Project
Inflammatory response time course, HUVEC (Wada, 2009)
Navigation
Downloads
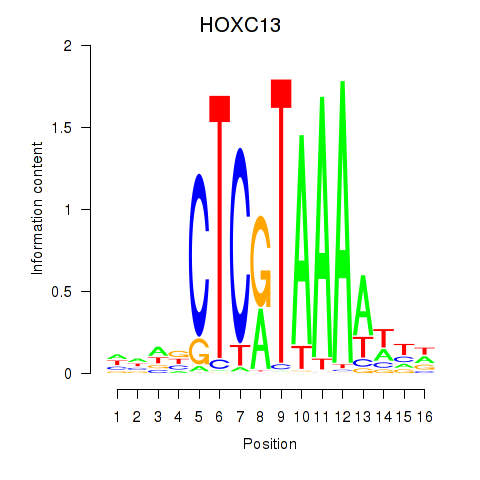
Results for HOXC13
Z-value: 0.58
Transcription factors associated with HOXC13
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HOXC13
|
ENSG00000123364.5 | HOXC13 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HOXC13 | hg38_v1_chr12_+_53938824_53938852 | 0.06 | 7.8e-01 | Click! |
Activity profile of HOXC13 motif
Sorted Z-values of HOXC13 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of HOXC13
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_150235972 | 1.54 |
ENST00000534220.1
|
ANP32E
|
acidic nuclear phosphoprotein 32 family member E |
| chr2_-_224947030 | 1.53 |
ENST00000409592.7
|
DOCK10
|
dedicator of cytokinesis 10 |
| chr6_+_127577168 | 1.46 |
ENST00000329722.8
|
C6orf58
|
chromosome 6 open reading frame 58 |
| chr3_-_123620496 | 1.24 |
ENST00000578202.1
|
MYLK
|
myosin light chain kinase |
| chr3_-_123620571 | 1.23 |
ENST00000583087.5
|
MYLK
|
myosin light chain kinase |
| chr6_-_11807045 | 1.08 |
ENST00000379415.6
|
ADTRP
|
androgen dependent TFPI regulating protein |
| chr11_-_5154757 | 1.01 |
ENST00000380367.3
|
OR52A1
|
olfactory receptor family 52 subfamily A member 1 |
| chr7_-_93148345 | 0.97 |
ENST00000437805.5
ENST00000446959.5 ENST00000439952.5 ENST00000414791.5 ENST00000446033.1 ENST00000411955.5 ENST00000318238.9 |
SAMD9L
|
sterile alpha motif domain containing 9 like |
| chr4_+_20251896 | 0.90 |
ENST00000504154.6
|
SLIT2
|
slit guidance ligand 2 |
| chr3_+_190615308 | 0.84 |
ENST00000412080.1
|
IL1RAP
|
interleukin 1 receptor accessory protein |
| chr3_+_155083889 | 0.83 |
ENST00000680282.1
|
MME
|
membrane metalloendopeptidase |
| chr15_+_70936487 | 0.76 |
ENST00000558456.5
ENST00000560158.6 ENST00000558808.5 ENST00000559806.5 ENST00000559069.1 |
LRRC49
|
leucine rich repeat containing 49 |
| chr1_-_89126066 | 0.75 |
ENST00000370466.4
|
GBP2
|
guanylate binding protein 2 |
| chr2_+_102418642 | 0.73 |
ENST00000264260.6
|
IL18RAP
|
interleukin 18 receptor accessory protein |
| chr3_+_155083523 | 0.67 |
ENST00000680057.1
|
MME
|
membrane metalloendopeptidase |
| chr2_+_167187364 | 0.65 |
ENST00000672671.1
|
XIRP2
|
xin actin binding repeat containing 2 |
| chr18_+_63887698 | 0.63 |
ENST00000457692.5
ENST00000299502.9 ENST00000413956.5 |
SERPINB2
|
serpin family B member 2 |
| chr2_+_161160299 | 0.63 |
ENST00000440506.5
ENST00000429217.5 ENST00000406287.5 ENST00000402568.5 |
TANK
|
TRAF family member associated NFKB activator |
| chr7_-_41703062 | 0.62 |
ENST00000242208.5
|
INHBA
|
inhibin subunit beta A |
| chr2_+_102337148 | 0.61 |
ENST00000311734.6
ENST00000409584.5 |
IL1RL1
|
interleukin 1 receptor like 1 |
| chr15_+_70892809 | 0.58 |
ENST00000260382.10
ENST00000560755.5 |
LRRC49
|
leucine rich repeat containing 49 |
| chr4_+_94757921 | 0.57 |
ENST00000515059.6
|
BMPR1B
|
bone morphogenetic protein receptor type 1B |
| chr22_+_18150162 | 0.56 |
ENST00000215794.8
|
USP18
|
ubiquitin specific peptidase 18 |
| chr5_-_59276109 | 0.54 |
ENST00000503258.5
|
PDE4D
|
phosphodiesterase 4D |
| chr15_+_70892443 | 0.54 |
ENST00000443425.6
ENST00000560369.5 |
LRRC49
|
leucine rich repeat containing 49 |
| chr1_-_150235943 | 0.54 |
ENST00000533654.5
|
ANP32E
|
acidic nuclear phosphoprotein 32 family member E |
| chr11_+_118304721 | 0.51 |
ENST00000361763.9
|
CD3E
|
CD3e molecule |
| chr1_-_150235995 | 0.51 |
ENST00000436748.6
|
ANP32E
|
acidic nuclear phosphoprotein 32 family member E |
| chr17_-_29930062 | 0.51 |
ENST00000579954.1
ENST00000269033.7 ENST00000540801.6 ENST00000590153.1 ENST00000582084.1 |
SSH2
|
slingshot protein phosphatase 2 |
| chr18_+_58196736 | 0.47 |
ENST00000675221.1
|
NEDD4L
|
NEDD4 like E3 ubiquitin protein ligase |
| chr9_-_2844058 | 0.44 |
ENST00000397885.3
|
PUM3
|
pumilio RNA binding family member 3 |
| chr7_-_16833411 | 0.44 |
ENST00000412973.1
|
AGR2
|
anterior gradient 2, protein disulphide isomerase family member |
| chr12_+_45216128 | 0.44 |
ENST00000681156.1
|
ANO6
|
anoctamin 6 |
| chr12_+_45216079 | 0.43 |
ENST00000423947.7
ENST00000680498.1 ENST00000320560.13 |
ANO6
|
anoctamin 6 |
| chr12_+_45216014 | 0.43 |
ENST00000425752.6
|
ANO6
|
anoctamin 6 |
| chr19_+_21082140 | 0.42 |
ENST00000616183.1
ENST00000596053.5 |
ZNF714
|
zinc finger protein 714 |
| chr14_+_73167829 | 0.41 |
ENST00000406768.1
|
PSEN1
|
presenilin 1 |
| chr10_-_77637902 | 0.39 |
ENST00000286627.10
ENST00000639486.1 ENST00000640523.1 |
KCNMA1
|
potassium calcium-activated channel subfamily M alpha 1 |
| chr1_-_150236064 | 0.39 |
ENST00000532744.2
ENST00000369114.9 ENST00000369115.3 ENST00000583931.6 |
ANP32E
|
acidic nuclear phosphoprotein 32 family member E |
| chr2_+_161136901 | 0.39 |
ENST00000259075.6
ENST00000432002.5 |
TANK
|
TRAF family member associated NFKB activator |
| chr4_+_121801311 | 0.37 |
ENST00000379663.7
ENST00000243498.10 ENST00000509800.5 |
EXOSC9
|
exosome component 9 |
| chr18_-_5544250 | 0.37 |
ENST00000540638.6
ENST00000342933.7 |
EPB41L3
|
erythrocyte membrane protein band 4.1 like 3 |
| chr5_-_161546970 | 0.36 |
ENST00000675303.1
|
GABRB2
|
gamma-aminobutyric acid type A receptor subunit beta2 |
| chr1_-_150236150 | 0.36 |
ENST00000629042.2
|
ANP32E
|
acidic nuclear phosphoprotein 32 family member E |
| chr8_-_27992624 | 0.35 |
ENST00000524352.5
|
SCARA5
|
scavenger receptor class A member 5 |
| chr1_+_213051229 | 0.35 |
ENST00000615329.4
ENST00000543354.5 ENST00000614059.4 ENST00000543470.5 ENST00000366960.8 ENST00000366959.4 |
RPS6KC1
|
ribosomal protein S6 kinase C1 |
| chr6_+_150721073 | 0.35 |
ENST00000358517.6
|
PLEKHG1
|
pleckstrin homology and RhoGEF domain containing G1 |
| chr11_+_62123991 | 0.35 |
ENST00000533896.5
ENST00000278849.4 ENST00000394818.8 |
INCENP
|
inner centromere protein |
| chr5_-_161546671 | 0.34 |
ENST00000517547.5
|
GABRB2
|
gamma-aminobutyric acid type A receptor subunit beta2 |
| chr17_-_41369807 | 0.34 |
ENST00000251646.8
|
KRT33B
|
keratin 33B |
| chr15_-_58279245 | 0.34 |
ENST00000558231.5
|
ALDH1A2
|
aldehyde dehydrogenase 1 family member A2 |
| chr12_-_16600703 | 0.34 |
ENST00000616247.4
|
LMO3
|
LIM domain only 3 |
| chr7_+_134866831 | 0.34 |
ENST00000435928.1
|
CALD1
|
caldesmon 1 |
| chr2_+_161160420 | 0.34 |
ENST00000392749.7
ENST00000405852.5 |
TANK
|
TRAF family member associated NFKB activator |
| chr5_+_169583636 | 0.33 |
ENST00000506574.5
ENST00000515224.5 ENST00000629457.2 ENST00000508247.5 ENST00000265295.9 ENST00000513941.5 |
SPDL1
|
spindle apparatus coiled-coil protein 1 |
| chr1_-_154608705 | 0.33 |
ENST00000649749.1
ENST00000648871.1 |
ADAR
|
adenosine deaminase RNA specific |
| chr19_+_3762665 | 0.32 |
ENST00000330133.5
|
MRPL54
|
mitochondrial ribosomal protein L54 |
| chr7_+_23710203 | 0.32 |
ENST00000422637.5
ENST00000355870.8 |
STK31
|
serine/threonine kinase 31 |
| chr19_+_21082224 | 0.31 |
ENST00000620627.1
|
ZNF714
|
zinc finger protein 714 |
| chr19_+_57128643 | 0.31 |
ENST00000598197.1
|
USP29
|
ubiquitin specific peptidase 29 |
| chr19_+_39655907 | 0.30 |
ENST00000392051.4
|
LGALS16
|
galectin 16 |
| chr7_+_23710326 | 0.30 |
ENST00000354639.7
ENST00000531170.5 ENST00000444333.2 |
STK31
|
serine/threonine kinase 31 |
| chr11_-_7830840 | 0.30 |
ENST00000641167.1
|
OR5P3
|
olfactory receptor family 5 subfamily P member 3 |
| chr11_+_30231000 | 0.30 |
ENST00000254122.8
ENST00000533718.2 ENST00000417547.1 |
FSHB
|
follicle stimulating hormone subunit beta |
| chr12_-_10998304 | 0.30 |
ENST00000538986.2
|
TAS2R20
|
taste 2 receptor member 20 |
| chr3_+_100709382 | 0.30 |
ENST00000620299.5
|
TFG
|
trafficking from ER to golgi regulator |
| chr3_+_100709473 | 0.29 |
ENST00000240851.9
ENST00000675243.1 ENST00000676111.1 ENST00000476228.5 ENST00000463568.6 ENST00000676395.1 |
TFG
|
trafficking from ER to golgi regulator |
| chr16_-_18896874 | 0.28 |
ENST00000565324.5
ENST00000561947.5 |
SMG1
|
SMG1 nonsense mediated mRNA decay associated PI3K related kinase |
| chr20_+_38317627 | 0.28 |
ENST00000417318.3
|
BPI
|
bactericidal permeability increasing protein |
| chr8_+_117135259 | 0.27 |
ENST00000519688.5
|
SLC30A8
|
solute carrier family 30 member 8 |
| chr7_-_30026617 | 0.27 |
ENST00000222803.10
|
FKBP14
|
FKBP prolyl isomerase 14 |
| chr10_-_77637789 | 0.26 |
ENST00000481070.1
ENST00000640969.1 ENST00000286628.14 ENST00000638991.1 ENST00000639913.1 ENST00000480683.2 |
KCNMA1
|
potassium calcium-activated channel subfamily M alpha 1 |
| chr1_+_50105666 | 0.26 |
ENST00000651347.1
|
ELAVL4
|
ELAV like RNA binding protein 4 |
| chr12_-_11134644 | 0.26 |
ENST00000539585.1
|
TAS2R30
|
taste 2 receptor member 30 |
| chr4_-_69961007 | 0.26 |
ENST00000353151.3
|
CSN2
|
casein beta |
| chr6_+_87407965 | 0.25 |
ENST00000369562.9
|
CFAP206
|
cilia and flagella associated protein 206 |
| chr3_-_71130557 | 0.25 |
ENST00000497355.7
|
FOXP1
|
forkhead box P1 |
| chr10_-_77637721 | 0.25 |
ENST00000638848.1
ENST00000639406.1 ENST00000618048.2 ENST00000639120.1 ENST00000640834.1 ENST00000639601.1 ENST00000638514.1 ENST00000457953.6 ENST00000639090.1 ENST00000639489.1 ENST00000372440.6 ENST00000404771.8 ENST00000638203.1 ENST00000638306.1 ENST00000638351.1 ENST00000638606.1 ENST00000639591.1 ENST00000640182.1 ENST00000640605.1 ENST00000640141.1 |
KCNMA1
|
potassium calcium-activated channel subfamily M alpha 1 |
| chr3_+_100709344 | 0.25 |
ENST00000418917.7
ENST00000675553.1 |
TFG
|
trafficking from ER to golgi regulator |
| chr1_+_50106265 | 0.25 |
ENST00000357083.8
|
ELAVL4
|
ELAV like RNA binding protein 4 |
| chr12_-_52814106 | 0.25 |
ENST00000551956.2
|
KRT4
|
keratin 4 |
| chr3_+_100709290 | 0.24 |
ENST00000675047.1
ENST00000490574.6 ENST00000675543.1 ENST00000676308.1 ENST00000675499.1 |
TFG
|
trafficking from ER to golgi regulator |
| chr2_-_189179754 | 0.24 |
ENST00000374866.9
ENST00000618828.1 |
COL5A2
|
collagen type V alpha 2 chain |
| chr12_+_130162456 | 0.24 |
ENST00000539839.1
ENST00000229030.5 |
FZD10
|
frizzled class receptor 10 |
| chr2_+_101252867 | 0.23 |
ENST00000289382.8
|
CNOT11
|
CCR4-NOT transcription complex subunit 11 |
| chr19_+_37594830 | 0.23 |
ENST00000589117.5
|
ZNF540
|
zinc finger protein 540 |
| chr3_+_101779425 | 0.23 |
ENST00000491511.6
|
NXPE3
|
neurexophilin and PC-esterase domain family member 3 |
| chr22_+_22758698 | 0.22 |
ENST00000390312.2
|
IGLV2-14
|
immunoglobulin lambda variable 2-14 |
| chr2_+_240560343 | 0.22 |
ENST00000405954.2
|
DUSP28
|
dual specificity phosphatase 28 |
| chr1_+_109114097 | 0.22 |
ENST00000457623.6
ENST00000369939.8 ENST00000529753.5 |
ELAPOR1
|
endosome-lysosome associated apoptosis and autophagy regulator 1 |
| chr4_-_68670648 | 0.22 |
ENST00000338206.6
|
UGT2B15
|
UDP glucuronosyltransferase family 2 member B15 |
| chr11_+_111245725 | 0.21 |
ENST00000280325.7
|
C11orf53
|
chromosome 11 open reading frame 53 |
| chr6_+_63521738 | 0.20 |
ENST00000648894.1
ENST00000639568.2 |
PTP4A1
|
protein tyrosine phosphatase 4A1 |
| chr20_+_45791978 | 0.20 |
ENST00000449078.5
ENST00000456939.5 |
DNTTIP1
|
deoxynucleotidyltransferase terminal interacting protein 1 |
| chr2_+_106063234 | 0.20 |
ENST00000409944.5
|
ECRG4
|
ECRG4 augurin precursor |
| chr22_-_32412207 | 0.19 |
ENST00000216038.6
|
RTCB
|
RNA 2',3'-cyclic phosphate and 5'-OH ligase |
| chr1_-_31919368 | 0.19 |
ENST00000457805.6
ENST00000602725.5 ENST00000679970.1 |
PTP4A2
ENSG00000288678.1
|
protein tyrosine phosphatase 4A2 novel protein |
| chr12_-_52434363 | 0.19 |
ENST00000252245.6
|
KRT75
|
keratin 75 |
| chr20_+_45791930 | 0.18 |
ENST00000372622.8
|
DNTTIP1
|
deoxynucleotidyltransferase terminal interacting protein 1 |
| chr14_+_96256194 | 0.18 |
ENST00000216629.11
ENST00000553356.1 |
BDKRB1
|
bradykinin receptor B1 |
| chr6_-_36547400 | 0.18 |
ENST00000229812.8
|
STK38
|
serine/threonine kinase 38 |
| chr10_+_17951906 | 0.18 |
ENST00000377371.3
ENST00000377369.7 |
SLC39A12
|
solute carrier family 39 member 12 |
| chr8_-_109680812 | 0.18 |
ENST00000528716.5
ENST00000527600.5 ENST00000531230.5 ENST00000532189.5 ENST00000534184.5 ENST00000408889.7 ENST00000533171.5 |
SYBU
|
syntabulin |
| chr5_-_79991237 | 0.18 |
ENST00000512560.5
ENST00000509852.6 ENST00000512528.3 ENST00000617335.4 |
MTX3
|
metaxin 3 |
| chr1_+_174159501 | 0.18 |
ENST00000251507.8
ENST00000681986.1 |
RABGAP1L
|
RAB GTPase activating protein 1 like |
| chr9_-_26892732 | 0.18 |
ENST00000333916.8
ENST00000520187.1 |
CAAP1
|
caspase activity and apoptosis inhibitor 1 |
| chr5_-_161546708 | 0.18 |
ENST00000393959.6
|
GABRB2
|
gamma-aminobutyric acid type A receptor subunit beta2 |
| chr4_-_82848843 | 0.18 |
ENST00000511338.1
|
SEC31A
|
SEC31 homolog A, COPII coat complex component |
| chr6_-_32816910 | 0.17 |
ENST00000447394.1
ENST00000438763.7 |
HLA-DOB
|
major histocompatibility complex, class II, DO beta |
| chr17_-_58692021 | 0.17 |
ENST00000240361.12
|
TEX14
|
testis expressed 14, intercellular bridge forming factor |
| chr16_-_72172135 | 0.17 |
ENST00000537465.5
ENST00000237353.15 |
PMFBP1
|
polyamine modulated factor 1 binding protein 1 |
| chr1_+_206684887 | 0.17 |
ENST00000367103.4
ENST00000294981.8 |
MAPKAPK2
|
MAPK activated protein kinase 2 |
| chr2_-_151289613 | 0.17 |
ENST00000243346.10
|
NMI
|
N-myc and STAT interactor |
| chr10_+_17951885 | 0.17 |
ENST00000377374.8
|
SLC39A12
|
solute carrier family 39 member 12 |
| chr15_-_89334794 | 0.17 |
ENST00000268124.11
ENST00000442287.6 |
POLG
|
DNA polymerase gamma, catalytic subunit |
| chr17_-_58692032 | 0.17 |
ENST00000349033.10
ENST00000389934.7 |
TEX14
|
testis expressed 14, intercellular bridge forming factor |
| chr17_-_41382298 | 0.17 |
ENST00000394001.3
|
KRT34
|
keratin 34 |
| chr19_+_8842593 | 0.17 |
ENST00000305625.2
|
MBD3L1
|
methyl-CpG binding domain protein 3 like 1 |
| chr1_+_34176900 | 0.16 |
ENST00000488417.2
|
C1orf94
|
chromosome 1 open reading frame 94 |
| chr10_+_17951825 | 0.16 |
ENST00000539911.5
|
SLC39A12
|
solute carrier family 39 member 12 |
| chr17_-_2511875 | 0.16 |
ENST00000263092.11
ENST00000576976.2 |
METTL16
|
methyltransferase like 16 |
| chr16_-_20352707 | 0.16 |
ENST00000396134.6
ENST00000573567.5 ENST00000570757.5 ENST00000396138.9 ENST00000571174.5 ENST00000576688.2 |
UMOD
|
uromodulin |
| chr1_+_174875505 | 0.16 |
ENST00000486220.5
|
RABGAP1L
|
RAB GTPase activating protein 1 like |
| chr19_-_20565769 | 0.16 |
ENST00000427401.9
|
ZNF737
|
zinc finger protein 737 |
| chr9_-_92482499 | 0.16 |
ENST00000375544.7
|
ASPN
|
asporin |
| chr6_+_133953210 | 0.16 |
ENST00000367869.1
ENST00000237264.9 ENST00000674115.1 |
TBPL1
ENSG00000288529.1
|
TATA-box binding protein like 1 novel protein |
| chr1_-_46941464 | 0.15 |
ENST00000462347.5
ENST00000371905.1 ENST00000310638.9 |
CYP4A11
|
cytochrome P450 family 4 subfamily A member 11 |
| chr20_+_45792498 | 0.15 |
ENST00000415790.5
|
DNTTIP1
|
deoxynucleotidyltransferase terminal interacting protein 1 |
| chr22_-_39532722 | 0.15 |
ENST00000334678.8
|
RPS19BP1
|
ribosomal protein S19 binding protein 1 |
| chr7_+_106865474 | 0.15 |
ENST00000359195.3
|
PIK3CG
|
phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit gamma |
| chr19_-_35228699 | 0.15 |
ENST00000324675.3
|
FAM187B
|
family with sequence similarity 187 member B |
| chr1_-_149861210 | 0.14 |
ENST00000579512.2
|
H4C15
|
H4 clustered histone 15 |
| chr14_-_67412112 | 0.14 |
ENST00000216446.9
|
PLEK2
|
pleckstrin 2 |
| chr6_-_85644043 | 0.14 |
ENST00000678930.1
ENST00000678355.1 |
SYNCRIP
|
synaptotagmin binding cytoplasmic RNA interacting protein |
| chr9_-_111330224 | 0.14 |
ENST00000302681.3
|
OR2K2
|
olfactory receptor family 2 subfamily K member 2 |
| chr17_-_3592877 | 0.13 |
ENST00000399756.8
|
TRPV1
|
transient receptor potential cation channel subfamily V member 1 |
| chr1_+_149477960 | 0.13 |
ENST00000369227.7
|
NBPF19
|
NBPF member 19 |
| chr20_+_38962299 | 0.13 |
ENST00000373325.6
ENST00000373323.8 ENST00000252011.8 ENST00000615559.1 |
DHX35
ENSG00000277581.1
|
DEAH-box helicase 35 novel transcript, sense intronic to DHX35 |
| chr15_+_78873723 | 0.13 |
ENST00000559690.5
ENST00000559158.5 |
MORF4L1
|
mortality factor 4 like 1 |
| chrX_+_49922605 | 0.13 |
ENST00000376088.7
|
CLCN5
|
chloride voltage-gated channel 5 |
| chr2_-_37157093 | 0.13 |
ENST00000681507.1
|
EIF2AK2
|
eukaryotic translation initiation factor 2 alpha kinase 2 |
| chr1_-_244862381 | 0.13 |
ENST00000640001.1
ENST00000639628.1 |
HNRNPU
|
heterogeneous nuclear ribonucleoprotein U |
| chr2_-_37156942 | 0.13 |
ENST00000680273.1
ENST00000233057.9 ENST00000679979.1 ENST00000679507.1 ENST00000681463.1 ENST00000395127.6 |
EIF2AK2
|
eukaryotic translation initiation factor 2 alpha kinase 2 |
| chr9_-_32526185 | 0.13 |
ENST00000379883.3
ENST00000379868.6 ENST00000679859.1 |
DDX58
|
DExD/H-box helicase 58 |
| chr22_+_39520553 | 0.12 |
ENST00000674920.3
ENST00000679776.1 ENST00000675582.2 ENST00000337304.2 ENST00000676346.2 ENST00000396680.3 ENST00000680446.1 ENST00000674568.2 ENST00000680748.1 ENST00000674835.2 |
ATF4
|
activating transcription factor 4 |
| chr10_-_67696115 | 0.12 |
ENST00000433211.7
|
CTNNA3
|
catenin alpha 3 |
| chr2_-_86105839 | 0.12 |
ENST00000263857.11
|
POLR1A
|
RNA polymerase I subunit A |
| chr5_+_111073309 | 0.12 |
ENST00000379706.4
|
TSLP
|
thymic stromal lymphopoietin |
| chr19_+_21082190 | 0.12 |
ENST00000618422.1
ENST00000618008.4 ENST00000425625.5 ENST00000456283.7 |
ZNF714
|
zinc finger protein 714 |
| chr4_+_69280472 | 0.12 |
ENST00000335568.10
ENST00000511240.1 |
UGT2B28
|
UDP glucuronosyltransferase family 2 member B28 |
| chr1_-_85708382 | 0.12 |
ENST00000370574.4
ENST00000431532.6 |
ZNHIT6
|
zinc finger HIT-type containing 6 |
| chr17_+_50373214 | 0.12 |
ENST00000393271.6
ENST00000338165.9 ENST00000511519.6 |
EME1
|
essential meiotic structure-specific endonuclease 1 |
| chr4_-_154612635 | 0.11 |
ENST00000407946.5
ENST00000405164.5 ENST00000336098.8 ENST00000393846.6 ENST00000404648.7 ENST00000443553.5 |
FGG
|
fibrinogen gamma chain |
| chr12_-_54295748 | 0.11 |
ENST00000540264.2
ENST00000312156.8 |
NFE2
|
nuclear factor, erythroid 2 |
| chr10_+_24449426 | 0.11 |
ENST00000307544.10
|
KIAA1217
|
KIAA1217 |
| chr19_-_6737565 | 0.11 |
ENST00000601716.1
ENST00000264080.11 |
GPR108
|
G protein-coupled receptor 108 |
| chr11_+_89924064 | 0.11 |
ENST00000623787.3
|
TRIM49D2
|
tripartite motif containing 49D2 |
| chr3_+_98463201 | 0.11 |
ENST00000642057.1
|
OR5K1
|
olfactory receptor family 5 subfamily K member 1 |
| chr12_-_7751605 | 0.11 |
ENST00000354629.9
|
CLEC4C
|
C-type lectin domain family 4 member C |
| chr12_+_96489569 | 0.11 |
ENST00000524981.9
|
CFAP54
|
cilia and flagella associated protein 54 |
| chr7_+_57450171 | 0.11 |
ENST00000420713.2
|
ZNF716
|
zinc finger protein 716 |
| chr16_-_84617547 | 0.11 |
ENST00000567786.2
|
COTL1
|
coactosin like F-actin binding protein 1 |
| chr1_+_99970430 | 0.11 |
ENST00000370153.6
|
SLC35A3
|
solute carrier family 35 member A3 |
| chr11_-_114559866 | 0.10 |
ENST00000534921.2
|
NXPE1
|
neurexophilin and PC-esterase domain family member 1 |
| chr19_+_3762705 | 0.10 |
ENST00000589174.1
|
MRPL54
|
mitochondrial ribosomal protein L54 |
| chr6_-_76072654 | 0.10 |
ENST00000369950.8
ENST00000611179.4 ENST00000369963.5 |
IMPG1
|
interphotoreceptor matrix proteoglycan 1 |
| chr17_-_41489907 | 0.10 |
ENST00000328119.11
|
KRT36
|
keratin 36 |
| chr1_+_67307360 | 0.10 |
ENST00000262345.5
ENST00000544434.5 |
IL12RB2
|
interleukin 12 receptor subunit beta 2 |
| chrX_+_49922559 | 0.10 |
ENST00000376091.8
|
CLCN5
|
chloride voltage-gated channel 5 |
| chr2_+_61145068 | 0.10 |
ENST00000426997.5
ENST00000464909.2 ENST00000489686.5 |
C2orf74
|
chromosome 2 open reading frame 74 |
| chr4_+_87799546 | 0.10 |
ENST00000226284.7
|
IBSP
|
integrin binding sialoprotein |
| chr9_+_114611206 | 0.10 |
ENST00000374049.4
ENST00000288502.9 |
TMEM268
|
transmembrane protein 268 |
| chr10_+_45972482 | 0.10 |
ENST00000580018.4
|
TIMM23
|
translocase of inner mitochondrial membrane 23 |
| chr8_-_108787563 | 0.09 |
ENST00000297459.4
|
TMEM74
|
transmembrane protein 74 |
| chr12_-_4538440 | 0.09 |
ENST00000261250.8
ENST00000541014.5 ENST00000545746.5 ENST00000542080.5 |
C12orf4
|
chromosome 12 open reading frame 4 |
| chr10_-_67838019 | 0.09 |
ENST00000483798.6
|
DNAJC12
|
DnaJ heat shock protein family (Hsp40) member C12 |
| chr2_-_222656067 | 0.09 |
ENST00000281828.8
|
FARSB
|
phenylalanyl-tRNA synthetase subunit beta |
| chr4_+_118034480 | 0.09 |
ENST00000296499.6
|
NDST3
|
N-deacetylase and N-sulfotransferase 3 |
| chr5_+_57175803 | 0.09 |
ENST00000511209.5
|
GPBP1
|
GC-rich promoter binding protein 1 |
| chr6_-_52803807 | 0.09 |
ENST00000334575.6
|
GSTA1
|
glutathione S-transferase alpha 1 |
| chr7_+_23680130 | 0.09 |
ENST00000409192.7
ENST00000409653.5 ENST00000409994.3 ENST00000344962.9 |
FAM221A
|
family with sequence similarity 221 member A |
| chr1_-_109113818 | 0.09 |
ENST00000369949.8
|
C1orf194
|
chromosome 1 open reading frame 194 |
| chr2_+_191245185 | 0.09 |
ENST00000418908.5
ENST00000339514.8 ENST00000392318.8 |
MYO1B
|
myosin IB |
| chr22_+_17628828 | 0.09 |
ENST00000399782.5
|
BCL2L13
|
BCL2 like 13 |
| chr9_+_68780029 | 0.09 |
ENST00000394264.7
|
PABIR1
|
PP2A Aalpha (PPP2R1A) and B55A (PPP2R2A) interacting phosphatase regulator 1 |
| chr12_+_69825273 | 0.09 |
ENST00000547771.6
|
MYRFL
|
myelin regulatory factor like |
| chr1_-_111563934 | 0.09 |
ENST00000443498.5
|
TMIGD3
|
transmembrane and immunoglobulin domain containing 3 |
| chr11_-_114559847 | 0.08 |
ENST00000251921.6
|
NXPE1
|
neurexophilin and PC-esterase domain family member 1 |
| chr12_-_118359639 | 0.08 |
ENST00000541786.5
ENST00000419821.6 ENST00000541878.5 |
TAOK3
|
TAO kinase 3 |
| chr12_+_64780465 | 0.08 |
ENST00000542120.6
|
TBC1D30
|
TBC1 domain family member 30 |
| chr17_-_10469558 | 0.08 |
ENST00000255381.2
|
MYH4
|
myosin heavy chain 4 |
| chr10_-_77638369 | 0.08 |
ENST00000372443.6
|
KCNMA1
|
potassium calcium-activated channel subfamily M alpha 1 |
| chr1_+_149832647 | 0.08 |
ENST00000578186.2
|
H4C14
|
H4 clustered histone 14 |
| chr6_-_127900958 | 0.07 |
ENST00000434358.3
ENST00000630369.2 ENST00000368248.4 |
THEMIS
|
thymocyte selection associated |
| chrX_+_114584037 | 0.07 |
ENST00000371951.5
ENST00000371950.3 ENST00000276198.6 |
HTR2C
|
5-hydroxytryptamine receptor 2C |
| chr5_-_65624288 | 0.07 |
ENST00000381018.7
ENST00000274327.11 ENST00000231524.14 |
TRIM23
|
tripartite motif containing 23 |
| chr14_-_67533720 | 0.07 |
ENST00000554278.6
|
TMEM229B
|
transmembrane protein 229B |
| chr4_+_71062642 | 0.07 |
ENST00000649996.1
|
SLC4A4
|
solute carrier family 4 member 4 |
| chr1_+_219173861 | 0.07 |
ENST00000366927.3
ENST00000366928.10 |
LYPLAL1
|
lysophospholipase like 1 |
| chr12_+_69825221 | 0.07 |
ENST00000552032.7
|
MYRFL
|
myelin regulatory factor like |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 2.5 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.3 | 1.3 | GO:0097045 | phosphatidylserine exposure on blood platelet(GO:0097045) |
| 0.3 | 0.9 | GO:0071676 | negative regulation of mononuclear cell migration(GO:0071676) |
| 0.2 | 1.3 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 0.2 | 1.5 | GO:0038172 | interleukin-33-mediated signaling pathway(GO:0038172) |
| 0.2 | 0.6 | GO:0060279 | positive regulation of ovulation(GO:0060279) |
| 0.1 | 1.5 | GO:0071492 | cellular response to UV-A(GO:0071492) |
| 0.1 | 0.5 | GO:0002913 | positive regulation of T cell anergy(GO:0002669) positive regulation of lymphocyte anergy(GO:0002913) |
| 0.1 | 0.4 | GO:0034476 | U1 snRNA 3'-end processing(GO:0034473) U5 snRNA 3'-end processing(GO:0034476) |
| 0.1 | 1.5 | GO:0002315 | marginal zone B cell differentiation(GO:0002315) |
| 0.1 | 0.4 | GO:0060480 | lung goblet cell differentiation(GO:0060480) |
| 0.1 | 0.3 | GO:1903487 | regulation of lactation(GO:1903487) |
| 0.1 | 0.4 | GO:0016080 | synaptic vesicle targeting(GO:0016080) |
| 0.1 | 0.6 | GO:0048165 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) negative regulation of chondrocyte proliferation(GO:1902731) |
| 0.1 | 1.0 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.1 | 0.2 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.1 | 1.1 | GO:0060083 | smooth muscle contraction involved in micturition(GO:0060083) |
| 0.1 | 0.2 | GO:0002581 | negative regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002581) |
| 0.1 | 0.1 | GO:2000097 | regulation of smooth muscle cell-matrix adhesion(GO:2000097) |
| 0.1 | 0.2 | GO:1902524 | positive regulation of protein K48-linked ubiquitination(GO:1902524) |
| 0.1 | 0.3 | GO:1900369 | negative regulation of RNA interference(GO:1900369) |
| 0.1 | 0.5 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.1 | 3.3 | GO:0043486 | histone exchange(GO:0043486) |
| 0.1 | 0.2 | GO:0072233 | thick ascending limb development(GO:0072023) metanephric thick ascending limb development(GO:0072233) |
| 0.0 | 0.3 | GO:2000324 | positive regulation of glucocorticoid receptor signaling pathway(GO:2000324) |
| 0.0 | 0.4 | GO:0010835 | regulation of protein ADP-ribosylation(GO:0010835) |
| 0.0 | 0.3 | GO:0060011 | progesterone biosynthetic process(GO:0006701) Sertoli cell proliferation(GO:0060011) |
| 0.0 | 0.4 | GO:0002175 | protein localization to paranode region of axon(GO:0002175) |
| 0.0 | 0.1 | GO:0061394 | regulation of transcription from RNA polymerase II promoter in response to arsenic-containing substance(GO:0061394) |
| 0.0 | 0.5 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.0 | 0.1 | GO:0072429 | response to intra-S DNA damage checkpoint signaling(GO:0072429) |
| 0.0 | 0.3 | GO:0042905 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.0 | 0.3 | GO:0052564 | response to immune response of other organism involved in symbiotic interaction(GO:0052564) response to host immune response(GO:0052572) |
| 0.0 | 0.2 | GO:0070171 | negative regulation of tooth mineralization(GO:0070171) |
| 0.0 | 0.1 | GO:1990569 | UDP-N-acetylglucosamine transport(GO:0015788) UDP-N-acetylglucosamine transmembrane transport(GO:1990569) |
| 0.0 | 0.3 | GO:0032466 | negative regulation of cytokinesis(GO:0032466) |
| 0.0 | 0.9 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.0 | 0.5 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.0 | 0.2 | GO:0038031 | non-canonical Wnt signaling pathway via JNK cascade(GO:0038031) |
| 0.0 | 0.2 | GO:0006287 | base-excision repair, gap-filling(GO:0006287) |
| 0.0 | 0.3 | GO:0034501 | protein localization to kinetochore(GO:0034501) |
| 0.0 | 0.1 | GO:0003095 | pressure natriuresis(GO:0003095) |
| 0.0 | 0.1 | GO:0034344 | type III interferon production(GO:0034343) regulation of type III interferon production(GO:0034344) |
| 0.0 | 0.1 | GO:0043397 | corticotropin-releasing hormone secretion(GO:0043396) regulation of corticotropin-releasing hormone secretion(GO:0043397) |
| 0.0 | 0.3 | GO:0061088 | regulation of sequestering of zinc ion(GO:0061088) |
| 0.0 | 0.2 | GO:0009212 | dTTP biosynthetic process(GO:0006235) pyrimidine deoxyribonucleoside triphosphate biosynthetic process(GO:0009212) |
| 0.0 | 0.7 | GO:0042730 | fibrinolysis(GO:0042730) |
| 0.0 | 0.1 | GO:1904749 | regulation of protein localization to nucleolus(GO:1904749) |
| 0.0 | 0.1 | GO:0003218 | cardiac left ventricle formation(GO:0003218) |
| 0.0 | 0.3 | GO:0070234 | positive regulation of T cell apoptotic process(GO:0070234) |
| 0.0 | 0.3 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.0 | 0.2 | GO:0080009 | rRNA base methylation(GO:0070475) mRNA methylation(GO:0080009) |
| 0.0 | 0.1 | GO:0052405 | negative regulation by host of symbiont molecular function(GO:0052405) |
| 0.0 | 0.1 | GO:0048254 | snoRNA localization(GO:0048254) |
| 0.0 | 0.1 | GO:0010897 | negative regulation of triglyceride catabolic process(GO:0010897) secretory granule localization(GO:0032252) |
| 0.0 | 0.1 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.0 | 0.1 | GO:0086073 | bundle of His cell-Purkinje myocyte adhesion involved in cell communication(GO:0086073) |
| 0.0 | 0.3 | GO:0032625 | interleukin-21 production(GO:0032625) interleukin-21 secretion(GO:0072619) |
| 0.0 | 0.6 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.0 | 0.4 | GO:0034755 | iron ion transmembrane transport(GO:0034755) |
| 0.0 | 0.1 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 | 0.2 | GO:0042535 | positive regulation of tumor necrosis factor biosynthetic process(GO:0042535) |
| 0.0 | 0.3 | GO:0032717 | negative regulation of interleukin-8 production(GO:0032717) |
| 0.0 | 0.2 | GO:0052695 | cellular glucuronidation(GO:0052695) |
| 0.0 | 0.2 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0043512 | inhibin complex(GO:0043511) inhibin A complex(GO:0043512) |
| 0.2 | 3.3 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.1 | 0.3 | GO:0097545 | radial spoke(GO:0001534) axonemal outer doublet(GO:0097545) |
| 0.1 | 0.2 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.1 | 0.2 | GO:0005760 | gamma DNA polymerase complex(GO:0005760) |
| 0.1 | 0.4 | GO:0000801 | central element(GO:0000801) |
| 0.0 | 0.3 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.0 | 0.5 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.0 | 0.4 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.0 | 1.2 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.0 | 0.1 | GO:1990590 | ATF1-ATF4 transcription factor complex(GO:1990590) |
| 0.0 | 0.6 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.0 | 2.5 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 0.4 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.0 | 0.1 | GO:0005944 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) |
| 0.0 | 0.1 | GO:0009328 | phenylalanine-tRNA ligase complex(GO:0009328) |
| 0.0 | 0.3 | GO:0030478 | actin cap(GO:0030478) |
| 0.0 | 0.2 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.0 | 0.9 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.0 | 0.3 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.0 | 1.3 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 0.2 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.0 | 0.4 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.0 | 0.3 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.0 | 0.0 | GO:0005753 | mitochondrial proton-transporting ATP synthase complex(GO:0005753) proton-transporting ATP synthase complex(GO:0045259) |
| 0.0 | 0.1 | GO:0070761 | pre-snoRNP complex(GO:0070761) |
| 0.0 | 0.0 | GO:0044609 | DBIRD complex(GO:0044609) |
| 0.0 | 0.2 | GO:0097433 | dense body(GO:0097433) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.1 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.2 | 1.3 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.2 | 1.5 | GO:0002114 | interleukin-33 receptor activity(GO:0002114) |
| 0.1 | 0.9 | GO:0048495 | laminin-1 binding(GO:0043237) Roundabout binding(GO:0048495) |
| 0.1 | 1.0 | GO:0060072 | large conductance calcium-activated potassium channel activity(GO:0060072) |
| 0.1 | 0.9 | GO:0005237 | inhibitory extracellular ligand-gated ion channel activity(GO:0005237) |
| 0.1 | 0.6 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.1 | 0.3 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.1 | 0.3 | GO:0030395 | lactose binding(GO:0030395) |
| 0.1 | 1.3 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.1 | 0.4 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.1 | 0.2 | GO:0008988 | rRNA (adenine-N6-)-methyltransferase activity(GO:0008988) mRNA (2'-O-methyladenosine-N6-)-methyltransferase activity(GO:0016422) |
| 0.0 | 0.6 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 0.3 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.0 | 3.3 | GO:0019212 | phosphatase inhibitor activity(GO:0019212) |
| 0.0 | 0.3 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.0 | 0.1 | GO:0005462 | UDP-N-acetylglucosamine transmembrane transporter activity(GO:0005462) |
| 0.0 | 0.3 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.0 | 0.3 | GO:0042608 | T cell receptor binding(GO:0042608) |
| 0.0 | 0.6 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.0 | 0.5 | GO:0019870 | potassium channel inhibitor activity(GO:0019870) |
| 0.0 | 0.2 | GO:0004727 | prenylated protein tyrosine phosphatase activity(GO:0004727) |
| 0.0 | 0.8 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.9 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.0 | 0.1 | GO:0097603 | temperature-gated ion channel activity(GO:0097603) |
| 0.0 | 0.5 | GO:0031698 | beta-2 adrenergic receptor binding(GO:0031698) |
| 0.0 | 0.1 | GO:0018685 | alkane 1-monooxygenase activity(GO:0018685) leukotriene-B4 20-monooxygenase activity(GO:0050051) |
| 0.0 | 0.1 | GO:0046848 | hydroxyapatite binding(GO:0046848) |
| 0.0 | 0.1 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.0 | 0.1 | GO:0042328 | heparan sulfate N-acetylglucosaminyltransferase activity(GO:0042328) |
| 0.0 | 0.1 | GO:0086083 | cell adhesive protein binding involved in bundle of His cell-Purkinje myocyte communication(GO:0086083) |
| 0.0 | 0.2 | GO:0016886 | ligase activity, forming phosphoric ester bonds(GO:0016886) |
| 0.0 | 0.9 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 0.2 | GO:0032395 | MHC class II receptor activity(GO:0032395) |
| 0.0 | 0.3 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 0.1 | GO:0015450 | P-P-bond-hydrolysis-driven protein transmembrane transporter activity(GO:0015450) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 2.8 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 1.2 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 1.5 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.4 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 0.8 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.0 | 0.6 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 0.7 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 0.6 | PID BMP PATHWAY | BMP receptor signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.0 | 2.7 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.9 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 1.5 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.0 | 0.4 | REACTOME REGULATED PROTEOLYSIS OF P75NTR | Genes involved in Regulated proteolysis of p75NTR |
| 0.0 | 0.6 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.0 | 0.4 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 0.5 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.0 | 0.9 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 0.2 | REACTOME DESTABILIZATION OF MRNA BY BRF1 | Genes involved in Destabilization of mRNA by Butyrate Response Factor 1 (BRF1) |
| 0.0 | 1.1 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 0.8 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.0 | 0.6 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.1 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 0.3 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |