Project
Inflammatory response time course, HUVEC (Wada, 2009)
Navigation
Downloads
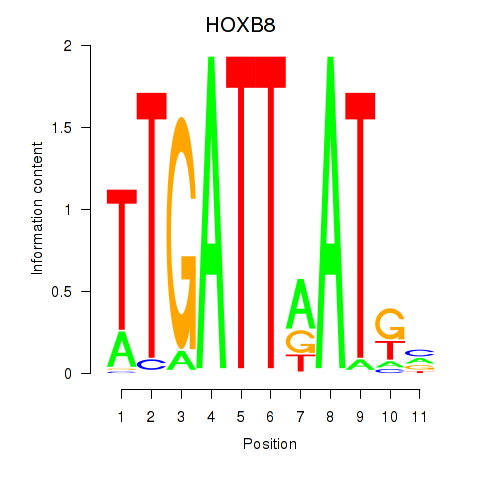
Results for HOXB8
Z-value: 0.87
Transcription factors associated with HOXB8
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HOXB8
|
ENSG00000120068.7 | HOXB8 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HOXB8 | hg38_v1_chr17_-_48613468_48613522 | -0.15 | 4.8e-01 | Click! |
Activity profile of HOXB8 motif
Sorted Z-values of HOXB8 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of HOXB8
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_-_122621011 | 1.69 |
ENST00000611104.2
ENST00000648588.1 |
IL21
|
interleukin 21 |
| chr5_+_157269317 | 1.35 |
ENST00000618329.4
|
CYFIP2
|
cytoplasmic FMR1 interacting protein 2 |
| chr1_+_86547070 | 1.16 |
ENST00000370563.3
|
CLCA4
|
chloride channel accessory 4 |
| chr9_-_83267230 | 1.14 |
ENST00000328788.5
|
FRMD3
|
FERM domain containing 3 |
| chr3_+_148827800 | 1.10 |
ENST00000282957.9
ENST00000468341.1 |
CPB1
|
carboxypeptidase B1 |
| chr15_+_76336755 | 1.04 |
ENST00000290759.9
|
ISL2
|
ISL LIM homeobox 2 |
| chr19_-_14835162 | 0.96 |
ENST00000322301.5
|
OR7A5
|
olfactory receptor family 7 subfamily A member 5 |
| chr10_-_88952763 | 0.95 |
ENST00000224784.10
|
ACTA2
|
actin alpha 2, smooth muscle |
| chr7_-_116030735 | 0.89 |
ENST00000393485.5
|
TFEC
|
transcription factor EC |
| chr20_-_56497608 | 0.86 |
ENST00000617620.1
|
GCNT7
|
glucosaminyl (N-acetyl) transferase family member 7 |
| chr2_-_201697993 | 0.85 |
ENST00000428900.6
|
MPP4
|
membrane palmitoylated protein 4 |
| chr3_+_151814102 | 0.82 |
ENST00000232892.12
|
AADAC
|
arylacetamide deacetylase |
| chr11_-_5324297 | 0.79 |
ENST00000624187.1
|
OR51B2
|
olfactory receptor family 51 subfamily B member 2 |
| chr11_+_5389377 | 0.79 |
ENST00000328611.5
|
OR51M1
|
olfactory receptor family 51 subfamily M member 1 |
| chr10_+_68106109 | 0.79 |
ENST00000540630.5
ENST00000354393.6 |
MYPN
|
myopalladin |
| chr7_-_116030750 | 0.76 |
ENST00000265440.12
ENST00000320239.11 |
TFEC
|
transcription factor EC |
| chr2_-_201698040 | 0.76 |
ENST00000396886.7
ENST00000409143.5 |
MPP4
|
membrane palmitoylated protein 4 |
| chr3_-_191282383 | 0.71 |
ENST00000427544.6
|
UTS2B
|
urotensin 2B |
| chr8_-_134510182 | 0.69 |
ENST00000521673.5
|
ZFAT
|
zinc finger and AT-hook domain containing |
| chr22_+_22704265 | 0.68 |
ENST00000390307.2
|
IGLV3-22
|
immunoglobulin lambda variable 3-22 |
| chr4_-_69214743 | 0.67 |
ENST00000446444.2
|
UGT2B11
|
UDP glucuronosyltransferase family 2 member B11 |
| chr3_+_155083889 | 0.66 |
ENST00000680282.1
|
MME
|
membrane metalloendopeptidase |
| chr6_-_52840843 | 0.66 |
ENST00000370989.6
|
GSTA5
|
glutathione S-transferase alpha 5 |
| chr6_-_132659178 | 0.66 |
ENST00000275216.3
|
TAAR1
|
trace amine associated receptor 1 |
| chr1_+_78620432 | 0.62 |
ENST00000370751.10
ENST00000459784.6 ENST00000680110.1 ENST00000680295.1 |
IFI44L
|
interferon induced protein 44 like |
| chr6_-_118710065 | 0.60 |
ENST00000392500.7
ENST00000368488.9 ENST00000434604.5 |
CEP85L
|
centrosomal protein 85 like |
| chr5_-_135954962 | 0.58 |
ENST00000522943.5
ENST00000514447.2 ENST00000274507.6 |
LECT2
|
leukocyte cell derived chemotaxin 2 |
| chr12_-_16600703 | 0.57 |
ENST00000616247.4
|
LMO3
|
LIM domain only 3 |
| chr11_-_102625332 | 0.57 |
ENST00000260228.3
|
MMP20
|
matrix metallopeptidase 20 |
| chr2_+_89884740 | 0.55 |
ENST00000509129.1
|
IGKV1D-37
|
immunoglobulin kappa variable 1D-37 (non-functional) |
| chr4_+_69280472 | 0.55 |
ENST00000335568.10
ENST00000511240.1 |
UGT2B28
|
UDP glucuronosyltransferase family 2 member B28 |
| chr21_-_10649835 | 0.55 |
ENST00000622028.1
|
IGHV1OR21-1
|
immunoglobulin heavy variable 1/OR21-1 (non-functional) |
| chr12_-_7444139 | 0.55 |
ENST00000416109.2
ENST00000313599.8 |
CD163L1
|
CD163 molecule like 1 |
| chr16_-_28623560 | 0.55 |
ENST00000350842.8
|
SULT1A1
|
sulfotransferase family 1A member 1 |
| chr1_-_247758680 | 0.54 |
ENST00000408896.4
|
OR1C1
|
olfactory receptor family 1 subfamily C member 1 |
| chr3_-_15440560 | 0.53 |
ENST00000595627.5
ENST00000597949.1 ENST00000494875.3 ENST00000595975.1 ENST00000598878.1 |
EAF1-AS1
METTL6
|
EAF1 antisense RNA 1 methyltransferase like 6 |
| chr5_-_59216826 | 0.53 |
ENST00000638939.1
|
PDE4D
|
phosphodiesterase 4D |
| chr19_-_10587219 | 0.53 |
ENST00000591240.5
ENST00000589684.5 ENST00000591676.1 ENST00000250244.11 ENST00000590923.5 |
AP1M2
|
adaptor related protein complex 1 subunit mu 2 |
| chr1_+_214603173 | 0.52 |
ENST00000366955.8
|
CENPF
|
centromere protein F |
| chr17_-_41140487 | 0.51 |
ENST00000345847.4
|
KRTAP4-6
|
keratin associated protein 4-6 |
| chr9_+_122370523 | 0.51 |
ENST00000643810.1
ENST00000540753.6 |
PTGS1
|
prostaglandin-endoperoxide synthase 1 |
| chr7_-_138679045 | 0.50 |
ENST00000419765.4
|
SVOPL
|
SVOP like |
| chr15_+_67125707 | 0.49 |
ENST00000540846.6
|
SMAD3
|
SMAD family member 3 |
| chr3_+_155083523 | 0.48 |
ENST00000680057.1
|
MME
|
membrane metalloendopeptidase |
| chr4_-_76023489 | 0.47 |
ENST00000306602.3
|
CXCL10
|
C-X-C motif chemokine ligand 10 |
| chr10_+_68109433 | 0.47 |
ENST00000613327.4
ENST00000358913.10 ENST00000373675.3 |
MYPN
|
myopalladin |
| chr3_-_112845950 | 0.47 |
ENST00000398214.5
|
CD200R1L
|
CD200 receptor 1 like |
| chr8_-_133060347 | 0.46 |
ENST00000427060.6
|
SLA
|
Src like adaptor |
| chr10_+_13100075 | 0.45 |
ENST00000378747.8
ENST00000378757.6 ENST00000378752.7 ENST00000378748.7 |
OPTN
|
optineurin |
| chr1_+_24319342 | 0.45 |
ENST00000361548.9
|
GRHL3
|
grainyhead like transcription factor 3 |
| chr4_-_176195563 | 0.44 |
ENST00000280191.7
|
SPATA4
|
spermatogenesis associated 4 |
| chr17_-_1229706 | 0.44 |
ENST00000574139.7
|
ABR
|
ABR activator of RhoGEF and GTPase |
| chr11_-_7796942 | 0.44 |
ENST00000329434.3
|
OR5P2
|
olfactory receptor family 5 subfamily P member 2 |
| chr12_-_49897056 | 0.43 |
ENST00000552863.5
|
FAIM2
|
Fas apoptotic inhibitory molecule 2 |
| chr10_+_13099440 | 0.43 |
ENST00000263036.9
|
OPTN
|
optineurin |
| chr4_+_122732652 | 0.42 |
ENST00000542236.5
ENST00000314218.8 |
BBS12
|
Bardet-Biedl syndrome 12 |
| chr5_+_129748091 | 0.41 |
ENST00000564719.2
|
MINAR2
|
membrane integral NOTCH2 associated receptor 2 |
| chr5_-_58999885 | 0.41 |
ENST00000317118.12
|
PDE4D
|
phosphodiesterase 4D |
| chr11_-_114595777 | 0.41 |
ENST00000375478.4
|
NXPE4
|
neurexophilin and PC-esterase domain family member 4 |
| chr1_-_111563956 | 0.40 |
ENST00000369717.8
|
TMIGD3
|
transmembrane and immunoglobulin domain containing 3 |
| chr3_-_12545499 | 0.40 |
ENST00000564146.4
|
MKRN2OS
|
MKRN2 opposite strand |
| chr1_+_244352627 | 0.40 |
ENST00000366537.5
ENST00000308105.5 |
C1orf100
|
chromosome 1 open reading frame 100 |
| chr4_+_73481737 | 0.39 |
ENST00000226355.5
|
AFM
|
afamin |
| chr11_-_102874974 | 0.39 |
ENST00000571244.3
|
MMP12
|
matrix metallopeptidase 12 |
| chr1_-_27914513 | 0.39 |
ENST00000313433.11
ENST00000373912.8 ENST00000444045.1 |
RPA2
|
replication protein A2 |
| chr3_+_98353854 | 0.38 |
ENST00000354924.2
|
OR5K4
|
olfactory receptor family 5 subfamily K member 4 |
| chr12_+_112978386 | 0.38 |
ENST00000342315.8
|
OAS2
|
2'-5'-oligoadenylate synthetase 2 |
| chr14_-_67412112 | 0.38 |
ENST00000216446.9
|
PLEK2
|
pleckstrin 2 |
| chr10_+_13099585 | 0.38 |
ENST00000378764.6
|
OPTN
|
optineurin |
| chr3_+_148791058 | 0.38 |
ENST00000491148.5
|
CPB1
|
carboxypeptidase B1 |
| chr3_-_122993232 | 0.38 |
ENST00000650207.1
ENST00000616742.4 ENST00000393583.6 |
SEMA5B
|
semaphorin 5B |
| chr12_+_21526287 | 0.37 |
ENST00000256969.7
|
SPX
|
spexin hormone |
| chr7_+_70596078 | 0.37 |
ENST00000644506.1
|
AUTS2
|
activator of transcription and developmental regulator AUTS2 |
| chr6_-_132763424 | 0.37 |
ENST00000532012.1
ENST00000525270.5 ENST00000530536.5 ENST00000524919.5 |
VNN2
|
vanin 2 |
| chr2_-_99255107 | 0.37 |
ENST00000333017.6
ENST00000626374.2 ENST00000409679.5 ENST00000423306.1 |
LYG2
|
lysozyme g2 |
| chr7_-_13986439 | 0.37 |
ENST00000443608.5
ENST00000438956.5 |
ETV1
|
ETS variant transcription factor 1 |
| chr11_+_96389985 | 0.36 |
ENST00000332349.5
|
JRKL
|
JRK like |
| chr7_-_80919017 | 0.36 |
ENST00000265361.8
|
SEMA3C
|
semaphorin 3C |
| chr16_-_67416420 | 0.36 |
ENST00000348579.6
ENST00000565726.3 |
ZDHHC1
|
zinc finger DHHC-type containing 1 |
| chr22_-_30246739 | 0.36 |
ENST00000403987.3
ENST00000249075.4 |
LIF
|
LIF interleukin 6 family cytokine |
| chr4_+_87832917 | 0.35 |
ENST00000395102.8
ENST00000497649.6 ENST00000540395.1 ENST00000560249.5 ENST00000511670.5 ENST00000361056.3 |
MEPE
|
matrix extracellular phosphoglycoprotein |
| chr5_+_122129533 | 0.35 |
ENST00000296600.5
ENST00000504912.1 ENST00000505843.1 |
ZNF474
|
zinc finger protein 474 |
| chr11_-_124315099 | 0.35 |
ENST00000641897.1
|
OR8D1
|
olfactory receptor family 8 subfamily D member 1 |
| chr8_-_100106034 | 0.35 |
ENST00000360863.11
ENST00000617334.1 |
RGS22
|
regulator of G protein signaling 22 |
| chr4_+_168092530 | 0.35 |
ENST00000359299.8
|
ANXA10
|
annexin A10 |
| chr9_+_72577369 | 0.35 |
ENST00000651183.1
|
TMC1
|
transmembrane channel like 1 |
| chr15_-_79971164 | 0.35 |
ENST00000335661.6
ENST00000267953.4 ENST00000677151.1 |
BCL2A1
|
BCL2 related protein A1 |
| chr17_+_41249687 | 0.35 |
ENST00000334109.3
|
KRTAP9-4
|
keratin associated protein 9-4 |
| chr3_-_108058361 | 0.34 |
ENST00000398258.7
|
CD47
|
CD47 molecule |
| chr20_+_33217325 | 0.34 |
ENST00000375452.3
ENST00000375454.8 |
BPIFA3
|
BPI fold containing family A member 3 |
| chr7_-_45111673 | 0.34 |
ENST00000461363.1
ENST00000258770.8 ENST00000495078.1 ENST00000494076.5 ENST00000478532.5 ENST00000361278.7 |
TBRG4
|
transforming growth factor beta regulator 4 |
| chr17_-_41009124 | 0.34 |
ENST00000391588.3
|
KRTAP3-1
|
keratin associated protein 3-1 |
| chr18_+_63887698 | 0.34 |
ENST00000457692.5
ENST00000299502.9 ENST00000413956.5 |
SERPINB2
|
serpin family B member 2 |
| chr2_+_167135901 | 0.33 |
ENST00000628543.2
|
XIRP2
|
xin actin binding repeat containing 2 |
| chr11_-_60243103 | 0.33 |
ENST00000651255.1
|
MS4A4E
|
membrane spanning 4-domains A4E |
| chr10_+_96129707 | 0.32 |
ENST00000316045.9
|
ZNF518A
|
zinc finger protein 518A |
| chr5_-_41213505 | 0.32 |
ENST00000337836.10
ENST00000433294.1 |
C6
|
complement C6 |
| chr1_-_154627945 | 0.32 |
ENST00000681683.1
ENST00000368471.8 ENST00000649042.1 ENST00000680270.1 ENST00000649022.2 ENST00000681056.1 ENST00000649724.1 |
ADAR
|
adenosine deaminase RNA specific |
| chr10_+_94683771 | 0.32 |
ENST00000339022.6
|
CYP2C18
|
cytochrome P450 family 2 subfamily C member 18 |
| chr14_+_22508602 | 0.32 |
ENST00000390504.1
|
TRAJ33
|
T cell receptor alpha joining 33 |
| chr10_+_114239245 | 0.32 |
ENST00000392982.8
|
VWA2
|
von Willebrand factor A domain containing 2 |
| chr17_-_40937641 | 0.32 |
ENST00000209718.8
|
KRT23
|
keratin 23 |
| chr9_+_108862255 | 0.32 |
ENST00000333999.5
|
ACTL7A
|
actin like 7A |
| chr11_-_18939493 | 0.31 |
ENST00000526914.1
|
MRGPRX1
|
MAS related GPR family member X1 |
| chrX_+_134990980 | 0.31 |
ENST00000330288.6
|
SMIM10
|
small integral membrane protein 10 |
| chr18_+_74534493 | 0.30 |
ENST00000358821.8
|
CNDP1
|
carnosine dipeptidase 1 |
| chr12_+_9827472 | 0.30 |
ENST00000617793.4
ENST00000617889.5 ENST00000354855.7 ENST00000279545.7 |
KLRF1
|
killer cell lectin like receptor F1 |
| chr3_+_130081831 | 0.30 |
ENST00000425059.1
|
ALG1L2
|
ALG1 chitobiosyldiphosphodolichol beta-mannosyltransferase like 2 |
| chr4_-_71784046 | 0.30 |
ENST00000513476.5
ENST00000273951.13 |
GC
|
GC vitamin D binding protein |
| chr19_-_52095704 | 0.30 |
ENST00000594440.6
ENST00000426391.6 |
ZNF841
|
zinc finger protein 841 |
| chr19_+_54803604 | 0.30 |
ENST00000359085.8
|
KIR2DL4
|
killer cell immunoglobulin like receptor, two Ig domains and long cytoplasmic tail 4 |
| chr7_-_105679089 | 0.30 |
ENST00000477775.5
|
ATXN7L1
|
ataxin 7 like 1 |
| chr14_+_75605462 | 0.30 |
ENST00000539311.5
|
FLVCR2
|
FLVCR heme transporter 2 |
| chr8_+_36784324 | 0.30 |
ENST00000523973.5
ENST00000399881.8 |
KCNU1
|
potassium calcium-activated channel subfamily U member 1 |
| chr10_+_92593112 | 0.29 |
ENST00000260731.5
|
KIF11
|
kinesin family member 11 |
| chr14_+_73591873 | 0.29 |
ENST00000326303.5
|
ACOT4
|
acyl-CoA thioesterase 4 |
| chr8_+_103880412 | 0.29 |
ENST00000436393.6
|
RIMS2
|
regulating synaptic membrane exocytosis 2 |
| chr6_+_26365176 | 0.29 |
ENST00000377708.7
|
BTN3A2
|
butyrophilin subfamily 3 member A2 |
| chr10_+_60778490 | 0.29 |
ENST00000448257.6
ENST00000614696.4 |
CDK1
|
cyclin dependent kinase 1 |
| chr3_-_58627596 | 0.29 |
ENST00000474531.5
ENST00000465970.1 |
FAM107A
|
family with sequence similarity 107 member A |
| chr6_+_26365215 | 0.29 |
ENST00000527422.5
ENST00000356386.6 ENST00000396948.5 |
BTN3A2
|
butyrophilin subfamily 3 member A2 |
| chr9_-_146140 | 0.29 |
ENST00000475990.5
|
CBWD1
|
COBW domain containing 1 |
| chr1_+_24319511 | 0.29 |
ENST00000356046.6
|
GRHL3
|
grainyhead like transcription factor 3 |
| chr18_+_74534594 | 0.28 |
ENST00000582365.1
|
CNDP1
|
carnosine dipeptidase 1 |
| chr1_-_200620729 | 0.28 |
ENST00000367350.5
|
KIF14
|
kinesin family member 14 |
| chr1_-_89198868 | 0.28 |
ENST00000355754.7
|
GBP4
|
guanylate binding protein 4 |
| chr7_+_120273129 | 0.28 |
ENST00000331113.9
|
KCND2
|
potassium voltage-gated channel subfamily D member 2 |
| chr5_+_162067858 | 0.27 |
ENST00000361925.9
|
GABRG2
|
gamma-aminobutyric acid type A receptor subunit gamma2 |
| chr10_-_78029487 | 0.27 |
ENST00000372371.8
|
POLR3A
|
RNA polymerase III subunit A |
| chr12_-_262828 | 0.27 |
ENST00000343164.9
ENST00000436453.1 ENST00000445055.6 ENST00000546319.5 |
SLC6A13
|
solute carrier family 6 member 13 |
| chr10_-_13099652 | 0.27 |
ENST00000378839.1
|
CCDC3
|
coiled-coil domain containing 3 |
| chr8_+_18391276 | 0.27 |
ENST00000286479.4
ENST00000520116.1 |
NAT2
|
N-acetyltransferase 2 |
| chr4_-_129093454 | 0.27 |
ENST00000281142.10
ENST00000511426.5 |
SCLT1
|
sodium channel and clathrin linker 1 |
| chr10_+_60778331 | 0.27 |
ENST00000519078.6
ENST00000316629.8 ENST00000395284.8 |
CDK1
|
cyclin dependent kinase 1 |
| chr16_-_28623330 | 0.27 |
ENST00000677940.1
|
ENSG00000288656.1
|
novel protein |
| chr8_+_85107172 | 0.26 |
ENST00000360375.8
|
LRRCC1
|
leucine rich repeat and coiled-coil centrosomal protein 1 |
| chr11_-_114595750 | 0.26 |
ENST00000424261.6
|
NXPE4
|
neurexophilin and PC-esterase domain family member 4 |
| chrX_-_111412162 | 0.26 |
ENST00000637570.1
ENST00000356220.8 ENST00000636035.2 ENST00000637453.1 ENST00000635795.1 |
DCX
|
doublecortin |
| chr8_-_100105955 | 0.26 |
ENST00000523437.5
|
RGS22
|
regulator of G protein signaling 22 |
| chr17_+_42773442 | 0.26 |
ENST00000253794.7
ENST00000590339.5 ENST00000589520.1 |
VPS25
|
vacuolar protein sorting 25 homolog |
| chr8_+_144477975 | 0.26 |
ENST00000435887.2
|
PPP1R16A
|
protein phosphatase 1 regulatory subunit 16A |
| chr2_-_89297785 | 0.26 |
ENST00000465170.1
|
IGKV1-37
|
immunoglobulin kappa variable 1-37 (non-functional) |
| chr1_+_248445512 | 0.26 |
ENST00000642130.1
ENST00000641925.2 |
OR2T2
|
olfactory receptor family 2 subfamily T member 2 |
| chr4_+_155903688 | 0.26 |
ENST00000536354.3
|
TDO2
|
tryptophan 2,3-dioxygenase |
| chr15_+_84235773 | 0.26 |
ENST00000510439.7
ENST00000422563.6 |
GOLGA6L4
|
golgin A6 family like 4 |
| chr14_+_51489112 | 0.26 |
ENST00000356218.8
|
FRMD6
|
FERM domain containing 6 |
| chr20_+_36876119 | 0.26 |
ENST00000602922.5
ENST00000217320.8 |
TLDC2
|
TBC/LysM-associated domain containing 2 |
| chr19_-_48513161 | 0.26 |
ENST00000673139.1
|
LMTK3
|
lemur tyrosine kinase 3 |
| chr11_+_57597563 | 0.26 |
ENST00000619430.2
ENST00000457869.1 ENST00000340687.10 ENST00000278407.9 ENST00000378323.8 ENST00000378324.6 ENST00000403558.1 |
SERPING1
|
serpin family G member 1 |
| chr10_+_55599041 | 0.25 |
ENST00000512524.4
|
MTRNR2L5
|
MT-RNR2 like 5 |
| chrX_-_152733715 | 0.25 |
ENST00000452779.3
ENST00000370287.7 |
CSAG1
|
chondrosarcoma associated gene 1 |
| chr1_-_111563934 | 0.25 |
ENST00000443498.5
|
TMIGD3
|
transmembrane and immunoglobulin domain containing 3 |
| chr4_-_70666492 | 0.25 |
ENST00000254801.9
ENST00000391614.7 |
JCHAIN
|
joining chain of multimeric IgA and IgM |
| chr7_+_97005538 | 0.25 |
ENST00000518156.3
|
DLX6
|
distal-less homeobox 6 |
| chr8_-_86743626 | 0.25 |
ENST00000320005.6
|
CNGB3
|
cyclic nucleotide gated channel subunit beta 3 |
| chr9_+_122371036 | 0.25 |
ENST00000619306.5
ENST00000426608.6 ENST00000223423.8 |
PTGS1
|
prostaglandin-endoperoxide synthase 1 |
| chr7_-_113086815 | 0.24 |
ENST00000424100.2
|
GPR85
|
G protein-coupled receptor 85 |
| chr14_+_19743571 | 0.24 |
ENST00000642117.2
|
OR4Q3
|
olfactory receptor family 4 subfamily Q member 3 |
| chr15_-_82349437 | 0.24 |
ENST00000621197.4
ENST00000610657.2 ENST00000619556.4 |
GOLGA6L10
|
golgin A6 family like 10 |
| chrX_+_96684712 | 0.24 |
ENST00000373049.8
|
DIAPH2
|
diaphanous related formin 2 |
| chr18_+_24113341 | 0.23 |
ENST00000540918.2
|
TTC39C
|
tetratricopeptide repeat domain 39C |
| chr8_-_7018295 | 0.23 |
ENST00000327857.7
|
DEFA3
|
defensin alpha 3 |
| chr6_+_52420992 | 0.23 |
ENST00000636954.1
ENST00000636566.1 ENST00000638075.1 |
EFHC1
|
EF-hand domain containing 1 |
| chr12_-_11022620 | 0.23 |
ENST00000390673.2
|
TAS2R19
|
taste 2 receptor member 19 |
| chr15_-_65115185 | 0.23 |
ENST00000559089.6
|
UBAP1L
|
ubiquitin associated protein 1 like |
| chr14_-_23155302 | 0.23 |
ENST00000529705.6
|
SLC7A8
|
solute carrier family 7 member 8 |
| chr11_+_67452392 | 0.23 |
ENST00000438189.6
|
CABP4
|
calcium binding protein 4 |
| chr16_+_8642375 | 0.23 |
ENST00000562973.1
|
METTL22
|
methyltransferase like 22 |
| chrX_+_56563569 | 0.23 |
ENST00000338222.7
|
UBQLN2
|
ubiquilin 2 |
| chr1_-_205121964 | 0.23 |
ENST00000264515.11
|
RBBP5
|
RB binding protein 5, histone lysine methyltransferase complex subunit |
| chr12_-_51324652 | 0.23 |
ENST00000544402.5
|
BIN2
|
bridging integrator 2 |
| chr9_-_92878018 | 0.23 |
ENST00000332591.6
ENST00000375495.8 ENST00000395505.6 ENST00000395506.7 |
ZNF484
|
zinc finger protein 484 |
| chr10_+_46375645 | 0.23 |
ENST00000622769.4
|
ANXA8L1
|
annexin A8 like 1 |
| chr1_+_31413187 | 0.23 |
ENST00000373709.8
|
SERINC2
|
serine incorporator 2 |
| chr1_+_84408230 | 0.23 |
ENST00000370662.3
|
DNASE2B
|
deoxyribonuclease 2 beta |
| chr21_-_14546297 | 0.23 |
ENST00000400566.6
ENST00000400564.5 |
SAMSN1
|
SAM domain, SH3 domain and nuclear localization signals 1 |
| chr3_-_149221811 | 0.23 |
ENST00000455472.3
ENST00000264613.11 |
CP
|
ceruloplasmin |
| chr10_+_46375619 | 0.23 |
ENST00000584982.7
ENST00000613703.4 |
ANXA8L1
|
annexin A8 like 1 |
| chr3_+_142723999 | 0.23 |
ENST00000476941.6
ENST00000273482.10 |
TRPC1
|
transient receptor potential cation channel subfamily C member 1 |
| chr3_+_108602776 | 0.22 |
ENST00000497905.5
ENST00000463306.1 |
DZIP3
|
DAZ interacting zinc finger protein 3 |
| chr3_-_179251615 | 0.22 |
ENST00000314235.9
ENST00000392685.6 |
KCNMB3
|
potassium calcium-activated channel subfamily M regulatory beta subunit 3 |
| chr12_+_18242955 | 0.22 |
ENST00000676171.1
|
PIK3C2G
|
phosphatidylinositol-4-phosphate 3-kinase catalytic subunit type 2 gamma |
| chr10_+_120851341 | 0.22 |
ENST00000263461.11
|
WDR11
|
WD repeat domain 11 |
| chr12_+_8509460 | 0.22 |
ENST00000382064.6
|
CLEC4D
|
C-type lectin domain family 4 member D |
| chr3_+_2892199 | 0.22 |
ENST00000397459.6
|
CNTN4
|
contactin 4 |
| chr12_+_80099535 | 0.22 |
ENST00000646859.1
ENST00000547103.7 |
OTOGL
|
otogelin like |
| chr11_-_123654939 | 0.22 |
ENST00000657191.1
|
SCN3B
|
sodium voltage-gated channel beta subunit 3 |
| chr11_-_35360050 | 0.22 |
ENST00000644868.1
ENST00000643454.1 ENST00000646080.1 |
SLC1A2
|
solute carrier family 1 member 2 |
| chrX_-_13817027 | 0.22 |
ENST00000493677.5
ENST00000355135.6 ENST00000316715.9 |
GPM6B
|
glycoprotein M6B |
| chr1_-_160579439 | 0.21 |
ENST00000368054.8
ENST00000368048.7 ENST00000311224.8 ENST00000368051.3 ENST00000534968.5 |
CD84
|
CD84 molecule |
| chr4_+_36281591 | 0.21 |
ENST00000639862.2
ENST00000357504.7 |
DTHD1
|
death domain containing 1 |
| chr3_+_102099244 | 0.21 |
ENST00000491959.5
|
ZPLD1
|
zona pellucida like domain containing 1 |
| chr2_-_96505345 | 0.21 |
ENST00000310865.7
ENST00000451794.6 |
NEURL3
|
neuralized E3 ubiquitin protein ligase 3 |
| chr5_-_139383284 | 0.21 |
ENST00000353963.7
ENST00000348729.8 |
SLC23A1
|
solute carrier family 23 member 1 |
| chr12_-_109021015 | 0.21 |
ENST00000546618.2
ENST00000610966.5 |
SVOP
|
SV2 related protein |
| chr20_-_56392131 | 0.21 |
ENST00000422322.5
ENST00000371356.6 ENST00000451915.1 ENST00000347343.6 ENST00000395911.5 ENST00000395915.8 ENST00000395907.5 ENST00000441357.5 ENST00000456249.5 ENST00000420474.5 ENST00000395914.5 ENST00000312783.10 ENST00000395913.7 |
AURKA
|
aurora kinase A |
| chr4_-_159035226 | 0.21 |
ENST00000434826.3
|
C4orf45
|
chromosome 4 open reading frame 45 |
| chr14_+_22052503 | 0.21 |
ENST00000390449.3
|
TRAV21
|
T cell receptor alpha variable 21 |
| chr9_+_74497308 | 0.21 |
ENST00000376896.8
|
RORB
|
RAR related orphan receptor B |
| chrX_-_154371210 | 0.21 |
ENST00000369856.8
ENST00000422373.6 ENST00000360319.9 |
FLNA
|
filamin A |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:0021524 | visceral motor neuron differentiation(GO:0021524) |
| 0.2 | 0.9 | GO:0090131 | mesenchyme migration(GO:0090131) |
| 0.2 | 1.3 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.2 | 0.5 | GO:0001868 | regulation of complement activation, lectin pathway(GO:0001868) negative regulation of complement activation, lectin pathway(GO:0001869) |
| 0.1 | 1.3 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 0.1 | 0.1 | GO:2000506 | negative regulation of energy homeostasis(GO:2000506) |
| 0.1 | 1.1 | GO:0071492 | cellular response to UV-A(GO:0071492) |
| 0.1 | 0.4 | GO:0043314 | negative regulation of neutrophil degranulation(GO:0043314) |
| 0.1 | 1.7 | GO:0045078 | positive regulation of interferon-gamma biosynthetic process(GO:0045078) |
| 0.1 | 0.4 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.1 | 0.1 | GO:0045349 | interferon-alpha biosynthetic process(GO:0045349) regulation of interferon-alpha biosynthetic process(GO:0045354) |
| 0.1 | 0.9 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.1 | 0.4 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.1 | 0.5 | GO:0061767 | negative regulation of lung blood pressure(GO:0061767) |
| 0.1 | 0.6 | GO:2000324 | positive regulation of glucocorticoid receptor signaling pathway(GO:2000324) |
| 0.1 | 0.3 | GO:0045917 | positive regulation of complement activation(GO:0045917) positive regulation of protein activation cascade(GO:2000259) |
| 0.1 | 0.6 | GO:0000270 | peptidoglycan metabolic process(GO:0000270) peptidoglycan catabolic process(GO:0009253) |
| 0.1 | 0.4 | GO:0003350 | pulmonary myocardium development(GO:0003350) |
| 0.1 | 0.2 | GO:0015882 | L-ascorbic acid transport(GO:0015882) transepithelial L-ascorbic acid transport(GO:0070904) |
| 0.1 | 0.2 | GO:0007057 | spindle assembly involved in female meiosis I(GO:0007057) positive regulation of oocyte maturation(GO:1900195) |
| 0.1 | 0.2 | GO:1905000 | regulation of membrane repolarization during atrial cardiac muscle cell action potential(GO:1905000) |
| 0.1 | 0.6 | GO:0014038 | regulation of Schwann cell differentiation(GO:0014038) Golgi disassembly(GO:0090166) |
| 0.1 | 0.2 | GO:0014707 | branchiomeric skeletal muscle development(GO:0014707) |
| 0.1 | 0.1 | GO:0042322 | negative regulation of circadian sleep/wake cycle, REM sleep(GO:0042322) |
| 0.1 | 0.1 | GO:0035744 | T-helper 1 cell cytokine production(GO:0035744) |
| 0.1 | 0.3 | GO:0010513 | positive regulation of phosphatidylinositol biosynthetic process(GO:0010513) |
| 0.1 | 0.8 | GO:0010898 | positive regulation of triglyceride catabolic process(GO:0010898) |
| 0.1 | 0.2 | GO:0021658 | rhombomere morphogenesis(GO:0021593) rhombomere 3 morphogenesis(GO:0021658) |
| 0.1 | 0.2 | GO:0002378 | immunoglobulin biosynthetic process(GO:0002378) |
| 0.1 | 0.2 | GO:1900110 | negative regulation of histone H3-K9 dimethylation(GO:1900110) regulation of TORC2 signaling(GO:1903939) |
| 0.1 | 0.7 | GO:0060339 | negative regulation of type I interferon-mediated signaling pathway(GO:0060339) |
| 0.1 | 0.2 | GO:0035691 | macrophage migration inhibitory factor signaling pathway(GO:0035691) |
| 0.1 | 0.2 | GO:1904021 | negative regulation of clathrin-mediated endocytosis(GO:1900186) negative regulation of G-protein coupled receptor internalization(GO:1904021) |
| 0.1 | 0.2 | GO:0019605 | butyrate metabolic process(GO:0019605) |
| 0.1 | 0.2 | GO:0006238 | CMP salvage(GO:0006238) CMP biosynthetic process(GO:0009224) CMP metabolic process(GO:0046035) |
| 0.1 | 0.9 | GO:0042487 | regulation of odontogenesis of dentin-containing tooth(GO:0042487) |
| 0.1 | 1.0 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.1 | 0.2 | GO:0032661 | regulation of interleukin-18 production(GO:0032661) |
| 0.1 | 0.1 | GO:0010645 | regulation of cell communication by chemical coupling(GO:0010645) positive regulation of cell communication by chemical coupling(GO:0010652) |
| 0.1 | 0.4 | GO:0060373 | regulation of ventricular cardiac muscle cell membrane depolarization(GO:0060373) |
| 0.1 | 0.3 | GO:0043328 | protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) |
| 0.1 | 0.2 | GO:0032304 | negative regulation of icosanoid secretion(GO:0032304) |
| 0.1 | 0.1 | GO:0035992 | tendon cell differentiation(GO:0035990) tendon formation(GO:0035992) |
| 0.1 | 0.2 | GO:0035915 | pore formation in membrane of other organism(GO:0035915) |
| 0.1 | 0.2 | GO:0048560 | establishment of anatomical structure orientation(GO:0048560) |
| 0.0 | 0.1 | GO:0044240 | multicellular organism lipid catabolic process(GO:0044240) |
| 0.0 | 0.3 | GO:0032487 | regulation of Rap protein signal transduction(GO:0032487) |
| 0.0 | 0.4 | GO:1904306 | regulation of gastro-intestinal system smooth muscle contraction(GO:1904304) positive regulation of gastro-intestinal system smooth muscle contraction(GO:1904306) |
| 0.0 | 0.2 | GO:1904222 | regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904217) positive regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904219) positive regulation of serine C-palmitoyltransferase activity(GO:1904222) |
| 0.0 | 0.3 | GO:0007161 | calcium-independent cell-matrix adhesion(GO:0007161) |
| 0.0 | 0.4 | GO:0038044 | transforming growth factor-beta secretion(GO:0038044) |
| 0.0 | 0.1 | GO:0045359 | interferon-beta biosynthetic process(GO:0045350) regulation of interferon-beta biosynthetic process(GO:0045357) positive regulation of interferon-beta biosynthetic process(GO:0045359) |
| 0.0 | 0.2 | GO:0007538 | primary sex determination(GO:0007538) |
| 0.0 | 0.1 | GO:0090675 | intermicrovillar adhesion(GO:0090675) |
| 0.0 | 0.5 | GO:0043950 | positive regulation of cAMP-mediated signaling(GO:0043950) |
| 0.0 | 0.2 | GO:0007206 | phospholipase C-activating G-protein coupled glutamate receptor signaling pathway(GO:0007206) |
| 0.0 | 0.3 | GO:0003383 | apical constriction(GO:0003383) |
| 0.0 | 0.1 | GO:0009720 | detection of hormone stimulus(GO:0009720) |
| 0.0 | 0.9 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.0 | 0.2 | GO:0000023 | maltose metabolic process(GO:0000023) |
| 0.0 | 0.4 | GO:2000620 | innate vocalization behavior(GO:0098582) positive regulation of histone H4-K16 acetylation(GO:2000620) |
| 0.0 | 0.2 | GO:0045590 | negative regulation of regulatory T cell differentiation(GO:0045590) |
| 0.0 | 0.2 | GO:0030035 | microspike assembly(GO:0030035) |
| 0.0 | 0.4 | GO:0019441 | tryptophan catabolic process to kynurenine(GO:0019441) |
| 0.0 | 0.2 | GO:0060040 | retinal bipolar neuron differentiation(GO:0060040) |
| 0.0 | 0.7 | GO:0060712 | spongiotrophoblast layer development(GO:0060712) |
| 0.0 | 0.7 | GO:0090179 | planar cell polarity pathway involved in neural tube closure(GO:0090179) |
| 0.0 | 0.3 | GO:0045040 | protein import into mitochondrial outer membrane(GO:0045040) |
| 0.0 | 0.2 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 0.0 | 0.2 | GO:2000393 | negative regulation of lamellipodium morphogenesis(GO:2000393) |
| 0.0 | 0.1 | GO:0045645 | regulation of eosinophil differentiation(GO:0045643) positive regulation of eosinophil differentiation(GO:0045645) |
| 0.0 | 0.1 | GO:0044837 | assembly of actomyosin apparatus involved in cytokinesis(GO:0000912) actomyosin contractile ring assembly(GO:0000915) actomyosin contractile ring organization(GO:0044837) |
| 0.0 | 0.1 | GO:0019087 | transformation of host cell by virus(GO:0019087) |
| 0.0 | 0.1 | GO:0046110 | xanthine metabolic process(GO:0046110) |
| 0.0 | 0.2 | GO:0006015 | 5-phosphoribose 1-diphosphate biosynthetic process(GO:0006015) 5-phosphoribose 1-diphosphate metabolic process(GO:0046391) |
| 0.0 | 0.1 | GO:0007499 | ectoderm and mesoderm interaction(GO:0007499) |
| 0.0 | 0.1 | GO:1903410 | lysine import(GO:0034226) L-lysine import(GO:0061461) L-lysine import into cell(GO:1903410) |
| 0.0 | 0.3 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.0 | 0.2 | GO:0044245 | polysaccharide digestion(GO:0044245) |
| 0.0 | 0.4 | GO:0018377 | protein myristoylation(GO:0018377) |
| 0.0 | 0.5 | GO:0002227 | innate immune response in mucosa(GO:0002227) |
| 0.0 | 0.2 | GO:0002322 | B cell proliferation involved in immune response(GO:0002322) |
| 0.0 | 0.1 | GO:0035698 | CD8-positive, alpha-beta cytotoxic T cell extravasation(GO:0035698) regulation of CD8-positive, alpha-beta cytotoxic T cell extravasation(GO:2000452) |
| 0.0 | 0.3 | GO:0060267 | positive regulation of respiratory burst(GO:0060267) |
| 0.0 | 0.2 | GO:0097498 | endothelial tube lumen extension(GO:0097498) |
| 0.0 | 0.1 | GO:0039022 | pronephric nephron morphogenesis(GO:0039007) pronephric nephron tubule morphogenesis(GO:0039008) pronephric duct development(GO:0039022) pronephric duct morphogenesis(GO:0039023) Kupffer's vesicle development(GO:0070121) |
| 0.0 | 0.3 | GO:0098814 | spontaneous neurotransmitter secretion(GO:0061669) spontaneous synaptic transmission(GO:0098814) |
| 0.0 | 0.1 | GO:0060437 | lung growth(GO:0060437) |
| 0.0 | 1.2 | GO:0008210 | estrogen metabolic process(GO:0008210) |
| 0.0 | 0.3 | GO:0072734 | response to staurosporine(GO:0072733) cellular response to staurosporine(GO:0072734) |
| 0.0 | 0.1 | GO:0036378 | calcitriol biosynthetic process from calciol(GO:0036378) |
| 0.0 | 0.1 | GO:0071395 | response to jasmonic acid(GO:0009753) cellular response to jasmonic acid stimulus(GO:0071395) |
| 0.0 | 0.1 | GO:1903070 | negative regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903070) |
| 0.0 | 0.1 | GO:0046900 | tetrahydrofolylpolyglutamate metabolic process(GO:0046900) |
| 0.0 | 0.3 | GO:0090292 | nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.0 | 0.1 | GO:1902093 | positive regulation of sperm motility(GO:1902093) |
| 0.0 | 0.2 | GO:0042796 | snRNA transcription from RNA polymerase III promoter(GO:0042796) |
| 0.0 | 0.1 | GO:0000350 | generation of catalytic spliceosome for second transesterification step(GO:0000350) |
| 0.0 | 0.2 | GO:0010989 | negative regulation of low-density lipoprotein particle clearance(GO:0010989) |
| 0.0 | 0.2 | GO:0000727 | double-strand break repair via break-induced replication(GO:0000727) |
| 0.0 | 0.4 | GO:0021702 | cerebellar Purkinje cell layer formation(GO:0021694) cerebellar Purkinje cell differentiation(GO:0021702) |
| 0.0 | 0.4 | GO:0015939 | pantothenate metabolic process(GO:0015939) |
| 0.0 | 0.1 | GO:0031296 | positive regulation of germinal center formation(GO:0002636) B cell costimulation(GO:0031296) |
| 0.0 | 0.2 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.0 | 0.5 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.0 | 0.2 | GO:0070562 | regulation of vitamin D receptor signaling pathway(GO:0070562) |
| 0.0 | 0.2 | GO:0045919 | positive regulation of cytolysis(GO:0045919) |
| 0.0 | 0.2 | GO:1903936 | cellular response to sodium arsenite(GO:1903936) |
| 0.0 | 0.1 | GO:0006663 | platelet activating factor biosynthetic process(GO:0006663) |
| 0.0 | 0.1 | GO:0035880 | embryonic nail plate morphogenesis(GO:0035880) establishment of body hair or bristle planar orientation(GO:0048104) establishment of body hair planar orientation(GO:0048105) |
| 0.0 | 0.1 | GO:0071231 | cellular response to folic acid(GO:0071231) |
| 0.0 | 0.1 | GO:0000961 | negative regulation of mitochondrial RNA catabolic process(GO:0000961) |
| 0.0 | 0.1 | GO:0051792 | short-chain fatty acid biosynthetic process(GO:0051790) medium-chain fatty acid biosynthetic process(GO:0051792) |
| 0.0 | 0.6 | GO:0072643 | interferon-gamma secretion(GO:0072643) |
| 0.0 | 0.1 | GO:0050955 | thermoception(GO:0050955) |
| 0.0 | 0.1 | GO:1904862 | inhibitory synapse assembly(GO:1904862) |
| 0.0 | 0.1 | GO:0046092 | deoxycytidine metabolic process(GO:0046092) |
| 0.0 | 0.2 | GO:0035881 | amacrine cell differentiation(GO:0035881) |
| 0.0 | 0.1 | GO:0060018 | astrocyte fate commitment(GO:0060018) |
| 0.0 | 0.1 | GO:0075732 | viral penetration into host nucleus(GO:0075732) multi-organism nuclear import(GO:1902594) |
| 0.0 | 0.3 | GO:0070779 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.0 | 0.1 | GO:0043128 | regulation of 1-phosphatidylinositol 4-kinase activity(GO:0043126) positive regulation of 1-phosphatidylinositol 4-kinase activity(GO:0043128) |
| 0.0 | 0.1 | GO:0046103 | adenosine catabolic process(GO:0006154) inosine biosynthetic process(GO:0046103) |
| 0.0 | 0.1 | GO:0010636 | positive regulation of mitochondrial fusion(GO:0010636) |
| 0.0 | 0.2 | GO:0042866 | pyruvate biosynthetic process(GO:0042866) |
| 0.0 | 0.2 | GO:0051918 | negative regulation of smooth muscle cell apoptotic process(GO:0034392) negative regulation of fibrinolysis(GO:0051918) |
| 0.0 | 0.2 | GO:0051612 | negative regulation of neurotransmitter uptake(GO:0051581) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.0 | 0.0 | GO:0048769 | sarcomerogenesis(GO:0048769) |
| 0.0 | 0.5 | GO:0010569 | regulation of double-strand break repair via homologous recombination(GO:0010569) |
| 0.0 | 0.2 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.0 | 0.1 | GO:0018094 | protein polyglycylation(GO:0018094) |
| 0.0 | 0.0 | GO:0007008 | outer mitochondrial membrane organization(GO:0007008) |
| 0.0 | 0.1 | GO:0042738 | exogenous drug catabolic process(GO:0042738) |
| 0.0 | 0.3 | GO:0015812 | gamma-aminobutyric acid transport(GO:0015812) |
| 0.0 | 0.1 | GO:1904252 | negative regulation of bile acid biosynthetic process(GO:0070858) negative regulation of bile acid metabolic process(GO:1904252) |
| 0.0 | 0.3 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.0 | 0.2 | GO:0098828 | positive regulation of inhibitory postsynaptic potential(GO:0097151) modulation of inhibitory postsynaptic potential(GO:0098828) |
| 0.0 | 0.1 | GO:0010430 | fatty acid omega-oxidation(GO:0010430) |
| 0.0 | 0.3 | GO:0045475 | locomotor rhythm(GO:0045475) |
| 0.0 | 0.1 | GO:0044026 | DNA hypermethylation(GO:0044026) |
| 0.0 | 0.5 | GO:0050908 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.0 | 0.1 | GO:0006568 | tryptophan metabolic process(GO:0006568) |
| 0.0 | 0.1 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.0 | 0.1 | GO:0051410 | detoxification of nitrogen compound(GO:0051410) cellular detoxification of nitrogen compound(GO:0070458) |
| 0.0 | 0.2 | GO:0002857 | positive regulation of natural killer cell mediated immune response to tumor cell(GO:0002857) positive regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002860) |
| 0.0 | 0.1 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.0 | 0.1 | GO:0071799 | response to prostaglandin D(GO:0071798) cellular response to prostaglandin D stimulus(GO:0071799) |
| 0.0 | 0.2 | GO:0006828 | manganese ion transport(GO:0006828) |
| 0.0 | 2.6 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 0.3 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.0 | 0.2 | GO:0072619 | interleukin-21 production(GO:0032625) interleukin-21 secretion(GO:0072619) |
| 0.0 | 0.4 | GO:0048935 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.0 | 0.2 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.0 | 0.2 | GO:0051451 | myoblast migration(GO:0051451) |
| 0.0 | 0.8 | GO:0051180 | vitamin transport(GO:0051180) |
| 0.0 | 0.2 | GO:0006702 | androgen biosynthetic process(GO:0006702) |
| 0.0 | 0.0 | GO:0035552 | tRNA wobble cytosine modification(GO:0002101) oxidative single-stranded DNA demethylation(GO:0035552) |
| 0.0 | 0.0 | GO:0032831 | regulation of CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation(GO:0032829) positive regulation of CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation(GO:0032831) |
| 0.0 | 0.1 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.0 | 0.3 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.0 | 0.2 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.0 | 0.4 | GO:0051131 | chaperone-mediated protein complex assembly(GO:0051131) |
| 0.0 | 0.2 | GO:0010764 | negative regulation of fibroblast migration(GO:0010764) |
| 0.0 | 0.0 | GO:0007079 | mitotic chromosome movement towards spindle pole(GO:0007079) |
| 0.0 | 0.1 | GO:0051315 | attachment of mitotic spindle microtubules to kinetochore(GO:0051315) |
| 0.0 | 0.4 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.0 | 0.2 | GO:0046602 | regulation of mitotic centrosome separation(GO:0046602) |
| 0.0 | 0.1 | GO:0003344 | pericardium morphogenesis(GO:0003344) |
| 0.0 | 0.1 | GO:0019918 | peptidyl-arginine methylation, to symmetrical-dimethyl arginine(GO:0019918) |
| 0.0 | 0.2 | GO:0019321 | pentose metabolic process(GO:0019321) |
| 0.0 | 0.1 | GO:0035502 | metanephric part of ureteric bud development(GO:0035502) |
| 0.0 | 0.4 | GO:0048846 | axon extension involved in axon guidance(GO:0048846) neuron projection extension involved in neuron projection guidance(GO:1902284) |
| 0.0 | 0.2 | GO:0008228 | opsonization(GO:0008228) |
| 0.0 | 0.0 | GO:1903251 | multi-ciliated epithelial cell differentiation(GO:1903251) |
| 0.0 | 0.1 | GO:0072656 | maintenance of protein location in mitochondrion(GO:0072656) |
| 0.0 | 0.3 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.0 | 0.0 | GO:0019348 | dolichol metabolic process(GO:0019348) |
| 0.0 | 0.1 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.0 | 0.2 | GO:0006450 | regulation of translational fidelity(GO:0006450) |
| 0.0 | 0.2 | GO:0033141 | positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) |
| 0.0 | 0.1 | GO:0015747 | urate transport(GO:0015747) |
| 0.0 | 0.3 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.0 | 0.6 | GO:0007190 | activation of adenylate cyclase activity(GO:0007190) |
| 0.0 | 0.2 | GO:0046784 | viral mRNA export from host cell nucleus(GO:0046784) |
| 0.0 | 0.2 | GO:0010804 | negative regulation of tumor necrosis factor-mediated signaling pathway(GO:0010804) |
| 0.0 | 0.1 | GO:1990928 | response to amino acid starvation(GO:1990928) |
| 0.0 | 0.2 | GO:0035721 | intraciliary retrograde transport(GO:0035721) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.9 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.1 | 0.6 | GO:0097125 | cyclin B1-CDK1 complex(GO:0097125) |
| 0.1 | 0.3 | GO:0001674 | female germ cell nucleus(GO:0001674) |
| 0.1 | 0.4 | GO:0034685 | integrin alphav-beta6 complex(GO:0034685) |
| 0.1 | 0.2 | GO:0002199 | zona pellucida receptor complex(GO:0002199) |
| 0.1 | 0.3 | GO:0000814 | ESCRT II complex(GO:0000814) |
| 0.1 | 0.3 | GO:0071750 | dimeric IgA immunoglobulin complex(GO:0071750) secretory dimeric IgA immunoglobulin complex(GO:0071752) IgM immunoglobulin complex(GO:0071753) IgM immunoglobulin complex, circulating(GO:0071754) pentameric IgM immunoglobulin complex(GO:0071756) |
| 0.1 | 0.1 | GO:0034686 | integrin alphav-beta8 complex(GO:0034686) |
| 0.1 | 0.2 | GO:0000811 | GINS complex(GO:0000811) |
| 0.1 | 0.5 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.0 | 0.3 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.0 | 0.2 | GO:0031523 | Myb complex(GO:0031523) |
| 0.0 | 0.3 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 0.2 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.0 | 0.1 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.0 | 0.4 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 0.7 | GO:0097539 | ciliary transition fiber(GO:0097539) |
| 0.0 | 0.2 | GO:0000835 | ER ubiquitin ligase complex(GO:0000835) Hrd1p ubiquitin ligase complex(GO:0000836) |
| 0.0 | 0.2 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.0 | 0.2 | GO:0043196 | varicosity(GO:0043196) |
| 0.0 | 0.5 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.0 | 0.3 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.0 | 0.1 | GO:0097149 | centralspindlin complex(GO:0097149) |
| 0.0 | 0.5 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.0 | 0.1 | GO:0009328 | phenylalanine-tRNA ligase complex(GO:0009328) |
| 0.0 | 0.2 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.0 | 0.1 | GO:0071008 | U2-type post-mRNA release spliceosomal complex(GO:0071008) |
| 0.0 | 0.1 | GO:0036195 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 0.0 | 0.2 | GO:0070761 | pre-snoRNP complex(GO:0070761) |
| 0.0 | 0.1 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 0.0 | 0.1 | GO:0044214 | spanning component of plasma membrane(GO:0044214) spanning component of membrane(GO:0089717) |
| 0.0 | 0.1 | GO:0043541 | UDP-N-acetylglucosamine transferase complex(GO:0043541) |
| 0.0 | 0.3 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.0 | 0.1 | GO:0030849 | X chromosome(GO:0000805) autosome(GO:0030849) |
| 0.0 | 0.3 | GO:0043073 | germ cell nucleus(GO:0043073) |
| 0.0 | 0.5 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.0 | 0.1 | GO:0071062 | alphav-beta3 integrin-vitronectin complex(GO:0071062) |
| 0.0 | 0.4 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 0.1 | GO:0071014 | post-mRNA release spliceosomal complex(GO:0071014) |
| 0.0 | 0.2 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.0 | 0.0 | GO:0033185 | dolichol-phosphate-mannose synthase complex(GO:0033185) |
| 0.0 | 1.3 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.0 | 0.3 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.1 | GO:0044194 | cytolytic granule(GO:0044194) |
| 0.0 | 0.2 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.0 | 0.1 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.0 | 1.1 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 0.1 | GO:0005587 | collagen type IV trimer(GO:0005587) |
| 0.0 | 0.2 | GO:0031045 | dense core granule(GO:0031045) |
| 0.0 | 0.1 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.0 | 0.1 | GO:0019907 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) |
| 0.0 | 0.1 | GO:0071203 | WASH complex(GO:0071203) |
| 0.0 | 0.1 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.0 | 0.0 | GO:0033565 | ESCRT-0 complex(GO:0033565) |
| 0.0 | 0.1 | GO:0030868 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 0.0 | 0.1 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.0 | 0.2 | GO:0034464 | BBSome(GO:0034464) |
| 0.0 | 0.1 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.0 | 0.2 | GO:0030897 | HOPS complex(GO:0030897) |
| 0.0 | 0.1 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.0 | 0.1 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.7 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.2 | 0.9 | GO:0004666 | prostaglandin-endoperoxide synthase activity(GO:0004666) |
| 0.1 | 0.4 | GO:0008431 | vitamin E binding(GO:0008431) |
| 0.1 | 0.6 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.1 | 0.7 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.1 | 0.3 | GO:1902271 | D3 vitamins binding(GO:1902271) |
| 0.1 | 0.5 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.1 | 0.4 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.1 | 0.5 | GO:0047894 | flavonol 3-sulfotransferase activity(GO:0047894) |
| 0.1 | 0.3 | GO:0004060 | arylamine N-acetyltransferase activity(GO:0004060) |
| 0.1 | 0.5 | GO:0031962 | mineralocorticoid receptor binding(GO:0031962) |
| 0.1 | 0.2 | GO:0004531 | deoxyribonuclease II activity(GO:0004531) |
| 0.1 | 0.2 | GO:0070890 | L-ascorbate:sodium symporter activity(GO:0008520) L-ascorbic acid transporter activity(GO:0015229) sodium-dependent L-ascorbate transmembrane transporter activity(GO:0070890) |
| 0.1 | 0.2 | GO:0050528 | acyloxyacyl hydrolase activity(GO:0050528) |
| 0.1 | 0.3 | GO:0004833 | tryptophan 2,3-dioxygenase activity(GO:0004833) |
| 0.1 | 0.3 | GO:0004558 | alpha-1,4-glucosidase activity(GO:0004558) |
| 0.1 | 0.2 | GO:0051431 | corticotropin-releasing hormone receptor 2 binding(GO:0051431) |
| 0.1 | 0.2 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.1 | 0.1 | GO:0047023 | androsterone dehydrogenase activity(GO:0047023) |
| 0.1 | 0.2 | GO:0005148 | prolactin receptor binding(GO:0005148) |
| 0.1 | 0.2 | GO:0097259 | tocopherol omega-hydroxylase activity(GO:0052870) alpha-tocopherol omega-hydroxylase activity(GO:0052871) 20-hydroxy-leukotriene B4 omega oxidase activity(GO:0097258) 20-aldehyde-leukotriene B4 20-monooxygenase activity(GO:0097259) |
| 0.1 | 0.3 | GO:0019862 | IgA binding(GO:0019862) |
| 0.0 | 0.2 | GO:0005223 | intracellular cGMP activated cation channel activity(GO:0005223) |
| 0.0 | 0.1 | GO:0050509 | N-acetylglucosaminyl-proteoglycan 4-beta-glucuronosyltransferase activity(GO:0050509) |
| 0.0 | 0.4 | GO:0086006 | voltage-gated sodium channel activity involved in cardiac muscle cell action potential(GO:0086006) |
| 0.0 | 0.3 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 0.0 | 0.2 | GO:0004447 | iodide peroxidase activity(GO:0004447) |
| 0.0 | 0.7 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.0 | 0.1 | GO:0035248 | alpha-1,4-N-acetylgalactosaminyltransferase activity(GO:0035248) |
| 0.0 | 0.2 | GO:0047888 | fatty acid peroxidase activity(GO:0047888) |
| 0.0 | 0.2 | GO:0034988 | Fc-gamma receptor I complex binding(GO:0034988) |
| 0.0 | 0.2 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.0 | 0.2 | GO:0004769 | steroid delta-isomerase activity(GO:0004769) |
| 0.0 | 0.7 | GO:0098505 | G-rich strand telomeric DNA binding(GO:0098505) |
| 0.0 | 0.1 | GO:0003845 | 11-beta-hydroxysteroid dehydrogenase [NAD(P)] activity(GO:0003845) |
| 0.0 | 0.3 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.0 | 0.1 | GO:0004315 | 3-oxoacyl-[acyl-carrier-protein] synthase activity(GO:0004315) |
| 0.0 | 1.3 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.0 | 0.2 | GO:0016211 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.0 | 0.1 | GO:0005137 | interleukin-5 receptor binding(GO:0005137) |
| 0.0 | 1.1 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.0 | 0.2 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.0 | 0.3 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.0 | 0.2 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.0 | 0.1 | GO:0004031 | aldehyde oxidase activity(GO:0004031) |
| 0.0 | 0.8 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 0.3 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.0 | 0.8 | GO:0035173 | histone kinase activity(GO:0035173) |
| 0.0 | 1.3 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.1 | GO:0034041 | sterol-transporting ATPase activity(GO:0034041) |
| 0.0 | 1.0 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.0 | 0.1 | GO:0070643 | vitamin D3 25-hydroxylase activity(GO:0030343) vitamin D 25-hydroxylase activity(GO:0070643) |
| 0.0 | 0.1 | GO:0005287 | high-affinity basic amino acid transmembrane transporter activity(GO:0005287) high-affinity arginine transmembrane transporter activity(GO:0005289) high-affinity lysine transmembrane transporter activity(GO:0005292) |
| 0.0 | 0.1 | GO:0004958 | prostaglandin F receptor activity(GO:0004958) |
| 0.0 | 0.2 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.0 | 0.4 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.0 | 0.3 | GO:0060072 | large conductance calcium-activated potassium channel activity(GO:0060072) |
| 0.0 | 0.6 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.0 | 0.5 | GO:0004000 | adenosine deaminase activity(GO:0004000) |
| 0.0 | 0.1 | GO:0000386 | second spliceosomal transesterification activity(GO:0000386) |
| 0.0 | 0.2 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.0 | 0.1 | GO:0046538 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 0.0 | 0.2 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.0 | 0.3 | GO:0043176 | amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 0.0 | 0.2 | GO:0001165 | RNA polymerase I upstream control element sequence-specific DNA binding(GO:0001165) |
| 0.0 | 0.3 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.0 | 0.4 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.0 | 0.1 | GO:0008466 | glycogenin glucosyltransferase activity(GO:0008466) |
| 0.0 | 0.3 | GO:0022833 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.0 | 0.7 | GO:0031698 | beta-2 adrenergic receptor binding(GO:0031698) |
| 0.0 | 0.8 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 0.1 | GO:0061714 | folic acid receptor activity(GO:0061714) |
| 0.0 | 0.6 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.0 | 0.3 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.0 | 0.1 | GO:0010698 | acetyltransferase activator activity(GO:0010698) |
| 0.0 | 0.1 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.0 | 0.2 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.0 | 0.1 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.0 | 0.7 | GO:0005549 | odorant binding(GO:0005549) |
| 0.0 | 0.9 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 0.2 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.1 | GO:0047631 | ADP-ribose diphosphatase activity(GO:0047631) |
| 0.0 | 0.3 | GO:0032393 | MHC class I receptor activity(GO:0032393) |
| 0.0 | 0.3 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.0 | 0.2 | GO:0019534 | toxin transporter activity(GO:0019534) |
| 0.0 | 0.2 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.0 | 0.3 | GO:0015266 | protein channel activity(GO:0015266) |
| 0.0 | 0.2 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.0 | 0.2 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.0 | 0.1 | GO:0016160 | amylase activity(GO:0016160) |
| 0.0 | 0.2 | GO:0004565 | beta-galactosidase activity(GO:0004565) |
| 0.0 | 0.2 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.1 | GO:0051429 | corticotropin-releasing hormone receptor binding(GO:0051429) corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.0 | 0.4 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 0.1 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.0 | 0.3 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.0 | 0.3 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.0 | 0.1 | GO:0004137 | deoxycytidine kinase activity(GO:0004137) thymidine kinase activity(GO:0004797) |
| 0.0 | 0.2 | GO:0002161 | aminoacyl-tRNA editing activity(GO:0002161) |
| 0.0 | 0.1 | GO:0035241 | protein-arginine omega-N monomethyltransferase activity(GO:0035241) |
| 0.0 | 0.1 | GO:0070735 | protein-glycine ligase activity(GO:0070735) |
| 0.0 | 0.1 | GO:1904288 | BAT3 complex binding(GO:1904288) |
| 0.0 | 0.3 | GO:0008391 | arachidonic acid monooxygenase activity(GO:0008391) arachidonic acid epoxygenase activity(GO:0008392) |
| 0.0 | 0.2 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.0 | 0.1 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.0 | 0.2 | GO:0015194 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.0 | 0.2 | GO:0043138 | 3'-5' DNA helicase activity(GO:0043138) |
| 0.0 | 0.1 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 0.0 | 0.0 | GO:0004582 | dolichyl-phosphate beta-D-mannosyltransferase activity(GO:0004582) |
| 0.0 | 0.1 | GO:0018636 | phenanthrene 9,10-monooxygenase activity(GO:0018636) ketosteroid monooxygenase activity(GO:0047086) |
| 0.0 | 0.0 | GO:0031685 | adenosine receptor binding(GO:0031685) |
| 0.0 | 0.5 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 0.0 | GO:0046848 | hydroxyapatite binding(GO:0046848) |
| 0.0 | 1.0 | GO:0008375 | acetylglucosaminyltransferase activity(GO:0008375) |
| 0.0 | 0.1 | GO:0086075 | gap junction channel activity involved in cardiac conduction electrical coupling(GO:0086075) |
| 0.0 | 1.9 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.0 | 0.2 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.0 | 0.1 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.0 | 0.2 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.0 | 0.5 | GO:0051393 | alpha-actinin binding(GO:0051393) |
| 0.0 | 0.1 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.0 | 0.5 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 0.2 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.0 | 0.4 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.0 | 0.0 | GO:0004583 | dolichyl-phosphate-glucose-glycolipid alpha-glucosyltransferase activity(GO:0004583) |
| 0.0 | 0.2 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) |
| 0.0 | 0.1 | GO:0035381 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.0 | 1.8 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.1 | GO:0008970 | phosphatidylcholine 1-acylhydrolase activity(GO:0008970) |
| 0.0 | 0.1 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.0 | 0.3 | GO:0001056 | RNA polymerase III activity(GO:0001056) |
| 0.0 | 0.1 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.0 | 0.1 | GO:0005221 | intracellular cyclic nucleotide activated cation channel activity(GO:0005221) cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.0 | 0.2 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.0 | 0.6 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 0.1 | GO:0004396 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.0 | 0.1 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.0 | 0.5 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.0 | 0.3 | GO:0000030 | mannosyltransferase activity(GO:0000030) |
| 0.0 | 0.1 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.0 | 0.2 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 0.0 | 0.3 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.0 | 0.2 | GO:0015250 | water channel activity(GO:0015250) |
| 0.0 | 0.1 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.0 | 0.1 | GO:1901702 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.0 | 0.0 | GO:0043734 | DNA-N1-methyladenine dioxygenase activity(GO:0043734) |
| 0.0 | 0.1 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 0.6 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 0.2 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.0 | 0.1 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 0.6 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 0.1 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 0.6 | PID BCR 5PATHWAY | BCR signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | REACTOME APC C CDH1 MEDIATED DEGRADATION OF CDC20 AND OTHER APC C CDH1 TARGETED PROTEINS IN LATE MITOSIS EARLY G1 | Genes involved in APC/C:Cdh1 mediated degradation of Cdc20 and other APC/C:Cdh1 targeted proteins in late mitosis/early G1 |
| 0.1 | 1.7 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.0 | 0.6 | REACTOME RAF MAP KINASE CASCADE | Genes involved in RAF/MAP kinase cascade |
| 0.0 | 0.5 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.0 | 0.5 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.0 | 0.4 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 1.3 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.0 | 0.4 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 2.9 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.0 | 1.2 | REACTOME PHASE1 FUNCTIONALIZATION OF COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |
| 0.0 | 0.3 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.0 | 0.7 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.0 | 0.4 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.0 | 0.5 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.5 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 0.5 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 3 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 3 Promoter |
| 0.0 | 0.2 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.0 | 0.1 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 0.2 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 0.0 | REACTOME TRANSPORT OF RIBONUCLEOPROTEINS INTO THE HOST NUCLEUS | Genes involved in Transport of Ribonucleoproteins into the Host Nucleus |
| 0.0 | 0.5 | REACTOME DEFENSINS | Genes involved in Defensins |
| 0.0 | 0.1 | REACTOME OPSINS | Genes involved in Opsins |
| 0.0 | 0.1 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |