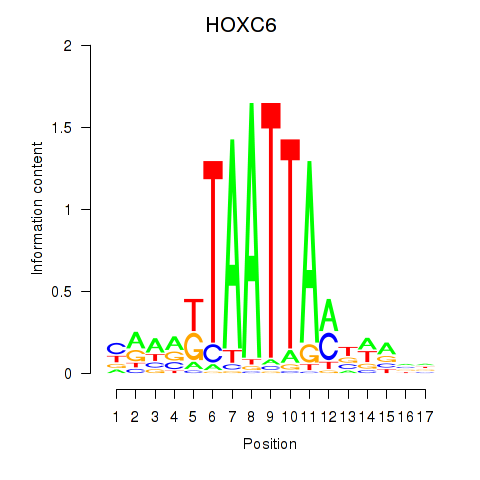
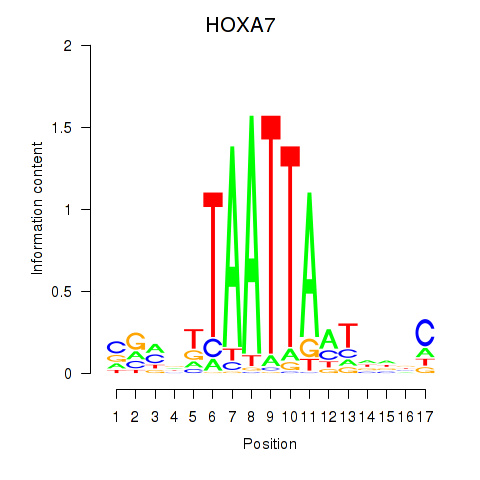
|
chr15_+_67125707
Show fit
|
3.15 |
ENST00000540846.6
|
SMAD3
|
SMAD family member 3
|
|
chr2_+_151357583
Show fit
|
1.02 |
ENST00000243347.5
|
TNFAIP6
|
TNF alpha induced protein 6
|
|
chr11_-_102780620
Show fit
|
0.96 |
ENST00000279441.9
ENST00000539681.1
|
MMP10
|
matrix metallopeptidase 10
|
|
chr19_-_14979848
Show fit
|
0.53 |
ENST00000594383.2
|
SLC1A6
|
solute carrier family 1 member 6
|
|
chr3_+_111998739
Show fit
|
0.52 |
ENST00000393917.6
ENST00000273368.8
|
TAGLN3
|
transgelin 3
|
|
chr1_-_120054225
Show fit
|
0.51 |
ENST00000602566.6
|
NOTCH2
|
notch receptor 2
|
|
chr19_-_14835162
Show fit
|
0.50 |
ENST00000322301.5
|
OR7A5
|
olfactory receptor family 7 subfamily A member 5
|
|
chr3_+_111999189
Show fit
|
0.48 |
ENST00000455401.6
|
TAGLN3
|
transgelin 3
|
|
chr1_-_150765785
Show fit
|
0.48 |
ENST00000680311.1
ENST00000681728.1
ENST00000680288.1
|
CTSS
|
cathepsin S
|
|
chr3_+_111998915
Show fit
|
0.48 |
ENST00000478951.6
|
TAGLN3
|
transgelin 3
|
|
chr6_+_113857333
Show fit
|
0.46 |
ENST00000612661.2
|
MARCKS
|
myristoylated alanine rich protein kinase C substrate
|
|
chr17_+_1771688
Show fit
|
0.41 |
ENST00000572048.1
ENST00000573763.1
|
SERPINF1
|
serpin family F member 1
|
|
chr12_-_9999176
Show fit
|
0.38 |
ENST00000298527.10
ENST00000348658.4
|
CLEC1B
|
C-type lectin domain family 1 member B
|
|
chr18_-_72544336
Show fit
|
0.36 |
ENST00000269503.9
|
CBLN2
|
cerebellin 2 precursor
|
|
chr1_+_205256189
Show fit
|
0.36 |
ENST00000329800.7
|
TMCC2
|
transmembrane and coiled-coil domain family 2
|
|
chr6_+_26365159
Show fit
|
0.35 |
ENST00000532865.5
ENST00000396934.7
ENST00000508906.6
ENST00000530653.5
ENST00000527417.5
|
BTN3A2
|
butyrophilin subfamily 3 member A2
|
|
chr12_-_89526253
Show fit
|
0.34 |
ENST00000547474.1
|
POC1B-GALNT4
|
POC1B-GALNT4 readthrough
|
|
chr1_+_160400543
Show fit
|
0.34 |
ENST00000368061.3
|
VANGL2
|
VANGL planar cell polarity protein 2
|
|
chr6_+_26365215
Show fit
|
0.33 |
ENST00000527422.5
ENST00000356386.6
ENST00000396948.5
|
BTN3A2
|
butyrophilin subfamily 3 member A2
|
|
chr6_+_26440472
Show fit
|
0.32 |
ENST00000494393.5
ENST00000482451.5
ENST00000471353.5
ENST00000361232.7
ENST00000487627.5
ENST00000496719.1
ENST00000244519.7
ENST00000490254.5
ENST00000487272.1
|
BTN3A3
|
butyrophilin subfamily 3 member A3
|
|
chr16_+_11965193
Show fit
|
0.31 |
ENST00000053243.6
ENST00000396495.3
|
TNFRSF17
|
TNF receptor superfamily member 17
|
|
chr8_+_53851786
Show fit
|
0.31 |
ENST00000297313.8
ENST00000344277.10
|
RGS20
|
regulator of G protein signaling 20
|
|
chr4_-_39977836
Show fit
|
0.30 |
ENST00000303538.13
ENST00000503396.5
|
PDS5A
|
PDS5 cohesin associated factor A
|
|
chr6_+_26365176
Show fit
|
0.30 |
ENST00000377708.7
|
BTN3A2
|
butyrophilin subfamily 3 member A2
|
|
chr12_-_89526164
Show fit
|
0.30 |
ENST00000548729.5
|
POC1B-GALNT4
|
POC1B-GALNT4 readthrough
|
|
chr3_-_191282383
Show fit
|
0.30 |
ENST00000427544.6
|
UTS2B
|
urotensin 2B
|
|
chr15_-_75455767
Show fit
|
0.28 |
ENST00000360439.8
|
SIN3A
|
SIN3 transcription regulator family member A
|
|
chr3_+_138621225
Show fit
|
0.27 |
ENST00000479848.1
|
FAIM
|
Fas apoptotic inhibitory molecule
|
|
chr6_+_26402237
Show fit
|
0.27 |
ENST00000476549.6
ENST00000450085.6
ENST00000425234.6
ENST00000427334.5
ENST00000506698.1
ENST00000289361.11
|
BTN3A1
|
butyrophilin subfamily 3 member A1
|
|
chr13_-_35476682
Show fit
|
0.25 |
ENST00000379919.6
|
MAB21L1
|
mab-21 like 1
|
|
chr3_+_138621207
Show fit
|
0.25 |
ENST00000464668.5
|
FAIM
|
Fas apoptotic inhibitory molecule
|
|
chr3_+_111999326
Show fit
|
0.25 |
ENST00000494932.1
|
TAGLN3
|
transgelin 3
|
|
chr8_-_30812867
Show fit
|
0.25 |
ENST00000518243.5
|
PPP2CB
|
protein phosphatase 2 catalytic subunit beta
|
|
chr15_+_64387828
Show fit
|
0.24 |
ENST00000261884.8
|
TRIP4
|
thyroid hormone receptor interactor 4
|
|
chr12_-_51028234
Show fit
|
0.24 |
ENST00000547688.7
ENST00000394904.9
|
SLC11A2
|
solute carrier family 11 member 2
|
|
chr5_+_67004618
Show fit
|
0.23 |
ENST00000261569.11
ENST00000436277.5
|
MAST4
|
microtubule associated serine/threonine kinase family member 4
|
|
chr12_-_91179355
Show fit
|
0.21 |
ENST00000550563.5
ENST00000546370.5
|
DCN
|
decorin
|
|
chr18_-_77127935
Show fit
|
0.21 |
ENST00000581878.5
|
MBP
|
myelin basic protein
|
|
chr9_+_122370523
Show fit
|
0.21 |
ENST00000643810.1
ENST00000540753.6
|
PTGS1
|
prostaglandin-endoperoxide synthase 1
|
|
chr10_+_113126670
Show fit
|
0.21 |
ENST00000369389.6
|
TCF7L2
|
transcription factor 7 like 2
|
|
chr13_+_31739542
Show fit
|
0.20 |
ENST00000380314.2
|
RXFP2
|
relaxin family peptide receptor 2
|
|
chr4_+_168497044
Show fit
|
0.20 |
ENST00000505667.6
|
PALLD
|
palladin, cytoskeletal associated protein
|
|
chr21_-_41926680
Show fit
|
0.20 |
ENST00000329623.11
|
C2CD2
|
C2 calcium dependent domain containing 2
|
|
chr2_+_168901290
Show fit
|
0.19 |
ENST00000429379.2
ENST00000375363.8
ENST00000421979.1
|
G6PC2
|
glucose-6-phosphatase catalytic subunit 2
|
|
chr4_+_168497066
Show fit
|
0.19 |
ENST00000261509.10
|
PALLD
|
palladin, cytoskeletal associated protein
|
|
chr17_-_41467386
Show fit
|
0.19 |
ENST00000225899.4
|
KRT32
|
keratin 32
|
|
chr4_-_119322128
Show fit
|
0.19 |
ENST00000274024.4
|
FABP2
|
fatty acid binding protein 2
|
|
chr4_-_8871817
Show fit
|
0.18 |
ENST00000400677.5
|
HMX1
|
H6 family homeobox 1
|
|
chr11_+_33039996
Show fit
|
0.17 |
ENST00000432887.5
ENST00000528898.1
ENST00000531632.6
|
TCP11L1
|
t-complex 11 like 1
|
|
chr9_+_122371036
Show fit
|
0.17 |
ENST00000619306.5
ENST00000426608.6
ENST00000223423.8
|
PTGS1
|
prostaglandin-endoperoxide synthase 1
|
|
chr1_-_113871665
Show fit
|
0.16 |
ENST00000528414.5
ENST00000460620.5
ENST00000359785.10
ENST00000420377.6
ENST00000525799.1
ENST00000538253.5
|
PTPN22
|
protein tyrosine phosphatase non-receptor type 22
|
|
chr6_-_17706852
Show fit
|
0.15 |
ENST00000262077.3
|
NUP153
|
nucleoporin 153
|
|
chr17_-_41168219
Show fit
|
0.15 |
ENST00000391356.4
|
KRTAP4-3
|
keratin associated protein 4-3
|
|
chr19_-_14835252
Show fit
|
0.15 |
ENST00000641666.1
ENST00000642030.1
ENST00000642000.1
|
OR7C1
|
olfactory receptor family 7 subfamily C member 1
|
|
chr3_-_157503574
Show fit
|
0.15 |
ENST00000494677.5
ENST00000468233.5
|
VEPH1
|
ventricular zone expressed PH domain containing 1
|
|
chr1_-_247760556
Show fit
|
0.14 |
ENST00000641256.1
|
OR1C1
|
olfactory receptor family 1 subfamily C member 1
|
|
chr2_+_181986015
Show fit
|
0.14 |
ENST00000409702.1
|
PPP1R1C
|
protein phosphatase 1 regulatory inhibitor subunit 1C
|
|
chr12_-_89526011
Show fit
|
0.14 |
ENST00000313546.8
|
POC1B
|
POC1 centriolar protein B
|
|
chr7_+_134843884
Show fit
|
0.14 |
ENST00000445569.6
|
CALD1
|
caldesmon 1
|
|
chr10_+_84424919
Show fit
|
0.14 |
ENST00000543283.2
ENST00000494586.5
|
CCSER2
|
coiled-coil serine rich protein 2
|
|
chr11_-_26572130
Show fit
|
0.13 |
ENST00000527569.1
|
MUC15
|
mucin 15, cell surface associated
|
|
chr18_+_58341038
Show fit
|
0.13 |
ENST00000679791.1
|
NEDD4L
|
NEDD4 like E3 ubiquitin protein ligase
|
|
chr5_-_16916400
Show fit
|
0.13 |
ENST00000513882.5
|
MYO10
|
myosin X
|
|
chr14_+_34993240
Show fit
|
0.13 |
ENST00000677647.1
|
SRP54
|
signal recognition particle 54
|
|
chr2_-_25168571
Show fit
|
0.13 |
ENST00000264708.7
ENST00000449220.1
ENST00000395826.7
|
POMC
|
proopiomelanocortin
|
|
chr7_-_44541318
Show fit
|
0.13 |
ENST00000381160.8
|
NPC1L1
|
NPC1 like intracellular cholesterol transporter 1
|
|
chr2_-_25168903
Show fit
|
0.13 |
ENST00000405623.5
|
POMC
|
proopiomelanocortin
|
|
chr2_-_25168690
Show fit
|
0.13 |
ENST00000380794.5
|
POMC
|
proopiomelanocortin
|
|
chr7_-_32299287
Show fit
|
0.12 |
ENST00000396193.5
|
PDE1C
|
phosphodiesterase 1C
|
|
chr14_-_106470788
Show fit
|
0.12 |
ENST00000434710.1
|
IGHV3-43
|
immunoglobulin heavy variable 3-43
|
|
chr4_+_70242583
Show fit
|
0.12 |
ENST00000304954.3
|
CSN3
|
casein kappa
|
|
chr6_+_29301701
Show fit
|
0.12 |
ENST00000641895.1
|
OR14J1
|
olfactory receptor family 14 subfamily J member 1
|
|
chr2_-_89143133
Show fit
|
0.12 |
ENST00000492167.1
|
IGKV3-20
|
immunoglobulin kappa variable 3-20
|
|
chr2_+_24793098
Show fit
|
0.12 |
ENST00000473706.5
|
CENPO
|
centromere protein O
|
|
chr5_+_75611182
Show fit
|
0.12 |
ENST00000672850.1
ENST00000672844.1
|
ANKDD1B
|
ankyrin repeat and death domain containing 1B
|
|
chr6_+_26402289
Show fit
|
0.11 |
ENST00000414912.2
|
BTN3A1
|
butyrophilin subfamily 3 member A1
|
|
chr4_+_107824555
Show fit
|
0.11 |
ENST00000394684.8
|
SGMS2
|
sphingomyelin synthase 2
|
|
chr1_-_160954801
Show fit
|
0.11 |
ENST00000368029.4
|
ITLN2
|
intelectin 2
|
|
chr2_-_40453438
Show fit
|
0.11 |
ENST00000455476.5
|
SLC8A1
|
solute carrier family 8 member A1
|
|
chr4_+_69931066
Show fit
|
0.10 |
ENST00000246891.9
|
CSN1S1
|
casein alpha s1
|
|
chr8_-_42768602
Show fit
|
0.10 |
ENST00000534622.5
|
CHRNA6
|
cholinergic receptor nicotinic alpha 6 subunit
|
|
chr6_-_34671918
Show fit
|
0.10 |
ENST00000374021.1
|
ILRUN
|
inflammation and lipid regulator with UBA-like and NBR1-like domains
|
|
chrX_+_83861126
Show fit
|
0.10 |
ENST00000621735.4
ENST00000329312.5
|
CYLC1
|
cylicin 1
|
|
chr14_+_94081278
Show fit
|
0.10 |
ENST00000555523.6
ENST00000393115.7
ENST00000554166.5
ENST00000556381.5
ENST00000553664.1
ENST00000555341.5
ENST00000557218.5
ENST00000554544.5
ENST00000557066.5
|
IFI27L1
|
interferon alpha inducible protein 27 like 1
|
|
chr3_-_165078480
Show fit
|
0.10 |
ENST00000264382.8
|
SI
|
sucrase-isomaltase
|
|
chr11_+_60056587
Show fit
|
0.10 |
ENST00000395032.6
ENST00000358152.6
|
MS4A3
|
membrane spanning 4-domains A3
|
|
chr20_+_15196834
Show fit
|
0.10 |
ENST00000402914.5
|
MACROD2
|
mono-ADP ribosylhydrolase 2
|
|
chr11_-_63144221
Show fit
|
0.09 |
ENST00000417740.5
ENST00000612278.4
ENST00000326192.5
|
SLC22A24
|
solute carrier family 22 member 24
|
|
chr10_-_110918934
Show fit
|
0.09 |
ENST00000605742.5
|
BBIP1
|
BBSome interacting protein 1
|
|
chr2_-_127526444
Show fit
|
0.09 |
ENST00000295321.9
|
IWS1
|
interacts with SUPT6H, CTD assembly factor 1
|
|
chr14_+_21997531
Show fit
|
0.09 |
ENST00000390445.2
|
TRAV17
|
T cell receptor alpha variable 17
|
|
chr16_-_28623560
Show fit
|
0.08 |
ENST00000350842.8
|
SULT1A1
|
sulfotransferase family 1A member 1
|
|
chr1_+_202348687
Show fit
|
0.08 |
ENST00000608999.6
ENST00000391959.5
ENST00000480184.5
|
PPP1R12B
|
protein phosphatase 1 regulatory subunit 12B
|
|
chr11_-_36598221
Show fit
|
0.08 |
ENST00000311485.8
ENST00000527033.5
ENST00000532616.1
ENST00000618712.4
|
RAG2
|
recombination activating 2
|
|
chr6_+_46793379
Show fit
|
0.08 |
ENST00000230588.9
ENST00000611727.2
|
MEP1A
|
meprin A subunit alpha
|
|
chr19_-_51417700
Show fit
|
0.08 |
ENST00000529627.1
ENST00000439889.6
|
SIGLEC10
|
sialic acid binding Ig like lectin 10
|
|
chr17_-_41149823
Show fit
|
0.08 |
ENST00000343246.6
|
KRTAP4-5
|
keratin associated protein 4-5
|
|
chr11_-_26572254
Show fit
|
0.08 |
ENST00000529533.6
|
MUC15
|
mucin 15, cell surface associated
|
|
chr8_+_91249307
Show fit
|
0.08 |
ENST00000309536.6
ENST00000276609.8
|
SLC26A7
|
solute carrier family 26 member 7
|
|
chr19_-_3557563
Show fit
|
0.07 |
ENST00000389395.7
ENST00000355415.7
|
MFSD12
|
major facilitator superfamily domain containing 12
|
|
chr11_+_60056653
Show fit
|
0.07 |
ENST00000278865.8
|
MS4A3
|
membrane spanning 4-domains A3
|
|
chr3_-_197184131
Show fit
|
0.07 |
ENST00000452595.5
|
DLG1
|
discs large MAGUK scaffold protein 1
|
|
chr11_-_107858777
Show fit
|
0.07 |
ENST00000525815.6
|
SLC35F2
|
solute carrier family 35 member F2
|
|
chr8_+_12108172
Show fit
|
0.07 |
ENST00000400078.3
|
ZNF705D
|
zinc finger protein 705D
|
|
chr12_-_91179517
Show fit
|
0.07 |
ENST00000551354.1
|
DCN
|
decorin
|
|
chr17_+_41084150
Show fit
|
0.07 |
ENST00000391417.5
ENST00000621138.1
|
KRTAP4-7
|
keratin associated protein 4-7
|
|
chr8_-_33567118
Show fit
|
0.07 |
ENST00000256257.2
|
RNF122
|
ring finger protein 122
|
|
chr1_+_202348727
Show fit
|
0.07 |
ENST00000356764.6
|
PPP1R12B
|
protein phosphatase 1 regulatory subunit 12B
|
|
chr9_+_69145463
Show fit
|
0.07 |
ENST00000636438.1
|
TJP2
|
tight junction protein 2
|
|
chr2_-_133568393
Show fit
|
0.07 |
ENST00000317721.10
ENST00000405974.7
ENST00000409261.6
ENST00000409213.5
|
NCKAP5
|
NCK associated protein 5
|
|
chr3_-_64019334
Show fit
|
0.06 |
ENST00000480205.5
|
PSMD6
|
proteasome 26S subunit, non-ATPase 6
|
|
chr4_-_46124046
Show fit
|
0.06 |
ENST00000295452.5
|
GABRG1
|
gamma-aminobutyric acid type A receptor subunit gamma1
|
|
chr6_+_30061231
Show fit
|
0.06 |
ENST00000376782.6
ENST00000359374.8
ENST00000376785.2
ENST00000332435.10
|
POLR1H
|
RNA polymerase I subunit H
|
|
chr17_-_5035418
Show fit
|
0.06 |
ENST00000254853.10
ENST00000424747.1
|
SLC52A1
|
solute carrier family 52 member 1
|
|
chr16_-_28623330
Show fit
|
0.06 |
ENST00000677940.1
|
ENSG00000288656.1
|
novel protein
|
|
chr2_+_26401909
Show fit
|
0.06 |
ENST00000288710.7
|
DRC1
|
dynein regulatory complex subunit 1
|
|
chrX_+_100584928
Show fit
|
0.06 |
ENST00000373031.5
|
TNMD
|
tenomodulin
|
|
chr1_+_117420597
Show fit
|
0.06 |
ENST00000449370.6
|
MAN1A2
|
mannosidase alpha class 1A member 2
|
|
chr1_-_242449478
Show fit
|
0.06 |
ENST00000427495.5
|
PLD5
|
phospholipase D family member 5
|
|
chr17_-_66229380
Show fit
|
0.06 |
ENST00000205948.11
|
APOH
|
apolipoprotein H
|
|
chr6_+_158649997
Show fit
|
0.06 |
ENST00000360448.8
ENST00000367081.7
ENST00000611299.5
|
SYTL3
|
synaptotagmin like 3
|
|
chr5_-_127073467
Show fit
|
0.05 |
ENST00000607731.5
ENST00000509733.7
ENST00000296662.10
ENST00000535381.6
|
C5orf63
|
chromosome 5 open reading frame 63
|
|
chr11_-_5301946
Show fit
|
0.05 |
ENST00000380224.2
|
OR51B4
|
olfactory receptor family 51 subfamily B member 4
|
|
chr7_-_111392915
Show fit
|
0.05 |
ENST00000450877.5
|
IMMP2L
|
inner mitochondrial membrane peptidase subunit 2
|
|
chr2_+_90172802
Show fit
|
0.05 |
ENST00000390277.3
|
IGKV3D-11
|
immunoglobulin kappa variable 3D-11
|
|
chr4_+_150582119
Show fit
|
0.05 |
ENST00000317605.6
|
MAB21L2
|
mab-21 like 2
|
|
chr9_-_5339874
Show fit
|
0.05 |
ENST00000223862.2
|
RLN1
|
relaxin 1
|
|
chr21_-_30166782
Show fit
|
0.05 |
ENST00000286808.5
|
CLDN17
|
claudin 17
|
|
chr17_-_74776323
Show fit
|
0.05 |
ENST00000582870.5
ENST00000581136.5
ENST00000579218.5
ENST00000583476.5
ENST00000580301.5
ENST00000583757.5
ENST00000357814.8
ENST00000582524.5
|
NAT9
|
N-acetyltransferase 9 (putative)
|
|
chr11_+_2384603
Show fit
|
0.05 |
ENST00000527343.5
ENST00000464784.6
|
CD81
|
CD81 molecule
|
|
chr8_+_24294107
Show fit
|
0.04 |
ENST00000437154.6
|
ADAM28
|
ADAM metallopeptidase domain 28
|
|
chr9_-_27005659
Show fit
|
0.04 |
ENST00000380055.6
|
LRRC19
|
leucine rich repeat containing 19
|
|
chr19_-_54313074
Show fit
|
0.04 |
ENST00000486742.2
ENST00000432233.8
|
LILRA5
|
leukocyte immunoglobulin like receptor A5
|
|
chr5_+_40909490
Show fit
|
0.04 |
ENST00000313164.10
|
C7
|
complement C7
|
|
chr20_+_45416551
Show fit
|
0.04 |
ENST00000639292.1
|
PIGT
|
phosphatidylinositol glycan anchor biosynthesis class T
|
|
chr10_+_97446137
Show fit
|
0.04 |
ENST00000370854.7
ENST00000393760.6
ENST00000414567.5
|
ZDHHC16
|
zinc finger DHHC-type palmitoyltransferase 16
|
|
chr3_+_98168700
Show fit
|
0.04 |
ENST00000383696.4
|
OR5H15
|
olfactory receptor family 5 subfamily H member 15
|
|
chr10_-_126670686
Show fit
|
0.04 |
ENST00000488181.3
|
C10orf90
|
chromosome 10 open reading frame 90
|
|
chr6_+_111087495
Show fit
|
0.04 |
ENST00000612036.4
ENST00000368851.10
|
SLC16A10
|
solute carrier family 16 member 10
|
|
chr12_-_86256299
Show fit
|
0.04 |
ENST00000552808.6
ENST00000547225.5
|
MGAT4C
|
MGAT4 family member C
|
|
chr11_-_13496018
Show fit
|
0.04 |
ENST00000529816.1
|
PTH
|
parathyroid hormone
|
|
chr20_-_10420737
Show fit
|
0.04 |
ENST00000649912.1
|
ENSG00000285723.1
|
novel protein
|
|
chr10_+_97446194
Show fit
|
0.04 |
ENST00000370846.8
ENST00000352634.8
ENST00000353979.7
ENST00000370842.6
ENST00000345745.9
|
ZDHHC16
|
zinc finger DHHC-type palmitoyltransferase 16
|
|
chr2_+_87338511
Show fit
|
0.04 |
ENST00000421835.2
|
IGKV3OR2-268
|
immunoglobulin kappa variable 3/OR2-268 (non-functional)
|
|
chr17_+_47651061
Show fit
|
0.04 |
ENST00000540627.5
|
KPNB1
|
karyopherin subunit beta 1
|
|
chrX_+_1591590
Show fit
|
0.04 |
ENST00000313871.9
ENST00000381261.8
|
AKAP17A
|
A-kinase anchoring protein 17A
|
|
chr7_+_150405668
Show fit
|
0.04 |
ENST00000642087.1
|
ENSG00000284691.1
|
novel zinc finger protein
|
|
chr12_+_80716906
Show fit
|
0.03 |
ENST00000228644.4
|
MYF5
|
myogenic factor 5
|
|
chr7_+_134986483
Show fit
|
0.03 |
ENST00000436302.6
ENST00000435976.6
ENST00000455283.2
|
AGBL3
|
ATP/GTP binding protein like 3
|
|
chr15_+_21579912
Show fit
|
0.03 |
ENST00000628444.1
|
LINC02203
|
long intergenic non-protein coding RNA 2203
|
|
chr2_-_142131004
Show fit
|
0.03 |
ENST00000434794.1
ENST00000389484.8
|
LRP1B
|
LDL receptor related protein 1B
|
|
chr11_-_26572102
Show fit
|
0.03 |
ENST00000455601.6
|
MUC15
|
mucin 15, cell surface associated
|
|
chr3_-_167474026
Show fit
|
0.03 |
ENST00000466903.1
ENST00000264677.8
|
SERPINI2
|
serpin family I member 2
|
|
chr4_+_112647059
Show fit
|
0.03 |
ENST00000511529.1
|
LARP7
|
La ribonucleoprotein 7, transcriptional regulator
|
|
chr17_+_59155726
Show fit
|
0.03 |
ENST00000578777.5
ENST00000577457.1
ENST00000582995.5
ENST00000262293.9
ENST00000614081.1
|
PRR11
|
proline rich 11
|
|
chr5_+_55853314
Show fit
|
0.03 |
ENST00000354961.8
ENST00000297015.7
|
IL31RA
|
interleukin 31 receptor A
|
|
chr18_+_34978244
Show fit
|
0.03 |
ENST00000436190.6
|
MAPRE2
|
microtubule associated protein RP/EB family member 2
|
|
chr16_+_28553908
Show fit
|
0.03 |
ENST00000317058.8
|
SGF29
|
SAGA complex associated factor 29
|
|
chr3_-_20012250
Show fit
|
0.03 |
ENST00000389050.5
|
PP2D1
|
protein phosphatase 2C like domain containing 1
|
|
chr7_-_122995700
Show fit
|
0.03 |
ENST00000249284.3
|
TAS2R16
|
taste 2 receptor member 16
|
|
chr7_+_150405146
Show fit
|
0.03 |
ENST00000498682.3
ENST00000641717.1
|
ENSG00000284691.1
|
novel zinc finger protein
|
|
chr8_-_41309434
Show fit
|
0.03 |
ENST00000220772.8
|
SFRP1
|
secreted frizzled related protein 1
|
|
chr10_-_110919146
Show fit
|
0.03 |
ENST00000652396.1
ENST00000423273.5
ENST00000436562.1
ENST00000651952.1
ENST00000448814.7
ENST00000652400.1
ENST00000447005.5
ENST00000454061.5
|
BBIP1
|
BBSome interacting protein 1
|
|
chr10_-_97445850
Show fit
|
0.03 |
ENST00000477692.6
ENST00000485122.6
ENST00000370886.9
ENST00000370885.8
ENST00000370902.8
ENST00000370884.5
|
EXOSC1
|
exosome component 1
|
|
chr12_+_106684696
Show fit
|
0.03 |
ENST00000229387.6
|
RFX4
|
regulatory factor X4
|
|
chr8_-_61689768
Show fit
|
0.03 |
ENST00000517847.6
ENST00000389204.8
ENST00000517661.5
ENST00000517903.5
ENST00000522603.5
ENST00000541428.5
ENST00000522349.5
ENST00000522835.5
ENST00000518306.5
|
ASPH
|
aspartate beta-hydroxylase
|
|
chr11_-_129024157
Show fit
|
0.03 |
ENST00000392657.7
|
ARHGAP32
|
Rho GTPase activating protein 32
|
|
chr1_+_15341744
Show fit
|
0.02 |
ENST00000444385.5
|
FHAD1
|
forkhead associated phosphopeptide binding domain 1
|
|
chr7_+_138460238
Show fit
|
0.02 |
ENST00000343526.9
|
TRIM24
|
tripartite motif containing 24
|
|
chr4_-_86360010
Show fit
|
0.02 |
ENST00000641911.1
ENST00000641072.1
ENST00000359221.8
ENST00000640490.1
|
MAPK10
|
mitogen-activated protein kinase 10
|
|
chr8_+_76681208
Show fit
|
0.02 |
ENST00000651372.2
|
ZFHX4
|
zinc finger homeobox 4
|
|
chr14_+_56117702
Show fit
|
0.02 |
ENST00000559044.5
|
PELI2
|
pellino E3 ubiquitin protein ligase family member 2
|
|
chr11_-_31369728
Show fit
|
0.02 |
ENST00000684477.1
ENST00000452803.1
|
DCDC1
|
doublecortin domain containing 1
|
|
chr10_+_18260715
Show fit
|
0.02 |
ENST00000615785.4
ENST00000617363.4
ENST00000396576.6
|
CACNB2
|
calcium voltage-gated channel auxiliary subunit beta 2
|
|
chr16_+_15395745
Show fit
|
0.02 |
ENST00000287594.7
ENST00000396385.4
ENST00000568766.1
|
MPV17L
ENSG00000261130.5
|
MPV17 mitochondrial inner membrane protein like
novel protein
|
|
chr4_-_86360071
Show fit
|
0.02 |
ENST00000641677.1
ENST00000639234.1
ENST00000641553.1
ENST00000641826.1
ENST00000641537.1
ENST00000395169.9
ENST00000641408.1
ENST00000638225.1
ENST00000641052.1
|
MAPK10
|
mitogen-activated protein kinase 10
|
|
chr2_-_175181663
Show fit
|
0.02 |
ENST00000392541.3
ENST00000284727.9
ENST00000409194.5
|
ATP5MC3
|
ATP synthase membrane subunit c locus 3
|
|
chr2_+_181985846
Show fit
|
0.02 |
ENST00000682840.1
ENST00000409137.7
ENST00000280295.7
|
PPP1R1C
|
protein phosphatase 1 regulatory inhibitor subunit 1C
|
|
chr11_+_73787853
Show fit
|
0.02 |
ENST00000310614.12
ENST00000497094.6
ENST00000411840.6
ENST00000535277.5
ENST00000398483.7
ENST00000542303.5
|
MRPL48
|
mitochondrial ribosomal protein L48
|
|
chr3_-_54928044
Show fit
|
0.02 |
ENST00000273286.6
|
LRTM1
|
leucine rich repeats and transmembrane domains 1
|
|
chr2_+_233917371
Show fit
|
0.02 |
ENST00000324695.9
ENST00000433712.6
|
TRPM8
|
transient receptor potential cation channel subfamily M member 8
|
|
chr4_-_86360039
Show fit
|
0.02 |
ENST00000515650.2
ENST00000641724.1
ENST00000641607.1
ENST00000641324.1
ENST00000641903.1
ENST00000395157.9
ENST00000641823.1
ENST00000641873.1
ENST00000641102.1
ENST00000641462.2
ENST00000641217.1
ENST00000642006.1
ENST00000641020.1
ENST00000641110.1
ENST00000639175.1
ENST00000641485.1
ENST00000641864.1
ENST00000641954.1
ENST00000641647.1
ENST00000641459.1
ENST00000641762.1
ENST00000641777.1
ENST00000641208.1
ENST00000642015.1
ENST00000641493.1
ENST00000642032.1
ENST00000641010.1
ENST00000641287.1
ENST00000641943.1
ENST00000642103.1
ENST00000641047.1
ENST00000641166.1
ENST00000641207.1
|
MAPK10
|
mitogen-activated protein kinase 10
|
|
chr2_+_206159580
Show fit
|
0.01 |
ENST00000236957.9
ENST00000392221.5
ENST00000445505.5
|
EEF1B2
|
eukaryotic translation elongation factor 1 beta 2
|
|
chr12_-_52367478
Show fit
|
0.01 |
ENST00000257901.7
|
KRT85
|
keratin 85
|
|
chr6_-_41734160
Show fit
|
0.01 |
ENST00000424495.2
ENST00000420312.6
|
TFEB
|
transcription factor EB
|
|
chr5_+_141245384
Show fit
|
0.01 |
ENST00000623671.1
ENST00000231173.6
|
PCDHB15
|
protocadherin beta 15
|
|
chr17_+_29941605
Show fit
|
0.01 |
ENST00000394835.7
|
EFCAB5
|
EF-hand calcium binding domain 5
|
|
chr2_+_90234809
Show fit
|
0.01 |
ENST00000443397.5
|
IGKV3D-7
|
immunoglobulin kappa variable 3D-7
|
|
chr2_-_206159509
Show fit
|
0.01 |
ENST00000423725.5
|
NDUFS1
|
NADH:ubiquinone oxidoreductase core subunit S1
|
|
chr4_-_46909235
Show fit
|
0.01 |
ENST00000505102.1
ENST00000355591.8
|
COX7B2
|
cytochrome c oxidase subunit 7B2
|
|
chr12_-_86256267
Show fit
|
0.01 |
ENST00000620241.4
|
MGAT4C
|
MGAT4 family member C
|
|
chrM_+_10055
Show fit
|
0.00 |
ENST00000361227.2
|
MT-ND3
|
mitochondrially encoded NADH:ubiquinone oxidoreductase core subunit 3
|
|
chr2_-_74392025
Show fit
|
0.00 |
ENST00000440727.1
ENST00000409240.5
|
DCTN1
|
dynactin subunit 1
|
|
chr14_+_71933116
Show fit
|
0.00 |
ENST00000553530.5
ENST00000556437.5
|
RGS6
|
regulator of G protein signaling 6
|
|
chr15_+_58138368
Show fit
|
0.00 |
ENST00000219919.9
ENST00000536493.1
|
AQP9
|
aquaporin 9
|
|
chr4_-_159035226
Show fit
|
0.00 |
ENST00000434826.3
|
C4orf45
|
chromosome 4 open reading frame 45
|
|
chr7_-_22822779
Show fit
|
0.00 |
ENST00000372879.8
|
TOMM7
|
translocase of outer mitochondrial membrane 7
|
|
chr5_+_163503075
Show fit
|
0.00 |
ENST00000280969.9
|
MAT2B
|
methionine adenosyltransferase 2B
|