Project
Inflammatory response time course, HUVEC (Wada, 2009)
Navigation
Downloads
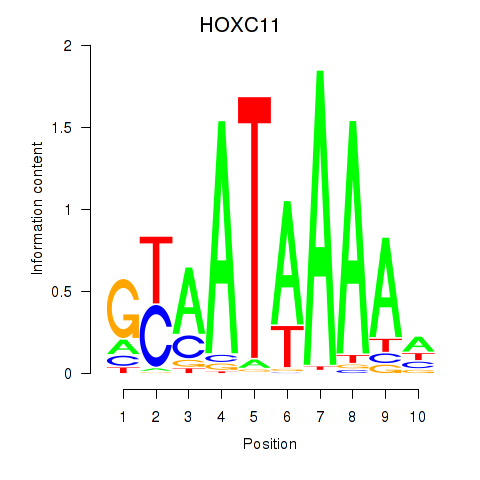
Results for HOXC11
Z-value: 0.41
Transcription factors associated with HOXC11
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HOXC11
|
ENSG00000123388.4 | HOXC11 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HOXC11 | hg38_v1_chr12_+_53973108_53973140 | 0.27 | 2.0e-01 | Click! |
Activity profile of HOXC11 motif
Sorted Z-values of HOXC11 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of HOXC11
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_+_61404076 | 1.21 |
ENST00000357977.5
|
NFIA
|
nuclear factor I A |
| chr18_+_75210789 | 0.86 |
ENST00000580243.3
|
TSHZ1
|
teashirt zinc finger homeobox 1 |
| chr18_+_75210755 | 0.85 |
ENST00000322038.5
|
TSHZ1
|
teashirt zinc finger homeobox 1 |
| chr9_-_122213903 | 0.84 |
ENST00000464484.3
|
LHX6
|
LIM homeobox 6 |
| chr9_-_122213874 | 0.83 |
ENST00000482062.1
|
LHX6
|
LIM homeobox 6 |
| chr5_-_88785493 | 0.78 |
ENST00000503554.4
|
MEF2C
|
myocyte enhancer factor 2C |
| chr4_-_185956652 | 0.71 |
ENST00000355634.9
|
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr4_-_185956348 | 0.62 |
ENST00000431902.5
ENST00000284776.11 ENST00000415274.5 |
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr9_+_2158487 | 0.58 |
ENST00000634706.1
ENST00000634338.1 ENST00000635688.1 ENST00000634435.1 |
SMARCA2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr7_+_22727147 | 0.56 |
ENST00000426291.5
ENST00000401651.5 ENST00000258743.10 ENST00000407492.5 ENST00000401630.7 ENST00000406575.1 |
IL6
|
interleukin 6 |
| chr17_-_48613468 | 0.55 |
ENST00000498634.2
|
HOXB8
|
homeobox B8 |
| chr9_-_122227525 | 0.54 |
ENST00000373755.6
ENST00000373754.6 |
LHX6
|
LIM homeobox 6 |
| chr5_-_111756245 | 0.52 |
ENST00000447165.6
|
NREP
|
neuronal regeneration related protein |
| chr12_-_95790755 | 0.51 |
ENST00000343702.9
ENST00000344911.8 |
NTN4
|
netrin 4 |
| chr21_-_30497160 | 0.50 |
ENST00000334058.3
|
KRTAP19-4
|
keratin associated protein 19-4 |
| chr8_+_76683779 | 0.50 |
ENST00000523885.2
|
ZFHX4
|
zinc finger homeobox 4 |
| chr22_+_35066136 | 0.43 |
ENST00000308700.6
ENST00000404699.7 |
ISX
|
intestine specific homeobox |
| chr7_-_11832190 | 0.43 |
ENST00000423059.9
ENST00000617773.1 |
THSD7A
|
thrombospondin type 1 domain containing 7A |
| chr18_-_48950960 | 0.43 |
ENST00000262158.8
|
SMAD7
|
SMAD family member 7 |
| chr15_+_66702219 | 0.41 |
ENST00000288840.10
|
SMAD6
|
SMAD family member 6 |
| chr7_-_20786879 | 0.39 |
ENST00000418710.3
ENST00000617581.4 ENST00000361443.4 |
SP8
|
Sp8 transcription factor |
| chr9_-_20622479 | 0.37 |
ENST00000380338.9
|
MLLT3
|
MLLT3 super elongation complex subunit |
| chr6_+_15246054 | 0.37 |
ENST00000341776.7
|
JARID2
|
jumonji and AT-rich interaction domain containing 2 |
| chr5_-_88823763 | 0.35 |
ENST00000635898.1
ENST00000626391.2 ENST00000628656.2 |
MEF2C
|
myocyte enhancer factor 2C |
| chr17_-_41489907 | 0.35 |
ENST00000328119.11
|
KRT36
|
keratin 36 |
| chr17_+_7558296 | 0.33 |
ENST00000438470.5
ENST00000436057.5 |
TNFSF13
|
TNF superfamily member 13 |
| chr8_+_38974212 | 0.32 |
ENST00000302495.5
|
HTRA4
|
HtrA serine peptidase 4 |
| chr7_-_27165517 | 0.32 |
ENST00000396345.1
ENST00000343483.7 |
HOXA9
|
homeobox A9 |
| chr1_+_66533575 | 0.30 |
ENST00000684751.1
ENST00000683291.1 ENST00000682054.1 ENST00000435165.3 ENST00000684539.1 ENST00000681971.1 ENST00000682476.1 ENST00000684168.1 ENST00000371039.5 |
SGIP1
|
SH3GL interacting endocytic adaptor 1 |
| chrX_-_10833643 | 0.27 |
ENST00000380785.5
ENST00000380787.5 |
MID1
|
midline 1 |
| chr12_+_25052512 | 0.27 |
ENST00000557489.5
ENST00000354454.7 ENST00000536173.5 |
IRAG2
|
inositol 1,4,5-triphosphate receptor associated 2 |
| chr1_+_92168915 | 0.27 |
ENST00000637221.2
|
BTBD8
|
BTB domain containing 8 |
| chr6_-_31777319 | 0.25 |
ENST00000375688.5
|
VWA7
|
von Willebrand factor A domain containing 7 |
| chr1_-_9069797 | 0.23 |
ENST00000473209.1
|
SLC2A5
|
solute carrier family 2 member 5 |
| chr16_+_30378312 | 0.23 |
ENST00000528032.5
ENST00000622647.3 |
ZNF48
|
zinc finger protein 48 |
| chr1_-_93681829 | 0.23 |
ENST00000260502.11
|
BCAR3
|
BCAR3 adaptor protein, NSP family member |
| chr6_-_166168700 | 0.23 |
ENST00000366871.7
|
TBXT
|
T-box transcription factor T |
| chr7_-_27152561 | 0.22 |
ENST00000467897.2
ENST00000612286.5 ENST00000498652.1 |
ENSG00000273433.1
HOXA3
ENSG00000272801.1
|
novel transcript homeobox A3 novel transcript |
| chr5_+_160229499 | 0.21 |
ENST00000402432.4
|
FABP6
|
fatty acid binding protein 6 |
| chr6_-_166168612 | 0.21 |
ENST00000296946.6
|
TBXT
|
T-box transcription factor T |
| chr2_-_55334529 | 0.21 |
ENST00000645860.1
ENST00000642563.1 ENST00000647396.1 |
CCDC88A
|
coiled-coil domain containing 88A |
| chr19_-_19515542 | 0.21 |
ENST00000585580.4
|
TSSK6
|
testis specific serine kinase 6 |
| chr17_+_81395469 | 0.20 |
ENST00000584436.7
|
BAHCC1
|
BAH domain and coiled-coil containing 1 |
| chr4_-_48780242 | 0.20 |
ENST00000507711.5
ENST00000358350.9 |
FRYL
|
FRY like transcription coactivator |
| chr15_-_43879835 | 0.20 |
ENST00000636859.1
|
FRMD5
|
FERM domain containing 5 |
| chr17_+_81395449 | 0.19 |
ENST00000675386.2
|
BAHCC1
|
BAH domain and coiled-coil containing 1 |
| chr11_+_34642468 | 0.19 |
ENST00000527935.1
|
EHF
|
ETS homologous factor |
| chr1_-_204166334 | 0.19 |
ENST00000272190.9
|
REN
|
renin |
| chr19_-_51082883 | 0.18 |
ENST00000650543.2
|
KLK14
|
kallikrein related peptidase 14 |
| chr22_+_40177917 | 0.18 |
ENST00000454349.7
ENST00000335727.13 |
TNRC6B
|
trinucleotide repeat containing adaptor 6B |
| chr10_+_61901678 | 0.18 |
ENST00000644638.1
ENST00000681100.1 ENST00000279873.12 |
ARID5B
|
AT-rich interaction domain 5B |
| chr3_+_148827800 | 0.17 |
ENST00000282957.9
ENST00000468341.1 |
CPB1
|
carboxypeptidase B1 |
| chr4_-_84499281 | 0.17 |
ENST00000295886.5
|
NKX6-1
|
NK6 homeobox 1 |
| chr12_+_130162456 | 0.17 |
ENST00000539839.1
ENST00000229030.5 |
FZD10
|
frizzled class receptor 10 |
| chr2_+_108588453 | 0.16 |
ENST00000393310.5
|
LIMS1
|
LIM zinc finger domain containing 1 |
| chr19_-_14090963 | 0.16 |
ENST00000269724.5
|
SAMD1
|
sterile alpha motif domain containing 1 |
| chr8_+_26293112 | 0.15 |
ENST00000523925.5
ENST00000315985.7 |
PPP2R2A
|
protein phosphatase 2 regulatory subunit Balpha |
| chr12_+_25052634 | 0.15 |
ENST00000548766.5
|
IRAG2
|
inositol 1,4,5-triphosphate receptor associated 2 |
| chr1_+_26159071 | 0.15 |
ENST00000374268.5
|
FAM110D
|
family with sequence similarity 110 member D |
| chr4_+_143433491 | 0.15 |
ENST00000512843.1
|
GAB1
|
GRB2 associated binding protein 1 |
| chr12_-_70754631 | 0.15 |
ENST00000440835.6
ENST00000549308.5 ENST00000550661.1 ENST00000378778.5 |
PTPRR
|
protein tyrosine phosphatase receptor type R |
| chr18_-_55321640 | 0.14 |
ENST00000637169.2
|
TCF4
|
transcription factor 4 |
| chr1_+_32362537 | 0.14 |
ENST00000373534.4
|
TSSK3
|
testis specific serine kinase 3 |
| chr9_+_79573162 | 0.14 |
ENST00000425506.5
|
TLE4
|
TLE family member 4, transcriptional corepressor |
| chr19_-_35742431 | 0.13 |
ENST00000592537.5
ENST00000246532.6 ENST00000588992.5 |
IGFLR1
|
IGF like family receptor 1 |
| chr19_-_14090695 | 0.13 |
ENST00000533683.7
|
SAMD1
|
sterile alpha motif domain containing 1 |
| chr19_+_49591170 | 0.13 |
ENST00000418929.7
|
PRR12
|
proline rich 12 |
| chr12_-_95116967 | 0.13 |
ENST00000551521.5
|
FGD6
|
FYVE, RhoGEF and PH domain containing 6 |
| chr3_-_20012250 | 0.13 |
ENST00000389050.5
|
PP2D1
|
protein phosphatase 2C like domain containing 1 |
| chr2_+_27217361 | 0.13 |
ENST00000264705.9
ENST00000403525.5 |
CAD
|
carbamoyl-phosphate synthetase 2, aspartate transcarbamylase, and dihydroorotase |
| chr18_+_32190015 | 0.13 |
ENST00000581447.1
|
MEP1B
|
meprin A subunit beta |
| chrX_+_108439866 | 0.13 |
ENST00000361603.7
|
COL4A5
|
collagen type IV alpha 5 chain |
| chr4_-_162163989 | 0.13 |
ENST00000306100.10
ENST00000427802.2 |
FSTL5
|
follistatin like 5 |
| chr6_+_156776020 | 0.13 |
ENST00000346085.10
|
ARID1B
|
AT-rich interaction domain 1B |
| chrX_-_19799751 | 0.13 |
ENST00000379698.8
|
SH3KBP1
|
SH3 domain containing kinase binding protein 1 |
| chr18_-_55321986 | 0.12 |
ENST00000570287.6
|
TCF4
|
transcription factor 4 |
| chr18_+_32190033 | 0.12 |
ENST00000269202.11
|
MEP1B
|
meprin A subunit beta |
| chr11_+_34642366 | 0.12 |
ENST00000532302.1
|
EHF
|
ETS homologous factor |
| chr13_-_46052712 | 0.12 |
ENST00000242848.8
ENST00000679008.1 ENST00000282007.7 |
ZC3H13
|
zinc finger CCCH-type containing 13 |
| chr4_-_25863537 | 0.12 |
ENST00000502949.5
ENST00000264868.9 ENST00000513691.1 ENST00000514872.1 |
SEL1L3
|
SEL1L family member 3 |
| chr11_-_62706304 | 0.12 |
ENST00000278893.11
ENST00000407022.7 ENST00000421906.5 |
BSCL2
|
BSCL2 lipid droplet biogenesis associated, seipin |
| chrX_+_108439779 | 0.12 |
ENST00000328300.11
|
COL4A5
|
collagen type IV alpha 5 chain |
| chr17_-_41528293 | 0.12 |
ENST00000455635.1
ENST00000361566.7 |
KRT19
|
keratin 19 |
| chr4_-_162163955 | 0.12 |
ENST00000379164.8
|
FSTL5
|
follistatin like 5 |
| chr18_-_55351977 | 0.12 |
ENST00000643689.1
|
TCF4
|
transcription factor 4 |
| chr5_+_102808057 | 0.12 |
ENST00000684043.1
ENST00000682407.1 |
PAM
|
peptidylglycine alpha-amidating monooxygenase |
| chr6_+_157036315 | 0.11 |
ENST00000637904.1
|
ARID1B
|
AT-rich interaction domain 1B |
| chr7_-_123199960 | 0.11 |
ENST00000194130.7
|
SLC13A1
|
solute carrier family 13 member 1 |
| chr1_-_241999091 | 0.11 |
ENST00000357246.4
|
MAP1LC3C
|
microtubule associated protein 1 light chain 3 gamma |
| chr20_+_34977625 | 0.11 |
ENST00000618182.6
|
MYH7B
|
myosin heavy chain 7B |
| chr3_-_98035295 | 0.11 |
ENST00000621172.4
|
GABRR3
|
gamma-aminobutyric acid type A receptor subunit rho3 |
| chr6_+_151690492 | 0.11 |
ENST00000404742.5
ENST00000440973.5 |
ESR1
|
estrogen receptor 1 |
| chr21_+_37367097 | 0.11 |
ENST00000644942.1
|
DYRK1A
|
dual specificity tyrosine phosphorylation regulated kinase 1A |
| chr12_-_53677397 | 0.11 |
ENST00000338662.6
ENST00000552242.5 |
ATP5MC2
|
ATP synthase membrane subunit c locus 2 |
| chr12_-_27971970 | 0.10 |
ENST00000395872.5
ENST00000201015.8 |
PTHLH
|
parathyroid hormone like hormone |
| chr19_+_4402615 | 0.10 |
ENST00000301280.10
|
CHAF1A
|
chromatin assembly factor 1 subunit A |
| chr11_-_62706234 | 0.10 |
ENST00000403550.5
|
BSCL2
|
BSCL2 lipid droplet biogenesis associated, seipin |
| chr5_-_34043205 | 0.10 |
ENST00000382065.8
ENST00000231338.7 |
C1QTNF3
|
C1q and TNF related 3 |
| chr7_-_81770122 | 0.10 |
ENST00000423064.7
|
HGF
|
hepatocyte growth factor |
| chr18_-_55322215 | 0.10 |
ENST00000457482.7
|
TCF4
|
transcription factor 4 |
| chr8_-_56211257 | 0.09 |
ENST00000316981.8
ENST00000423799.6 ENST00000429357.2 |
PLAG1
|
PLAG1 zinc finger |
| chr13_-_29850605 | 0.09 |
ENST00000380680.5
|
UBL3
|
ubiquitin like 3 |
| chr3_+_15003912 | 0.09 |
ENST00000406272.6
ENST00000617312.4 |
NR2C2
|
nuclear receptor subfamily 2 group C member 2 |
| chr7_+_129144691 | 0.09 |
ENST00000486685.3
|
TSPAN33
|
tetraspanin 33 |
| chr20_-_57710539 | 0.09 |
ENST00000395816.7
ENST00000347215.8 |
PMEPA1
|
prostate transmembrane protein, androgen induced 1 |
| chr8_+_69466617 | 0.09 |
ENST00000525061.5
ENST00000260128.8 ENST00000458141.6 |
SULF1
|
sulfatase 1 |
| chr18_+_3252267 | 0.08 |
ENST00000536605.1
ENST00000580887.5 |
MYL12A
|
myosin light chain 12A |
| chr9_+_12693327 | 0.08 |
ENST00000388918.10
|
TYRP1
|
tyrosinase related protein 1 |
| chr2_+_210556590 | 0.08 |
ENST00000233072.10
ENST00000619804.1 |
CPS1
|
carbamoyl-phosphate synthase 1 |
| chr7_-_93890160 | 0.08 |
ENST00000451238.1
|
TFPI2
|
tissue factor pathway inhibitor 2 |
| chr11_+_64234569 | 0.08 |
ENST00000309422.7
ENST00000426086.3 |
VEGFB
|
vascular endothelial growth factor B |
| chr3_+_101827982 | 0.08 |
ENST00000461724.5
ENST00000483180.5 ENST00000394054.6 |
NFKBIZ
|
NFKB inhibitor zeta |
| chr2_+_147845020 | 0.07 |
ENST00000241416.12
|
ACVR2A
|
activin A receptor type 2A |
| chr12_+_55549602 | 0.07 |
ENST00000641569.1
ENST00000641851.1 |
OR6C4
|
olfactory receptor family 6 subfamily C member 4 |
| chr2_+_190649062 | 0.07 |
ENST00000409581.5
ENST00000337386.10 |
NAB1
|
NGFI-A binding protein 1 |
| chr16_+_7303245 | 0.07 |
ENST00000674626.1
|
RBFOX1
|
RNA binding fox-1 homolog 1 |
| chr2_-_50347789 | 0.07 |
ENST00000628364.2
|
NRXN1
|
neurexin 1 |
| chr11_+_118530990 | 0.07 |
ENST00000411589.6
ENST00000359862.8 ENST00000442938.6 |
TMEM25
|
transmembrane protein 25 |
| chr2_-_40512361 | 0.07 |
ENST00000403092.5
|
SLC8A1
|
solute carrier family 8 member A1 |
| chr1_+_23019415 | 0.07 |
ENST00000465864.2
ENST00000356634.7 ENST00000400181.9 |
KDM1A
|
lysine demethylase 1A |
| chr19_-_45424364 | 0.07 |
ENST00000589165.5
|
ERCC1
|
ERCC excision repair 1, endonuclease non-catalytic subunit |
| chr16_+_14711689 | 0.07 |
ENST00000552140.5
|
NPIPA3
|
nuclear pore complex interacting protein family member A3 |
| chr2_+_209579399 | 0.07 |
ENST00000360351.8
|
MAP2
|
microtubule associated protein 2 |
| chr5_+_36606355 | 0.06 |
ENST00000681909.1
ENST00000513903.5 ENST00000681795.1 ENST00000680125.1 ENST00000612708.5 ENST00000680232.1 ENST00000681776.1 ENST00000681926.1 ENST00000679958.1 ENST00000265113.9 ENST00000504121.5 ENST00000512374.1 ENST00000613445.5 ENST00000679983.1 |
SLC1A3
|
solute carrier family 1 member 3 |
| chr8_+_75539862 | 0.06 |
ENST00000396423.4
|
HNF4G
|
hepatocyte nuclear factor 4 gamma |
| chr4_-_173530219 | 0.06 |
ENST00000359562.4
|
HAND2
|
heart and neural crest derivatives expressed 2 |
| chr17_+_59940908 | 0.06 |
ENST00000591035.1
|
ENSG00000267318.1
|
novel protein |
| chr2_-_55917699 | 0.06 |
ENST00000634374.1
|
EFEMP1
|
EGF containing fibulin extracellular matrix protein 1 |
| chr6_+_26156323 | 0.06 |
ENST00000304218.6
|
H1-4
|
H1.4 linker histone, cluster member |
| chr16_-_15378294 | 0.05 |
ENST00000360151.9
ENST00000543801.5 |
NPIPA5
|
nuclear pore complex interacting protein family member A5 |
| chr17_-_40665121 | 0.05 |
ENST00000394052.5
|
KRT222
|
keratin 222 |
| chr4_+_113292838 | 0.05 |
ENST00000672411.1
ENST00000673231.1 |
ANK2
|
ankyrin 2 |
| chr6_+_46793379 | 0.05 |
ENST00000230588.9
ENST00000611727.2 |
MEP1A
|
meprin A subunit alpha |
| chr7_-_108569453 | 0.05 |
ENST00000313516.5
|
THAP5
|
THAP domain containing 5 |
| chrX_-_108439472 | 0.05 |
ENST00000372216.8
|
COL4A6
|
collagen type IV alpha 6 chain |
| chr21_-_42350987 | 0.05 |
ENST00000291526.5
|
TFF2
|
trefoil factor 2 |
| chrX_-_134658450 | 0.05 |
ENST00000359237.9
|
PLAC1
|
placenta enriched 1 |
| chr12_+_25052732 | 0.05 |
ENST00000547044.5
|
IRAG2
|
inositol 1,4,5-triphosphate receptor associated 2 |
| chr11_-_5516690 | 0.04 |
ENST00000380184.2
|
UBQLNL
|
ubiquilin like |
| chr8_+_75539893 | 0.04 |
ENST00000674002.1
|
HNF4G
|
hepatocyte nuclear factor 4 gamma |
| chr20_+_4721901 | 0.04 |
ENST00000305817.3
|
PRND
|
prion like protein doppel |
| chr2_+_209579598 | 0.04 |
ENST00000445941.5
ENST00000673860.1 |
MAP2
|
microtubule associated protein 2 |
| chr12_+_1691011 | 0.04 |
ENST00000357103.5
|
ADIPOR2
|
adiponectin receptor 2 |
| chrX_+_66164210 | 0.04 |
ENST00000343002.7
ENST00000336279.9 |
HEPH
|
hephaestin |
| chr7_-_41703062 | 0.04 |
ENST00000242208.5
|
INHBA
|
inhibin subunit beta A |
| chr16_+_14937443 | 0.04 |
ENST00000328085.10
|
NPIPA1
|
nuclear pore complex interacting protein family member A1 |
| chr6_-_43053832 | 0.04 |
ENST00000265348.9
ENST00000674134.1 ENST00000674100.1 |
CUL7
|
cullin 7 |
| chr4_+_113292925 | 0.04 |
ENST00000673353.1
ENST00000505342.6 ENST00000672915.1 ENST00000509550.5 |
ANK2
|
ankyrin 2 |
| chr7_-_126533850 | 0.03 |
ENST00000444921.3
|
GRM8
|
glutamate metabotropic receptor 8 |
| chr16_+_16379055 | 0.03 |
ENST00000530217.2
|
NPIPA7
|
nuclear pore complex interacting protein family member A7 |
| chr8_-_21812320 | 0.03 |
ENST00000517328.5
|
GFRA2
|
GDNF family receptor alpha 2 |
| chr5_-_83720813 | 0.03 |
ENST00000515590.1
ENST00000274341.9 |
HAPLN1
|
hyaluronan and proteoglycan link protein 1 |
| chr1_-_9069608 | 0.03 |
ENST00000377424.9
|
SLC2A5
|
solute carrier family 2 member 5 |
| chr6_-_63319943 | 0.03 |
ENST00000622415.1
ENST00000370658.9 ENST00000485906.6 ENST00000370657.9 |
LGSN
|
lengsin, lens protein with glutamine synthetase domain |
| chr15_+_36594868 | 0.03 |
ENST00000566807.5
ENST00000643612.1 ENST00000567389.5 ENST00000562877.5 |
CDIN1
|
CDAN1 interacting nuclease 1 |
| chr4_-_73223082 | 0.03 |
ENST00000509867.6
|
ANKRD17
|
ankyrin repeat domain 17 |
| chr6_+_6588708 | 0.03 |
ENST00000230568.5
|
LY86
|
lymphocyte antigen 86 |
| chr4_+_119135825 | 0.03 |
ENST00000307128.6
|
MYOZ2
|
myozenin 2 |
| chrX_+_66164340 | 0.03 |
ENST00000441993.7
ENST00000419594.6 ENST00000425114.2 |
HEPH
|
hephaestin |
| chr12_-_23949642 | 0.03 |
ENST00000537393.5
ENST00000451604.7 ENST00000381381.6 |
SOX5
|
SRY-box transcription factor 5 |
| chr9_+_92974476 | 0.03 |
ENST00000337352.10
|
FGD3
|
FYVE, RhoGEF and PH domain containing 3 |
| chrM_+_4467 | 0.03 |
ENST00000361453.3
|
MT-ND2
|
mitochondrially encoded NADH:ubiquinone oxidoreductase core subunit 2 |
| chr1_-_9069572 | 0.03 |
ENST00000377414.7
|
SLC2A5
|
solute carrier family 2 member 5 |
| chr1_+_151766655 | 0.02 |
ENST00000400999.7
|
OAZ3
|
ornithine decarboxylase antizyme 3 |
| chr8_+_26577843 | 0.02 |
ENST00000311151.9
|
DPYSL2
|
dihydropyrimidinase like 2 |
| chr15_-_53733103 | 0.02 |
ENST00000559418.5
|
WDR72
|
WD repeat domain 72 |
| chr14_+_55661272 | 0.02 |
ENST00000555573.5
|
KTN1
|
kinectin 1 |
| chr4_-_99144238 | 0.02 |
ENST00000512499.5
ENST00000504125.1 ENST00000505590.5 ENST00000629236.2 ENST00000508393.5 ENST00000265512.12 |
ADH4
|
alcohol dehydrogenase 4 (class II), pi polypeptide |
| chr6_+_12290353 | 0.02 |
ENST00000379375.6
|
EDN1
|
endothelin 1 |
| chr2_-_175181663 | 0.02 |
ENST00000392541.3
ENST00000284727.9 ENST00000409194.5 |
ATP5MC3
|
ATP synthase membrane subunit c locus 3 |
| chr1_+_110451132 | 0.02 |
ENST00000271331.4
|
PROK1
|
prokineticin 1 |
| chr21_+_37366894 | 0.01 |
ENST00000647504.1
|
DYRK1A
|
dual specificity tyrosine phosphorylation regulated kinase 1A |
| chr10_+_113854610 | 0.01 |
ENST00000369301.3
|
NHLRC2
|
NHL repeat containing 2 |
| chr2_-_40512423 | 0.01 |
ENST00000402441.5
ENST00000448531.1 |
SLC8A1
|
solute carrier family 8 member A1 |
| chr8_-_18808837 | 0.01 |
ENST00000286485.12
|
PSD3
|
pleckstrin and Sec7 domain containing 3 |
| chr17_-_48728705 | 0.01 |
ENST00000290295.8
|
HOXB13
|
homeobox B13 |
| chr10_+_17229267 | 0.01 |
ENST00000224237.9
|
VIM
|
vimentin |
| chr19_-_39433703 | 0.00 |
ENST00000602153.5
|
RPS16
|
ribosomal protein S16 |
| chr19_+_926001 | 0.00 |
ENST00000263620.8
|
ARID3A
|
AT-rich interaction domain 3A |
| chr14_-_60486021 | 0.00 |
ENST00000555476.5
ENST00000321731.8 |
C14orf39
|
chromosome 14 open reading frame 39 |
| chr3_-_165078480 | 0.00 |
ENST00000264382.8
|
SI
|
sucrase-isomaltase |
| chr8_-_113436883 | 0.00 |
ENST00000455883.2
ENST00000297405.10 |
CSMD3
|
CUB and Sushi multiple domains 3 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.7 | GO:0060023 | soft palate development(GO:0060023) |
| 0.2 | 2.2 | GO:1904936 | cerebral cortex GABAergic interneuron migration(GO:0021853) interneuron migration(GO:1904936) |
| 0.1 | 1.1 | GO:0003172 | primary heart field specification(GO:0003138) sinoatrial valve development(GO:0003172) sinoatrial valve morphogenesis(GO:0003185) |
| 0.1 | 0.6 | GO:1990637 | hepatic immune response(GO:0002384) response to prolactin(GO:1990637) |
| 0.1 | 0.4 | GO:1904479 | negative regulation of intestinal absorption(GO:1904479) |
| 0.1 | 0.5 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
| 0.1 | 0.4 | GO:0007352 | zygotic specification of dorsal/ventral axis(GO:0007352) |
| 0.1 | 0.2 | GO:0021913 | glandular epithelial cell maturation(GO:0002071) regulation of transcription from RNA polymerase II promoter involved in ventral spinal cord interneuron specification(GO:0021913) positive regulation of type B pancreatic cell development(GO:2000078) |
| 0.1 | 0.2 | GO:0010159 | specification of organ position(GO:0010159) |
| 0.1 | 0.4 | GO:2000320 | regulation of ventricular cardiac muscle cell membrane depolarization(GO:0060373) negative regulation of T-helper 17 type immune response(GO:2000317) negative regulation of T-helper 17 cell differentiation(GO:2000320) |
| 0.0 | 0.3 | GO:0097319 | fructose transport(GO:0015755) fructose import(GO:0032445) carbohydrate import into cell(GO:0097319) carbohydrate import across plasma membrane(GO:0098704) fructose import across plasma membrane(GO:1990539) |
| 0.0 | 0.1 | GO:0044205 | 'de novo' UMP biosynthetic process(GO:0044205) |
| 0.0 | 1.2 | GO:0072189 | ureter development(GO:0072189) |
| 0.0 | 0.2 | GO:0002018 | renin-angiotensin regulation of aldosterone production(GO:0002018) |
| 0.0 | 1.3 | GO:0061049 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.0 | 0.2 | GO:1903566 | positive regulation of protein localization to cilium(GO:1903566) |
| 0.0 | 0.2 | GO:0097026 | dendritic cell dendrite assembly(GO:0097026) |
| 0.0 | 0.1 | GO:0018032 | protein amidation(GO:0018032) |
| 0.0 | 0.1 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) |
| 0.0 | 0.1 | GO:0070408 | carbamoyl phosphate metabolic process(GO:0070408) carbamoyl phosphate biosynthetic process(GO:0070409) response to ammonia(GO:1903717) cellular response to ammonia(GO:1903718) |
| 0.0 | 0.4 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.0 | 0.2 | GO:0038031 | non-canonical Wnt signaling pathway via JNK cascade(GO:0038031) |
| 0.0 | 0.1 | GO:1903385 | regulation of homophilic cell adhesion(GO:1903385) |
| 0.0 | 0.4 | GO:0051574 | positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.0 | 0.3 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) |
| 0.0 | 0.6 | GO:0021516 | dorsal spinal cord development(GO:0021516) |
| 0.0 | 0.3 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.0 | 0.4 | GO:0031507 | heterochromatin assembly(GO:0031507) |
| 0.0 | 0.3 | GO:0007320 | insemination(GO:0007320) |
| 0.0 | 0.1 | GO:0060750 | epithelial cell proliferation involved in mammary gland duct elongation(GO:0060750) |
| 0.0 | 0.2 | GO:0035092 | sperm chromatin condensation(GO:0035092) |
| 0.0 | 0.2 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) |
| 0.0 | 0.1 | GO:0033184 | positive regulation of histone ubiquitination(GO:0033184) |
| 0.0 | 0.0 | GO:0060279 | positive regulation of ovulation(GO:0060279) |
| 0.0 | 0.2 | GO:0048298 | positive regulation of isotype switching to IgA isotypes(GO:0048298) |
| 0.0 | 0.1 | GO:0006335 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 | 0.1 | GO:0061032 | cardiac right ventricle formation(GO:0003219) norepinephrine biosynthetic process(GO:0042421) visceral serous pericardium development(GO:0061032) |
| 0.0 | 0.1 | GO:0000720 | pyrimidine dimer repair by nucleotide-excision repair(GO:0000720) |
| 0.0 | 0.3 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.0 | 0.1 | GO:0070162 | adiponectin secretion(GO:0070162) regulation of adiponectin secretion(GO:0070163) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.0 | 0.3 | GO:0005587 | collagen type IV trimer(GO:0005587) |
| 0.0 | 0.4 | GO:0005677 | chromatin silencing complex(GO:0005677) |
| 0.0 | 0.1 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.0 | 0.1 | GO:1990357 | terminal web(GO:1990357) |
| 0.0 | 0.8 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 0.0 | 0.1 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.0 | 0.4 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 0.1 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.0 | 0.0 | GO:0043512 | inhibin complex(GO:0043511) inhibin A complex(GO:0043512) |
| 0.0 | 0.1 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.0 | 0.3 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.2 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.1 | 0.5 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.1 | 0.6 | GO:0005138 | interleukin-6 receptor binding(GO:0005138) |
| 0.1 | 0.2 | GO:0004088 | carbamoyl-phosphate synthase (ammonia) activity(GO:0004087) carbamoyl-phosphate synthase (glutamine-hydrolyzing) activity(GO:0004088) |
| 0.0 | 0.8 | GO:0034713 | type I transforming growth factor beta receptor binding(GO:0034713) |
| 0.0 | 0.3 | GO:0005353 | fructose transmembrane transporter activity(GO:0005353) |
| 0.0 | 1.3 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.1 | GO:0004504 | peptidylglycine monooxygenase activity(GO:0004504) peptidylamidoglycolate lyase activity(GO:0004598) |
| 0.0 | 0.5 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.0 | 0.1 | GO:0001093 | TFIIB-class transcription factor binding(GO:0001093) |
| 0.0 | 0.3 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.0 | 0.1 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 0.1 | GO:0061752 | telomeric repeat-containing RNA binding(GO:0061752) |
| 0.0 | 0.1 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.0 | 0.0 | GO:0055100 | adiponectin binding(GO:0055100) |
| 0.0 | 0.1 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.0 | 0.2 | GO:0032052 | bile acid binding(GO:0032052) |
| 0.0 | 0.1 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.0 | 0.5 | GO:0032452 | histone demethylase activity(GO:0032452) |
| 0.0 | 0.3 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | ST STAT3 PATHWAY | STAT3 Pathway |
| 0.0 | 1.4 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.0 | 0.8 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 1.0 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 0.5 | PID ALK1 PATHWAY | ALK1 signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 1.1 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.0 | 0.1 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.0 | 0.3 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 0.5 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |