Project
Inflammatory response time course, HUVEC (Wada, 2009)
Navigation
Downloads
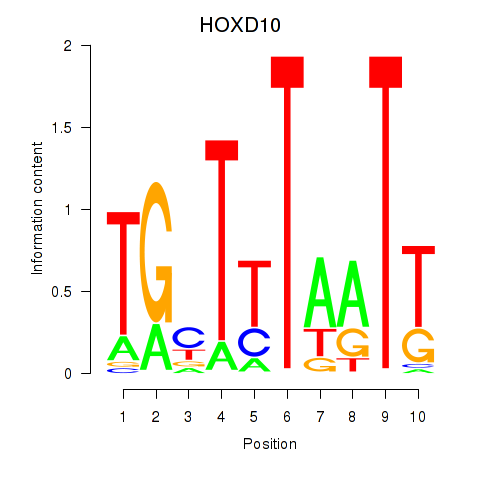
Results for HOXD10
Z-value: 0.72
Transcription factors associated with HOXD10
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HOXD10
|
ENSG00000128710.6 | HOXD10 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HOXD10 | hg38_v1_chr2_+_176116768_176116794 | 0.10 | 6.3e-01 | Click! |
Activity profile of HOXD10 motif
Sorted Z-values of HOXD10 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of HOXD10
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_-_76023489 | 4.87 |
ENST00000306602.3
|
CXCL10
|
C-X-C motif chemokine ligand 10 |
| chr3_-_127736329 | 1.48 |
ENST00000398101.7
|
MGLL
|
monoglyceride lipase |
| chr2_+_167248638 | 1.37 |
ENST00000295237.10
|
XIRP2
|
xin actin binding repeat containing 2 |
| chr1_-_111200633 | 1.34 |
ENST00000357640.9
|
DENND2D
|
DENN domain containing 2D |
| chr1_+_78649818 | 1.29 |
ENST00000370747.9
ENST00000438486.1 |
IFI44
|
interferon induced protein 44 |
| chr4_+_99574812 | 1.27 |
ENST00000422897.6
ENST00000265517.10 |
MTTP
|
microsomal triglyceride transfer protein |
| chr1_-_89270751 | 1.17 |
ENST00000370459.7
|
GBP5
|
guanylate binding protein 5 |
| chr5_-_169980474 | 1.09 |
ENST00000377365.4
|
INSYN2B
|
inhibitory synaptic factor family member 2B |
| chr6_-_134318097 | 1.06 |
ENST00000367858.10
ENST00000533224.1 |
SGK1
|
serum/glucocorticoid regulated kinase 1 |
| chr12_-_76423256 | 1.06 |
ENST00000546946.5
|
OSBPL8
|
oxysterol binding protein like 8 |
| chr4_-_76007501 | 1.02 |
ENST00000264888.6
|
CXCL9
|
C-X-C motif chemokine ligand 9 |
| chr3_+_130850585 | 0.93 |
ENST00000505330.5
ENST00000504381.5 ENST00000507488.6 |
ATP2C1
|
ATPase secretory pathway Ca2+ transporting 1 |
| chr4_-_152411734 | 0.91 |
ENST00000603841.1
|
FBXW7
|
F-box and WD repeat domain containing 7 |
| chr11_+_54706832 | 0.90 |
ENST00000319760.8
|
OR4A5
|
olfactory receptor family 4 subfamily A member 5 |
| chr12_+_55362975 | 0.90 |
ENST00000641576.1
|
OR6C75
|
olfactory receptor family 6 subfamily C member 75 |
| chr12_-_9999176 | 0.89 |
ENST00000298527.10
ENST00000348658.4 |
CLEC1B
|
C-type lectin domain family 1 member B |
| chr2_-_201698628 | 0.89 |
ENST00000602867.1
ENST00000409474.8 |
MPP4
|
membrane palmitoylated protein 4 |
| chr3_-_116444983 | 0.76 |
ENST00000333617.8
|
LSAMP
|
limbic system associated membrane protein |
| chr5_-_139482285 | 0.75 |
ENST00000652110.1
|
STING1
|
stimulator of interferon response cGAMP interactor 1 |
| chr7_-_115968302 | 0.71 |
ENST00000457268.5
|
TFEC
|
transcription factor EC |
| chr5_-_139482341 | 0.69 |
ENST00000651699.1
|
STING1
|
stimulator of interferon response cGAMP interactor 1 |
| chr15_+_71096941 | 0.66 |
ENST00000355327.7
|
THSD4
|
thrombospondin type 1 domain containing 4 |
| chr10_-_122845850 | 0.66 |
ENST00000392790.6
|
CUZD1
|
CUB and zona pellucida like domains 1 |
| chr10_+_13099440 | 0.66 |
ENST00000263036.9
|
OPTN
|
optineurin |
| chr4_-_47981535 | 0.65 |
ENST00000402813.9
|
CNGA1
|
cyclic nucleotide gated channel subunit alpha 1 |
| chr10_+_13099585 | 0.64 |
ENST00000378764.6
|
OPTN
|
optineurin |
| chr10_+_94683771 | 0.63 |
ENST00000339022.6
|
CYP2C18
|
cytochrome P450 family 2 subfamily C member 18 |
| chr2_+_222861423 | 0.62 |
ENST00000681383.1
ENST00000413316.1 ENST00000679558.1 ENST00000680684.1 |
ACSL3
|
acyl-CoA synthetase long chain family member 3 |
| chr12_-_16606795 | 0.61 |
ENST00000447609.5
|
LMO3
|
LIM domain only 3 |
| chr4_-_67883987 | 0.61 |
ENST00000283916.11
|
TMPRSS11D
|
transmembrane serine protease 11D |
| chr6_+_26365176 | 0.58 |
ENST00000377708.7
|
BTN3A2
|
butyrophilin subfamily 3 member A2 |
| chr14_+_36657560 | 0.58 |
ENST00000402703.6
|
PAX9
|
paired box 9 |
| chr10_-_60733467 | 0.57 |
ENST00000373827.6
|
ANK3
|
ankyrin 3 |
| chr3_+_101827982 | 0.57 |
ENST00000461724.5
ENST00000483180.5 ENST00000394054.6 |
NFKBIZ
|
NFKB inhibitor zeta |
| chr6_+_26365215 | 0.56 |
ENST00000527422.5
ENST00000356386.6 ENST00000396948.5 |
BTN3A2
|
butyrophilin subfamily 3 member A2 |
| chr15_+_64387828 | 0.54 |
ENST00000261884.8
|
TRIP4
|
thyroid hormone receptor interactor 4 |
| chr13_-_46897021 | 0.54 |
ENST00000542664.4
ENST00000543956.5 |
HTR2A
|
5-hydroxytryptamine receptor 2A |
| chr3_+_190615308 | 0.54 |
ENST00000412080.1
|
IL1RAP
|
interleukin 1 receptor accessory protein |
| chr12_-_21774688 | 0.54 |
ENST00000240662.3
|
KCNJ8
|
potassium inwardly rectifying channel subfamily J member 8 |
| chr9_-_127755243 | 0.54 |
ENST00000629203.2
ENST00000420366.5 |
SH2D3C
|
SH2 domain containing 3C |
| chr15_+_76336755 | 0.53 |
ENST00000290759.9
|
ISL2
|
ISL LIM homeobox 2 |
| chrX_+_15749848 | 0.51 |
ENST00000479740.5
ENST00000454127.2 |
CA5B
|
carbonic anhydrase 5B |
| chr6_-_29087313 | 0.50 |
ENST00000377173.4
|
OR2B3
|
olfactory receptor family 2 subfamily B member 3 |
| chr4_-_68670648 | 0.50 |
ENST00000338206.6
|
UGT2B15
|
UDP glucuronosyltransferase family 2 member B15 |
| chr4_+_71062642 | 0.50 |
ENST00000649996.1
|
SLC4A4
|
solute carrier family 4 member 4 |
| chr3_+_169911566 | 0.49 |
ENST00000428432.6
ENST00000335556.7 |
SAMD7
|
sterile alpha motif domain containing 7 |
| chr18_-_22417910 | 0.49 |
ENST00000391403.4
|
CTAGE1
|
cutaneous T cell lymphoma-associated antigen 1 |
| chr1_-_154627945 | 0.49 |
ENST00000681683.1
ENST00000368471.8 ENST00000649042.1 ENST00000680270.1 ENST00000649022.2 ENST00000681056.1 ENST00000649724.1 |
ADAR
|
adenosine deaminase RNA specific |
| chr6_-_118710065 | 0.48 |
ENST00000392500.7
ENST00000368488.9 ENST00000434604.5 |
CEP85L
|
centrosomal protein 85 like |
| chr1_+_45913647 | 0.48 |
ENST00000674079.1
|
MAST2
|
microtubule associated serine/threonine kinase 2 |
| chr12_+_64497968 | 0.48 |
ENST00000676593.1
ENST00000677093.1 |
TBK1
ENSG00000288665.1
|
TANK binding kinase 1 novel transcript |
| chr2_-_162152404 | 0.47 |
ENST00000375497.3
|
GCG
|
glucagon |
| chr2_-_29074515 | 0.47 |
ENST00000331664.6
|
PCARE
|
photoreceptor cilium actin regulator |
| chr5_+_31193739 | 0.47 |
ENST00000514738.5
|
CDH6
|
cadherin 6 |
| chr12_+_8157034 | 0.47 |
ENST00000396570.7
|
ZNF705A
|
zinc finger protein 705A |
| chr5_-_135954962 | 0.46 |
ENST00000522943.5
ENST00000514447.2 ENST00000274507.6 |
LECT2
|
leukocyte cell derived chemotaxin 2 |
| chr5_-_59586393 | 0.46 |
ENST00000505453.1
ENST00000360047.9 |
PDE4D
|
phosphodiesterase 4D |
| chr6_-_25830557 | 0.45 |
ENST00000468082.1
|
SLC17A1
|
solute carrier family 17 member 1 |
| chr11_+_93746433 | 0.45 |
ENST00000526335.1
|
C11orf54
|
chromosome 11 open reading frame 54 |
| chr15_-_55270280 | 0.45 |
ENST00000564609.5
|
RAB27A
|
RAB27A, member RAS oncogene family |
| chr7_-_138627444 | 0.44 |
ENST00000463557.1
|
SVOPL
|
SVOP like |
| chr5_+_168486462 | 0.43 |
ENST00000231572.8
ENST00000626454.1 |
RARS1
|
arginyl-tRNA synthetase 1 |
| chr8_+_131939865 | 0.43 |
ENST00000520362.5
ENST00000519656.1 |
EFR3A
|
EFR3 homolog A |
| chr2_+_101839815 | 0.42 |
ENST00000421882.5
|
MAP4K4
|
mitogen-activated protein kinase kinase kinase kinase 4 |
| chr4_+_99511008 | 0.42 |
ENST00000514652.5
ENST00000326581.9 |
C4orf17
|
chromosome 4 open reading frame 17 |
| chr10_-_73591330 | 0.42 |
ENST00000451492.5
ENST00000681793.1 ENST00000680396.1 ENST00000413442.5 |
USP54
|
ubiquitin specific peptidase 54 |
| chr11_-_30586866 | 0.42 |
ENST00000528686.2
|
MPPED2
|
metallophosphoesterase domain containing 2 |
| chr9_+_100427123 | 0.41 |
ENST00000395067.7
|
MSANTD3
|
Myb/SANT DNA binding domain containing 3 |
| chr6_-_130222813 | 0.40 |
ENST00000437477.6
ENST00000439090.7 |
SAMD3
|
sterile alpha motif domain containing 3 |
| chr2_-_206218024 | 0.39 |
ENST00000407325.6
ENST00000612892.4 ENST00000411719.1 |
GPR1
|
G protein-coupled receptor 1 |
| chr3_+_98130721 | 0.39 |
ENST00000641874.1
|
OR5H1
|
olfactory receptor family 5 subfamily H member 1 |
| chr8_+_123072667 | 0.39 |
ENST00000519418.5
ENST00000287380.6 ENST00000327098.9 ENST00000522420.5 ENST00000521676.5 |
TBC1D31
|
TBC1 domain family member 31 |
| chr4_-_122456725 | 0.38 |
ENST00000226730.5
|
IL2
|
interleukin 2 |
| chr2_+_119679184 | 0.38 |
ENST00000445518.1
ENST00000272521.7 ENST00000409951.1 |
TMEM177
|
transmembrane protein 177 |
| chr3_+_98168700 | 0.37 |
ENST00000383696.4
|
OR5H15
|
olfactory receptor family 5 subfamily H member 15 |
| chr3_+_98463201 | 0.37 |
ENST00000642057.1
|
OR5K1
|
olfactory receptor family 5 subfamily K member 1 |
| chr2_+_218710931 | 0.36 |
ENST00000442769.5
ENST00000424644.1 |
TTLL4
|
tubulin tyrosine ligase like 4 |
| chr5_+_122129533 | 0.35 |
ENST00000296600.5
ENST00000504912.1 ENST00000505843.1 |
ZNF474
|
zinc finger protein 474 |
| chrX_+_15790446 | 0.35 |
ENST00000380308.7
ENST00000307771.8 |
ZRSR2
|
zinc finger CCCH-type, RNA binding motif and serine/arginine rich 2 |
| chr18_+_616711 | 0.34 |
ENST00000579494.1
|
CLUL1
|
clusterin like 1 |
| chr6_+_150866333 | 0.34 |
ENST00000618312.4
ENST00000423867.2 |
MTHFD1L
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1 like |
| chr1_-_94925759 | 0.34 |
ENST00000415017.1
ENST00000545882.5 |
CNN3
|
calponin 3 |
| chr4_-_69961007 | 0.33 |
ENST00000353151.3
|
CSN2
|
casein beta |
| chr3_+_171843337 | 0.33 |
ENST00000334567.9
ENST00000619900.4 ENST00000450693.1 |
TMEM212
|
transmembrane protein 212 |
| chr20_-_18413216 | 0.33 |
ENST00000480488.2
|
DZANK1
|
double zinc ribbon and ankyrin repeat domains 1 |
| chr5_+_162067990 | 0.33 |
ENST00000641017.1
|
GABRG2
|
gamma-aminobutyric acid type A receptor subunit gamma2 |
| chr2_-_213151590 | 0.32 |
ENST00000374319.8
ENST00000457361.5 ENST00000451136.6 ENST00000434687.6 |
IKZF2
|
IKAROS family zinc finger 2 |
| chr5_+_31193678 | 0.32 |
ENST00000265071.3
|
CDH6
|
cadherin 6 |
| chr16_-_9943182 | 0.32 |
ENST00000535259.6
|
GRIN2A
|
glutamate ionotropic receptor NMDA type subunit 2A |
| chr2_+_68734773 | 0.32 |
ENST00000409202.8
|
ARHGAP25
|
Rho GTPase activating protein 25 |
| chr1_+_74235377 | 0.32 |
ENST00000326637.8
|
TNNI3K
|
TNNI3 interacting kinase |
| chr2_+_165469647 | 0.32 |
ENST00000421875.5
ENST00000314499.11 ENST00000409664.5 ENST00000651982.1 |
CSRNP3
|
cysteine and serine rich nuclear protein 3 |
| chr4_-_99435396 | 0.32 |
ENST00000209665.8
|
ADH7
|
alcohol dehydrogenase 7 (class IV), mu or sigma polypeptide |
| chr4_+_154563003 | 0.32 |
ENST00000302068.9
ENST00000509493.1 |
FGB
|
fibrinogen beta chain |
| chr3_+_109136707 | 0.31 |
ENST00000622536.6
|
C3orf85
|
chromosome 3 open reading frame 85 |
| chr5_+_157180816 | 0.31 |
ENST00000422843.8
|
ITK
|
IL2 inducible T cell kinase |
| chr11_+_123902167 | 0.30 |
ENST00000641687.1
|
OR8D4
|
olfactory receptor family 8 subfamily D member 4 |
| chr4_-_71784046 | 0.30 |
ENST00000513476.5
ENST00000273951.13 |
GC
|
GC vitamin D binding protein |
| chr7_+_95485898 | 0.29 |
ENST00000428113.5
|
ASB4
|
ankyrin repeat and SOCS box containing 4 |
| chr17_-_10549612 | 0.29 |
ENST00000532183.6
ENST00000397183.6 ENST00000420805.1 |
MYH2
|
myosin heavy chain 2 |
| chr17_-_10549694 | 0.29 |
ENST00000622564.4
|
MYH2
|
myosin heavy chain 2 |
| chr11_+_55827219 | 0.29 |
ENST00000378397.1
|
OR5L2
|
olfactory receptor family 5 subfamily L member 2 |
| chr17_-_10549652 | 0.29 |
ENST00000245503.10
|
MYH2
|
myosin heavy chain 2 |
| chr18_-_28036585 | 0.29 |
ENST00000399380.7
|
CDH2
|
cadherin 2 |
| chr14_+_51489112 | 0.29 |
ENST00000356218.8
|
FRMD6
|
FERM domain containing 6 |
| chr3_-_37174578 | 0.29 |
ENST00000336686.9
|
LRRFIP2
|
LRR binding FLII interacting protein 2 |
| chr8_-_28490220 | 0.28 |
ENST00000517673.5
ENST00000380254.7 ENST00000518734.5 ENST00000346498.6 |
FBXO16
|
F-box protein 16 |
| chr11_+_121102666 | 0.28 |
ENST00000264037.2
|
TECTA
|
tectorin alpha |
| chr1_+_40247926 | 0.28 |
ENST00000372766.4
|
TMCO2
|
transmembrane and coiled-coil domains 2 |
| chr1_-_162376841 | 0.28 |
ENST00000367935.10
|
SPATA46
|
spermatogenesis associated 46 |
| chr8_-_100706931 | 0.28 |
ENST00000520868.5
|
PABPC1
|
poly(A) binding protein cytoplasmic 1 |
| chr7_+_95485934 | 0.28 |
ENST00000325885.6
|
ASB4
|
ankyrin repeat and SOCS box containing 4 |
| chr17_-_65561137 | 0.28 |
ENST00000580513.1
|
AXIN2
|
axin 2 |
| chr1_-_86383078 | 0.28 |
ENST00000460698.6
|
ODF2L
|
outer dense fiber of sperm tails 2 like |
| chr17_+_62370218 | 0.27 |
ENST00000450662.7
|
EFCAB3
|
EF-hand calcium binding domain 3 |
| chr3_+_63967738 | 0.27 |
ENST00000484332.1
|
ATXN7
|
ataxin 7 |
| chr6_+_28267107 | 0.27 |
ENST00000621053.1
ENST00000617168.4 ENST00000421553.7 ENST00000611552.2 ENST00000623276.3 |
ENSG00000276302.1
ZSCAN26
|
novel protein zinc finger and SCAN domain containing 26 |
| chr18_+_616672 | 0.27 |
ENST00000338387.11
|
CLUL1
|
clusterin like 1 |
| chr19_-_47886308 | 0.27 |
ENST00000222002.4
|
SULT2A1
|
sulfotransferase family 2A member 1 |
| chr10_-_59362584 | 0.27 |
ENST00000618427.4
ENST00000611933.4 |
FAM13C
|
family with sequence similarity 13 member C |
| chr10_-_113854368 | 0.26 |
ENST00000369305.1
|
DCLRE1A
|
DNA cross-link repair 1A |
| chr12_+_53097656 | 0.26 |
ENST00000301464.4
|
IGFBP6
|
insulin like growth factor binding protein 6 |
| chr6_-_83193207 | 0.26 |
ENST00000652468.1
ENST00000512866.5 ENST00000616566.5 ENST00000510258.1 ENST00000650642.1 ENST00000513973.6 ENST00000503094.1 ENST00000283977.9 ENST00000509219.2 ENST00000651425.1 ENST00000508748.5 |
PGM3
|
phosphoglucomutase 3 |
| chr2_-_89085787 | 0.26 |
ENST00000390252.2
|
IGKV3-15
|
immunoglobulin kappa variable 3-15 |
| chr11_+_56027654 | 0.26 |
ENST00000641320.1
|
OR5AS1
|
olfactory receptor family 5 subfamily AS member 1 |
| chr6_+_158312459 | 0.26 |
ENST00000367097.8
|
TULP4
|
TUB like protein 4 |
| chr7_+_144000320 | 0.26 |
ENST00000641698.1
|
OR6B1
|
olfactory receptor family 6 subfamily B member 1 |
| chr1_-_100894818 | 0.25 |
ENST00000370114.8
|
EXTL2
|
exostosin like glycosyltransferase 2 |
| chr1_-_100894775 | 0.25 |
ENST00000416479.1
ENST00000370113.7 |
EXTL2
|
exostosin like glycosyltransferase 2 |
| chr2_-_135047432 | 0.25 |
ENST00000392915.7
ENST00000637841.1 ENST00000414343.1 |
MAP3K19
|
mitogen-activated protein kinase kinase kinase 19 |
| chr20_+_59163810 | 0.25 |
ENST00000371030.4
|
ZNF831
|
zinc finger protein 831 |
| chr11_-_64935882 | 0.25 |
ENST00000532246.1
ENST00000279168.7 |
GPHA2
|
glycoprotein hormone subunit alpha 2 |
| chr3_+_35679690 | 0.25 |
ENST00000413378.5
ENST00000417925.5 |
ARPP21
|
cAMP regulated phosphoprotein 21 |
| chr10_-_103153609 | 0.25 |
ENST00000675985.1
|
NT5C2
|
5'-nucleotidase, cytosolic II |
| chr12_+_41437680 | 0.24 |
ENST00000649474.1
ENST00000539469.6 ENST00000298919.7 |
PDZRN4
|
PDZ domain containing ring finger 4 |
| chr12_-_18738006 | 0.24 |
ENST00000266505.12
ENST00000543242.5 ENST00000539072.5 ENST00000541966.5 ENST00000648272.1 |
PLCZ1
|
phospholipase C zeta 1 |
| chr18_+_34976928 | 0.24 |
ENST00000591734.5
ENST00000413393.5 ENST00000589180.5 ENST00000587359.5 |
MAPRE2
|
microtubule associated protein RP/EB family member 2 |
| chr2_-_202870862 | 0.24 |
ENST00000454326.5
ENST00000432273.5 ENST00000450143.5 ENST00000411681.5 |
ICA1L
|
islet cell autoantigen 1 like |
| chr2_-_212124901 | 0.24 |
ENST00000402597.6
|
ERBB4
|
erb-b2 receptor tyrosine kinase 4 |
| chr4_+_119135825 | 0.24 |
ENST00000307128.6
|
MYOZ2
|
myozenin 2 |
| chr17_+_42773442 | 0.24 |
ENST00000253794.7
ENST00000590339.5 ENST00000589520.1 |
VPS25
|
vacuolar protein sorting 25 homolog |
| chr17_+_19533818 | 0.23 |
ENST00000436810.6
ENST00000270570.8 ENST00000575023.5 ENST00000395585.5 |
SLC47A1
|
solute carrier family 47 member 1 |
| chr15_+_94297939 | 0.23 |
ENST00000357742.9
|
MCTP2
|
multiple C2 and transmembrane domain containing 2 |
| chr6_+_131637296 | 0.23 |
ENST00000358229.6
ENST00000357639.8 |
ENPP3
|
ectonucleotide pyrophosphatase/phosphodiesterase 3 |
| chr3_+_98147479 | 0.23 |
ENST00000641380.1
|
OR5H14
|
olfactory receptor family 5 subfamily H member 14 |
| chr3_-_190449782 | 0.23 |
ENST00000354905.3
|
TMEM207
|
transmembrane protein 207 |
| chr6_+_26224192 | 0.23 |
ENST00000634733.1
|
H3C6
|
H3 clustered histone 6 |
| chr9_-_101435760 | 0.22 |
ENST00000647789.2
ENST00000616752.1 |
ALDOB
|
aldolase, fructose-bisphosphate B |
| chr2_+_218710821 | 0.22 |
ENST00000392102.6
ENST00000457313.5 ENST00000415717.5 |
TTLL4
|
tubulin tyrosine ligase like 4 |
| chr6_+_12717660 | 0.22 |
ENST00000674637.1
|
PHACTR1
|
phosphatase and actin regulator 1 |
| chr17_+_75721327 | 0.22 |
ENST00000579662.5
|
ITGB4
|
integrin subunit beta 4 |
| chr3_+_46370854 | 0.22 |
ENST00000292303.4
|
CCR5
|
C-C motif chemokine receptor 5 |
| chr3_+_98149326 | 0.22 |
ENST00000437310.1
|
OR5H14
|
olfactory receptor family 5 subfamily H member 14 |
| chr1_+_236395394 | 0.21 |
ENST00000359362.6
|
EDARADD
|
EDAR associated death domain |
| chr20_-_56497608 | 0.21 |
ENST00000617620.1
|
GCNT7
|
glucosaminyl (N-acetyl) transferase family member 7 |
| chr6_-_15586006 | 0.21 |
ENST00000462989.6
|
DTNBP1
|
dystrobrevin binding protein 1 |
| chr2_-_162152239 | 0.21 |
ENST00000418842.7
|
GCG
|
glucagon |
| chr10_+_94762673 | 0.21 |
ENST00000480405.2
ENST00000371321.9 |
CYP2C19
|
cytochrome P450 family 2 subfamily C member 19 |
| chr7_+_130207847 | 0.21 |
ENST00000297819.4
|
SSMEM1
|
serine rich single-pass membrane protein 1 |
| chr14_+_22304051 | 0.21 |
ENST00000390466.1
|
TRAV39
|
T cell receptor alpha variable 39 |
| chrX_-_15315615 | 0.21 |
ENST00000380470.7
ENST00000480796.6 |
ASB11
|
ankyrin repeat and SOCS box containing 11 |
| chr1_+_218285283 | 0.21 |
ENST00000366932.4
|
RRP15
|
ribosomal RNA processing 15 homolog |
| chr6_+_28267044 | 0.20 |
ENST00000316606.10
|
ZSCAN26
|
zinc finger and SCAN domain containing 26 |
| chr14_-_67412112 | 0.20 |
ENST00000216446.9
|
PLEK2
|
pleckstrin 2 |
| chr12_-_118359105 | 0.20 |
ENST00000541186.5
ENST00000539872.5 |
TAOK3
|
TAO kinase 3 |
| chr12_-_118359639 | 0.20 |
ENST00000541786.5
ENST00000419821.6 ENST00000541878.5 |
TAOK3
|
TAO kinase 3 |
| chr10_-_60572599 | 0.20 |
ENST00000503366.5
|
ANK3
|
ankyrin 3 |
| chr10_+_122980448 | 0.20 |
ENST00000405485.2
|
PSTK
|
phosphoseryl-tRNA kinase |
| chr10_+_96304425 | 0.19 |
ENST00000371174.5
|
DNTT
|
DNA nucleotidylexotransferase |
| chr11_-_27722021 | 0.19 |
ENST00000314915.6
|
BDNF
|
brain derived neurotrophic factor |
| chr3_-_190862688 | 0.19 |
ENST00000442080.6
|
GMNC
|
geminin coiled-coil domain containing |
| chr2_+_184598520 | 0.19 |
ENST00000302277.7
|
ZNF804A
|
zinc finger protein 804A |
| chr3_-_108953870 | 0.19 |
ENST00000261047.8
|
GUCA1C
|
guanylate cyclase activator 1C |
| chr12_-_48157583 | 0.19 |
ENST00000535988.3
ENST00000536953.5 ENST00000535055.5 ENST00000536549.5 |
ASB8
|
ankyrin repeat and SOCS box containing 8 |
| chr1_+_162069674 | 0.19 |
ENST00000361897.10
|
NOS1AP
|
nitric oxide synthase 1 adaptor protein |
| chr10_+_96304392 | 0.19 |
ENST00000630152.1
|
DNTT
|
DNA nucleotidylexotransferase |
| chr4_+_15339818 | 0.19 |
ENST00000397700.6
ENST00000295297.4 |
C1QTNF7
|
C1q and TNF related 7 |
| chr6_+_63521738 | 0.18 |
ENST00000648894.1
ENST00000639568.2 |
PTP4A1
|
protein tyrosine phosphatase 4A1 |
| chr2_-_189179754 | 0.18 |
ENST00000374866.9
ENST00000618828.1 |
COL5A2
|
collagen type V alpha 2 chain |
| chr13_-_36370171 | 0.18 |
ENST00000355182.8
|
SPART
|
spartin |
| chr5_+_122129597 | 0.18 |
ENST00000514925.1
|
ENSG00000250803.6
|
novel zinc finger protein |
| chr1_-_197067234 | 0.18 |
ENST00000367412.2
|
F13B
|
coagulation factor XIII B chain |
| chr1_-_229434086 | 0.18 |
ENST00000366684.7
ENST00000684723.1 ENST00000366683.4 |
ACTA1
|
actin alpha 1, skeletal muscle |
| chr7_+_114414997 | 0.18 |
ENST00000462331.5
ENST00000393491.7 ENST00000403559.8 ENST00000408937.7 ENST00000393498.6 ENST00000393495.7 ENST00000378237.7 |
FOXP2
|
forkhead box P2 |
| chr3_+_35679614 | 0.18 |
ENST00000474696.5
ENST00000412048.5 ENST00000396482.6 ENST00000432682.5 |
ARPP21
|
cAMP regulated phosphoprotein 21 |
| chr2_-_86105839 | 0.18 |
ENST00000263857.11
|
POLR1A
|
RNA polymerase I subunit A |
| chr10_-_59362460 | 0.18 |
ENST00000422313.6
ENST00000435852.6 ENST00000614220.4 ENST00000618804.5 ENST00000621119.4 |
FAM13C
|
family with sequence similarity 13 member C |
| chr6_-_135054785 | 0.17 |
ENST00000367820.6
ENST00000314674.7 ENST00000524715.5 ENST00000415177.6 ENST00000367826.6 ENST00000367837.10 |
HBS1L
|
HBS1 like translational GTPase |
| chr7_+_138076453 | 0.17 |
ENST00000242375.8
ENST00000411726.6 |
AKR1D1
|
aldo-keto reductase family 1 member D1 |
| chr17_-_7080275 | 0.17 |
ENST00000571664.1
ENST00000254868.8 |
CLEC10A
|
C-type lectin domain containing 10A |
| chr10_+_94683722 | 0.17 |
ENST00000285979.11
|
CYP2C18
|
cytochrome P450 family 2 subfamily C member 18 |
| chr11_+_118077009 | 0.17 |
ENST00000616579.4
ENST00000534111.5 |
TMPRSS4
|
transmembrane serine protease 4 |
| chr11_-_26572102 | 0.17 |
ENST00000455601.6
|
MUC15
|
mucin 15, cell surface associated |
| chr3_+_98166696 | 0.16 |
ENST00000641450.1
|
OR5H15
|
olfactory receptor family 5 subfamily H member 15 |
| chr5_+_136132772 | 0.16 |
ENST00000545279.6
ENST00000507118.5 ENST00000511116.5 ENST00000545620.5 ENST00000509297.6 |
SMAD5
|
SMAD family member 5 |
| chrX_-_49043345 | 0.16 |
ENST00000315869.8
|
TFE3
|
transcription factor binding to IGHM enhancer 3 |
| chr1_+_9588860 | 0.16 |
ENST00000340381.11
ENST00000340305.9 |
TMEM201
|
transmembrane protein 201 |
| chr9_-_21482313 | 0.16 |
ENST00000448696.4
|
IFNE
|
interferon epsilon |
| chr3_+_184300564 | 0.16 |
ENST00000435761.5
ENST00000439383.5 |
PSMD2
|
proteasome 26S subunit, non-ATPase 2 |
| chr2_+_68734861 | 0.15 |
ENST00000467265.5
|
ARHGAP25
|
Rho GTPase activating protein 25 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 5.7 | GO:0043950 | positive regulation of cAMP-mediated signaling(GO:0043950) |
| 0.2 | 0.9 | GO:2000638 | regulation of SREBP signaling pathway(GO:2000638) negative regulation of SREBP signaling pathway(GO:2000639) |
| 0.2 | 1.4 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.2 | 0.8 | GO:1900827 | maintenance of protein location in membrane(GO:0072658) maintenance of protein location in plasma membrane(GO:0072660) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.2 | 0.5 | GO:0021524 | visceral motor neuron differentiation(GO:0021524) |
| 0.2 | 1.5 | GO:2000124 | regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.2 | 0.9 | GO:0032468 | Golgi calcium ion homeostasis(GO:0032468) |
| 0.2 | 0.6 | GO:2001247 | positive regulation of phosphatidylcholine biosynthetic process(GO:2001247) |
| 0.1 | 0.4 | GO:0030573 | bile acid catabolic process(GO:0030573) |
| 0.1 | 1.2 | GO:0032621 | interleukin-18 production(GO:0032621) |
| 0.1 | 0.7 | GO:0035948 | positive regulation of gluconeogenesis by positive regulation of transcription from RNA polymerase II promoter(GO:0035948) |
| 0.1 | 1.1 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 0.1 | 0.3 | GO:1903487 | regulation of lactation(GO:1903487) |
| 0.1 | 0.5 | GO:0010513 | positive regulation of phosphatidylinositol biosynthetic process(GO:0010513) |
| 0.1 | 1.3 | GO:0034378 | chylomicron assembly(GO:0034378) |
| 0.1 | 0.5 | GO:0044565 | dendritic cell proliferation(GO:0044565) |
| 0.1 | 0.7 | GO:0006931 | substrate-dependent cell migration, cell attachment to substrate(GO:0006931) |
| 0.1 | 1.1 | GO:0090204 | protein localization to nuclear pore(GO:0090204) |
| 0.1 | 0.6 | GO:2000324 | positive regulation of glucocorticoid receptor signaling pathway(GO:2000324) |
| 0.1 | 0.3 | GO:0009257 | 10-formyltetrahydrofolate biosynthetic process(GO:0009257) |
| 0.1 | 0.5 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.1 | 0.5 | GO:1900369 | negative regulation of RNA interference(GO:1900369) |
| 0.1 | 0.3 | GO:0010430 | fatty acid omega-oxidation(GO:0010430) |
| 0.1 | 0.2 | GO:0061026 | cardiac muscle tissue regeneration(GO:0061026) |
| 0.1 | 0.5 | GO:0038172 | interleukin-33-mediated signaling pathway(GO:0038172) |
| 0.1 | 0.2 | GO:0030505 | inorganic diphosphate transport(GO:0030505) |
| 0.1 | 0.4 | GO:0046013 | regulation of T cell homeostatic proliferation(GO:0046013) |
| 0.1 | 0.4 | GO:1903435 | positive regulation of constitutive secretory pathway(GO:1903435) |
| 0.1 | 0.3 | GO:0003383 | apical constriction(GO:0003383) |
| 0.1 | 0.2 | GO:0031550 | positive regulation of brain-derived neurotrophic factor receptor signaling pathway(GO:0031550) |
| 0.1 | 0.3 | GO:0033058 | directional locomotion(GO:0033058) |
| 0.1 | 0.2 | GO:1903947 | positive regulation of voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization(GO:1903762) positive regulation of ventricular cardiac muscle cell action potential(GO:1903947) positive regulation of membrane repolarization during ventricular cardiac muscle cell action potential(GO:1905026) positive regulation of membrane repolarization during cardiac muscle cell action potential(GO:1905033) |
| 0.1 | 0.2 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.1 | 0.2 | GO:0021758 | caudate nucleus development(GO:0021757) putamen development(GO:0021758) |
| 0.1 | 0.3 | GO:0097056 | selenocysteinyl-tRNA(Sec) biosynthetic process(GO:0097056) |
| 0.1 | 0.1 | GO:0052047 | interaction with other organism via secreted substance involved in symbiotic interaction(GO:0052047) |
| 0.1 | 0.7 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.0 | 0.6 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.0 | 0.3 | GO:0061181 | regulation of chondrocyte development(GO:0061181) |
| 0.0 | 0.2 | GO:0007343 | egg activation(GO:0007343) |
| 0.0 | 0.2 | GO:0043328 | protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) |
| 0.0 | 0.3 | GO:0090238 | positive regulation of arachidonic acid secretion(GO:0090238) |
| 0.0 | 0.5 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.0 | 0.1 | GO:0060366 | subpallium cell proliferation in forebrain(GO:0022012) lateral ganglionic eminence cell proliferation(GO:0022018) lambdoid suture morphogenesis(GO:0060366) sagittal suture morphogenesis(GO:0060367) anterior semicircular canal development(GO:0060873) lateral semicircular canal development(GO:0060875) |
| 0.0 | 0.3 | GO:1904749 | regulation of protein localization to nucleolus(GO:1904749) |
| 0.0 | 0.3 | GO:0019255 | glucosamine metabolic process(GO:0006041) glucose 1-phosphate metabolic process(GO:0019255) |
| 0.0 | 1.0 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.0 | 0.6 | GO:2001214 | positive regulation of vasculogenesis(GO:2001214) |
| 0.0 | 0.5 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.0 | 1.1 | GO:0072643 | interferon-gamma secretion(GO:0072643) |
| 0.0 | 0.5 | GO:0042090 | interleukin-12 biosynthetic process(GO:0042090) regulation of interleukin-12 biosynthetic process(GO:0045075) |
| 0.0 | 0.5 | GO:1990573 | potassium ion import across plasma membrane(GO:1990573) |
| 0.0 | 0.1 | GO:1903575 | cornified envelope assembly(GO:1903575) |
| 0.0 | 0.2 | GO:0090131 | mesenchyme migration(GO:0090131) |
| 0.0 | 0.1 | GO:0001978 | regulation of systemic arterial blood pressure by carotid sinus baroreceptor feedback(GO:0001978) baroreceptor response to increased systemic arterial blood pressure(GO:0001983) |
| 0.0 | 0.1 | GO:0034414 | tRNA 3'-trailer cleavage, endonucleolytic(GO:0034414) tRNA 3'-trailer cleavage(GO:0042779) |
| 0.0 | 0.1 | GO:1905000 | regulation of membrane repolarization during atrial cardiac muscle cell action potential(GO:1905000) |
| 0.0 | 0.3 | GO:1902897 | regulation of postsynaptic density protein 95 clustering(GO:1902897) |
| 0.0 | 0.2 | GO:0061624 | fructose catabolic process(GO:0006001) fructose catabolic process to hydroxyacetone phosphate and glyceraldehyde-3-phosphate(GO:0061624) |
| 0.0 | 0.4 | GO:2000622 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.0 | 0.6 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.0 | 0.1 | GO:0002378 | immunoglobulin biosynthetic process(GO:0002378) |
| 0.0 | 5.3 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 0.1 | GO:0002528 | regulation of vascular permeability involved in acute inflammatory response(GO:0002528) |
| 0.0 | 0.3 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.0 | 0.3 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.0 | 0.2 | GO:0001880 | Mullerian duct regression(GO:0001880) |
| 0.0 | 0.1 | GO:0090134 | mesendoderm migration(GO:0090133) cell migration involved in mesendoderm migration(GO:0090134) |
| 0.0 | 0.2 | GO:0046618 | drug export(GO:0046618) |
| 0.0 | 0.5 | GO:0086069 | bundle of His cell to Purkinje myocyte communication(GO:0086069) |
| 0.0 | 0.1 | GO:0035854 | regulation of primitive erythrocyte differentiation(GO:0010725) eosinophil fate commitment(GO:0035854) |
| 0.0 | 0.7 | GO:0035641 | locomotory exploration behavior(GO:0035641) |
| 0.0 | 0.2 | GO:0044800 | fusion of virus membrane with host plasma membrane(GO:0019064) membrane fusion involved in viral entry into host cell(GO:0039663) multi-organism membrane fusion(GO:0044800) |
| 0.0 | 0.1 | GO:0010645 | positive regulation of glomerular filtration(GO:0003104) regulation of cell communication by chemical coupling(GO:0010645) positive regulation of cell communication by chemical coupling(GO:0010652) |
| 0.0 | 0.1 | GO:0070105 | positive regulation of interleukin-6-mediated signaling pathway(GO:0070105) |
| 0.0 | 0.3 | GO:0031848 | protection from non-homologous end joining at telomere(GO:0031848) |
| 0.0 | 0.8 | GO:0022400 | regulation of rhodopsin mediated signaling pathway(GO:0022400) |
| 0.0 | 0.1 | GO:0072268 | specification of loop of Henle identity(GO:0072086) pattern specification involved in metanephros development(GO:0072268) |
| 0.0 | 0.1 | GO:0000023 | maltose metabolic process(GO:0000023) |
| 0.0 | 0.1 | GO:0070171 | negative regulation of tooth mineralization(GO:0070171) |
| 0.0 | 0.9 | GO:0030220 | platelet formation(GO:0030220) |
| 0.0 | 0.0 | GO:1902159 | regulation of cyclic nucleotide-gated ion channel activity(GO:1902159) |
| 0.0 | 0.1 | GO:2001107 | negative regulation of establishment of T cell polarity(GO:1903904) negative regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001107) |
| 0.0 | 0.6 | GO:0042481 | regulation of odontogenesis(GO:0042481) |
| 0.0 | 0.3 | GO:0060159 | regulation of dopamine receptor signaling pathway(GO:0060159) |
| 0.0 | 0.3 | GO:0052695 | cellular glucuronidation(GO:0052695) |
| 0.0 | 0.1 | GO:0015722 | canalicular bile acid transport(GO:0015722) |
| 0.0 | 0.1 | GO:1904381 | Golgi apparatus mannose trimming(GO:1904381) |
| 0.0 | 0.1 | GO:0090080 | positive regulation of MAPKKK cascade by fibroblast growth factor receptor signaling pathway(GO:0090080) |
| 0.0 | 0.1 | GO:0010897 | negative regulation of triglyceride catabolic process(GO:0010897) |
| 0.0 | 0.1 | GO:0046598 | positive regulation of viral entry into host cell(GO:0046598) |
| 0.0 | 0.1 | GO:0006620 | posttranslational protein targeting to membrane(GO:0006620) |
| 0.0 | 1.1 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 0.3 | GO:0042359 | vitamin D metabolic process(GO:0042359) |
| 0.0 | 0.3 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.0 | 0.2 | GO:0035878 | nail development(GO:0035878) |
| 0.0 | 0.1 | GO:0021859 | pyramidal neuron differentiation(GO:0021859) |
| 0.0 | 0.1 | GO:2000809 | positive regulation of synaptic vesicle clustering(GO:2000809) |
| 0.0 | 1.4 | GO:0055008 | cardiac muscle tissue morphogenesis(GO:0055008) |
| 0.0 | 0.1 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.0 | 0.4 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 0.0 | GO:0061300 | cerebellum vasculature development(GO:0061300) |
| 0.0 | 0.2 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.0 | 0.1 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.0 | 0.3 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.0 | 0.0 | GO:1902544 | regulation of DNA N-glycosylase activity(GO:1902544) |
| 0.0 | 0.1 | GO:1901409 | positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.0 | 0.1 | GO:0051967 | negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.0 | 0.2 | GO:0032625 | interleukin-21 production(GO:0032625) interleukin-21 secretion(GO:0072619) |
| 0.0 | 0.1 | GO:0009253 | peptidoglycan metabolic process(GO:0000270) peptidoglycan catabolic process(GO:0009253) |
| 0.0 | 0.2 | GO:0000478 | endonucleolytic cleavage involved in rRNA processing(GO:0000478) |
| 0.0 | 0.3 | GO:0007342 | fusion of sperm to egg plasma membrane(GO:0007342) |
| 0.0 | 0.3 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.0 | 0.1 | GO:0007060 | male meiosis chromosome segregation(GO:0007060) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.1 | 0.9 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.1 | 0.2 | GO:0000814 | ESCRT II complex(GO:0000814) |
| 0.1 | 0.5 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.1 | 0.4 | GO:0089701 | U2AF(GO:0089701) |
| 0.1 | 0.2 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.1 | 0.5 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.1 | 0.6 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.1 | 0.2 | GO:0034515 | proteasome storage granule(GO:0034515) |
| 0.0 | 0.5 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.0 | 0.1 | GO:1990038 | glial cytoplasmic inclusion(GO:0097409) classical Lewy body(GO:0097414) Lewy neurite(GO:0097462) Lewy body corona(GO:1990038) |
| 0.0 | 0.2 | GO:1902937 | inward rectifier potassium channel complex(GO:1902937) |
| 0.0 | 0.7 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.0 | 0.4 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.0 | 0.8 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.0 | 0.6 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.0 | 0.7 | GO:0043205 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.0 | 0.3 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.3 | GO:0036454 | insulin-like growth factor binding protein complex(GO:0016942) growth factor complex(GO:0036454) |
| 0.0 | 1.5 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 0.3 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.5 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.0 | 0.1 | GO:0005944 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) |
| 0.0 | 0.1 | GO:0060171 | stereocilium membrane(GO:0060171) |
| 0.0 | 0.0 | GO:0072536 | interleukin-23 receptor complex(GO:0072536) |
| 0.0 | 0.3 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.0 | 5.9 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 0.3 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 0.4 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 1.1 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.0 | 0.2 | GO:0033162 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.0 | 0.1 | GO:0000308 | cytoplasmic cyclin-dependent protein kinase holoenzyme complex(GO:0000308) |
| 0.0 | 0.1 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) |
| 0.0 | 0.1 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.0 | 0.2 | GO:0030056 | hemidesmosome(GO:0030056) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 5.9 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.2 | 0.5 | GO:0035248 | alpha-1,4-N-acetylgalactosaminyltransferase activity(GO:0035248) |
| 0.1 | 0.9 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.1 | 0.3 | GO:1902271 | D3 vitamins binding(GO:1902271) |
| 0.1 | 0.5 | GO:0017098 | sulfonylurea receptor binding(GO:0017098) |
| 0.1 | 2.7 | GO:0005549 | odorant binding(GO:0005549) |
| 0.1 | 0.3 | GO:0004329 | formate-tetrahydrofolate ligase activity(GO:0004329) |
| 0.1 | 0.2 | GO:0050146 | nucleoside phosphotransferase activity(GO:0050146) |
| 0.1 | 0.3 | GO:0004031 | aldehyde oxidase activity(GO:0004031) |
| 0.1 | 0.5 | GO:0002114 | interleukin-33 receptor activity(GO:0002114) |
| 0.1 | 0.2 | GO:0061609 | fructose-1-phosphate aldolase activity(GO:0061609) |
| 0.1 | 1.5 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.1 | 0.6 | GO:0070740 | tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.1 | 0.4 | GO:0043208 | glycosphingolipid binding(GO:0043208) |
| 0.1 | 0.4 | GO:0030628 | pre-mRNA 3'-splice site binding(GO:0030628) |
| 0.1 | 0.3 | GO:0031531 | thyrotropin-releasing hormone receptor binding(GO:0031531) |
| 0.1 | 0.2 | GO:0047787 | delta4-3-oxosteroid 5beta-reductase activity(GO:0047787) |
| 0.1 | 0.5 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.1 | 0.4 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.1 | 0.6 | GO:0051378 | amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 0.1 | 0.6 | GO:0005221 | intracellular cyclic nucleotide activated cation channel activity(GO:0005221) cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.1 | 1.0 | GO:0008392 | arachidonic acid monooxygenase activity(GO:0008391) arachidonic acid epoxygenase activity(GO:0008392) |
| 0.0 | 0.2 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 0.0 | 0.2 | GO:0035529 | NADH pyrophosphatase activity(GO:0035529) |
| 0.0 | 0.5 | GO:0015321 | sodium-dependent phosphate transmembrane transporter activity(GO:0015321) |
| 0.0 | 1.3 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.0 | 0.2 | GO:0015307 | drug:proton antiporter activity(GO:0015307) |
| 0.0 | 1.1 | GO:0017081 | chloride channel regulator activity(GO:0017081) |
| 0.0 | 0.3 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.0 | 0.1 | GO:0042781 | 3'-tRNA processing endoribonuclease activity(GO:0042781) |
| 0.0 | 0.3 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.0 | 0.1 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.0 | 0.2 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.0 | 0.3 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.0 | 0.9 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.3 | GO:0043008 | ATP-dependent protein binding(GO:0043008) |
| 0.0 | 0.5 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.1 | GO:0031716 | calcitonin receptor binding(GO:0031716) |
| 0.0 | 0.6 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.0 | 2.2 | GO:0005548 | phospholipid transporter activity(GO:0005548) |
| 0.0 | 0.5 | GO:0031994 | insulin-like growth factor I binding(GO:0031994) |
| 0.0 | 0.3 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.0 | 0.2 | GO:0008048 | calcium sensitive guanylate cyclase activator activity(GO:0008048) |
| 0.0 | 0.1 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.0 | 0.5 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.1 | GO:0032450 | maltose alpha-glucosidase activity(GO:0032450) |
| 0.0 | 0.0 | GO:0004558 | alpha-1,4-glucosidase activity(GO:0004558) |
| 0.0 | 0.9 | GO:0000146 | microfilament motor activity(GO:0000146) |
| 0.0 | 0.1 | GO:0034988 | Fc-gamma receptor I complex binding(GO:0034988) |
| 0.0 | 0.1 | GO:0005436 | sodium:phosphate symporter activity(GO:0005436) |
| 0.0 | 1.4 | GO:0051393 | alpha-actinin binding(GO:0051393) |
| 0.0 | 0.3 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.0 | 0.3 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) |
| 0.0 | 1.4 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 2.6 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.0 | 0.2 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.0 | 0.5 | GO:0031698 | beta-2 adrenergic receptor binding(GO:0031698) |
| 0.0 | 0.4 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.0 | 0.1 | GO:0086083 | cell adhesive protein binding involved in bundle of His cell-Purkinje myocyte communication(GO:0086083) |
| 0.0 | 0.1 | GO:0030882 | lipid antigen binding(GO:0030882) endogenous lipid antigen binding(GO:0030883) exogenous lipid antigen binding(GO:0030884) |
| 0.0 | 0.0 | GO:0043739 | G/U mismatch-specific uracil-DNA glycosylase activity(GO:0043739) |
| 0.0 | 0.1 | GO:0004704 | NF-kappaB-inducing kinase activity(GO:0004704) |
| 0.0 | 0.2 | GO:0004716 | receptor signaling protein tyrosine kinase activity(GO:0004716) |
| 0.0 | 0.1 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.0 | 0.3 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.0 | 0.5 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.0 | 0.2 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.0 | 0.3 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 0.1 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.0 | 0.3 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.0 | 0.4 | GO:0042923 | neuropeptide binding(GO:0042923) |
| 0.0 | 0.4 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 0.1 | GO:0003963 | RNA-3'-phosphate cyclase activity(GO:0003963) |
| 0.0 | 0.1 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.3 | GO:0030275 | LRR domain binding(GO:0030275) |
| 0.0 | 0.1 | GO:0086075 | gap junction channel activity involved in cardiac conduction electrical coupling(GO:0086075) |
| 0.0 | 0.1 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.0 | 0.3 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.0 | 0.4 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.4 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 0.0 | GO:0050571 | 1,5-anhydro-D-fructose reductase activity(GO:0050571) |
| 0.0 | 0.1 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.0 | 0.3 | GO:0001972 | retinoic acid binding(GO:0001972) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 5.9 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.0 | 0.8 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.0 | 0.9 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.0 | 1.4 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.0 | 0.9 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 0.4 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 0.4 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 0.2 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 0.5 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.0 | 0.3 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 6.1 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.1 | 1.0 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.0 | 1.1 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 0.5 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.0 | 0.3 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF CAMKII | Genes involved in CREB phosphorylation through the activation of CaMKII |
| 0.0 | 1.5 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.7 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.0 | 0.5 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.0 | 0.5 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.0 | 1.1 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.3 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.0 | 3.2 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.0 | 0.5 | REACTOME TRAF3 DEPENDENT IRF ACTIVATION PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |
| 0.0 | 0.8 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.6 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 0.9 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.7 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.3 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.0 | 0.2 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 0.2 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.6 | REACTOME INWARDLY RECTIFYING K CHANNELS | Genes involved in Inwardly rectifying K+ channels |
| 0.0 | 0.1 | REACTOME DOWNSTREAM TCR SIGNALING | Genes involved in Downstream TCR signaling |
| 0.0 | 1.2 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.0 | 0.4 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |