0061484 | hematopoietic stem cell homeostasis(GO:0061484) |
| 0.0 | 0.3 | GO:0033235 | positive regulation of protein sumoylation(GO:0033235) |
| 0.0 | 0.3 | GO:0031293 | membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.0 | 0.4 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.0 | 0.0 | GO:0060694 | regulation of cholesterol transporter activity(GO:0060694) |
| 0.0 | 0.3 | GO:0006228 | UTP biosynthetic process(GO:0006228) |
| 0.0 | 1.2 | GO:0045600 | positive regulation of fat cell differentiation(GO:0045600) |
| 0.0 | 0.5 | GO:0031954 | positive regulation of protein autophosphorylation(GO:0031954) |
| 0.0 | 0.2 | GO:0032119 | sequestering of zinc ion(GO:0032119) regulation of sequestering of zinc ion(GO:0061088) |
| 0.0 | 0.4 | GO:0035338 | long-chain fatty-acyl-CoA biosynthetic process(GO:0035338) |
| 0.0 | 0.2 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.0 | 0.1 | GO:0060012 | synaptic transmission, glycinergic(GO:0060012) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | GO:0044214 | spanning component of plasma membrane(GO:0044214) spanning component of membrane(GO:0089717) |
| 0.1 | 0.2 | GO:0033185 | dolichol-phosphate-mannose synthase complex(GO:0033185) |
| 0.1 | 0.3 | GO:0071458 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) |
| 0.0 | 0.7 | GO:0033202 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 0.0 | 0.5 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.0 | 0.2 | GO:0005688 | U6 snRNP(GO:0005688) |
| 0.0 | 0.7 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 0.5 | GO:0071339 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.0 | 0.1 | GO:0098536 | deuterosome(GO:0098536) |
| 0.0 | 0.3 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.5 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.1 | 0.5 | GO:0008453 | alanine-glyoxylate transaminase activity(GO:0008453) |
| 0.1 | 0.7 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.1 | 0.4 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) intracellular cGMP activated cation channel activity(GO:0005223) |
| 0.1 | 0.2 | GO:0004582 | dolichyl-phosphate beta-D-mannosyltransferase activity(GO:0004582) |
| 0.1 | 0.5 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.0 | 0.7 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.0 | 0.5 | GO:0005237 | inhibitory extracellular ligand-gated ion channel activity(GO:0005237) |
| 0.0 | 0.3 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.0 | 0.2 | GO:0030197 | extracellular matrix constituent, lubricant activity(GO:0030197) |
| 0.0 | 0.2 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.0 | 0.2 | GO:0046923 | A-type (transient outward) potassium channel activity(GO:0005250) ER retention sequence binding(GO:0046923) |
| 0.0 | 0.1 | GO:0030395 | lactose binding(GO:0030395) |
| 0.0 | 0.1 | GO:0004337 | dimethylallyltranstransferase activity(GO:0004161) geranyltranstransferase activity(GO:0004337) |
| 0.0 | 0.1 | GO:0070576 | vitamin D 24-hydroxylase activity(GO:0070576) |
| 0.0 | 0.4 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.0 | 0.2 | GO:0030621 | U4 snRNA binding(GO:0030621) |
| 0.0 | 0.1 | GO:0031811 | G-protein coupled nucleotide receptor binding(GO:0031811) P2Y1 nucleotide receptor binding(GO:0031812) |
| 0.0 | 0.2 | GO:0008504 | monoamine transmembrane transporter activity(GO:0008504) |
| 0.0 | 0.0 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 0.0 | 0.4 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.5 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.5 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 0.5 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |