Project
Inflammatory response time course, HUVEC (Wada, 2009)
Navigation
Downloads
Results for HOXB6_PRRX2
Z-value: 2.31
Transcription factors associated with HOXB6_PRRX2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
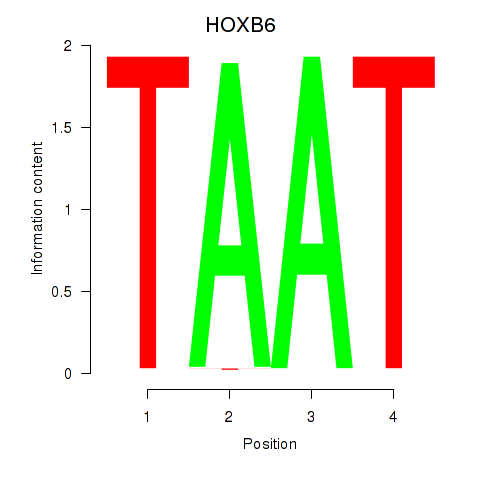
HOXB6
|
ENSG00000108511.10 | HOXB6 |
|
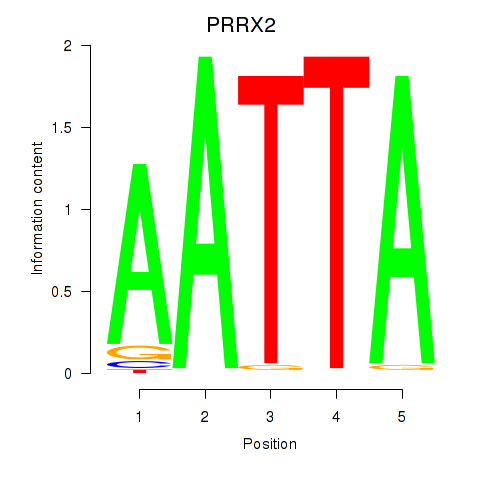
PRRX2
|
ENSG00000167157.11 | PRRX2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HOXB6 | hg38_v1_chr17_-_48604959_48605000 | -0.32 | 1.2e-01 | Click! |
| PRRX2 | hg38_v1_chr9_+_129665603_129665672 | -0.32 | 1.2e-01 | Click! |
Activity profile of HOXB6_PRRX2 motif
Sorted Z-values of HOXB6_PRRX2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of HOXB6_PRRX2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.0 | 47.8 | GO:0003185 | primary heart field specification(GO:0003138) sinoatrial valve development(GO:0003172) sinoatrial valve morphogenesis(GO:0003185) |
| 5.1 | 15.3 | GO:0060435 | bronchiole development(GO:0060435) |
| 4.0 | 12.0 | GO:0072097 | apoptotic process involved in endocardial cushion morphogenesis(GO:0003277) intermediate mesoderm morphogenesis(GO:0048390) intermediate mesoderm formation(GO:0048391) intermediate mesodermal cell differentiation(GO:0048392) regulation of cardiac muscle fiber development(GO:0055018) positive regulation of cardiac muscle fiber development(GO:0055020) bud dilation involved in lung branching(GO:0060503) BMP signaling pathway involved in ureter morphogenesis(GO:0061149) renal system segmentation(GO:0061150) BMP signaling pathway involved in renal system segmentation(GO:0061151) pulmonary artery endothelial tube morphogenesis(GO:0061155) regulation of transcription from RNA polymerase II promoter involved in mesonephros development(GO:0061216) BMP signaling pathway involved in nephric duct formation(GO:0071893) negative regulation of branch elongation involved in ureteric bud branching(GO:0072096) negative regulation of branch elongation involved in ureteric bud branching by BMP signaling pathway(GO:0072097) anterior/posterior pattern specification involved in ureteric bud development(GO:0072099) specification of ureteric bud anterior/posterior symmetry(GO:0072100) specification of ureteric bud anterior/posterior symmetry by BMP signaling pathway(GO:0072101) ureter epithelial cell differentiation(GO:0072192) negative regulation of mesenchymal cell proliferation involved in ureter development(GO:0072200) positive regulation of cell proliferation involved in outflow tract morphogenesis(GO:1901964) cardiac jelly development(GO:1905072) regulation of metanephric S-shaped body morphogenesis(GO:2000004) negative regulation of metanephric S-shaped body morphogenesis(GO:2000005) regulation of metanephric comma-shaped body morphogenesis(GO:2000006) negative regulation of metanephric comma-shaped body morphogenesis(GO:2000007) |
| 3.9 | 42.5 | GO:2000795 | negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 3.8 | 26.7 | GO:0097338 | response to clozapine(GO:0097338) |
| 3.0 | 18.2 | GO:0060023 | soft palate development(GO:0060023) |
| 3.0 | 9.0 | GO:0098746 | fast, calcium ion-dependent exocytosis of neurotransmitter(GO:0098746) |
| 2.1 | 8.2 | GO:0009956 | radial pattern formation(GO:0009956) |
| 1.9 | 5.7 | GO:0002302 | CD8-positive, alpha-beta T cell differentiation involved in immune response(GO:0002302) |
| 1.9 | 7.4 | GO:1903758 | regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903756) negative regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903758) |
| 1.8 | 7.3 | GO:0010159 | specification of organ position(GO:0010159) |
| 1.8 | 52.0 | GO:0072189 | ureter development(GO:0072189) |
| 1.7 | 5.2 | GO:0021658 | rhombomere morphogenesis(GO:0021593) rhombomere 3 morphogenesis(GO:0021658) |
| 1.7 | 6.9 | GO:0072137 | condensed mesenchymal cell proliferation(GO:0072137) |
| 1.7 | 1.7 | GO:0021569 | rhombomere 3 development(GO:0021569) |
| 1.5 | 5.8 | GO:1904428 | negative regulation of tubulin deacetylation(GO:1904428) |
| 1.4 | 12.4 | GO:0060244 | negative regulation of cell proliferation involved in contact inhibition(GO:0060244) |
| 1.1 | 6.8 | GO:2000504 | positive regulation of blood vessel remodeling(GO:2000504) |
| 1.1 | 15.6 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 1.0 | 11.7 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.9 | 5.7 | GO:1903566 | positive regulation of protein localization to cilium(GO:1903566) |
| 0.9 | 18.7 | GO:0060044 | negative regulation of cardiac muscle cell proliferation(GO:0060044) |
| 0.9 | 3.5 | GO:0061528 | negative regulation of gamma-aminobutyric acid secretion(GO:0014053) aspartate secretion(GO:0061528) regulation of aspartate secretion(GO:1904448) positive regulation of aspartate secretion(GO:1904450) |
| 0.9 | 3.5 | GO:0061357 | positive regulation of Wnt protein secretion(GO:0061357) |
| 0.9 | 5.1 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 0.8 | 2.5 | GO:0002194 | hepatocyte cell migration(GO:0002194) otic placode formation(GO:0043049) branching involved in pancreas morphogenesis(GO:0061114) acinar cell differentiation(GO:0090425) positive regulation of forebrain neuron differentiation(GO:2000979) |
| 0.8 | 2.4 | GO:0035026 | leading edge cell differentiation(GO:0035026) |
| 0.8 | 3.9 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.8 | 2.3 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) |
| 0.7 | 5.1 | GO:0090325 | regulation of locomotion involved in locomotory behavior(GO:0090325) |
| 0.7 | 25.2 | GO:0003298 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.7 | 2.0 | GO:0043973 | histone H3-K4 acetylation(GO:0043973) |
| 0.7 | 2.0 | GO:0060366 | subpallium cell proliferation in forebrain(GO:0022012) lateral ganglionic eminence cell proliferation(GO:0022018) lambdoid suture morphogenesis(GO:0060366) sagittal suture morphogenesis(GO:0060367) anterior semicircular canal development(GO:0060873) lateral semicircular canal development(GO:0060875) |
| 0.6 | 1.3 | GO:0061386 | closure of optic fissure(GO:0061386) |
| 0.6 | 1.8 | GO:0060823 | canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060823) |
| 0.6 | 4.7 | GO:0035583 | sequestering of TGFbeta in extracellular matrix(GO:0035583) |
| 0.6 | 0.6 | GO:0034724 | DNA replication-independent nucleosome organization(GO:0034724) |
| 0.5 | 2.1 | GO:0010157 | response to chlorate(GO:0010157) |
| 0.5 | 1.5 | GO:0015917 | aminophospholipid transport(GO:0015917) |
| 0.5 | 1.5 | GO:0031550 | positive regulation of brain-derived neurotrophic factor receptor signaling pathway(GO:0031550) |
| 0.5 | 6.4 | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.5 | 9.8 | GO:0040015 | negative regulation of multicellular organism growth(GO:0040015) |
| 0.5 | 1.5 | GO:0016103 | diterpenoid catabolic process(GO:0016103) retinoic acid catabolic process(GO:0034653) |
| 0.5 | 96.7 | GO:0007286 | spermatid development(GO:0007286) |
| 0.5 | 2.8 | GO:0060399 | positive regulation of growth hormone receptor signaling pathway(GO:0060399) |
| 0.5 | 1.4 | GO:0001869 | regulation of complement activation, lectin pathway(GO:0001868) negative regulation of complement activation, lectin pathway(GO:0001869) |
| 0.5 | 4.1 | GO:0048539 | bone marrow development(GO:0048539) |
| 0.5 | 1.8 | GO:0061113 | pancreas morphogenesis(GO:0061113) |
| 0.5 | 9.9 | GO:0034391 | smooth muscle cell apoptotic process(GO:0034390) regulation of smooth muscle cell apoptotic process(GO:0034391) |
| 0.4 | 3.4 | GO:0097026 | dendritic cell dendrite assembly(GO:0097026) |
| 0.4 | 5.0 | GO:0003402 | planar cell polarity pathway involved in axis elongation(GO:0003402) |
| 0.4 | 1.3 | GO:0046005 | positive regulation of circadian sleep/wake cycle, REM sleep(GO:0046005) |
| 0.4 | 1.2 | GO:0002752 | cell surface pattern recognition receptor signaling pathway(GO:0002752) |
| 0.4 | 1.2 | GO:0035470 | positive regulation of vascular wound healing(GO:0035470) |
| 0.4 | 22.4 | GO:0045599 | negative regulation of fat cell differentiation(GO:0045599) |
| 0.4 | 3.9 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.4 | 1.9 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.4 | 6.1 | GO:0090190 | positive regulation of branching involved in ureteric bud morphogenesis(GO:0090190) |
| 0.4 | 5.3 | GO:0043983 | histone H4-K12 acetylation(GO:0043983) |
| 0.4 | 3.4 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) |
| 0.4 | 1.8 | GO:0007352 | zygotic specification of dorsal/ventral axis(GO:0007352) |
| 0.4 | 2.9 | GO:0048014 | Tie signaling pathway(GO:0048014) |
| 0.4 | 1.4 | GO:0061552 | vestibulocochlear nerve structural organization(GO:0021649) neuropilin signaling pathway(GO:0038189) VEGF-activated neuropilin signaling pathway(GO:0038190) positive regulation of cytokine activity(GO:0060301) ganglion morphogenesis(GO:0061552) positive regulation of retinal ganglion cell axon guidance(GO:1902336) VEGF-activated neuropilin signaling pathway involved in axon guidance(GO:1902378) dorsal root ganglion morphogenesis(GO:1904835) otic placode development(GO:1905040) |
| 0.3 | 1.4 | GO:0051799 | negative regulation of hair follicle development(GO:0051799) |
| 0.3 | 2.0 | GO:0050916 | sensory perception of sweet taste(GO:0050916) sensory perception of umami taste(GO:0050917) |
| 0.3 | 1.0 | GO:0021913 | regulation of transcription from RNA polymerase II promoter involved in ventral spinal cord interneuron specification(GO:0021913) |
| 0.3 | 3.3 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.3 | 4.4 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.3 | 1.2 | GO:0048859 | formation of anatomical boundary(GO:0048859) |
| 0.3 | 5.9 | GO:0042789 | mRNA transcription from RNA polymerase II promoter(GO:0042789) |
| 0.3 | 0.9 | GO:0032877 | positive regulation of DNA endoreduplication(GO:0032877) |
| 0.3 | 1.2 | GO:0018032 | protein amidation(GO:0018032) |
| 0.3 | 1.4 | GO:0035022 | positive regulation of Rac protein signal transduction(GO:0035022) |
| 0.3 | 0.8 | GO:2000174 | pro-T cell differentiation(GO:0002572) regulation of pro-T cell differentiation(GO:2000174) positive regulation of pro-T cell differentiation(GO:2000176) |
| 0.3 | 3.6 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.3 | 2.8 | GO:0042985 | negative regulation of amyloid precursor protein biosynthetic process(GO:0042985) |
| 0.3 | 0.8 | GO:1903676 | regulation of cap-dependent translational initiation(GO:1903674) positive regulation of cap-dependent translational initiation(GO:1903676) |
| 0.3 | 3.0 | GO:1904936 | cerebral cortex GABAergic interneuron migration(GO:0021853) interneuron migration(GO:1904936) |
| 0.3 | 2.1 | GO:0003406 | retinal pigment epithelium development(GO:0003406) |
| 0.3 | 1.3 | GO:0099590 | neurotransmitter receptor internalization(GO:0099590) |
| 0.3 | 1.3 | GO:0003150 | muscular septum morphogenesis(GO:0003150) |
| 0.3 | 24.6 | GO:0043268 | positive regulation of potassium ion transport(GO:0043268) |
| 0.3 | 3.1 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 0.3 | 2.8 | GO:0034350 | regulation of glial cell apoptotic process(GO:0034350) negative regulation of glial cell apoptotic process(GO:0034351) |
| 0.3 | 1.0 | GO:0038044 | transforming growth factor-beta secretion(GO:0038044) |
| 0.3 | 1.5 | GO:0010746 | regulation of plasma membrane long-chain fatty acid transport(GO:0010746) negative regulation of plasma membrane long-chain fatty acid transport(GO:0010748) |
| 0.3 | 1.5 | GO:0046543 | development of secondary female sexual characteristics(GO:0046543) |
| 0.3 | 3.1 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.3 | 3.3 | GO:0007351 | tripartite regional subdivision(GO:0007351) anterior/posterior axis specification, embryo(GO:0008595) |
| 0.2 | 1.0 | GO:0002270 | plasmacytoid dendritic cell activation(GO:0002270) regulation of restriction endodeoxyribonuclease activity(GO:0032072) |
| 0.2 | 0.7 | GO:0036233 | glycine import(GO:0036233) |
| 0.2 | 0.7 | GO:0018315 | molybdenum incorporation into molybdenum-molybdopterin complex(GO:0018315) metal incorporation into metallo-molybdopterin complex(GO:0042040) glycine receptor clustering(GO:0072579) |
| 0.2 | 1.9 | GO:2000332 | response to linoleic acid(GO:0070543) blood microparticle formation(GO:0072564) regulation of blood microparticle formation(GO:2000332) positive regulation of blood microparticle formation(GO:2000334) |
| 0.2 | 1.2 | GO:0009099 | branched-chain amino acid biosynthetic process(GO:0009082) leucine biosynthetic process(GO:0009098) valine biosynthetic process(GO:0009099) |
| 0.2 | 4.4 | GO:0097090 | presynaptic membrane organization(GO:0097090) |
| 0.2 | 1.4 | GO:0010166 | wax biosynthetic process(GO:0010025) wax metabolic process(GO:0010166) |
| 0.2 | 2.5 | GO:0014877 | response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.2 | 11.2 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.2 | 3.5 | GO:0007379 | segment specification(GO:0007379) |
| 0.2 | 62.7 | GO:0065004 | protein-DNA complex assembly(GO:0065004) |
| 0.2 | 0.4 | GO:1903671 | negative regulation of sprouting angiogenesis(GO:1903671) |
| 0.2 | 1.1 | GO:0046013 | regulation of T cell homeostatic proliferation(GO:0046013) |
| 0.2 | 1.7 | GO:0055059 | asymmetric neuroblast division(GO:0055059) |
| 0.2 | 0.8 | GO:0010982 | regulation of high-density lipoprotein particle clearance(GO:0010982) |
| 0.2 | 0.2 | GO:0048686 | regulation of sprouting of injured axon(GO:0048686) regulation of axon extension involved in regeneration(GO:0048690) |
| 0.2 | 0.4 | GO:0090214 | spongiotrophoblast layer developmental growth(GO:0090214) |
| 0.2 | 1.2 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.2 | 1.1 | GO:0001692 | histamine metabolic process(GO:0001692) |
| 0.2 | 0.6 | GO:0002416 | IgG immunoglobulin transcytosis in epithelial cells mediated by FcRn immunoglobulin receptor(GO:0002416) |
| 0.2 | 0.7 | GO:0042427 | serotonin biosynthetic process(GO:0042427) |
| 0.2 | 1.4 | GO:0071386 | cellular response to corticosterone stimulus(GO:0071386) |
| 0.2 | 2.7 | GO:0090091 | positive regulation of extracellular matrix disassembly(GO:0090091) |
| 0.2 | 0.5 | GO:0052047 | interaction with other organism via secreted substance involved in symbiotic interaction(GO:0052047) |
| 0.2 | 0.9 | GO:1900245 | positive regulation of MDA-5 signaling pathway(GO:1900245) |
| 0.2 | 1.1 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.2 | 0.2 | GO:0033092 | positive regulation of immature T cell proliferation(GO:0033091) positive regulation of immature T cell proliferation in thymus(GO:0033092) |
| 0.2 | 4.8 | GO:0060065 | uterus development(GO:0060065) |
| 0.2 | 1.3 | GO:0022027 | interkinetic nuclear migration(GO:0022027) |
| 0.2 | 0.5 | GO:0016999 | antibiotic metabolic process(GO:0016999) detoxification of mercury ion(GO:0050787) |
| 0.2 | 0.3 | GO:0035910 | ascending aorta development(GO:0035905) ascending aorta morphogenesis(GO:0035910) |
| 0.2 | 1.1 | GO:0060242 | contact inhibition(GO:0060242) |
| 0.2 | 2.2 | GO:0001780 | neutrophil homeostasis(GO:0001780) |
| 0.2 | 0.8 | GO:0006537 | glutamate biosynthetic process(GO:0006537) |
| 0.2 | 0.6 | GO:2000342 | negative regulation of chemokine (C-X-C motif) ligand 2 production(GO:2000342) |
| 0.2 | 1.1 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.2 | 0.5 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.1 | 4.0 | GO:0002230 | positive regulation of defense response to virus by host(GO:0002230) |
| 0.1 | 2.2 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.1 | 1.9 | GO:0009642 | response to light intensity(GO:0009642) |
| 0.1 | 0.7 | GO:0014886 | transition between slow and fast fiber(GO:0014886) |
| 0.1 | 2.2 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.1 | 0.4 | GO:0097156 | fasciculation of motor neuron axon(GO:0097156) |
| 0.1 | 0.6 | GO:0001507 | acetylcholine catabolic process in synaptic cleft(GO:0001507) acetylcholine catabolic process(GO:0006581) |
| 0.1 | 2.0 | GO:0021514 | ventral spinal cord interneuron differentiation(GO:0021514) |
| 0.1 | 0.4 | GO:0021759 | globus pallidus development(GO:0021759) |
| 0.1 | 0.3 | GO:0055095 | lipoprotein particle mediated signaling(GO:0055095) low-density lipoprotein particle mediated signaling(GO:0055096) |
| 0.1 | 0.8 | GO:0061343 | cell adhesion involved in heart morphogenesis(GO:0061343) |
| 0.1 | 2.9 | GO:0035162 | embryonic hemopoiesis(GO:0035162) |
| 0.1 | 0.8 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.1 | 1.4 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.1 | 0.4 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.1 | 0.6 | GO:0008616 | queuosine biosynthetic process(GO:0008616) queuosine metabolic process(GO:0046116) |
| 0.1 | 0.5 | GO:0070345 | negative regulation of fat cell proliferation(GO:0070345) |
| 0.1 | 0.5 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.1 | 0.6 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.1 | 0.9 | GO:0033227 | dsRNA transport(GO:0033227) |
| 0.1 | 0.7 | GO:0060087 | relaxation of vascular smooth muscle(GO:0060087) |
| 0.1 | 0.3 | GO:1904844 | response to L-glutamine(GO:1904844) cellular response to L-glutamine(GO:1904845) |
| 0.1 | 4.3 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.1 | 0.9 | GO:0010727 | negative regulation of hydrogen peroxide metabolic process(GO:0010727) |
| 0.1 | 0.5 | GO:0018076 | N-terminal peptidyl-lysine acetylation(GO:0018076) |
| 0.1 | 0.4 | GO:0052651 | monoacylglycerol metabolic process(GO:0046462) monoacylglycerol catabolic process(GO:0052651) |
| 0.1 | 0.3 | GO:0097119 | postsynaptic density protein 95 clustering(GO:0097119) |
| 0.1 | 1.6 | GO:0045656 | negative regulation of monocyte differentiation(GO:0045656) |
| 0.1 | 1.7 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 0.1 | 0.6 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.1 | 0.7 | GO:0038165 | oncostatin-M-mediated signaling pathway(GO:0038165) |
| 0.1 | 1.8 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.1 | 0.9 | GO:0002725 | negative regulation of T cell cytokine production(GO:0002725) |
| 0.1 | 0.4 | GO:0042905 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.1 | 1.1 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.1 | 0.8 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.1 | 3.3 | GO:0006625 | protein targeting to peroxisome(GO:0006625) protein localization to peroxisome(GO:0072662) establishment of protein localization to peroxisome(GO:0072663) |
| 0.1 | 0.8 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.1 | 1.6 | GO:0042420 | dopamine catabolic process(GO:0042420) |
| 0.1 | 1.6 | GO:1900364 | negative regulation of mRNA polyadenylation(GO:1900364) |
| 0.1 | 8.9 | GO:0048207 | vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) |
| 0.1 | 4.6 | GO:0048384 | retinoic acid receptor signaling pathway(GO:0048384) |
| 0.1 | 0.8 | GO:0090160 | Golgi to lysosome transport(GO:0090160) |
| 0.1 | 1.0 | GO:0070307 | lens fiber cell development(GO:0070307) |
| 0.1 | 0.5 | GO:1902896 | terminal web assembly(GO:1902896) |
| 0.1 | 0.4 | GO:0048294 | negative regulation of isotype switching to IgE isotypes(GO:0048294) |
| 0.1 | 0.3 | GO:0060591 | chondroblast differentiation(GO:0060591) |
| 0.1 | 1.4 | GO:1990118 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.1 | 0.4 | GO:0072502 | cellular phosphate ion homeostasis(GO:0030643) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.1 | 0.4 | GO:0021615 | glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.1 | 0.2 | GO:0035526 | retrograde transport, plasma membrane to Golgi(GO:0035526) |
| 0.1 | 0.3 | GO:0090362 | positive regulation of platelet-derived growth factor production(GO:0090362) |
| 0.1 | 0.7 | GO:0009299 | mRNA transcription(GO:0009299) |
| 0.1 | 0.6 | GO:0010835 | regulation of protein ADP-ribosylation(GO:0010835) nitric oxide homeostasis(GO:0033484) |
| 0.1 | 0.9 | GO:0070315 | G1 to G0 transition involved in cell differentiation(GO:0070315) |
| 0.1 | 1.0 | GO:0071313 | cellular response to caffeine(GO:0071313) |
| 0.1 | 1.8 | GO:0043537 | negative regulation of blood vessel endothelial cell migration(GO:0043537) |
| 0.1 | 0.3 | GO:0033306 | phytol metabolic process(GO:0033306) fatty alcohol metabolic process(GO:1903173) |
| 0.1 | 0.4 | GO:0006196 | AMP catabolic process(GO:0006196) |
| 0.1 | 0.7 | GO:2000353 | positive regulation of endothelial cell apoptotic process(GO:2000353) |
| 0.1 | 1.8 | GO:0010569 | regulation of double-strand break repair via homologous recombination(GO:0010569) |
| 0.1 | 0.6 | GO:0006848 | pyruvate transport(GO:0006848) pyruvate transmembrane transport(GO:1901475) |
| 0.1 | 0.4 | GO:0048050 | post-embryonic eye morphogenesis(GO:0048050) |
| 0.1 | 8.3 | GO:1990830 | response to leukemia inhibitory factor(GO:1990823) cellular response to leukemia inhibitory factor(GO:1990830) |
| 0.1 | 1.2 | GO:0030043 | actin filament fragmentation(GO:0030043) |
| 0.1 | 1.0 | GO:0042226 | interleukin-6 biosynthetic process(GO:0042226) |
| 0.1 | 1.3 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.1 | 0.3 | GO:2000393 | negative regulation of lamellipodium morphogenesis(GO:2000393) |
| 0.1 | 0.1 | GO:0036292 | DNA rewinding(GO:0036292) |
| 0.1 | 0.3 | GO:0030070 | insulin processing(GO:0030070) |
| 0.1 | 1.9 | GO:0043555 | regulation of translation in response to stress(GO:0043555) |
| 0.1 | 3.9 | GO:0007566 | embryo implantation(GO:0007566) |
| 0.1 | 0.3 | GO:0007161 | calcium-independent cell-matrix adhesion(GO:0007161) regulation of collagen catabolic process(GO:0010710) |
| 0.1 | 0.1 | GO:0034241 | macrophage fusion(GO:0034238) regulation of macrophage fusion(GO:0034239) positive regulation of macrophage fusion(GO:0034241) |
| 0.1 | 1.3 | GO:0010801 | negative regulation of peptidyl-threonine phosphorylation(GO:0010801) |
| 0.1 | 0.3 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.1 | 0.2 | GO:0023016 | signal transduction by trans-phosphorylation(GO:0023016) |
| 0.1 | 0.4 | GO:0070459 | prolactin secretion(GO:0070459) |
| 0.1 | 0.2 | GO:0030576 | Cajal body organization(GO:0030576) |
| 0.1 | 0.7 | GO:0033601 | positive regulation of mammary gland epithelial cell proliferation(GO:0033601) endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.1 | 0.4 | GO:0050955 | thermoception(GO:0050955) |
| 0.1 | 0.4 | GO:0035865 | cellular response to potassium ion(GO:0035865) |
| 0.1 | 1.2 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.1 | 0.9 | GO:0035728 | response to hepatocyte growth factor(GO:0035728) |
| 0.1 | 0.2 | GO:0035627 | ceramide transport(GO:0035627) |
| 0.1 | 0.5 | GO:2000189 | positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.1 | 0.5 | GO:0051451 | myoblast migration(GO:0051451) |
| 0.1 | 1.0 | GO:0043116 | negative regulation of vascular permeability(GO:0043116) |
| 0.1 | 13.6 | GO:0008360 | regulation of cell shape(GO:0008360) |
| 0.1 | 1.1 | GO:0051412 | response to corticosterone(GO:0051412) |
| 0.1 | 0.1 | GO:0021562 | vestibulocochlear nerve development(GO:0021562) |
| 0.1 | 0.1 | GO:0072501 | cellular divalent inorganic anion homeostasis(GO:0072501) |
| 0.1 | 0.3 | GO:0043418 | homocysteine catabolic process(GO:0043418) |
| 0.1 | 2.1 | GO:0019933 | cAMP-mediated signaling(GO:0019933) |
| 0.1 | 12.0 | GO:0031032 | actomyosin structure organization(GO:0031032) |
| 0.1 | 1.0 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.1 | 1.3 | GO:1902036 | regulation of hematopoietic stem cell differentiation(GO:1902036) |
| 0.0 | 1.8 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.0 | 0.3 | GO:0048570 | notochord morphogenesis(GO:0048570) |
| 0.0 | 0.1 | GO:0031117 | positive regulation of microtubule depolymerization(GO:0031117) |
| 0.0 | 0.2 | GO:0006127 | glycerophosphate shuttle(GO:0006127) |
| 0.0 | 0.2 | GO:0031274 | positive regulation of pseudopodium assembly(GO:0031274) |
| 0.0 | 1.9 | GO:0046329 | negative regulation of JNK cascade(GO:0046329) |
| 0.0 | 1.1 | GO:2000249 | regulation of actin cytoskeleton reorganization(GO:2000249) |
| 0.0 | 0.4 | GO:0071638 | negative regulation of monocyte chemotactic protein-1 production(GO:0071638) |
| 0.0 | 0.6 | GO:0042340 | keratan sulfate catabolic process(GO:0042340) |
| 0.0 | 3.3 | GO:0032781 | positive regulation of ATPase activity(GO:0032781) |
| 0.0 | 0.7 | GO:0060575 | intestinal epithelial cell differentiation(GO:0060575) |
| 0.0 | 0.3 | GO:0032927 | positive regulation of activin receptor signaling pathway(GO:0032927) |
| 0.0 | 0.5 | GO:0097094 | craniofacial suture morphogenesis(GO:0097094) |
| 0.0 | 1.1 | GO:0006337 | nucleosome disassembly(GO:0006337) |
| 0.0 | 0.8 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.0 | 0.8 | GO:0019321 | pentose metabolic process(GO:0019321) |
| 0.0 | 0.7 | GO:0034214 | protein hexamerization(GO:0034214) |
| 0.0 | 1.1 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 0.3 | GO:0071477 | hypotonic salinity response(GO:0042539) cellular hypotonic salinity response(GO:0071477) |
| 0.0 | 1.0 | GO:0006004 | fucose metabolic process(GO:0006004) |
| 0.0 | 0.4 | GO:0032815 | negative regulation of natural killer cell activation(GO:0032815) |
| 0.0 | 0.8 | GO:0032225 | regulation of synaptic transmission, dopaminergic(GO:0032225) |
| 0.0 | 0.1 | GO:1903367 | positive regulation of fear response(GO:1903367) positive regulation of behavioral fear response(GO:2000987) |
| 0.0 | 0.4 | GO:0034384 | high-density lipoprotein particle clearance(GO:0034384) |
| 0.0 | 0.7 | GO:0075522 | IRES-dependent viral translational initiation(GO:0075522) |
| 0.0 | 0.3 | GO:1903593 | regulation of histamine secretion by mast cell(GO:1903593) |
| 0.0 | 0.3 | GO:0050957 | equilibrioception(GO:0050957) |
| 0.0 | 0.3 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.0 | 0.6 | GO:0040032 | post-embryonic body morphogenesis(GO:0040032) |
| 0.0 | 0.5 | GO:0060287 | epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 0.0 | 2.5 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.0 | 1.1 | GO:0031529 | ruffle organization(GO:0031529) |
| 0.0 | 2.1 | GO:0043966 | histone H3 acetylation(GO:0043966) |
| 0.0 | 1.6 | GO:0034260 | negative regulation of GTPase activity(GO:0034260) |
| 0.0 | 1.5 | GO:0038084 | vascular endothelial growth factor signaling pathway(GO:0038084) |
| 0.0 | 0.7 | GO:0042417 | dopamine metabolic process(GO:0042417) |
| 0.0 | 0.4 | GO:0006657 | CDP-choline pathway(GO:0006657) |
| 0.0 | 0.7 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.0 | 0.6 | GO:0097151 | positive regulation of inhibitory postsynaptic potential(GO:0097151) modulation of inhibitory postsynaptic potential(GO:0098828) |
| 0.0 | 0.6 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.0 | 0.3 | GO:0035020 | regulation of Rac protein signal transduction(GO:0035020) |
| 0.0 | 0.1 | GO:0051005 | negative regulation of lipoprotein lipase activity(GO:0051005) |
| 0.0 | 0.1 | GO:0021965 | spinal cord ventral commissure morphogenesis(GO:0021965) |
| 0.0 | 0.2 | GO:0046440 | L-lysine catabolic process to acetyl-CoA(GO:0019474) L-lysine catabolic process(GO:0019477) L-lysine metabolic process(GO:0046440) |
| 0.0 | 0.8 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 0.6 | GO:1901386 | negative regulation of voltage-gated calcium channel activity(GO:1901386) |
| 0.0 | 0.3 | GO:0050966 | detection of mechanical stimulus involved in sensory perception of pain(GO:0050966) |
| 0.0 | 0.1 | GO:1900738 | positive regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900738) |
| 0.0 | 0.3 | GO:0031115 | negative regulation of microtubule polymerization(GO:0031115) |
| 0.0 | 0.3 | GO:0000447 | endonucleolytic cleavage in ITS1 to separate SSU-rRNA from 5.8S rRNA and LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000447) |
| 0.0 | 0.3 | GO:1903025 | regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903025) |
| 0.0 | 0.5 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.0 | 0.2 | GO:0042670 | retinal cone cell differentiation(GO:0042670) retinal cone cell development(GO:0046549) |
| 0.0 | 0.6 | GO:0050909 | sensory perception of taste(GO:0050909) |
| 0.0 | 0.5 | GO:0031297 | replication fork processing(GO:0031297) |
| 0.0 | 0.4 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.0 | 0.3 | GO:0001556 | oocyte maturation(GO:0001556) |
| 0.0 | 0.6 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.0 | 2.8 | GO:0090175 | Wnt signaling pathway, planar cell polarity pathway(GO:0060071) regulation of establishment of planar polarity(GO:0090175) |
| 0.0 | 0.2 | GO:0051988 | regulation of attachment of spindle microtubules to kinetochore(GO:0051988) |
| 0.0 | 0.1 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.0 | 0.0 | GO:0002740 | negative regulation of cytokine secretion involved in immune response(GO:0002740) |
| 0.0 | 0.3 | GO:0090312 | positive regulation of protein deacetylation(GO:0090312) |
| 0.0 | 0.1 | GO:0042362 | fat-soluble vitamin biosynthetic process(GO:0042362) |
| 0.0 | 1.0 | GO:0032007 | negative regulation of TOR signaling(GO:0032007) |
| 0.0 | 1.0 | GO:1901998 | toxin transport(GO:1901998) |
| 0.0 | 0.8 | GO:0060612 | adipose tissue development(GO:0060612) |
| 0.0 | 0.2 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.0 | 0.0 | GO:0097535 | lymphoid lineage cell migration(GO:0097534) lymphoid lineage cell migration into thymus(GO:0097535) |
| 0.0 | 0.4 | GO:0032793 | positive regulation of CREB transcription factor activity(GO:0032793) |
| 0.0 | 0.1 | GO:2000628 | regulation of miRNA metabolic process(GO:2000628) |
| 0.0 | 1.4 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.0 | 0.9 | GO:1901661 | quinone metabolic process(GO:1901661) |
| 0.0 | 0.4 | GO:0048873 | homeostasis of number of cells within a tissue(GO:0048873) |
| 0.0 | 0.8 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.0 | 0.3 | GO:0042359 | vitamin D metabolic process(GO:0042359) |
| 0.0 | 0.1 | GO:0031339 | negative regulation of vesicle fusion(GO:0031339) |
| 0.0 | 0.7 | GO:0006699 | bile acid biosynthetic process(GO:0006699) |
| 0.0 | 0.2 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.0 | 0.1 | GO:0033183 | negative regulation of histone ubiquitination(GO:0033183) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) |
| 0.0 | 0.0 | GO:0097211 | response to gonadotropin-releasing hormone(GO:0097210) cellular response to gonadotropin-releasing hormone(GO:0097211) |
| 0.0 | 0.1 | GO:0060736 | prostate gland growth(GO:0060736) |
| 0.0 | 0.5 | GO:0010761 | fibroblast migration(GO:0010761) |
| 0.0 | 2.2 | GO:0002576 | platelet degranulation(GO:0002576) |
| 0.0 | 1.0 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.7 | GO:0061001 | regulation of dendritic spine morphogenesis(GO:0061001) |
| 0.0 | 0.5 | GO:0000245 | spliceosomal complex assembly(GO:0000245) |
| 0.0 | 0.1 | GO:0044314 | protein K27-linked ubiquitination(GO:0044314) |
| 0.0 | 0.4 | GO:0030513 | positive regulation of BMP signaling pathway(GO:0030513) |
| 0.0 | 0.5 | GO:0007625 | grooming behavior(GO:0007625) |
| 0.0 | 0.2 | GO:0071285 | cellular response to lithium ion(GO:0071285) |
| 0.0 | 0.2 | GO:0007220 | Notch receptor processing(GO:0007220) |
| 0.0 | 0.0 | GO:0044537 | regulation of circulating fibrinogen levels(GO:0044537) |
| 0.0 | 0.1 | GO:0061298 | retina vasculature development in camera-type eye(GO:0061298) |
| 0.0 | 0.7 | GO:0032006 | regulation of TOR signaling(GO:0032006) |
| 0.0 | 0.7 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.0 | 1.2 | GO:0001578 | microtubule bundle formation(GO:0001578) |
| 0.0 | 0.7 | GO:0046677 | response to antibiotic(GO:0046677) |
| 0.0 | 0.1 | GO:2000427 | detection of molecule of bacterial origin(GO:0032490) positive regulation of apoptotic cell clearance(GO:2000427) |
| 0.0 | 0.5 | GO:0031110 | regulation of microtubule polymerization or depolymerization(GO:0031110) |
| 0.0 | 0.2 | GO:0036065 | fucosylation(GO:0036065) |
| 0.0 | 0.2 | GO:0010818 | T cell chemotaxis(GO:0010818) |
| 0.0 | 0.2 | GO:0071447 | cellular response to hydroperoxide(GO:0071447) |
| 0.0 | 0.1 | GO:1903393 | positive regulation of focal adhesion assembly(GO:0051894) positive regulation of adherens junction organization(GO:1903393) |
| 0.0 | 0.1 | GO:2000620 | positive regulation of histone H4 acetylation(GO:0090240) positive regulation of histone H4-K16 acetylation(GO:2000620) |
| 0.0 | 0.5 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.0 | 0.1 | GO:0046784 | viral mRNA export from host cell nucleus(GO:0046784) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.5 | 91.3 | GO:0071564 | npBAF complex(GO:0071564) |
| 2.3 | 20.9 | GO:0071953 | elastic fiber(GO:0071953) |
| 1.8 | 9.0 | GO:0060201 | clathrin-sculpted acetylcholine transport vesicle(GO:0060200) clathrin-sculpted acetylcholine transport vesicle membrane(GO:0060201) |
| 1.6 | 42.1 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 1.2 | 26.7 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 1.0 | 3.9 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.9 | 3.5 | GO:0032144 | 4-aminobutyrate transaminase complex(GO:0032144) |
| 0.6 | 4.6 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.6 | 2.2 | GO:0044307 | dendritic branch(GO:0044307) |
| 0.5 | 1.5 | GO:0097233 | lamellar body membrane(GO:0097232) alveolar lamellar body membrane(GO:0097233) |
| 0.4 | 9.2 | GO:0043220 | Schmidt-Lanterman incisure(GO:0043220) |
| 0.4 | 2.4 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.4 | 5.5 | GO:0016013 | syntrophin complex(GO:0016013) |
| 0.4 | 3.4 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 0.4 | 1.1 | GO:0071148 | TEAD-1-YAP complex(GO:0071148) |
| 0.4 | 5.0 | GO:0036057 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.3 | 15.1 | GO:0005720 | nuclear heterochromatin(GO:0005720) |
| 0.3 | 1.0 | GO:0034685 | integrin alphav-beta6 complex(GO:0034685) |
| 0.2 | 2.1 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.2 | 2.3 | GO:0031527 | filopodium membrane(GO:0031527) |
| 0.2 | 1.6 | GO:0035867 | alphav-beta3 integrin-IGF-1-IGF1R complex(GO:0035867) |
| 0.2 | 2.5 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.2 | 0.9 | GO:0016342 | catenin complex(GO:0016342) |
| 0.2 | 1.6 | GO:0042584 | chromaffin granule membrane(GO:0042584) |
| 0.2 | 3.7 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.2 | 2.6 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.2 | 39.9 | GO:0032993 | protein-DNA complex(GO:0032993) |
| 0.2 | 1.0 | GO:0032311 | angiogenin-PRI complex(GO:0032311) |
| 0.2 | 3.2 | GO:0012510 | trans-Golgi network transport vesicle membrane(GO:0012510) |
| 0.2 | 6.6 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.2 | 1.9 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.1 | 0.4 | GO:0005592 | collagen type XI trimer(GO:0005592) |
| 0.1 | 0.6 | GO:0070032 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) |
| 0.1 | 0.3 | GO:0071006 | U2-type catalytic step 1 spliceosome(GO:0071006) |
| 0.1 | 0.5 | GO:0071821 | FANCM-MHF complex(GO:0071821) |
| 0.1 | 64.0 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.1 | 2.1 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.1 | 20.8 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.1 | 1.7 | GO:0010369 | chromocenter(GO:0010369) |
| 0.1 | 0.3 | GO:0034677 | integrin alpha7-beta1 complex(GO:0034677) |
| 0.1 | 0.3 | GO:0045259 | mitochondrial proton-transporting ATP synthase complex(GO:0005753) proton-transporting ATP synthase complex(GO:0045259) |
| 0.1 | 1.8 | GO:0045180 | basal cortex(GO:0045180) |
| 0.1 | 2.9 | GO:0000145 | exocyst(GO:0000145) |
| 0.1 | 0.7 | GO:0000308 | cytoplasmic cyclin-dependent protein kinase holoenzyme complex(GO:0000308) |
| 0.1 | 2.4 | GO:0090545 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.1 | 33.9 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.1 | 0.6 | GO:0032280 | symmetric synapse(GO:0032280) |
| 0.1 | 2.4 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.1 | 34.1 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.1 | 0.4 | GO:0035841 | new growing cell tip(GO:0035841) |
| 0.1 | 2.7 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.1 | 0.5 | GO:0098839 | postsynaptic density membrane(GO:0098839) |
| 0.1 | 1.2 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.1 | 0.5 | GO:1990357 | terminal web(GO:1990357) |
| 0.1 | 1.6 | GO:0001527 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.1 | 1.4 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.1 | 0.8 | GO:0097470 | ribbon synapse(GO:0097470) |
| 0.1 | 0.4 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.1 | 0.3 | GO:0030905 | retromer, tubulation complex(GO:0030905) |
| 0.1 | 2.4 | GO:0000792 | heterochromatin(GO:0000792) |
| 0.1 | 5.2 | GO:0000123 | histone acetyltransferase complex(GO:0000123) |
| 0.1 | 13.1 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.1 | 4.2 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.1 | 1.4 | GO:0030128 | clathrin coat of endocytic vesicle(GO:0030128) |
| 0.1 | 1.7 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.5 | GO:0090544 | BAF-type complex(GO:0090544) |
| 0.0 | 2.1 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 1.4 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.0 | 0.5 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 1.4 | GO:0031231 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.0 | 0.2 | GO:0072534 | perineuronal net(GO:0072534) |
| 0.0 | 0.2 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.0 | 0.3 | GO:0097433 | dense body(GO:0097433) |
| 0.0 | 0.3 | GO:0097209 | epidermal lamellar body(GO:0097209) |
| 0.0 | 0.6 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.2 | GO:0097149 | centralspindlin complex(GO:0097149) |
| 0.0 | 2.1 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.0 | 2.2 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 0.4 | GO:0071439 | clathrin complex(GO:0071439) |
| 0.0 | 0.4 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.0 | 0.7 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.0 | 0.3 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 1.3 | GO:0030660 | Golgi-associated vesicle membrane(GO:0030660) |
| 0.0 | 0.4 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.0 | 0.4 | GO:0001673 | male germ cell nucleus(GO:0001673) |
| 0.0 | 5.2 | GO:0000922 | spindle pole(GO:0000922) |
| 0.0 | 0.2 | GO:0031673 | H zone(GO:0031673) |
| 0.0 | 0.2 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.0 | 1.5 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.0 | 2.0 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.4 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 0.7 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 2.8 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 1.1 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 0.1 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.0 | 1.1 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 0.4 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.0 | 1.3 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.0 | 7.8 | GO:0016604 | nuclear body(GO:0016604) |
| 0.0 | 2.3 | GO:0070821 | tertiary granule membrane(GO:0070821) |
| 0.0 | 1.8 | GO:0045335 | phagocytic vesicle(GO:0045335) |
| 0.0 | 0.1 | GO:0000839 | Hrd1p ubiquitin ligase ERAD-L complex(GO:0000839) |
| 0.0 | 0.5 | GO:0005657 | replication fork(GO:0005657) |
| 0.0 | 2.8 | GO:0005795 | Golgi stack(GO:0005795) |
| 0.0 | 1.9 | GO:0030496 | midbody(GO:0030496) |
| 0.0 | 0.2 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 0.5 | GO:0030669 | clathrin-coated endocytic vesicle membrane(GO:0030669) |
| 0.0 | 0.3 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 0.1 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.0 | 1.6 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.0 | 1.0 | GO:0031903 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 0.1 | GO:0000791 | euchromatin(GO:0000791) nuclear euchromatin(GO:0005719) |
| 0.0 | 0.2 | GO:0097546 | ciliary base(GO:0097546) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.7 | 57.4 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 2.3 | 43.3 | GO:0003680 | AT DNA binding(GO:0003680) |
| 1.7 | 10.3 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 1.7 | 26.7 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 1.5 | 8.7 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 1.3 | 10.2 | GO:0008269 | JAK pathway signal transduction adaptor activity(GO:0008269) |
| 1.1 | 93.1 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 1.1 | 4.3 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 1.0 | 3.9 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.9 | 4.7 | GO:0050436 | microfibril binding(GO:0050436) |
| 0.9 | 11.8 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.9 | 3.5 | GO:0016524 | latrotoxin receptor activity(GO:0016524) |
| 0.9 | 3.5 | GO:0003867 | 4-aminobutyrate transaminase activity(GO:0003867) succinate-semialdehyde dehydrogenase binding(GO:0032145) (S)-3-amino-2-methylpropionate transaminase activity(GO:0047298) |
| 0.9 | 5.1 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.8 | 13.4 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.8 | 24.9 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.7 | 2.9 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.7 | 2.1 | GO:0035375 | zymogen binding(GO:0035375) |
| 0.7 | 6.9 | GO:0070696 | transmembrane receptor protein serine/threonine kinase binding(GO:0070696) |
| 0.6 | 2.9 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) |
| 0.6 | 1.7 | GO:0008511 | sodium:potassium:chloride symporter activity(GO:0008511) |
| 0.5 | 3.3 | GO:0004699 | calcium-independent protein kinase C activity(GO:0004699) |
| 0.5 | 14.1 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.5 | 1.5 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.5 | 1.4 | GO:0019959 | interleukin-8 binding(GO:0019959) |
| 0.5 | 2.3 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.4 | 12.7 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.4 | 1.3 | GO:0004796 | thromboxane-A synthase activity(GO:0004796) 12-hydroxyheptadecatrienoic acid synthase activity(GO:0036134) |
| 0.4 | 2.1 | GO:0005119 | smoothened binding(GO:0005119) hedgehog family protein binding(GO:0097108) |
| 0.4 | 1.3 | GO:0016608 | growth hormone-releasing hormone activity(GO:0016608) |
| 0.4 | 2.7 | GO:0016715 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced ascorbate as one donor, and incorporation of one atom of oxygen(GO:0016715) |
| 0.4 | 1.5 | GO:0030226 | apolipoprotein receptor activity(GO:0030226) |
| 0.4 | 1.5 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 0.3 | 1.4 | GO:0047391 | alkylglycerophosphoethanolamine phosphodiesterase activity(GO:0047391) |
| 0.3 | 5.7 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.3 | 1.3 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.3 | 12.1 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.3 | 1.6 | GO:0008240 | tripeptidyl-peptidase activity(GO:0008240) |
| 0.3 | 8.8 | GO:0001190 | transcriptional activator activity, RNA polymerase II transcription factor binding(GO:0001190) transcriptional repressor activity, RNA polymerase II activating transcription factor binding(GO:0098811) |
| 0.3 | 0.9 | GO:0090555 | phosphatidylethanolamine-translocating ATPase activity(GO:0090555) |
| 0.3 | 1.7 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.3 | 1.4 | GO:0080019 | fatty-acyl-CoA reductase (alcohol-forming) activity(GO:0080019) |
| 0.3 | 3.7 | GO:0008381 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.3 | 7.3 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.3 | 8.2 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.3 | 0.5 | GO:1990763 | arrestin family protein binding(GO:1990763) |
| 0.2 | 2.5 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.2 | 111.1 | GO:0001077 | transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) |
| 0.2 | 0.7 | GO:0061599 | nitrate reductase activity(GO:0008940) molybdopterin adenylyltransferase activity(GO:0061598) molybdopterin molybdotransferase activity(GO:0061599) |
| 0.2 | 1.9 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 0.2 | 5.7 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.2 | 1.2 | GO:0052654 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.2 | 0.7 | GO:0005174 | CD40 receptor binding(GO:0005174) |
| 0.2 | 3.3 | GO:0019966 | interleukin-1 binding(GO:0019966) |
| 0.2 | 0.6 | GO:0005477 | pyruvate secondary active transmembrane transporter activity(GO:0005477) |
| 0.2 | 0.7 | GO:0004923 | leukemia inhibitory factor receptor activity(GO:0004923) |
| 0.2 | 0.6 | GO:0004556 | alpha-amylase activity(GO:0004556) |
| 0.2 | 1.2 | GO:0016714 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced pteridine as one donor, and incorporation of one atom of oxygen(GO:0016714) |
| 0.2 | 1.0 | GO:0004522 | ribonuclease A activity(GO:0004522) |
| 0.2 | 2.8 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.2 | 0.5 | GO:0004531 | deoxyribonuclease II activity(GO:0004531) |
| 0.2 | 2.0 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.2 | 1.9 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.2 | 3.4 | GO:0031432 | titin binding(GO:0031432) |
| 0.2 | 2.1 | GO:0046965 | retinoid X receptor binding(GO:0046965) |
| 0.2 | 4.1 | GO:0031852 | mu-type opioid receptor binding(GO:0031852) |
| 0.1 | 2.5 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.1 | 1.0 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.1 | 0.7 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.1 | 1.7 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.1 | 1.1 | GO:0047522 | 13-prostaglandin reductase activity(GO:0036132) 15-oxoprostaglandin 13-oxidase activity(GO:0047522) |
| 0.1 | 0.6 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.1 | 0.7 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) |
| 0.1 | 0.9 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.1 | 0.8 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.1 | 1.2 | GO:1904315 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.1 | 0.5 | GO:0015375 | glycine:sodium symporter activity(GO:0015375) |
| 0.1 | 0.7 | GO:0055077 | gap junction hemi-channel activity(GO:0055077) |
| 0.1 | 2.6 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.1 | 1.0 | GO:0016403 | dimethylargininase activity(GO:0016403) |
| 0.1 | 1.0 | GO:0010858 | calcium-dependent protein kinase regulator activity(GO:0010858) |
| 0.1 | 0.9 | GO:0003960 | NADPH:quinone reductase activity(GO:0003960) |
| 0.1 | 1.4 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.1 | 0.7 | GO:0022821 | potassium ion antiporter activity(GO:0022821) |
| 0.1 | 0.3 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.1 | 0.8 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.1 | 1.3 | GO:0072542 | protein phosphatase activator activity(GO:0072542) |
| 0.1 | 0.8 | GO:0030267 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 0.1 | 6.4 | GO:0001191 | transcriptional repressor activity, RNA polymerase II transcription factor binding(GO:0001191) |
| 0.1 | 0.4 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) |
| 0.1 | 0.4 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.1 | 3.3 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.1 | 2.5 | GO:0098748 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.1 | 0.3 | GO:0008670 | 2,4-dienoyl-CoA reductase (NADPH) activity(GO:0008670) trans-2-enoyl-CoA reductase (NADPH) activity(GO:0019166) |
| 0.1 | 1.1 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 0.1 | 0.5 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.1 | 0.3 | GO:0032093 | SAM domain binding(GO:0032093) |
| 0.1 | 0.3 | GO:0055100 | adiponectin binding(GO:0055100) |
| 0.1 | 0.6 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 0.1 | 0.7 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 0.1 | 3.0 | GO:0001530 | lipopolysaccharide binding(GO:0001530) |
| 0.1 | 2.3 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.1 | 1.8 | GO:0031690 | adrenergic receptor binding(GO:0031690) |
| 0.1 | 2.7 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.1 | 0.3 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.1 | 2.6 | GO:0019198 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.1 | 2.9 | GO:0032452 | histone demethylase activity(GO:0032452) |
| 0.1 | 1.8 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.1 | 3.2 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.1 | 2.0 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.1 | 0.4 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.1 | 2.1 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.1 | 20.1 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.1 | 2.8 | GO:0001158 | enhancer sequence-specific DNA binding(GO:0001158) |
| 0.1 | 2.9 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.1 | 1.5 | GO:0005542 | folic acid binding(GO:0005542) |
| 0.1 | 0.9 | GO:0034713 | type I transforming growth factor beta receptor binding(GO:0034713) |
| 0.1 | 21.4 | GO:0008022 | protein C-terminus binding(GO:0008022) |
| 0.1 | 0.5 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.1 | 1.6 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.1 | 11.6 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.1 | 0.8 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.1 | 1.4 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.1 | 0.2 | GO:0052590 | sn-glycerol-3-phosphate:ubiquinone oxidoreductase activity(GO:0052590) sn-glycerol-3-phosphate:ubiquinone-8 oxidoreductase activity(GO:0052591) |
| 0.1 | 0.2 | GO:0034191 | apolipoprotein A-I receptor binding(GO:0034191) |
| 0.1 | 2.1 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.1 | 0.4 | GO:0042974 | retinoic acid receptor binding(GO:0042974) |
| 0.0 | 2.1 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.4 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 1.0 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.0 | 0.8 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.0 | 1.6 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.0 | 0.5 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 0.0 | 1.7 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 0.5 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.0 | 1.5 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 2.7 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.0 | 0.4 | GO:0044213 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) intronic transcription regulatory region DNA binding(GO:0044213) |
| 0.0 | 0.2 | GO:0005502 | 11-cis retinal binding(GO:0005502) |
| 0.0 | 0.3 | GO:0098639 | collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.0 | 18.7 | GO:0003682 | chromatin binding(GO:0003682) |
| 0.0 | 0.3 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 0.0 | 0.8 | GO:0016595 | glutamate binding(GO:0016595) |
| 0.0 | 0.2 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.0 | 0.8 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.0 | 0.6 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.0 | 0.2 | GO:0016647 | oxidoreductase activity, acting on the CH-NH group of donors, oxygen as acceptor(GO:0016647) |
| 0.0 | 0.7 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.0 | 1.3 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.3 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.0 | 0.4 | GO:0047035 | testosterone dehydrogenase (NAD+) activity(GO:0047035) |
| 0.0 | 2.1 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.0 | 0.4 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 0.4 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.0 | 1.8 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.0 | 0.3 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.0 | 2.1 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.0 | 0.1 | GO:0015321 | sodium-dependent phosphate transmembrane transporter activity(GO:0015321) |
| 0.0 | 0.1 | GO:0071253 | connexin binding(GO:0071253) |
| 0.0 | 0.9 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 3.4 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 0.3 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.0 | 0.2 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.0 | 0.4 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.0 | 0.1 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.0 | 0.2 | GO:0019870 | potassium channel inhibitor activity(GO:0019870) |
| 0.0 | 0.2 | GO:0016929 | SUMO-specific protease activity(GO:0016929) |
| 0.0 | 0.8 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.5 | GO:0000339 | RNA cap binding(GO:0000339) |
| 0.0 | 0.8 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.0 | 0.3 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.0 | 0.5 | GO:0071949 | FAD binding(GO:0071949) |
| 0.0 | 1.3 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 0.2 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.3 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.0 | 0.5 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.7 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.1 | GO:0097617 | annealing helicase activity(GO:0036310) annealing activity(GO:0097617) |
| 0.0 | 0.3 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.0 | 0.1 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.0 | 1.0 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.4 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.0 | 0.7 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.0 | 0.1 | GO:0005113 | patched binding(GO:0005113) |
| 0.0 | 0.7 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 0.1 | GO:0016274 | arginine N-methyltransferase activity(GO:0016273) protein-arginine N-methyltransferase activity(GO:0016274) |
| 0.0 | 0.4 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.0 | 0.0 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.0 | 0.1 | GO:0030169 | low-density lipoprotein particle binding(GO:0030169) |
| 0.0 | 0.1 | GO:0050733 | RS domain binding(GO:0050733) |
| 0.0 | 0.4 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.1 | GO:0050664 | oxidoreductase activity, acting on NAD(P)H, oxygen as acceptor(GO:0050664) |
| 0.0 | 0.1 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 90.2 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 1.0 | 93.2 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.8 | 47.7 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.4 | 23.7 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.4 | 50.4 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.2 | 8.8 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.2 | 11.0 | PID IL2 1PATHWAY | IL2-mediated signaling events |
| 0.2 | 11.1 | PID BMP PATHWAY | BMP receptor signaling |
| 0.2 | 4.5 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.2 | 2.6 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.2 | 12.2 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.1 | 8.2 | PID SHP2 PATHWAY | SHP2 signaling |
| 0.1 | 12.0 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.1 | 2.5 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.1 | 3.8 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.1 | 3.7 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.1 | 5.6 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.1 | 2.2 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.1 | 0.9 | PID LYMPH ANGIOGENESIS PATHWAY | VEGFR3 signaling in lymphatic endothelium |
| 0.1 | 0.7 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.1 | 3.6 | PID TXA2PATHWAY | Thromboxane A2 receptor signaling |
| 0.1 | 0.9 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.1 | 0.3 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.1 | 5.1 | PID E2F PATHWAY | E2F transcription factor network |
| 0.1 | 1.4 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.1 | 0.8 | PID VEGFR1 PATHWAY | VEGFR1 specific signals |
| 0.0 | 2.4 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 1.0 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 2.0 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 1.4 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 1.0 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.0 | 5.2 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 1.4 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 1.0 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 1.0 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.0 | 0.4 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 0.9 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 0.1 | PID S1P S1P1 PATHWAY | S1P1 pathway |
| 0.0 | 0.4 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 1.2 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.0 | 1.0 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 5.0 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.4 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 0.8 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 0.4 | PID FOXO PATHWAY | FoxO family signaling |
| 0.0 | 0.2 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.0 | 0.8 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 95.6 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 1.1 | 42.5 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.8 | 30.3 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.5 | 11.8 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.3 | 4.2 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.2 | 11.1 | REACTOME GROWTH HORMONE RECEPTOR SIGNALING | Genes involved in Growth hormone receptor signaling |
| 0.2 | 4.5 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.2 | 10.0 | REACTOME CA DEPENDENT EVENTS | Genes involved in Ca-dependent events |
| 0.2 | 1.9 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.2 | 0.3 | REACTOME FGFR1 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR1 ligand binding and activation |
| 0.2 | 0.9 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.2 | 10.0 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.1 | 13.6 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.1 | 3.4 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.1 | 3.5 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.1 | 2.6 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.1 | 2.8 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.1 | 2.3 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.1 | 10.0 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.1 | 0.6 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.1 | 2.6 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.1 | 12.7 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.1 | 3.2 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.1 | 1.5 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.1 | 1.0 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.0 | 1.0 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 1.2 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.0 | 4.1 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 2.8 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.0 | 0.9 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.6 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 1.4 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 0.8 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.0 | 0.7 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.0 | 1.2 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 0.7 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 3.0 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 0.6 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.0 | 1.3 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.0 | 0.8 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.0 | 1.0 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 0.3 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.0 | 0.6 | REACTOME ELONGATION ARREST AND RECOVERY | Genes involved in Elongation arrest and recovery |
| 0.0 | 0.4 | REACTOME G BETA GAMMA SIGNALLING THROUGH PI3KGAMMA | Genes involved in G beta:gamma signalling through PI3Kgamma |
| 0.0 | 0.5 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.0 | 0.9 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.0 | 0.3 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 0.5 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 0.2 | REACTOME REGULATED PROTEOLYSIS OF P75NTR | Genes involved in Regulated proteolysis of p75NTR |
| 0.0 | 0.4 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.0 | 0.1 | REACTOME SOS MEDIATED SIGNALLING | Genes involved in SOS-mediated signalling |
| 0.0 | 0.4 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 0.2 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.0 | 0.4 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.0 | 0.0 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 0.2 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.0 | 1.0 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.0 | 0.3 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 0.3 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 0.5 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 1.8 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |