Project
Inflammatory response time course, HUVEC (Wada, 2009)
Navigation
Downloads
Results for HOXD9
Z-value: 0.88
Transcription factors associated with HOXD9
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HOXD9
|
ENSG00000128709.13 | HOXD9 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HOXD9 | hg38_v1_chr2_+_176122712_176122727 | -0.51 | 9.0e-03 | Click! |
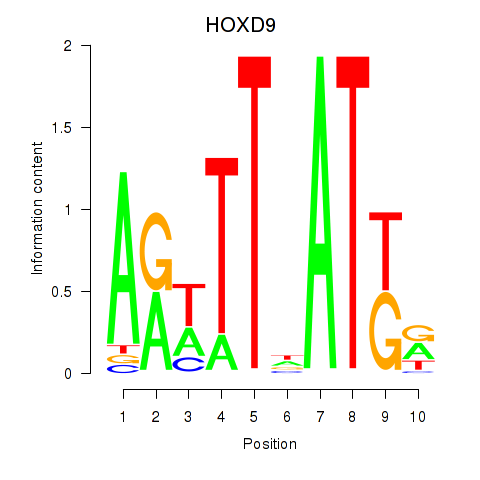
Activity profile of HOXD9 motif
Sorted Z-values of HOXD9 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of HOXD9
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_-_76023489 | 7.30 |
ENST00000306602.3
|
CXCL10
|
C-X-C motif chemokine ligand 10 |
| chr6_+_32844789 | 5.16 |
ENST00000414474.5
|
PSMB9
|
proteasome 20S subunit beta 9 |
| chr1_-_173050931 | 3.46 |
ENST00000404377.5
|
TNFSF18
|
TNF superfamily member 18 |
| chr18_+_63887698 | 2.91 |
ENST00000457692.5
ENST00000299502.9 ENST00000413956.5 |
SERPINB2
|
serpin family B member 2 |
| chr1_+_100719734 | 2.79 |
ENST00000370119.8
ENST00000294728.7 ENST00000347652.6 ENST00000370115.1 |
VCAM1
|
vascular cell adhesion molecule 1 |
| chr2_-_201697993 | 2.37 |
ENST00000428900.6
|
MPP4
|
membrane palmitoylated protein 4 |
| chr4_+_73740541 | 2.29 |
ENST00000401931.1
ENST00000307407.8 |
CXCL8
|
C-X-C motif chemokine ligand 8 |
| chr2_-_201698040 | 2.27 |
ENST00000396886.7
ENST00000409143.5 |
MPP4
|
membrane palmitoylated protein 4 |
| chr2_-_231125032 | 2.07 |
ENST00000258400.4
|
HTR2B
|
5-hydroxytryptamine receptor 2B |
| chr22_-_41286168 | 1.63 |
ENST00000356244.8
|
RANGAP1
|
Ran GTPase activating protein 1 |
| chr3_-_108058361 | 1.62 |
ENST00000398258.7
|
CD47
|
CD47 molecule |
| chr3_-_116444983 | 1.53 |
ENST00000333617.8
|
LSAMP
|
limbic system associated membrane protein |
| chr19_-_49896868 | 1.49 |
ENST00000593956.5
ENST00000391826.7 |
IL4I1
|
interleukin 4 induced 1 |
| chr11_-_105035113 | 1.27 |
ENST00000526568.5
ENST00000531166.5 ENST00000534497.5 ENST00000527979.5 ENST00000533400.6 ENST00000528974.1 ENST00000525825.5 ENST00000353247.9 ENST00000446369.5 ENST00000436863.7 |
CASP1
|
caspase 1 |
| chr18_-_5396265 | 1.27 |
ENST00000579951.2
|
EPB41L3
|
erythrocyte membrane protein band 4.1 like 3 |
| chr17_-_42112674 | 1.23 |
ENST00000251642.8
ENST00000591220.5 |
DHX58
|
DExH-box helicase 58 |
| chr11_-_128587551 | 1.16 |
ENST00000392668.8
|
ETS1
|
ETS proto-oncogene 1, transcription factor |
| chr12_-_57078784 | 1.15 |
ENST00000300128.9
|
NEMP1
|
nuclear envelope integral membrane protein 1 |
| chr2_+_142877653 | 1.11 |
ENST00000375773.6
ENST00000409512.5 ENST00000264170.9 ENST00000410015.6 |
KYNU
|
kynureninase |
| chr12_-_88580459 | 1.06 |
ENST00000552044.1
ENST00000644744.1 ENST00000357116.4 |
KITLG
|
KIT ligand |
| chr4_-_68670648 | 1.05 |
ENST00000338206.6
|
UGT2B15
|
UDP glucuronosyltransferase family 2 member B15 |
| chr22_-_30246739 | 1.05 |
ENST00000403987.3
ENST00000249075.4 |
LIF
|
LIF interleukin 6 family cytokine |
| chr7_-_122699108 | 1.01 |
ENST00000340112.3
|
RNF133
|
ring finger protein 133 |
| chr12_+_55681711 | 1.01 |
ENST00000394252.4
|
METTL7B
|
methyltransferase like 7B |
| chr2_+_143129379 | 1.00 |
ENST00000295095.11
|
ARHGAP15
|
Rho GTPase activating protein 15 |
| chr3_-_27456743 | 0.97 |
ENST00000295736.9
ENST00000428386.5 ENST00000428179.1 |
SLC4A7
|
solute carrier family 4 member 7 |
| chr16_-_18876305 | 0.96 |
ENST00000563235.5
|
SMG1
|
SMG1 nonsense mediated mRNA decay associated PI3K related kinase |
| chr7_-_108240049 | 0.94 |
ENST00000379022.8
|
NRCAM
|
neuronal cell adhesion molecule |
| chr4_-_76007501 | 0.94 |
ENST00000264888.6
|
CXCL9
|
C-X-C motif chemokine ligand 9 |
| chr15_-_55270383 | 0.93 |
ENST00000396307.6
|
RAB27A
|
RAB27A, member RAS oncogene family |
| chr2_-_174847765 | 0.93 |
ENST00000443238.6
|
CHN1
|
chimerin 1 |
| chr20_+_59835853 | 0.88 |
ENST00000492611.5
|
PHACTR3
|
phosphatase and actin regulator 3 |
| chr2_-_174847525 | 0.80 |
ENST00000295497.12
ENST00000652036.1 ENST00000444394.6 ENST00000650731.1 |
CHN1
|
chimerin 1 |
| chr17_-_10549694 | 0.80 |
ENST00000622564.4
|
MYH2
|
myosin heavy chain 2 |
| chrX_-_111270474 | 0.76 |
ENST00000324068.2
|
CAPN6
|
calpain 6 |
| chr3_-_167474026 | 0.75 |
ENST00000466903.1
ENST00000264677.8 |
SERPINI2
|
serpin family I member 2 |
| chr11_+_62190212 | 0.73 |
ENST00000306238.3
|
SCGB1D1
|
secretoglobin family 1D member 1 |
| chr1_-_94925759 | 0.72 |
ENST00000415017.1
ENST00000545882.5 |
CNN3
|
calponin 3 |
| chr4_+_186069144 | 0.71 |
ENST00000513189.1
ENST00000296795.8 |
TLR3
|
toll like receptor 3 |
| chr4_-_67883987 | 0.70 |
ENST00000283916.11
|
TMPRSS11D
|
transmembrane serine protease 11D |
| chr18_+_58196736 | 0.70 |
ENST00000675221.1
|
NEDD4L
|
NEDD4 like E3 ubiquitin protein ligase |
| chr9_-_92482350 | 0.69 |
ENST00000375543.2
|
ASPN
|
asporin |
| chr6_+_45422485 | 0.69 |
ENST00000359524.7
|
RUNX2
|
RUNX family transcription factor 2 |
| chr6_+_122996227 | 0.64 |
ENST00000275162.10
|
CLVS2
|
clavesin 2 |
| chr6_+_45422564 | 0.63 |
ENST00000625924.1
|
RUNX2
|
RUNX family transcription factor 2 |
| chr6_+_29306626 | 0.62 |
ENST00000377160.4
|
OR14J1
|
olfactory receptor family 14 subfamily J member 1 |
| chr3_-_157503574 | 0.61 |
ENST00000494677.5
ENST00000468233.5 |
VEPH1
|
ventricular zone expressed PH domain containing 1 |
| chr17_-_10549652 | 0.60 |
ENST00000245503.10
|
MYH2
|
myosin heavy chain 2 |
| chr17_-_10549612 | 0.60 |
ENST00000532183.6
ENST00000397183.6 ENST00000420805.1 |
MYH2
|
myosin heavy chain 2 |
| chr9_+_94084458 | 0.59 |
ENST00000620992.5
ENST00000288976.3 |
PTPDC1
|
protein tyrosine phosphatase domain containing 1 |
| chr1_-_100178215 | 0.59 |
ENST00000370138.1
ENST00000370137.6 ENST00000342895.7 ENST00000620882.4 |
LRRC39
|
leucine rich repeat containing 39 |
| chr3_+_156120572 | 0.57 |
ENST00000389636.9
ENST00000490337.6 |
KCNAB1
|
potassium voltage-gated channel subfamily A member regulatory beta subunit 1 |
| chr1_-_165445220 | 0.57 |
ENST00000619224.1
|
RXRG
|
retinoid X receptor gamma |
| chr4_+_174918400 | 0.57 |
ENST00000404450.8
ENST00000514159.1 |
ADAM29
|
ADAM metallopeptidase domain 29 |
| chr17_+_21126947 | 0.56 |
ENST00000579303.5
|
DHRS7B
|
dehydrogenase/reductase 7B |
| chr15_+_66504959 | 0.55 |
ENST00000535141.6
ENST00000613446.4 ENST00000446801.6 |
ZWILCH
|
zwilch kinetochore protein |
| chr1_-_165445088 | 0.54 |
ENST00000359842.10
|
RXRG
|
retinoid X receptor gamma |
| chr3_-_197260369 | 0.53 |
ENST00000658155.1
ENST00000453607.5 |
DLG1
|
discs large MAGUK scaffold protein 1 |
| chr10_-_113854368 | 0.53 |
ENST00000369305.1
|
DCLRE1A
|
DNA cross-link repair 1A |
| chr15_+_66505289 | 0.49 |
ENST00000565627.5
ENST00000564179.5 ENST00000307897.10 |
ZWILCH
|
zwilch kinetochore protein |
| chr8_+_103880412 | 0.47 |
ENST00000436393.6
|
RIMS2
|
regulating synaptic membrane exocytosis 2 |
| chrX_-_15314543 | 0.46 |
ENST00000344384.8
|
ASB11
|
ankyrin repeat and SOCS box containing 11 |
| chr2_-_165203870 | 0.46 |
ENST00000639244.1
ENST00000409101.7 ENST00000668657.1 |
SCN3A
|
sodium voltage-gated channel alpha subunit 3 |
| chr7_+_144000320 | 0.45 |
ENST00000641698.1
|
OR6B1
|
olfactory receptor family 6 subfamily B member 1 |
| chr4_-_167234552 | 0.45 |
ENST00000512648.5
|
SPOCK3
|
SPARC (osteonectin), cwcv and kazal like domains proteoglycan 3 |
| chr10_+_27532521 | 0.45 |
ENST00000683924.1
|
RAB18
|
RAB18, member RAS oncogene family |
| chr8_-_20183090 | 0.43 |
ENST00000265808.11
ENST00000522513.5 |
SLC18A1
|
solute carrier family 18 member A1 |
| chr6_+_13272709 | 0.43 |
ENST00000379335.8
|
PHACTR1
|
phosphatase and actin regulator 1 |
| chr10_+_84424919 | 0.43 |
ENST00000543283.2
ENST00000494586.5 |
CCSER2
|
coiled-coil serine rich protein 2 |
| chr1_+_171557845 | 0.42 |
ENST00000644916.1
|
PRRC2C
|
proline rich coiled-coil 2C |
| chr8_+_12104389 | 0.42 |
ENST00000400085.7
|
ZNF705D
|
zinc finger protein 705D |
| chr2_+_161136901 | 0.41 |
ENST00000259075.6
ENST00000432002.5 |
TANK
|
TRAF family member associated NFKB activator |
| chr12_-_84892120 | 0.40 |
ENST00000680379.1
|
SLC6A15
|
solute carrier family 6 member 15 |
| chr21_+_34364003 | 0.40 |
ENST00000290310.4
|
KCNE2
|
potassium voltage-gated channel subfamily E regulatory subunit 2 |
| chr5_+_141364231 | 0.40 |
ENST00000611914.1
|
PCDHGA5
|
protocadherin gamma subfamily A, 5 |
| chr8_-_20183127 | 0.39 |
ENST00000276373.10
ENST00000519026.5 ENST00000440926.3 ENST00000437980.3 |
SLC18A1
|
solute carrier family 18 member A1 |
| chr6_-_31665185 | 0.39 |
ENST00000456540.1
ENST00000445768.5 |
GPANK1
|
G-patch domain and ankyrin repeats 1 |
| chr15_-_66386668 | 0.38 |
ENST00000568216.5
ENST00000562124.5 ENST00000570251.1 |
TIPIN
|
TIMELESS interacting protein |
| chr20_+_6007245 | 0.38 |
ENST00000378868.4
|
CRLS1
|
cardiolipin synthase 1 |
| chr8_-_85341705 | 0.38 |
ENST00000517618.5
|
CA1
|
carbonic anhydrase 1 |
| chr1_+_86547070 | 0.37 |
ENST00000370563.3
|
CLCA4
|
chloride channel accessory 4 |
| chr3_-_112845950 | 0.36 |
ENST00000398214.5
|
CD200R1L
|
CD200 receptor 1 like |
| chr5_+_95391361 | 0.36 |
ENST00000283357.10
|
FAM81B
|
family with sequence similarity 81 member B |
| chr3_-_146528750 | 0.36 |
ENST00000483300.5
|
PLSCR1
|
phospholipid scramblase 1 |
| chr22_-_32255344 | 0.35 |
ENST00000266086.6
|
SLC5A4
|
solute carrier family 5 member 4 |
| chrX_-_131289412 | 0.35 |
ENST00000652189.1
ENST00000651556.1 |
IGSF1
|
immunoglobulin superfamily member 1 |
| chr2_-_227379297 | 0.35 |
ENST00000304568.4
|
TM4SF20
|
transmembrane 4 L six family member 20 |
| chr6_+_131637296 | 0.34 |
ENST00000358229.6
ENST00000357639.8 |
ENPP3
|
ectonucleotide pyrophosphatase/phosphodiesterase 3 |
| chr12_-_9999176 | 0.34 |
ENST00000298527.10
ENST00000348658.4 |
CLEC1B
|
C-type lectin domain family 1 member B |
| chr1_+_50105666 | 0.34 |
ENST00000651347.1
|
ELAVL4
|
ELAV like RNA binding protein 4 |
| chr6_+_10528326 | 0.33 |
ENST00000379597.7
|
GCNT2
|
glucosaminyl (N-acetyl) transferase 2 (I blood group) |
| chr19_-_56314788 | 0.33 |
ENST00000592509.5
ENST00000592679.5 ENST00000683990.1 ENST00000588442.5 ENST00000593106.5 ENST00000587492.5 |
ZSCAN5A
|
zinc finger and SCAN domain containing 5A |
| chr2_+_100974849 | 0.32 |
ENST00000450763.1
|
NPAS2
|
neuronal PAS domain protein 2 |
| chr11_+_117203135 | 0.32 |
ENST00000529622.1
|
TAGLN
|
transgelin |
| chr20_+_13221257 | 0.32 |
ENST00000262487.5
|
ISM1
|
isthmin 1 |
| chr9_-_121050264 | 0.32 |
ENST00000223642.3
|
C5
|
complement C5 |
| chr6_-_17706852 | 0.32 |
ENST00000262077.3
|
NUP153
|
nucleoporin 153 |
| chr11_-_95910748 | 0.31 |
ENST00000675933.1
|
MTMR2
|
myotubularin related protein 2 |
| chr22_-_35840218 | 0.31 |
ENST00000414461.6
ENST00000416721.6 ENST00000449924.6 ENST00000262829.11 ENST00000397305.3 |
RBFOX2
|
RNA binding fox-1 homolog 2 |
| chr14_-_60724300 | 0.31 |
ENST00000556952.3
ENST00000216513.5 |
SIX4
|
SIX homeobox 4 |
| chr1_+_44405164 | 0.31 |
ENST00000355387.6
|
RNF220
|
ring finger protein 220 |
| chr3_+_109136707 | 0.30 |
ENST00000622536.6
|
C3orf85
|
chromosome 3 open reading frame 85 |
| chr4_-_167234579 | 0.30 |
ENST00000502330.5
ENST00000357154.7 ENST00000421836.6 |
SPOCK3
|
SPARC (osteonectin), cwcv and kazal like domains proteoglycan 3 |
| chr11_-_95910824 | 0.30 |
ENST00000674528.1
ENST00000675477.1 ENST00000675636.1 |
MTMR2
|
myotubularin related protein 2 |
| chr21_-_18403754 | 0.30 |
ENST00000284885.8
|
TMPRSS15
|
transmembrane serine protease 15 |
| chr3_-_132684685 | 0.30 |
ENST00000512094.5
ENST00000632629.1 |
NPHP3
NPHP3-ACAD11
|
nephrocystin 3 NPHP3-ACAD11 readthrough (NMD candidate) |
| chr10_+_5364955 | 0.30 |
ENST00000380433.5
|
UCN3
|
urocortin 3 |
| chr2_+_89947508 | 0.30 |
ENST00000491977.1
|
IGKV2D-29
|
immunoglobulin kappa variable 2D-29 |
| chr12_+_11649666 | 0.30 |
ENST00000396373.9
|
ETV6
|
ETS variant transcription factor 6 |
| chr11_-_95910665 | 0.29 |
ENST00000674610.1
ENST00000675660.1 |
MTMR2
|
myotubularin related protein 2 |
| chr2_-_135837170 | 0.29 |
ENST00000264162.7
|
LCT
|
lactase |
| chr2_-_174597728 | 0.29 |
ENST00000409891.5
ENST00000410117.5 |
WIPF1
|
WAS/WASL interacting protein family member 1 |
| chr12_-_124567464 | 0.29 |
ENST00000458234.5
|
NCOR2
|
nuclear receptor corepressor 2 |
| chr17_-_41168219 | 0.29 |
ENST00000391356.4
|
KRTAP4-3
|
keratin associated protein 4-3 |
| chr17_+_62370218 | 0.29 |
ENST00000450662.7
|
EFCAB3
|
EF-hand calcium binding domain 3 |
| chr10_+_17752185 | 0.28 |
ENST00000377495.2
|
TMEM236
|
transmembrane protein 236 |
| chr14_+_23185316 | 0.28 |
ENST00000399910.5
ENST00000492621.5 |
RNF212B
|
ring finger protein 212B |
| chr10_-_50885656 | 0.28 |
ENST00000374001.6
ENST00000395489.6 ENST00000282641.6 ENST00000395495.5 ENST00000373995.7 ENST00000414883.1 |
A1CF
|
APOBEC1 complementation factor |
| chr8_+_54616078 | 0.28 |
ENST00000220676.2
|
RP1
|
RP1 axonemal microtubule associated |
| chr4_+_87799546 | 0.27 |
ENST00000226284.7
|
IBSP
|
integrin binding sialoprotein |
| chr7_+_93906557 | 0.27 |
ENST00000248572.10
ENST00000429473.1 ENST00000430875.1 ENST00000428834.1 |
GNGT1
|
G protein subunit gamma transducin 1 |
| chrX_+_136305988 | 0.27 |
ENST00000394141.1
|
ADGRG4
|
adhesion G protein-coupled receptor G4 |
| chr9_+_12693327 | 0.27 |
ENST00000388918.10
|
TYRP1
|
tyrosinase related protein 1 |
| chr4_-_167234426 | 0.27 |
ENST00000541354.5
ENST00000509854.5 ENST00000512681.5 ENST00000357545.9 ENST00000510741.5 ENST00000510403.5 |
SPOCK3
|
SPARC (osteonectin), cwcv and kazal like domains proteoglycan 3 |
| chr2_-_162838728 | 0.27 |
ENST00000328032.8
ENST00000332142.10 |
KCNH7
|
potassium voltage-gated channel subfamily H member 7 |
| chr12_+_14973020 | 0.26 |
ENST00000266395.3
|
PDE6H
|
phosphodiesterase 6H |
| chr4_+_70383123 | 0.26 |
ENST00000304915.8
|
SMR3B
|
submaxillary gland androgen regulated protein 3B |
| chrX_+_48761743 | 0.26 |
ENST00000303227.11
|
GLOD5
|
glyoxalase domain containing 5 |
| chr8_+_24294107 | 0.26 |
ENST00000437154.6
|
ADAM28
|
ADAM metallopeptidase domain 28 |
| chr2_-_95484731 | 0.26 |
ENST00000639673.2
|
TRIM43B
|
tripartite motif containing 43B |
| chr12_+_75334675 | 0.26 |
ENST00000312442.2
|
GLIPR1L1
|
GLIPR1 like 1 |
| chr3_+_63443076 | 0.26 |
ENST00000295894.9
|
SYNPR
|
synaptoporin |
| chr7_+_114416286 | 0.26 |
ENST00000635534.1
|
FOXP2
|
forkhead box P2 |
| chr5_+_141364153 | 0.25 |
ENST00000518069.2
|
PCDHGA5
|
protocadherin gamma subfamily A, 5 |
| chr15_+_58138368 | 0.25 |
ENST00000219919.9
ENST00000536493.1 |
AQP9
|
aquaporin 9 |
| chr22_+_50170720 | 0.25 |
ENST00000159647.9
ENST00000395842.3 |
PANX2
|
pannexin 2 |
| chr14_-_67533720 | 0.25 |
ENST00000554278.6
|
TMEM229B
|
transmembrane protein 229B |
| chr10_-_88851809 | 0.25 |
ENST00000371930.5
|
ANKRD22
|
ankyrin repeat domain 22 |
| chr11_+_112175526 | 0.24 |
ENST00000532612.5
ENST00000438022.5 |
BCO2
|
beta-carotene oxygenase 2 |
| chr1_+_197268222 | 0.24 |
ENST00000367400.8
ENST00000638467.1 ENST00000367399.6 |
CRB1
|
crumbs cell polarity complex component 1 |
| chr1_+_181088692 | 0.24 |
ENST00000367577.7
|
IER5
|
immediate early response 5 |
| chr10_-_50885619 | 0.24 |
ENST00000373997.8
|
A1CF
|
APOBEC1 complementation factor |
| chr12_+_53954870 | 0.24 |
ENST00000243103.4
|
HOXC12
|
homeobox C12 |
| chr5_+_141412979 | 0.23 |
ENST00000612503.1
ENST00000398610.3 |
PCDHGA10
|
protocadherin gamma subfamily A, 10 |
| chr2_-_178702479 | 0.23 |
ENST00000414766.5
|
TTN
|
titin |
| chr2_+_36696686 | 0.23 |
ENST00000379242.7
ENST00000389975.7 |
VIT
|
vitrin |
| chr9_-_28670285 | 0.23 |
ENST00000379992.6
ENST00000308675.5 ENST00000613945.3 |
LINGO2
|
leucine rich repeat and Ig domain containing 2 |
| chr10_-_104085847 | 0.23 |
ENST00000648076.2
|
COL17A1
|
collagen type XVII alpha 1 chain |
| chr8_+_106726115 | 0.22 |
ENST00000521592.5
|
OXR1
|
oxidation resistance 1 |
| chr3_-_167653916 | 0.22 |
ENST00000488012.5
ENST00000682715.1 ENST00000647816.1 |
WDR49
|
WD repeat domain 49 |
| chr11_-_16408853 | 0.22 |
ENST00000528252.5
|
SOX6
|
SRY-box transcription factor 6 |
| chr1_-_86383078 | 0.22 |
ENST00000460698.6
|
ODF2L
|
outer dense fiber of sperm tails 2 like |
| chr11_-_84720876 | 0.21 |
ENST00000648622.1
|
DLG2
|
discs large MAGUK scaffold protein 2 |
| chr7_+_90383672 | 0.21 |
ENST00000416322.5
|
CLDN12
|
claudin 12 |
| chr8_-_58500158 | 0.20 |
ENST00000301645.4
|
CYP7A1
|
cytochrome P450 family 7 subfamily A member 1 |
| chr19_+_15082211 | 0.20 |
ENST00000641398.1
|
OR1I1
|
olfactory receptor family 1 subfamily I member 1 |
| chr1_+_197268204 | 0.20 |
ENST00000535699.5
ENST00000538660.5 |
CRB1
|
crumbs cell polarity complex component 1 |
| chr5_-_79512794 | 0.20 |
ENST00000282260.10
ENST00000508576.5 ENST00000535690.1 |
HOMER1
|
homer scaffold protein 1 |
| chr17_+_43483949 | 0.20 |
ENST00000540306.5
ENST00000262415.8 |
DHX8
|
DEAH-box helicase 8 |
| chr17_+_59155726 | 0.19 |
ENST00000578777.5
ENST00000577457.1 ENST00000582995.5 ENST00000262293.9 ENST00000614081.1 |
PRR11
|
proline rich 11 |
| chr16_-_66730216 | 0.19 |
ENST00000569320.5
|
DYNC1LI2
|
dynein cytoplasmic 1 light intermediate chain 2 |
| chr1_+_192158448 | 0.19 |
ENST00000367460.4
|
RGS18
|
regulator of G protein signaling 18 |
| chr3_-_167653952 | 0.19 |
ENST00000466760.5
ENST00000479765.5 |
WDR49
|
WD repeat domain 49 |
| chr17_+_58169401 | 0.19 |
ENST00000641866.1
|
OR4D2
|
olfactory receptor family 4 subfamily D member 2 |
| chr6_-_49866453 | 0.19 |
ENST00000507853.5
|
CRISP1
|
cysteine rich secretory protein 1 |
| chr2_+_24793098 | 0.19 |
ENST00000473706.5
|
CENPO
|
centromere protein O |
| chr18_+_616672 | 0.19 |
ENST00000338387.11
|
CLUL1
|
clusterin like 1 |
| chr3_+_178558700 | 0.19 |
ENST00000432997.5
ENST00000455865.5 |
KCNMB2
|
potassium calcium-activated channel subfamily M regulatory beta subunit 2 |
| chr11_-_13496018 | 0.19 |
ENST00000529816.1
|
PTH
|
parathyroid hormone |
| chr1_-_148152272 | 0.19 |
ENST00000682118.1
ENST00000615281.4 |
NBPF11
|
NBPF member 11 |
| chr9_+_133376334 | 0.18 |
ENST00000371957.4
|
STKLD1
|
serine/threonine kinase like domain containing 1 |
| chr1_+_240014319 | 0.18 |
ENST00000447095.5
|
FMN2
|
formin 2 |
| chr18_-_77017042 | 0.18 |
ENST00000359645.7
ENST00000397875.7 ENST00000397869.7 ENST00000578193.5 ENST00000578873.5 ENST00000397866.8 ENST00000528160.1 ENST00000527041.1 ENST00000526111.5 ENST00000397865.9 ENST00000382582.7 |
MBP
|
myelin basic protein |
| chr1_+_160400543 | 0.17 |
ENST00000368061.3
|
VANGL2
|
VANGL planar cell polarity protein 2 |
| chr9_-_21482313 | 0.17 |
ENST00000448696.4
|
IFNE
|
interferon epsilon |
| chr2_+_127701490 | 0.17 |
ENST00000310981.6
|
SFT2D3
|
SFT2 domain containing 3 |
| chr7_+_45027634 | 0.17 |
ENST00000381112.7
ENST00000474617.1 |
CCM2
|
CCM2 scaffold protein |
| chr4_-_82844418 | 0.17 |
ENST00000503937.5
|
SEC31A
|
SEC31 homolog A, COPII coat complex component |
| chr8_-_673547 | 0.17 |
ENST00000522893.1
|
ERICH1
|
glutamate rich 1 |
| chr3_-_197226351 | 0.17 |
ENST00000656428.1
|
DLG1
|
discs large MAGUK scaffold protein 1 |
| chr9_-_21351378 | 0.17 |
ENST00000380210.1
|
IFNA6
|
interferon alpha 6 |
| chr6_+_151809105 | 0.17 |
ENST00000427531.6
|
ESR1
|
estrogen receptor 1 |
| chr18_-_72865680 | 0.16 |
ENST00000397929.5
|
NETO1
|
neuropilin and tolloid like 1 |
| chr17_-_41489907 | 0.16 |
ENST00000328119.11
|
KRT36
|
keratin 36 |
| chr8_+_36784324 | 0.16 |
ENST00000523973.5
ENST00000399881.8 |
KCNU1
|
potassium calcium-activated channel subfamily U member 1 |
| chr5_-_177780633 | 0.16 |
ENST00000513554.5
ENST00000440605.7 |
FAM153A
|
family with sequence similarity 153 member A |
| chr22_+_31754862 | 0.16 |
ENST00000382111.6
ENST00000645407.1 ENST00000646701.1 |
DEPDC5
ENSG00000285404.1
|
DEP domain containing 5, GATOR1 subcomplex subunit novel protein, DEPDC5-YWHAH readthrough |
| chr1_-_197067234 | 0.16 |
ENST00000367412.2
|
F13B
|
coagulation factor XIII B chain |
| chr4_+_48483324 | 0.15 |
ENST00000273861.5
|
SLC10A4
|
solute carrier family 10 member 4 |
| chr18_+_616711 | 0.15 |
ENST00000579494.1
|
CLUL1
|
clusterin like 1 |
| chr6_+_29111560 | 0.15 |
ENST00000377169.2
|
OR2J3
|
olfactory receptor family 2 subfamily J member 3 |
| chr1_+_197413827 | 0.15 |
ENST00000367397.1
ENST00000681519.1 |
CRB1
|
crumbs cell polarity complex component 1 |
| chr6_-_49866527 | 0.15 |
ENST00000335847.9
|
CRISP1
|
cysteine rich secretory protein 1 |
| chr6_-_106975616 | 0.14 |
ENST00000610952.1
|
CD24
|
CD24 molecule |
| chr5_+_136160986 | 0.14 |
ENST00000507637.1
|
SMAD5
|
SMAD family member 5 |
| chr2_-_208146150 | 0.14 |
ENST00000260988.5
|
CRYGB
|
crystallin gamma B |
| chr7_-_28180735 | 0.14 |
ENST00000283928.10
|
JAZF1
|
JAZF zinc finger 1 |
| chr4_-_99321362 | 0.14 |
ENST00000625860.2
ENST00000305046.13 ENST00000506651.5 |
ADH1B
|
alcohol dehydrogenase 1B (class I), beta polypeptide |
| chr6_+_132538290 | 0.14 |
ENST00000434551.2
|
TAAR9
|
trace amine associated receptor 9 |
| chr3_-_139678011 | 0.14 |
ENST00000646611.1
ENST00000645290.1 ENST00000647257.1 |
NMNAT3
|
nicotinamide nucleotide adenylyltransferase 3 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 3.5 | GO:2000329 | negative regulation of T-helper 17 cell lineage commitment(GO:2000329) |
| 0.7 | 8.2 | GO:0043950 | positive regulation of cAMP-mediated signaling(GO:0043950) |
| 0.4 | 1.3 | GO:0050717 | positive regulation of interleukin-1 alpha secretion(GO:0050717) |
| 0.4 | 2.1 | GO:0010513 | positive regulation of phosphatidylinositol biosynthetic process(GO:0010513) |
| 0.4 | 1.6 | GO:1904116 | response to vasopressin(GO:1904116) cellular response to vasopressin(GO:1904117) |
| 0.4 | 2.8 | GO:0022614 | membrane to membrane docking(GO:0022614) |
| 0.4 | 1.1 | GO:0033023 | mast cell homeostasis(GO:0033023) mast cell apoptotic process(GO:0033024) regulation of mast cell apoptotic process(GO:0033025) mast cell proliferation(GO:0070662) |
| 0.3 | 1.2 | GO:0039534 | negative regulation of MDA-5 signaling pathway(GO:0039534) |
| 0.3 | 1.0 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.2 | 1.1 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.2 | 2.3 | GO:2000535 | regulation of entry of bacterium into host cell(GO:2000535) |
| 0.2 | 0.4 | GO:1901979 | regulation of inward rectifier potassium channel activity(GO:1901979) |
| 0.2 | 0.9 | GO:1903435 | positive regulation of constitutive secretory pathway(GO:1903435) |
| 0.1 | 1.3 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 0.1 | 0.7 | GO:0034344 | type III interferon production(GO:0034343) regulation of type III interferon production(GO:0034344) positive regulation of interferon-alpha biosynthetic process(GO:0045356) |
| 0.1 | 1.3 | GO:0002175 | protein localization to paranode region of axon(GO:0002175) |
| 0.1 | 1.2 | GO:1904996 | positive regulation of leukocyte adhesion to vascular endothelial cell(GO:1904996) |
| 0.1 | 0.7 | GO:0070171 | negative regulation of tooth mineralization(GO:0070171) |
| 0.1 | 0.3 | GO:0030505 | inorganic diphosphate transport(GO:0030505) |
| 0.1 | 0.7 | GO:0015842 | aminergic neurotransmitter loading into synaptic vesicle(GO:0015842) |
| 0.1 | 0.3 | GO:1902594 | viral penetration into host nucleus(GO:0075732) multi-organism nuclear import(GO:1902594) |
| 0.1 | 0.3 | GO:0072365 | regulation of cellular ketone metabolic process by negative regulation of transcription from RNA polymerase II promoter(GO:0072365) |
| 0.1 | 0.3 | GO:0016116 | tetraterpenoid metabolic process(GO:0016108) carotenoid metabolic process(GO:0016116) carotene catabolic process(GO:0016121) xanthophyll metabolic process(GO:0016122) terpene catabolic process(GO:0046247) |
| 0.1 | 1.5 | GO:1902221 | L-phenylalanine metabolic process(GO:0006558) L-phenylalanine catabolic process(GO:0006559) erythrose 4-phosphate/phosphoenolpyruvate family amino acid metabolic process(GO:1902221) erythrose 4-phosphate/phosphoenolpyruvate family amino acid catabolic process(GO:1902222) |
| 0.1 | 0.3 | GO:0021757 | caudate nucleus development(GO:0021757) putamen development(GO:0021758) |
| 0.1 | 2.9 | GO:0042730 | fibrinolysis(GO:0042730) |
| 0.1 | 0.9 | GO:2000645 | negative regulation of receptor catabolic process(GO:2000645) |
| 0.1 | 0.3 | GO:0090290 | positive regulation of osteoclast proliferation(GO:0090290) |
| 0.1 | 0.3 | GO:0061055 | myotome development(GO:0061055) |
| 0.1 | 1.6 | GO:0008228 | opsonization(GO:0008228) |
| 0.1 | 0.5 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.1 | 0.4 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 0.1 | 0.3 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.1 | 0.9 | GO:0045162 | neuronal action potential propagation(GO:0019227) clustering of voltage-gated sodium channels(GO:0045162) action potential propagation(GO:0098870) |
| 0.1 | 0.3 | GO:0010760 | negative regulation of macrophage chemotaxis(GO:0010760) |
| 0.1 | 0.3 | GO:1904823 | canalicular bile acid transport(GO:0015722) pyrimidine nucleobase transport(GO:0015855) purine nucleobase transmembrane transport(GO:1904823) |
| 0.1 | 0.1 | GO:0003099 | positive regulation of the force of heart contraction by chemical signal(GO:0003099) |
| 0.1 | 0.2 | GO:0019355 | nicotinamide nucleotide biosynthetic process from aspartate(GO:0019355) 'de novo' NAD biosynthetic process(GO:0034627) 'de novo' NAD biosynthetic process from aspartate(GO:0034628) |
| 0.1 | 0.7 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.1 | 0.2 | GO:0061341 | non-canonical Wnt signaling pathway involved in heart development(GO:0061341) planar cell polarity pathway involved in heart morphogenesis(GO:0061346) |
| 0.1 | 1.5 | GO:0035641 | locomotory exploration behavior(GO:0035641) |
| 0.1 | 0.5 | GO:0098814 | spontaneous neurotransmitter secretion(GO:0061669) spontaneous synaptic transmission(GO:0098814) |
| 0.1 | 0.8 | GO:1903760 | regulation of voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization(GO:1903760) regulation of potassium ion export across plasma membrane(GO:1903764) |
| 0.1 | 0.2 | GO:0007206 | phospholipase C-activating G-protein coupled glutamate receptor signaling pathway(GO:0007206) |
| 0.0 | 0.3 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.0 | 5.2 | GO:0006521 | regulation of cellular amino acid metabolic process(GO:0006521) |
| 0.0 | 0.2 | GO:0030241 | skeletal muscle myosin thick filament assembly(GO:0030241) |
| 0.0 | 0.2 | GO:0051758 | homologous chromosome movement towards spindle pole involved in homologous chromosome segregation(GO:0051758) |
| 0.0 | 0.5 | GO:0031848 | protection from non-homologous end joining at telomere(GO:0031848) |
| 0.0 | 0.2 | GO:0021780 | oligodendrocyte cell fate specification(GO:0021778) oligodendrocyte cell fate commitment(GO:0021779) glial cell fate specification(GO:0021780) |
| 0.0 | 0.4 | GO:1904386 | response to thyroxine(GO:0097068) response to L-phenylalanine derivative(GO:1904386) |
| 0.0 | 0.3 | GO:0044245 | polysaccharide digestion(GO:0044245) |
| 0.0 | 0.3 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.0 | 0.4 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) |
| 0.0 | 0.4 | GO:0035524 | proline transmembrane transport(GO:0035524) |
| 0.0 | 1.7 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.0 | 0.1 | GO:0032597 | B cell receptor transport within lipid bilayer(GO:0032595) B cell receptor transport into membrane raft(GO:0032597) protein transport out of membrane raft(GO:0032599) chemokine receptor transport out of membrane raft(GO:0032600) negative regulation of transforming growth factor beta3 production(GO:0032913) chemokine receptor transport within lipid bilayer(GO:0033606) |
| 0.0 | 0.6 | GO:1903817 | negative regulation of delayed rectifier potassium channel activity(GO:1902260) negative regulation of voltage-gated potassium channel activity(GO:1903817) |
| 0.0 | 0.7 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.0 | 0.2 | GO:2000312 | regulation of kainate selective glutamate receptor activity(GO:2000312) |
| 0.0 | 0.7 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.0 | 0.3 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.0 | 0.3 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.0 | 0.2 | GO:1904207 | maintenance of blood-brain barrier(GO:0035633) regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904207) positive regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904209) |
| 0.0 | 0.0 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.0 | 0.3 | GO:1902857 | positive regulation of nonmotile primary cilium assembly(GO:1902857) |
| 0.0 | 0.1 | GO:0021888 | hypothalamus gonadotrophin-releasing hormone neuron differentiation(GO:0021886) hypothalamus gonadotrophin-releasing hormone neuron development(GO:0021888) |
| 0.0 | 0.2 | GO:0060750 | epithelial cell proliferation involved in mammary gland duct elongation(GO:0060750) |
| 0.0 | 0.1 | GO:2000354 | regulation of ovarian follicle development(GO:2000354) |
| 0.0 | 0.2 | GO:0048478 | replication fork protection(GO:0048478) |
| 0.0 | 0.1 | GO:0000414 | regulation of histone H3-K36 methylation(GO:0000414) |
| 0.0 | 0.1 | GO:2000563 | positive regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000563) |
| 0.0 | 0.1 | GO:1905225 | thyroid-stimulating hormone signaling pathway(GO:0038194) cellular response to glycoprotein(GO:1904588) response to thyrotropin-releasing hormone(GO:1905225) cellular response to thyrotropin-releasing hormone(GO:1905229) |
| 0.0 | 0.3 | GO:0034214 | protein hexamerization(GO:0034214) |
| 0.0 | 1.5 | GO:0030049 | muscle filament sliding(GO:0030049) actin-myosin filament sliding(GO:0033275) |
| 0.0 | 0.2 | GO:0070857 | regulation of bile acid biosynthetic process(GO:0070857) |
| 0.0 | 0.1 | GO:0070309 | lens fiber cell morphogenesis(GO:0070309) |
| 0.0 | 0.3 | GO:0048021 | regulation of melanin biosynthetic process(GO:0048021) positive regulation of melanin biosynthetic process(GO:0048023) regulation of secondary metabolite biosynthetic process(GO:1900376) positive regulation of secondary metabolite biosynthetic process(GO:1900378) |
| 0.0 | 0.1 | GO:0035022 | positive regulation of Rac protein signal transduction(GO:0035022) |
| 0.0 | 0.3 | GO:0051775 | response to redox state(GO:0051775) |
| 0.0 | 0.4 | GO:0071786 | endoplasmic reticulum tubular network organization(GO:0071786) |
| 0.0 | 0.3 | GO:0045838 | positive regulation of membrane potential(GO:0045838) |
| 0.0 | 0.9 | GO:0042462 | eye photoreceptor cell development(GO:0042462) |
| 0.0 | 1.3 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.0 | 0.1 | GO:0060054 | positive regulation of epithelial cell proliferation involved in wound healing(GO:0060054) |
| 0.0 | 0.1 | GO:0035989 | tendon development(GO:0035989) |
| 0.0 | 0.0 | GO:0061300 | cerebellum vasculature development(GO:0061300) |
| 0.0 | 0.3 | GO:0052695 | cellular glucuronidation(GO:0052695) |
| 0.0 | 0.0 | GO:0090310 | negative regulation of methylation-dependent chromatin silencing(GO:0090310) |
| 0.0 | 0.2 | GO:0048845 | venous blood vessel morphogenesis(GO:0048845) |
| 0.0 | 0.1 | GO:0008050 | female courtship behavior(GO:0008050) |
| 0.0 | 0.3 | GO:0007342 | fusion of sperm to egg plasma membrane(GO:0007342) |
| 0.0 | 0.5 | GO:1901522 | positive regulation of transcription from RNA polymerase II promoter involved in cellular response to chemical stimulus(GO:1901522) |
| 0.0 | 0.8 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 0.6 | GO:0019228 | neuronal action potential(GO:0019228) |
| 0.0 | 0.2 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 0.1 | GO:0043615 | astrocyte cell migration(GO:0043615) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 2.8 | GO:0071065 | alpha9-beta1 integrin-vascular cell adhesion molecule-1 complex(GO:0071065) |
| 0.6 | 5.2 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.5 | 1.6 | GO:1990723 | cytoplasmic periphery of the nuclear pore complex(GO:1990723) |
| 0.3 | 1.3 | GO:0097179 | protease inhibitor complex(GO:0097179) |
| 0.3 | 2.0 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.3 | 1.0 | GO:1990423 | RZZ complex(GO:1990423) |
| 0.1 | 0.5 | GO:0030895 | apolipoprotein B mRNA editing enzyme complex(GO:0030895) |
| 0.1 | 0.3 | GO:0097543 | ciliary inversin compartment(GO:0097543) |
| 0.1 | 0.8 | GO:0070083 | clathrin-sculpted monoamine transport vesicle(GO:0070081) clathrin-sculpted monoamine transport vesicle membrane(GO:0070083) |
| 0.1 | 0.6 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.1 | 0.9 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.1 | 1.4 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.1 | 0.2 | GO:0060187 | cell pole(GO:0060187) |
| 0.0 | 0.8 | GO:0043219 | lateral loop(GO:0043219) |
| 0.0 | 0.3 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 0.9 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.0 | 0.3 | GO:0043196 | varicosity(GO:0043196) |
| 0.0 | 0.7 | GO:0036020 | endolysosome membrane(GO:0036020) |
| 0.0 | 0.5 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.0 | 0.2 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.0 | 0.1 | GO:0005592 | collagen type XI trimer(GO:0005592) |
| 0.0 | 0.3 | GO:0045009 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.0 | 0.2 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.0 | 0.2 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.0 | 0.3 | GO:0044615 | nuclear pore nuclear basket(GO:0044615) |
| 0.0 | 6.3 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 1.0 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 1.6 | GO:0070821 | tertiary granule membrane(GO:0070821) |
| 0.0 | 0.4 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.0 | 0.3 | GO:0098563 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 0.0 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.0 | 0.2 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.6 | GO:0030669 | clathrin-coated endocytic vesicle membrane(GO:0030669) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 8.2 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.5 | 1.5 | GO:0001716 | L-amino-acid oxidase activity(GO:0001716) |
| 0.3 | 2.8 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.3 | 1.1 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.3 | 1.1 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.2 | 2.3 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.2 | 1.1 | GO:0016822 | hydrolase activity, acting on acid carbon-carbon bonds(GO:0016822) hydrolase activity, acting on acid carbon-carbon bonds, in ketonic substances(GO:0016823) |
| 0.2 | 0.8 | GO:0015222 | serotonin transmembrane transporter activity(GO:0015222) |
| 0.2 | 5.2 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.2 | 2.1 | GO:0051378 | amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 0.1 | 1.0 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.1 | 0.3 | GO:0005275 | amine transmembrane transporter activity(GO:0005275) |
| 0.1 | 1.3 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.1 | 0.3 | GO:0051431 | corticotropin-releasing hormone receptor 2 binding(GO:0051431) |
| 0.1 | 0.4 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.1 | 0.3 | GO:0035529 | NADH pyrophosphatase activity(GO:0035529) |
| 0.1 | 0.4 | GO:0005298 | proline:sodium symporter activity(GO:0005298) |
| 0.1 | 0.8 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.1 | 0.2 | GO:0000309 | nicotinamide-nucleotide adenylyltransferase activity(GO:0000309) |
| 0.1 | 0.9 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.1 | 0.3 | GO:0055077 | gap junction hemi-channel activity(GO:0055077) |
| 0.0 | 0.3 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.0 | 0.5 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.0 | 0.3 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.0 | 0.4 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.0 | 2.0 | GO:0000146 | microfilament motor activity(GO:0000146) |
| 0.0 | 0.8 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.0 | 0.9 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.0 | 0.1 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.0 | 1.0 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 3.2 | GO:0032813 | tumor necrosis factor receptor superfamily binding(GO:0032813) |
| 0.0 | 0.4 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.0 | 1.7 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.9 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 0.2 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.0 | 0.7 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.7 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.0 | 0.4 | GO:0047144 | 2-acylglycerol-3-phosphate O-acyltransferase activity(GO:0047144) |
| 0.0 | 1.3 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.4 | GO:1902282 | voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization(GO:1902282) |
| 0.0 | 0.2 | GO:0038052 | RNA polymerase II transcription factor activity, estrogen-activated sequence-specific DNA binding(GO:0038052) |
| 0.0 | 3.7 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.7 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.0 | 0.1 | GO:0015173 | aromatic amino acid transmembrane transporter activity(GO:0015173) |
| 0.0 | 0.1 | GO:1902271 | D3 vitamins binding(GO:1902271) |
| 0.0 | 0.6 | GO:0070402 | NADPH binding(GO:0070402) |
| 0.0 | 0.4 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.0 | 0.1 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.0 | 0.2 | GO:0031433 | telethonin binding(GO:0031433) |
| 0.0 | 0.6 | GO:0005549 | odorant binding(GO:0005549) |
| 0.0 | 1.1 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.0 | 0.8 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 1.0 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.0 | 0.2 | GO:0060072 | large conductance calcium-activated potassium channel activity(GO:0060072) |
| 0.0 | 0.3 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 0.1 | GO:0004024 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) |
| 0.0 | 0.3 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.1 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.0 | 0.5 | GO:1905030 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.0 | 0.4 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.0 | 0.2 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 0.0 | 0.1 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.0 | 0.1 | GO:0004996 | thyroid-stimulating hormone receptor activity(GO:0004996) |
| 0.0 | 0.2 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.3 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.0 | 0.4 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.0 | 0.3 | GO:0005112 | Notch binding(GO:0005112) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 4.1 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.1 | 8.2 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.1 | 1.6 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 2.0 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.0 | 1.9 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.0 | 1.5 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 1.5 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 1.3 | PID FGF PATHWAY | FGF signaling pathway |
| 0.0 | 1.2 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 7.6 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 0.3 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.0 | 0.3 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 1.1 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 0.8 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.0 | 3.8 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.1 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.1 | 8.2 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.1 | 1.1 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.1 | 5.2 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.1 | 1.3 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.1 | 1.1 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.1 | 0.9 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.1 | 1.6 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.3 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 0.7 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.0 | 1.2 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 1.3 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.9 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 2.8 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.0 | 0.3 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.0 | 0.8 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.9 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 2.5 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 0.3 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 1.4 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.8 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.3 | REACTOME NEP NS2 INTERACTS WITH THE CELLULAR EXPORT MACHINERY | Genes involved in NEP/NS2 Interacts with the Cellular Export Machinery |
| 0.0 | 0.2 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.0 | 0.2 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.0 | 0.1 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |