Project
Inflammatory response time course, HUVEC (Wada, 2009)
Navigation
Downloads
Results for HSF1
Z-value: 0.66
Transcription factors associated with HSF1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HSF1
|
ENSG00000185122.11 | HSF1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HSF1 | hg38_v1_chr8_+_144291581_144291617 | -0.16 | 4.6e-01 | Click! |
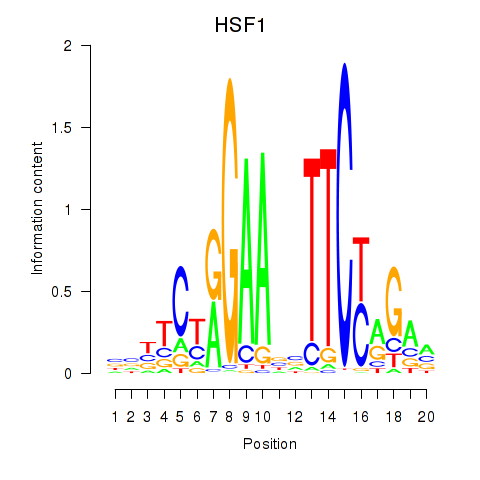
Activity profile of HSF1 motif
Sorted Z-values of HSF1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of HSF1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_+_150567347 | 1.36 |
ENST00000461940.5
|
GIMAP4
|
GTPase, IMAP family member 4 |
| chr7_+_150737382 | 1.23 |
ENST00000358647.5
|
GIMAP5
|
GTPase, IMAP family member 5 |
| chr7_+_150567382 | 1.23 |
ENST00000255945.4
ENST00000479232.1 |
GIMAP4
|
GTPase, IMAP family member 4 |
| chr20_-_59007807 | 1.18 |
ENST00000680386.1
|
CTSZ
|
cathepsin Z |
| chr8_-_80171106 | 1.15 |
ENST00000519303.6
|
TPD52
|
tumor protein D52 |
| chr1_-_160954801 | 1.10 |
ENST00000368029.4
|
ITLN2
|
intelectin 2 |
| chr4_+_41359599 | 1.08 |
ENST00000513024.5
|
LIMCH1
|
LIM and calponin homology domains 1 |
| chr10_-_114684457 | 1.05 |
ENST00000392955.7
|
ABLIM1
|
actin binding LIM protein 1 |
| chr10_-_114684612 | 0.96 |
ENST00000533213.6
ENST00000369252.8 |
ABLIM1
|
actin binding LIM protein 1 |
| chr2_-_191013955 | 0.93 |
ENST00000409465.5
|
STAT1
|
signal transducer and activator of transcription 1 |
| chr2_-_237590660 | 0.91 |
ENST00000409576.1
|
RAB17
|
RAB17, member RAS oncogene family |
| chr16_-_9943182 | 0.88 |
ENST00000535259.6
|
GRIN2A
|
glutamate ionotropic receptor NMDA type subunit 2A |
| chr4_+_54657918 | 0.84 |
ENST00000412167.6
ENST00000288135.6 |
KIT
|
KIT proto-oncogene, receptor tyrosine kinase |
| chr17_-_69141878 | 0.79 |
ENST00000590645.1
ENST00000284425.7 |
ABCA6
|
ATP binding cassette subfamily A member 6 |
| chr9_+_124291935 | 0.75 |
ENST00000546191.5
|
NEK6
|
NIMA related kinase 6 |
| chr2_-_191014137 | 0.72 |
ENST00000673777.1
ENST00000673942.1 ENST00000424722.6 ENST00000392322.7 ENST00000361099.8 ENST00000392323.6 ENST00000673816.1 ENST00000673847.1 ENST00000673952.1 ENST00000540176.6 ENST00000673841.1 |
STAT1
|
signal transducer and activator of transcription 1 |
| chr2_-_27628975 | 0.72 |
ENST00000324364.4
|
CCDC121
|
coiled-coil domain containing 121 |
| chrX_-_126552801 | 0.67 |
ENST00000371126.3
|
DCAF12L1
|
DDB1 and CUL4 associated factor 12 like 1 |
| chr14_-_22644352 | 0.65 |
ENST00000540461.2
|
OR6J1
|
olfactory receptor family 6 subfamily J member 1 |
| chr1_+_47023659 | 0.64 |
ENST00000371901.4
|
CYP4X1
|
cytochrome P450 family 4 subfamily X member 1 |
| chr8_-_80171496 | 0.64 |
ENST00000379096.9
ENST00000518937.6 |
TPD52
|
tumor protein D52 |
| chr19_+_34926892 | 0.64 |
ENST00000303586.11
ENST00000601142.2 ENST00000439785.5 ENST00000601540.5 ENST00000601957.5 |
ZNF30
|
zinc finger protein 30 |
| chr14_+_63204436 | 0.63 |
ENST00000316754.8
|
RHOJ
|
ras homolog family member J |
| chrX_+_54920796 | 0.62 |
ENST00000442098.5
ENST00000430420.5 ENST00000453081.5 ENST00000319167.12 ENST00000622017.4 ENST00000375022.8 ENST00000399736.5 ENST00000440072.5 ENST00000173898.12 ENST00000431115.5 ENST00000440759.5 ENST00000375041.6 |
TRO
|
trophinin |
| chr1_+_220879434 | 0.61 |
ENST00000366903.8
|
HLX
|
H2.0 like homeobox |
| chr10_-_96358989 | 0.61 |
ENST00000371172.8
|
OPALIN
|
oligodendrocytic myelin paranodal and inner loop protein |
| chr9_+_117704168 | 0.61 |
ENST00000472304.2
ENST00000394487.5 |
TLR4
|
toll like receptor 4 |
| chr8_+_142834247 | 0.60 |
ENST00000522728.5
|
GML
|
glycosylphosphatidylinositol anchored molecule like |
| chr6_+_116511626 | 0.59 |
ENST00000368599.4
|
CALHM5
|
calcium homeostasis modulator family member 5 |
| chr7_-_138663152 | 0.55 |
ENST00000288513.9
|
SVOPL
|
SVOP like |
| chr1_+_152663378 | 0.54 |
ENST00000368784.2
|
LCE2D
|
late cornified envelope 2D |
| chr9_-_13279407 | 0.54 |
ENST00000546205.5
|
MPDZ
|
multiple PDZ domain crumbs cell polarity complex component |
| chr5_-_43043170 | 0.54 |
ENST00000314890.3
|
ANXA2R
|
annexin A2 receptor |
| chr6_+_32637419 | 0.53 |
ENST00000374949.2
|
HLA-DQA1
|
major histocompatibility complex, class II, DQ alpha 1 |
| chr5_+_122311740 | 0.53 |
ENST00000506272.5
ENST00000508681.5 ENST00000509154.6 |
SNCAIP
|
synuclein alpha interacting protein |
| chr3_+_132597260 | 0.52 |
ENST00000249887.3
|
ACKR4
|
atypical chemokine receptor 4 |
| chr13_-_48444653 | 0.51 |
ENST00000378434.8
|
LPAR6
|
lysophosphatidic acid receptor 6 |
| chr19_+_17406099 | 0.51 |
ENST00000634942.2
ENST00000634568.1 ENST00000600514.5 |
BISPR
MVB12A
|
BST2 interferon stimulated positive regulator multivesicular body subunit 12A |
| chr1_-_205994439 | 0.51 |
ENST00000617991.4
|
RAB7B
|
RAB7B, member RAS oncogene family |
| chr9_+_117704382 | 0.49 |
ENST00000646089.1
ENST00000355622.8 |
ENSG00000285082.2
TLR4
|
novel protein toll like receptor 4 |
| chr19_+_48019726 | 0.49 |
ENST00000593413.1
|
ELSPBP1
|
epididymal sperm binding protein 1 |
| chr4_+_75556048 | 0.49 |
ENST00000616557.1
ENST00000435974.2 ENST00000311623.9 |
ODAPH
|
odontogenesis associated phosphoprotein |
| chr1_-_60073733 | 0.49 |
ENST00000450089.6
|
C1orf87
|
chromosome 1 open reading frame 87 |
| chr1_-_60073750 | 0.48 |
ENST00000371201.3
|
C1orf87
|
chromosome 1 open reading frame 87 |
| chr12_+_93378550 | 0.47 |
ENST00000550056.5
ENST00000549992.5 ENST00000548662.5 ENST00000547014.5 |
NUDT4
|
nudix hydrolase 4 |
| chr6_+_32637396 | 0.47 |
ENST00000395363.5
ENST00000496318.5 ENST00000343139.11 |
HLA-DQA1
|
major histocompatibility complex, class II, DQ alpha 1 |
| chrX_+_87517784 | 0.47 |
ENST00000373119.9
ENST00000373114.4 |
KLHL4
|
kelch like family member 4 |
| chr12_+_93378625 | 0.46 |
ENST00000546925.1
|
NUDT4
|
nudix hydrolase 4 |
| chr3_-_46464868 | 0.46 |
ENST00000417439.5
ENST00000231751.9 ENST00000431944.1 |
LTF
|
lactotransferrin |
| chrX_-_52517057 | 0.46 |
ENST00000375613.7
|
XAGE1B
|
X antigen family member 1B |
| chr5_+_150778733 | 0.46 |
ENST00000526627.2
|
SMIM3
|
small integral membrane protein 3 |
| chr3_-_39192584 | 0.46 |
ENST00000340369.4
ENST00000421646.1 ENST00000396251.1 |
XIRP1
|
xin actin binding repeat containing 1 |
| chr20_-_59007210 | 0.46 |
ENST00000681175.1
ENST00000681416.1 ENST00000680753.1 ENST00000680995.1 ENST00000680206.1 ENST00000680879.1 ENST00000217131.6 ENST00000680738.1 ENST00000679948.1 ENST00000680880.1 ENST00000681877.1 |
CTSZ
|
cathepsin Z |
| chr12_-_10674013 | 0.46 |
ENST00000535345.5
ENST00000542562.5 ENST00000075503.8 |
STYK1
|
serine/threonine/tyrosine kinase 1 |
| chr2_-_189762755 | 0.44 |
ENST00000520350.1
ENST00000521630.1 ENST00000264151.10 ENST00000517895.5 |
OSGEPL1
|
O-sialoglycoprotein endopeptidase like 1 |
| chr11_+_59713403 | 0.44 |
ENST00000641815.1
|
STX3
|
syntaxin 3 |
| chr6_-_28252246 | 0.44 |
ENST00000377294.3
|
ZKSCAN4
|
zinc finger with KRAB and SCAN domains 4 |
| chrX_+_48521788 | 0.43 |
ENST00000651615.1
ENST00000495186.6 |
ENSG00000286268.1
EBP
|
novel protein EBP cholestenol delta-isomerase |
| chr2_-_96740034 | 0.43 |
ENST00000264963.9
ENST00000377079.8 |
LMAN2L
|
lectin, mannose binding 2 like |
| chr7_+_80646305 | 0.43 |
ENST00000426978.5
ENST00000432207.5 |
CD36
|
CD36 molecule |
| chr11_+_124115404 | 0.42 |
ENST00000361352.9
ENST00000449321.5 ENST00000392748.5 ENST00000392744.4 ENST00000456829.7 |
VWA5A
|
von Willebrand factor A domain containing 5A |
| chr7_+_80646436 | 0.42 |
ENST00000419819.2
|
CD36
|
CD36 molecule |
| chr16_+_31483002 | 0.41 |
ENST00000569576.5
ENST00000330498.4 |
SLC5A2
|
solute carrier family 5 member 2 |
| chr5_+_150778781 | 0.41 |
ENST00000648745.1
|
SMIM3
|
small integral membrane protein 3 |
| chrX_+_52495791 | 0.40 |
ENST00000375602.2
ENST00000375600.5 |
XAGE1A
|
X antigen family member 1A |
| chr16_+_68085861 | 0.40 |
ENST00000570212.5
ENST00000562926.5 |
NFATC3
|
nuclear factor of activated T cells 3 |
| chrX_-_153673778 | 0.39 |
ENST00000340888.8
|
PNCK
|
pregnancy up-regulated nonubiquitous CaM kinase |
| chr17_-_35986649 | 0.39 |
ENST00000622526.1
ENST00000620991.1 ENST00000618404.5 |
CCL14
|
C-C motif chemokine ligand 14 |
| chr19_-_54941610 | 0.39 |
ENST00000328092.9
ENST00000590030.5 |
NLRP7
|
NLR family pyrin domain containing 7 |
| chr8_-_30082948 | 0.39 |
ENST00000521265.5
|
SARAF
|
store-operated calcium entry associated regulatory factor |
| chr8_-_30083053 | 0.38 |
ENST00000256255.11
ENST00000523761.1 |
SARAF
|
store-operated calcium entry associated regulatory factor |
| chr3_-_184017863 | 0.38 |
ENST00000427120.6
ENST00000334444.11 ENST00000392579.6 ENST00000382494.6 ENST00000265586.10 ENST00000446941.2 |
ABCC5
|
ATP binding cassette subfamily C member 5 |
| chr8_-_30083110 | 0.38 |
ENST00000545648.2
|
SARAF
|
store-operated calcium entry associated regulatory factor |
| chr1_-_147760592 | 0.38 |
ENST00000579774.3
|
GJA5
|
gap junction protein alpha 5 |
| chr5_-_123036664 | 0.38 |
ENST00000306442.5
|
PPIC
|
peptidylprolyl isomerase C |
| chr1_-_24143112 | 0.37 |
ENST00000270800.2
|
IL22RA1
|
interleukin 22 receptor subunit alpha 1 |
| chr19_-_14518383 | 0.37 |
ENST00000254322.3
ENST00000595139.2 |
DNAJB1
|
DnaJ heat shock protein family (Hsp40) member B1 |
| chr19_-_14517425 | 0.37 |
ENST00000676577.1
ENST00000677204.1 ENST00000598235.2 |
DNAJB1
|
DnaJ heat shock protein family (Hsp40) member B1 |
| chr4_-_47914569 | 0.37 |
ENST00000507489.2
|
NFXL1
|
nuclear transcription factor, X-box binding like 1 |
| chr1_+_220786853 | 0.37 |
ENST00000366910.10
|
MTARC1
|
mitochondrial amidoxime reducing component 1 |
| chrX_-_153673678 | 0.37 |
ENST00000370150.5
|
PNCK
|
pregnancy up-regulated nonubiquitous CaM kinase |
| chr10_-_96359273 | 0.37 |
ENST00000393871.5
ENST00000419479.5 ENST00000393870.3 |
OPALIN
|
oligodendrocytic myelin paranodal and inner loop protein |
| chr6_+_46652914 | 0.36 |
ENST00000411689.6
|
SLC25A27
|
solute carrier family 25 member 27 |
| chr4_-_47914596 | 0.36 |
ENST00000381538.7
ENST00000329043.7 |
NFXL1
|
nuclear transcription factor, X-box binding like 1 |
| chr6_+_47698538 | 0.36 |
ENST00000327753.7
|
ADGRF4
|
adhesion G protein-coupled receptor F4 |
| chr9_-_21202205 | 0.35 |
ENST00000239347.3
|
IFNA7
|
interferon alpha 7 |
| chr20_-_25585517 | 0.35 |
ENST00000422516.5
ENST00000278886.11 |
NINL
|
ninein like |
| chr17_+_50834581 | 0.35 |
ENST00000426127.1
|
WFIKKN2
|
WAP, follistatin/kazal, immunoglobulin, kunitz and netrin domain containing 2 |
| chr1_+_152675295 | 0.35 |
ENST00000368783.1
|
LCE2C
|
late cornified envelope 2C |
| chr6_+_47698574 | 0.35 |
ENST00000283303.3
|
ADGRF4
|
adhesion G protein-coupled receptor F4 |
| chr2_+_202634960 | 0.34 |
ENST00000392238.3
|
FAM117B
|
family with sequence similarity 117 member B |
| chr13_+_39038347 | 0.34 |
ENST00000379599.6
ENST00000379600.8 |
NHLRC3
|
NHL repeat containing 3 |
| chr13_+_39038292 | 0.34 |
ENST00000470258.5
|
NHLRC3
|
NHL repeat containing 3 |
| chr9_+_36036899 | 0.34 |
ENST00000377966.4
|
RECK
|
reversion inducing cysteine rich protein with kazal motifs |
| chr6_-_170291053 | 0.34 |
ENST00000366756.4
|
DLL1
|
delta like canonical Notch ligand 1 |
| chr2_+_189857393 | 0.34 |
ENST00000452382.1
|
PMS1
|
PMS1 homolog 1, mismatch repair system component |
| chr2_+_108786738 | 0.34 |
ENST00000412964.6
ENST00000295124.9 |
CCDC138
|
coiled-coil domain containing 138 |
| chr6_+_154995258 | 0.33 |
ENST00000682666.1
|
TIAM2
|
TIAM Rac1 associated GEF 2 |
| chrX_-_112840815 | 0.33 |
ENST00000304758.5
ENST00000371959.9 |
AMOT
|
angiomotin |
| chr13_-_112588125 | 0.33 |
ENST00000375669.7
ENST00000464139.5 ENST00000261965.8 |
TUBGCP3
|
tubulin gamma complex associated protein 3 |
| chr19_+_36851645 | 0.32 |
ENST00000331800.8
ENST00000612719.4 ENST00000586646.1 |
ZNF345
|
zinc finger protein 345 |
| chr22_-_40856565 | 0.32 |
ENST00000620312.4
ENST00000216218.8 |
ST13
|
ST13 Hsp70 interacting protein |
| chr19_-_4559663 | 0.32 |
ENST00000586582.6
|
SEMA6B
|
semaphorin 6B |
| chr9_-_94640130 | 0.32 |
ENST00000414122.1
|
FBP1
|
fructose-bisphosphatase 1 |
| chr12_+_51238854 | 0.32 |
ENST00000549732.6
ENST00000604900.5 |
DAZAP2
|
DAZ associated protein 2 |
| chr7_+_107660819 | 0.31 |
ENST00000644269.2
|
SLC26A4
|
solute carrier family 26 member 4 |
| chr19_-_17334844 | 0.30 |
ENST00000159087.7
|
ANO8
|
anoctamin 8 |
| chr22_+_26621952 | 0.30 |
ENST00000354760.4
|
CRYBA4
|
crystallin beta A4 |
| chr5_+_54517706 | 0.30 |
ENST00000326277.5
ENST00000381410.5 ENST00000343017.11 |
SNX18
|
sorting nexin 18 |
| chr1_-_113759464 | 0.30 |
ENST00000357783.6
ENST00000369604.6 |
PHTF1
|
putative homeodomain transcription factor 1 |
| chr9_+_124853417 | 0.30 |
ENST00000613760.4
ENST00000618744.4 ENST00000373574.2 |
WDR38
|
WD repeat domain 38 |
| chr19_-_7040179 | 0.30 |
ENST00000381394.9
|
MBD3L4
|
methyl-CpG binding domain protein 3 like 4 |
| chr13_-_44437214 | 0.29 |
ENST00000622051.1
|
TSC22D1
|
TSC22 domain family member 1 |
| chr6_+_35737028 | 0.29 |
ENST00000373869.7
ENST00000373866.4 |
ARMC12
|
armadillo repeat containing 12 |
| chr8_+_142834801 | 0.29 |
ENST00000220940.2
|
GML
|
glycosylphosphatidylinositol anchored molecule like |
| chr12_-_76348404 | 0.29 |
ENST00000650064.2
|
BBS10
|
Bardet-Biedl syndrome 10 |
| chr2_-_58241259 | 0.29 |
ENST00000481670.2
ENST00000403676.5 ENST00000427708.6 ENST00000403295.7 ENST00000233741.9 ENST00000446381.5 ENST00000417361.1 ENST00000402135.7 ENST00000449070.5 |
FANCL
|
FA complementation group L |
| chr1_-_206003385 | 0.29 |
ENST00000617070.5
|
RAB7B
|
RAB7B, member RAS oncogene family |
| chr7_+_101817601 | 0.29 |
ENST00000292535.12
ENST00000546411.7 ENST00000549414.6 ENST00000550008.6 ENST00000556210.1 |
CUX1
|
cut like homeobox 1 |
| chr6_+_46652968 | 0.28 |
ENST00000371347.10
|
SLC25A27
|
solute carrier family 25 member 27 |
| chr19_+_1354931 | 0.28 |
ENST00000591337.7
|
PWWP3A
|
PWWP domain containing 3A, DNA repair factor |
| chr3_-_36739791 | 0.28 |
ENST00000416516.2
|
DCLK3
|
doublecortin like kinase 3 |
| chr19_-_42302292 | 0.28 |
ENST00000594989.5
|
PAFAH1B3
|
platelet activating factor acetylhydrolase 1b catalytic subunit 3 |
| chr19_-_2051224 | 0.28 |
ENST00000309340.11
ENST00000589534.2 ENST00000250896.9 ENST00000589509.5 |
MKNK2
|
MAPK interacting serine/threonine kinase 2 |
| chr2_+_216659600 | 0.28 |
ENST00000456764.1
|
IGFBP2
|
insulin like growth factor binding protein 2 |
| chr6_+_35737078 | 0.28 |
ENST00000288065.6
|
ARMC12
|
armadillo repeat containing 12 |
| chr16_-_88703611 | 0.28 |
ENST00000541206.6
|
RNF166
|
ring finger protein 166 |
| chr13_-_44436801 | 0.28 |
ENST00000261489.6
|
TSC22D1
|
TSC22 domain family member 1 |
| chr14_-_106335613 | 0.28 |
ENST00000603660.1
|
IGHV3-30
|
immunoglobulin heavy variable 3-30 |
| chr4_-_83109843 | 0.28 |
ENST00000411416.6
|
PLAC8
|
placenta associated 8 |
| chr7_+_150323239 | 0.27 |
ENST00000323078.7
ENST00000493307.1 ENST00000359623.9 |
LRRC61
|
leucine rich repeat containing 61 |
| chr1_+_14929734 | 0.27 |
ENST00000376028.8
ENST00000400798.6 |
KAZN
|
kazrin, periplakin interacting protein |
| chr1_-_153963462 | 0.27 |
ENST00000368621.5
ENST00000368623.7 ENST00000621013.4 |
SLC39A1
|
solute carrier family 39 member 1 |
| chr12_+_52274610 | 0.27 |
ENST00000423955.7
|
KRT86
|
keratin 86 |
| chr11_+_86302211 | 0.27 |
ENST00000533986.5
ENST00000278483.8 |
HIKESHI
|
heat shock protein nuclear import factor hikeshi |
| chr2_-_191847068 | 0.26 |
ENST00000304141.5
|
CAVIN2
|
caveolae associated protein 2 |
| chr1_-_145095528 | 0.26 |
ENST00000612199.4
ENST00000641863.1 |
SRGAP2B
|
SLIT-ROBO Rho GTPase activating protein 2B |
| chr19_+_917287 | 0.26 |
ENST00000592648.1
ENST00000234371.10 |
KISS1R
|
KISS1 receptor |
| chr13_-_77975513 | 0.26 |
ENST00000646948.1
ENST00000435281.2 |
EDNRB
ENSG00000233379.2
|
endothelin receptor type B novel transcript |
| chrX_-_153674325 | 0.26 |
ENST00000370142.5
ENST00000447676.6 |
PNCK
|
pregnancy up-regulated nonubiquitous CaM kinase |
| chr19_+_1249870 | 0.26 |
ENST00000591446.6
|
MIDN
|
midnolin |
| chr21_-_32771712 | 0.26 |
ENST00000331923.9
|
PAXBP1
|
PAX3 and PAX7 binding protein 1 |
| chr1_-_235866916 | 0.26 |
ENST00000389794.7
|
LYST
|
lysosomal trafficking regulator |
| chr9_-_21207143 | 0.26 |
ENST00000357374.2
|
IFNA10
|
interferon alpha 10 |
| chrX_+_136197039 | 0.25 |
ENST00000370683.6
|
FHL1
|
four and a half LIM domains 1 |
| chr21_-_30372265 | 0.25 |
ENST00000399889.4
|
KRTAP13-2
|
keratin associated protein 13-2 |
| chr12_+_103930332 | 0.25 |
ENST00000681861.1
ENST00000550595.2 ENST00000680762.1 ENST00000614327.2 ENST00000681949.1 ENST00000299767.10 |
HSP90B1
|
heat shock protein 90 beta family member 1 |
| chr19_+_13151975 | 0.25 |
ENST00000588173.1
|
IER2
|
immediate early response 2 |
| chr4_-_103099811 | 0.25 |
ENST00000504285.5
ENST00000296424.9 |
BDH2
|
3-hydroxybutyrate dehydrogenase 2 |
| chr19_-_54364908 | 0.25 |
ENST00000391742.7
|
LAIR1
|
leukocyte associated immunoglobulin like receptor 1 |
| chr20_+_33283205 | 0.25 |
ENST00000253354.2
|
BPIFB1
|
BPI fold containing family B member 1 |
| chr1_-_153963505 | 0.25 |
ENST00000356205.9
ENST00000537590.5 |
SLC39A1
|
solute carrier family 39 member 1 |
| chr4_-_83485065 | 0.24 |
ENST00000515303.2
ENST00000321945.12 ENST00000503217.2 |
ABRAXAS1
|
abraxas 1, BRCA1 A complex subunit |
| chr6_+_87472925 | 0.24 |
ENST00000369556.7
ENST00000369557.9 ENST00000369552.9 |
SLC35A1
|
solute carrier family 35 member A1 |
| chr6_-_84763437 | 0.24 |
ENST00000606784.5
ENST00000606325.5 |
TBX18
|
T-box transcription factor 18 |
| chr19_-_7058640 | 0.24 |
ENST00000333843.8
|
MBD3L3
|
methyl-CpG binding domain protein 3 like 3 |
| chr20_+_64066211 | 0.24 |
ENST00000458442.1
|
TCEA2
|
transcription elongation factor A2 |
| chr17_+_80220406 | 0.24 |
ENST00000573809.5
ENST00000361193.8 ENST00000574967.5 ENST00000576126.5 ENST00000411502.7 ENST00000546047.6 ENST00000572725.5 |
SLC26A11
|
solute carrier family 26 member 11 |
| chr10_-_99913971 | 0.24 |
ENST00000543621.6
|
DNMBP
|
dynamin binding protein |
| chrX_-_154295085 | 0.24 |
ENST00000617225.4
ENST00000619903.4 |
TEX28
|
testis expressed 28 |
| chr18_-_35290184 | 0.24 |
ENST00000589178.5
ENST00000592278.1 ENST00000333206.10 ENST00000592211.1 ENST00000420878.7 ENST00000610712.1 ENST00000586922.2 |
ZSCAN30
ENSG00000268573.1
|
zinc finger and SCAN domain containing 30 novel transcript |
| chr9_-_21368057 | 0.24 |
ENST00000449498.2
|
IFNA13
|
interferon alpha 13 |
| chr11_-_102452758 | 0.23 |
ENST00000398136.7
ENST00000361236.7 |
TMEM123
|
transmembrane protein 123 |
| chr2_-_127642131 | 0.23 |
ENST00000426981.5
|
LIMS2
|
LIM zinc finger domain containing 2 |
| chr1_-_151372683 | 0.23 |
ENST00000458566.5
ENST00000447402.7 ENST00000426705.6 ENST00000368868.10 |
SELENBP1
|
selenium binding protein 1 |
| chr2_+_87748087 | 0.23 |
ENST00000359481.9
|
PLGLB2
|
plasminogen like B2 |
| chr10_+_133453936 | 0.23 |
ENST00000640237.1
|
SCART1
|
scavenger receptor family member expressed on T cells 1 |
| chr9_-_34710069 | 0.23 |
ENST00000378792.1
ENST00000259607.7 |
CCL21
|
C-C motif chemokine ligand 21 |
| chr19_-_51020019 | 0.23 |
ENST00000309958.7
|
KLK10
|
kallikrein related peptidase 10 |
| chr2_-_87021844 | 0.23 |
ENST00000355705.4
ENST00000409310.6 |
PLGLB1
|
plasminogen like B1 |
| chr19_-_7702124 | 0.23 |
ENST00000597921.6
|
FCER2
|
Fc fragment of IgE receptor II |
| chr17_-_61928005 | 0.23 |
ENST00000444766.7
|
INTS2
|
integrator complex subunit 2 |
| chr6_-_38639898 | 0.23 |
ENST00000481247.6
|
BTBD9
|
BTB domain containing 9 |
| chr4_+_89111521 | 0.23 |
ENST00000603357.3
|
TIGD2
|
tigger transposable element derived 2 |
| chr14_+_75522427 | 0.22 |
ENST00000286639.8
|
BATF
|
basic leucine zipper ATF-like transcription factor |
| chrX_-_30308333 | 0.22 |
ENST00000378963.1
|
NR0B1
|
nuclear receptor subfamily 0 group B member 1 |
| chr2_-_218002988 | 0.22 |
ENST00000682258.1
ENST00000446903.5 |
TNS1
|
tensin 1 |
| chr19_-_42302690 | 0.22 |
ENST00000596265.5
|
PAFAH1B3
|
platelet activating factor acetylhydrolase 1b catalytic subunit 3 |
| chr8_+_22995831 | 0.22 |
ENST00000522948.5
|
RHOBTB2
|
Rho related BTB domain containing 2 |
| chr11_+_61752603 | 0.22 |
ENST00000278836.10
|
MYRF
|
myelin regulatory factor |
| chr19_+_1248553 | 0.22 |
ENST00000586757.5
ENST00000300952.6 ENST00000682408.1 |
MIDN
|
midnolin |
| chr22_+_40857076 | 0.22 |
ENST00000614001.1
ENST00000357137.9 |
XPNPEP3
|
X-prolyl aminopeptidase 3 |
| chr1_-_235866867 | 0.22 |
ENST00000389793.7
|
LYST
|
lysosomal trafficking regulator |
| chr8_+_95024977 | 0.22 |
ENST00000396124.9
|
NDUFAF6
|
NADH:ubiquinone oxidoreductase complex assembly factor 6 |
| chr2_+_102473219 | 0.22 |
ENST00000295269.5
|
SLC9A4
|
solute carrier family 9 member A4 |
| chr2_-_240025299 | 0.22 |
ENST00000404554.5
ENST00000407129.3 ENST00000307300.8 ENST00000678455.1 ENST00000444548.6 ENST00000252711.7 ENST00000620965.5 ENST00000443626.5 |
NDUFA10
|
NADH:ubiquinone oxidoreductase subunit A10 |
| chr2_-_200864561 | 0.22 |
ENST00000434813.3
|
CLK1
|
CDC like kinase 1 |
| chr19_-_42302576 | 0.22 |
ENST00000262890.8
|
PAFAH1B3
|
platelet activating factor acetylhydrolase 1b catalytic subunit 3 |
| chr1_+_7961894 | 0.22 |
ENST00000377491.5
ENST00000377488.5 |
PARK7
|
Parkinsonism associated deglycase |
| chr14_+_39265278 | 0.22 |
ENST00000554392.5
ENST00000555716.5 ENST00000341749.7 ENST00000557038.5 |
MIA2
|
MIA SH3 domain ER export factor 2 |
| chr20_+_3888915 | 0.21 |
ENST00000316562.9
ENST00000495692.5 |
PANK2
|
pantothenate kinase 2 |
| chr12_+_119668109 | 0.21 |
ENST00000229328.10
ENST00000630317.1 |
PRKAB1
|
protein kinase AMP-activated non-catalytic subunit beta 1 |
| chr11_-_59810750 | 0.21 |
ENST00000300151.5
|
MRPL16
|
mitochondrial ribosomal protein L16 |
| chr3_+_119703001 | 0.21 |
ENST00000273390.9
ENST00000463700.1 ENST00000648112.1 |
CFAP91
ENSG00000285585.1
|
cilia and flagella associated protein 91 novel protein |
| chrX_-_75523011 | 0.21 |
ENST00000373367.8
|
ZDHHC15
|
zinc finger DHHC-type palmitoyltransferase 15 |
| chr12_+_119667859 | 0.21 |
ENST00000541640.5
|
PRKAB1
|
protein kinase AMP-activated non-catalytic subunit beta 1 |
| chr17_-_78577383 | 0.21 |
ENST00000389840.7
|
DNAH17
|
dynein axonemal heavy chain 17 |
| chr15_-_72197772 | 0.21 |
ENST00000309731.12
|
GRAMD2A
|
GRAM domain containing 2A |
| chr20_+_63235883 | 0.21 |
ENST00000342412.10
|
BIRC7
|
baculoviral IAP repeat containing 7 |
| chr22_-_26565362 | 0.21 |
ENST00000398110.6
|
TPST2
|
tyrosylprotein sulfotransferase 2 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.6 | GO:0072308 | negative regulation by virus of viral protein levels in host cell(GO:0046725) negative regulation of metanephric nephron tubule epithelial cell differentiation(GO:0072308) |
| 0.4 | 1.1 | GO:0032707 | negative regulation of interleukin-23 production(GO:0032707) |
| 0.3 | 0.9 | GO:0002415 | immunoglobulin transcytosis in epithelial cells mediated by polymeric immunoglobulin receptor(GO:0002415) |
| 0.3 | 1.1 | GO:1904349 | regulation of small intestine smooth muscle contraction(GO:1904347) positive regulation of small intestine smooth muscle contraction(GO:1904349) small intestine smooth muscle contraction(GO:1990770) |
| 0.2 | 1.6 | GO:0010757 | negative regulation of plasminogen activation(GO:0010757) |
| 0.2 | 0.8 | GO:0034164 | negative regulation of toll-like receptor 9 signaling pathway(GO:0034164) |
| 0.2 | 0.9 | GO:0033058 | directional locomotion(GO:0033058) |
| 0.2 | 0.6 | GO:0051562 | negative regulation of mitochondrial calcium ion concentration(GO:0051562) |
| 0.2 | 0.5 | GO:0033364 | mast cell secretory granule organization(GO:0033364) |
| 0.2 | 0.9 | GO:0015961 | diadenosine polyphosphate catabolic process(GO:0015961) diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 0.1 | 0.4 | GO:0002949 | tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.1 | 0.6 | GO:0005986 | sucrose biosynthetic process(GO:0005986) |
| 0.1 | 0.4 | GO:0035922 | pulmonary valve formation(GO:0003193) foramen ovale closure(GO:0035922) regulation of bundle of His cell action potential(GO:0098905) |
| 0.1 | 0.5 | GO:1900191 | biofilm formation(GO:0042710) single-species biofilm formation(GO:0044010) single-species biofilm formation in or on host organism(GO:0044407) regulation of single-species biofilm formation(GO:1900190) negative regulation of single-species biofilm formation(GO:1900191) regulation of single-species biofilm formation in or on host organism(GO:1900228) negative regulation of single-species biofilm formation in or on host organism(GO:1900229) positive regulation of chondrocyte proliferation(GO:1902732) |
| 0.1 | 0.2 | GO:2000547 | regulation of dendritic cell dendrite assembly(GO:2000547) |
| 0.1 | 0.3 | GO:0045608 | negative regulation of auditory receptor cell differentiation(GO:0045608) |
| 0.1 | 0.3 | GO:1903181 | enzyme active site formation(GO:0018307) neuron intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:0036482) detoxification of mercury ion(GO:0050787) positive regulation of mitochondrial electron transport, NADH to ubiquinone(GO:1902958) regulation of protein K48-linked deubiquitination(GO:1903093) negative regulation of protein K48-linked deubiquitination(GO:1903094) regulation of dopamine biosynthetic process(GO:1903179) positive regulation of dopamine biosynthetic process(GO:1903181) regulation of hydrogen peroxide-induced neuron intrinsic apoptotic signaling pathway(GO:1903383) negative regulation of hydrogen peroxide-induced neuron intrinsic apoptotic signaling pathway(GO:1903384) negative regulation of ubiquitin-specific protease activity(GO:2000157) |
| 0.1 | 0.5 | GO:0019075 | virus maturation(GO:0019075) |
| 0.1 | 0.8 | GO:2000332 | response to linoleic acid(GO:0070543) blood microparticle formation(GO:0072564) regulation of blood microparticle formation(GO:2000332) positive regulation of blood microparticle formation(GO:2000334) |
| 0.1 | 0.5 | GO:0033490 | cholesterol biosynthetic process via desmosterol(GO:0033489) cholesterol biosynthetic process via lathosterol(GO:0033490) |
| 0.1 | 0.3 | GO:0031247 | actin rod assembly(GO:0031247) |
| 0.1 | 0.3 | GO:1902283 | negative regulation of primary amine oxidase activity(GO:1902283) |
| 0.1 | 0.2 | GO:0060823 | canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060823) |
| 0.1 | 0.6 | GO:0045627 | positive regulation of T-helper 1 cell differentiation(GO:0045627) |
| 0.1 | 0.4 | GO:0002925 | positive regulation of humoral immune response mediated by circulating immunoglobulin(GO:0002925) |
| 0.1 | 1.2 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.1 | 0.2 | GO:0006478 | peptidyl-tyrosine sulfation(GO:0006478) |
| 0.1 | 0.2 | GO:0061357 | positive regulation of Wnt protein secretion(GO:0061357) |
| 0.1 | 0.5 | GO:1903300 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.1 | 0.3 | GO:0061083 | regulation of protein refolding(GO:0061083) negative regulation of protein refolding(GO:0061084) |
| 0.1 | 0.2 | GO:0002368 | B cell cytokine production(GO:0002368) protein import into mitochondrial intermembrane space(GO:0045041) |
| 0.1 | 0.2 | GO:0061386 | closure of optic fissure(GO:0061386) negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.1 | 0.2 | GO:0002892 | type IIa hypersensitivity(GO:0001794) regulation of type IIa hypersensitivity(GO:0001796) type II hypersensitivity(GO:0002445) regulation of type II hypersensitivity(GO:0002892) |
| 0.1 | 0.3 | GO:0035645 | enteric smooth muscle cell differentiation(GO:0035645) |
| 0.1 | 0.2 | GO:0046368 | GDP-L-fucose metabolic process(GO:0046368) |
| 0.1 | 0.1 | GO:0001812 | regulation of type I hypersensitivity(GO:0001810) positive regulation of type I hypersensitivity(GO:0001812) type I hypersensitivity(GO:0016068) |
| 0.0 | 0.1 | GO:0045715 | negative regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045715) |
| 0.0 | 0.1 | GO:0070676 | intralumenal vesicle formation(GO:0070676) negative regulation of early endosome to late endosome transport(GO:2000642) |
| 0.0 | 0.1 | GO:0071603 | endothelial cell-cell adhesion(GO:0071603) |
| 0.0 | 0.1 | GO:0040009 | regulation of growth rate(GO:0040009) |
| 0.0 | 0.2 | GO:1900226 | negative regulation of NLRP3 inflammasome complex assembly(GO:1900226) |
| 0.0 | 0.3 | GO:0090245 | axis elongation involved in somitogenesis(GO:0090245) |
| 0.0 | 0.1 | GO:1903015 | regulation of exo-alpha-sialidase activity(GO:1903015) |
| 0.0 | 0.4 | GO:0098881 | exocytic insertion of neurotransmitter receptor to plasma membrane(GO:0098881) exocytic insertion of neurotransmitter receptor to postsynaptic membrane(GO:0098967) |
| 0.0 | 0.6 | GO:0061299 | retina vasculature morphogenesis in camera-type eye(GO:0061299) |
| 0.0 | 0.3 | GO:0051418 | interphase microtubule nucleation by interphase microtubule organizing center(GO:0051415) microtubule nucleation by microtubule organizing center(GO:0051418) |
| 0.0 | 0.2 | GO:0034670 | chemotaxis to arachidonic acid(GO:0034670) response to arachidonic acid(GO:1904550) |
| 0.0 | 0.2 | GO:0030576 | Cajal body organization(GO:0030576) |
| 0.0 | 0.1 | GO:1901389 | regulation of transforming growth factor beta activation(GO:1901388) negative regulation of transforming growth factor beta activation(GO:1901389) |
| 0.0 | 0.3 | GO:1904684 | negative regulation of metalloendopeptidase activity(GO:1904684) |
| 0.0 | 0.2 | GO:0002803 | positive regulation of antimicrobial peptide production(GO:0002225) positive regulation of antibacterial peptide production(GO:0002803) |
| 0.0 | 0.9 | GO:0097201 | negative regulation of transcription from RNA polymerase II promoter in response to stress(GO:0097201) |
| 0.0 | 0.3 | GO:0015705 | iodide transport(GO:0015705) |
| 0.0 | 0.1 | GO:0030573 | bile acid catabolic process(GO:0030573) |
| 0.0 | 0.1 | GO:1903575 | cornified envelope assembly(GO:1903575) |
| 0.0 | 0.1 | GO:0019355 | nicotinamide nucleotide biosynthetic process from aspartate(GO:0019355) 'de novo' NAD biosynthetic process from aspartate(GO:0034628) |
| 0.0 | 0.1 | GO:0018032 | protein amidation(GO:0018032) |
| 0.0 | 0.1 | GO:0034395 | regulation of transcription from RNA polymerase II promoter in response to iron(GO:0034395) |
| 0.0 | 0.1 | GO:2000612 | thyroid-stimulating hormone secretion(GO:0070460) regulation of thyroid-stimulating hormone secretion(GO:2000612) |
| 0.0 | 0.3 | GO:0032286 | central nervous system myelin maintenance(GO:0032286) |
| 0.0 | 0.1 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.0 | 0.1 | GO:0031959 | mineralocorticoid receptor signaling pathway(GO:0031959) positive regulation of cardiac vascular smooth muscle cell differentiation(GO:2000724) |
| 0.0 | 0.1 | GO:0099640 | axo-dendritic protein transport(GO:0099640) |
| 0.0 | 0.1 | GO:0038178 | complement component C5a signaling pathway(GO:0038178) |
| 0.0 | 0.3 | GO:0042048 | olfactory behavior(GO:0042048) |
| 0.0 | 0.1 | GO:0052314 | phthalate metabolic process(GO:0018963) epinephrine biosynthetic process(GO:0042418) phytoalexin metabolic process(GO:0052314) |
| 0.0 | 0.1 | GO:0071947 | protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) |
| 0.0 | 0.3 | GO:0035973 | aggrephagy(GO:0035973) |
| 0.0 | 0.6 | GO:0033141 | positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) |
| 0.0 | 0.2 | GO:2000346 | negative regulation of hepatocyte proliferation(GO:2000346) |
| 0.0 | 0.2 | GO:0048295 | positive regulation of isotype switching to IgE isotypes(GO:0048295) |
| 0.0 | 0.1 | GO:0032072 | plasmacytoid dendritic cell activation(GO:0002270) regulation of restriction endodeoxyribonuclease activity(GO:0032072) |
| 0.0 | 0.2 | GO:0070943 | neutrophil mediated cytotoxicity(GO:0070942) neutrophil mediated killing of symbiont cell(GO:0070943) neutrophil mediated killing of bacterium(GO:0070944) |
| 0.0 | 0.1 | GO:0043091 | L-arginine import(GO:0043091) arginine import(GO:0090467) |
| 0.0 | 0.1 | GO:0009443 | pyridoxal 5'-phosphate salvage(GO:0009443) |
| 0.0 | 0.1 | GO:0042796 | snRNA transcription from RNA polymerase III promoter(GO:0042796) |
| 0.0 | 0.2 | GO:0042428 | serotonin metabolic process(GO:0042428) |
| 0.0 | 0.1 | GO:0048936 | peripheral nervous system neuron axonogenesis(GO:0048936) |
| 0.0 | 0.1 | GO:0046668 | regulation of retinal cell programmed cell death(GO:0046668) |
| 0.0 | 0.1 | GO:0006121 | mitochondrial electron transport, succinate to ubiquinone(GO:0006121) |
| 0.0 | 0.1 | GO:0006532 | aspartate biosynthetic process(GO:0006532) |
| 0.0 | 0.3 | GO:0036089 | cleavage furrow formation(GO:0036089) |
| 0.0 | 0.1 | GO:0036100 | leukotriene catabolic process(GO:0036100) leukotriene B4 catabolic process(GO:0036101) leukotriene B4 metabolic process(GO:0036102) icosanoid catabolic process(GO:1901523) fatty acid derivative catabolic process(GO:1901569) |
| 0.0 | 0.3 | GO:0015939 | pantothenate metabolic process(GO:0015939) |
| 0.0 | 0.4 | GO:1990001 | inhibition of cysteine-type endopeptidase activity involved in apoptotic process(GO:1990001) |
| 0.0 | 0.2 | GO:0034144 | negative regulation of toll-like receptor 4 signaling pathway(GO:0034144) |
| 0.0 | 0.6 | GO:0006346 | methylation-dependent chromatin silencing(GO:0006346) |
| 0.0 | 0.4 | GO:0070307 | lens fiber cell development(GO:0070307) |
| 0.0 | 0.2 | GO:0015781 | pyrimidine nucleotide-sugar transport(GO:0015781) |
| 0.0 | 0.1 | GO:0048669 | microglia differentiation(GO:0014004) microglia development(GO:0014005) collateral sprouting in absence of injury(GO:0048669) smooth endoplasmic reticulum calcium ion homeostasis(GO:0051563) cellular response to norepinephrine stimulus(GO:0071874) |
| 0.0 | 0.1 | GO:0052405 | negative regulation by host of symbiont molecular function(GO:0052405) |
| 0.0 | 0.3 | GO:0042074 | cell migration involved in gastrulation(GO:0042074) |
| 0.0 | 0.1 | GO:0051970 | negative regulation of transmission of nerve impulse(GO:0051970) |
| 0.0 | 0.2 | GO:2000580 | regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.0 | 0.1 | GO:1901475 | pyruvate transport(GO:0006848) pyruvate transmembrane transport(GO:1901475) |
| 0.0 | 0.3 | GO:0030213 | hyaluronan biosynthetic process(GO:0030213) |
| 0.0 | 0.1 | GO:0071442 | positive regulation of histone H3-K14 acetylation(GO:0071442) |
| 0.0 | 0.2 | GO:2000288 | positive regulation of myoblast proliferation(GO:2000288) |
| 0.0 | 0.3 | GO:0046951 | ketone body biosynthetic process(GO:0046951) |
| 0.0 | 0.2 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.0 | 0.1 | GO:0032380 | regulation of intracellular lipid transport(GO:0032377) regulation of intracellular sterol transport(GO:0032380) regulation of intracellular cholesterol transport(GO:0032383) |
| 0.0 | 0.1 | GO:1902630 | regulation of membrane hyperpolarization(GO:1902630) |
| 0.0 | 0.1 | GO:2000653 | chromatin reprogramming in the zygote(GO:0044725) regulation of DNA demethylation(GO:1901535) negative regulation of DNA demethylation(GO:1901536) regulation of genetic imprinting(GO:2000653) |
| 0.0 | 0.0 | GO:2000298 | regulation of Rho-dependent protein serine/threonine kinase activity(GO:2000298) |
| 0.0 | 0.3 | GO:2000394 | positive regulation of lamellipodium morphogenesis(GO:2000394) |
| 0.0 | 0.1 | GO:2000048 | negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 0.0 | 1.0 | GO:0031572 | G2 DNA damage checkpoint(GO:0031572) |
| 0.0 | 0.0 | GO:0050760 | negative regulation of thymidylate synthase biosynthetic process(GO:0050760) |
| 0.0 | 0.5 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.0 | 0.1 | GO:0003409 | optic cup structural organization(GO:0003409) |
| 0.0 | 0.2 | GO:0006307 | DNA dealkylation involved in DNA repair(GO:0006307) |
| 0.0 | 0.4 | GO:1902894 | negative regulation of pri-miRNA transcription from RNA polymerase II promoter(GO:1902894) |
| 0.0 | 0.1 | GO:0015798 | myo-inositol transport(GO:0015798) |
| 0.0 | 0.1 | GO:0015811 | L-cystine transport(GO:0015811) basic amino acid transmembrane transport(GO:1990822) |
| 0.0 | 0.2 | GO:0034472 | snRNA 3'-end processing(GO:0034472) |
| 0.0 | 0.0 | GO:2000452 | CD8-positive, alpha-beta cytotoxic T cell extravasation(GO:0035698) regulation of CD8-positive, alpha-beta cytotoxic T cell extravasation(GO:2000452) |
| 0.0 | 0.2 | GO:0033327 | Leydig cell differentiation(GO:0033327) |
| 0.0 | 0.2 | GO:0016075 | rRNA catabolic process(GO:0016075) |
| 0.0 | 0.9 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.0 | 0.1 | GO:0032202 | telomere assembly(GO:0032202) |
| 0.0 | 0.5 | GO:0051482 | positive regulation of cytosolic calcium ion concentration involved in phospholipase C-activating G-protein coupled signaling pathway(GO:0051482) |
| 0.0 | 0.2 | GO:0050713 | negative regulation of interleukin-1 beta secretion(GO:0050713) |
| 0.0 | 0.1 | GO:0007198 | adenylate cyclase-inhibiting serotonin receptor signaling pathway(GO:0007198) |
| 0.0 | 0.9 | GO:0007566 | embryo implantation(GO:0007566) |
| 0.0 | 1.5 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.0 | 0.1 | GO:0006196 | AMP catabolic process(GO:0006196) |
| 0.0 | 0.2 | GO:1990118 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.0 | 0.1 | GO:0045023 | G0 to G1 transition(GO:0045023) |
| 0.0 | 0.3 | GO:0040015 | negative regulation of multicellular organism growth(GO:0040015) |
| 0.0 | 0.4 | GO:0060285 | cilium-dependent cell motility(GO:0060285) |
| 0.0 | 0.1 | GO:0016139 | glycoside catabolic process(GO:0016139) |
| 0.0 | 0.1 | GO:0035608 | protein deglutamylation(GO:0035608) |
| 0.0 | 0.1 | GO:0006348 | chromatin silencing at telomere(GO:0006348) |
| 0.0 | 0.0 | GO:0002752 | cell surface pattern recognition receptor signaling pathway(GO:0002752) |
| 0.0 | 0.0 | GO:0002304 | gamma-delta intraepithelial T cell differentiation(GO:0002304) CD8-positive, gamma-delta intraepithelial T cell differentiation(GO:0002305) |
| 0.0 | 0.0 | GO:1904154 | positive regulation of retrograde protein transport, ER to cytosol(GO:1904154) |
| 0.0 | 0.4 | GO:1904659 | hexose transmembrane transport(GO:0035428) glucose transmembrane transport(GO:1904659) |
| 0.0 | 0.5 | GO:0071577 | zinc II ion transmembrane transport(GO:0071577) |
| 0.0 | 0.2 | GO:1902018 | negative regulation of cilium assembly(GO:1902018) |
| 0.0 | 0.2 | GO:0016188 | synaptic vesicle maturation(GO:0016188) |
| 0.0 | 0.1 | GO:0016198 | axon choice point recognition(GO:0016198) |
| 0.0 | 0.0 | GO:1903644 | regulation of chaperone-mediated protein folding(GO:1903644) |
| 0.0 | 0.4 | GO:0043304 | regulation of mast cell activation involved in immune response(GO:0033006) regulation of mast cell degranulation(GO:0043304) |
| 0.0 | 0.2 | GO:0072540 | T-helper 17 cell lineage commitment(GO:0072540) |
| 0.0 | 0.2 | GO:0003094 | glomerular filtration(GO:0003094) |
| 0.0 | 0.3 | GO:0043303 | mast cell activation involved in immune response(GO:0002279) mast cell degranulation(GO:0043303) |
| 0.0 | 0.3 | GO:0010226 | response to lithium ion(GO:0010226) |
| 0.0 | 0.1 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.0 | 0.2 | GO:0090161 | Golgi ribbon formation(GO:0090161) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.3 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.1 | 0.3 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 0.1 | 0.4 | GO:1990796 | photoreceptor cell terminal bouton(GO:1990796) |
| 0.1 | 0.5 | GO:0044279 | other organism cell membrane(GO:0044218) other organism membrane(GO:0044279) |
| 0.1 | 0.2 | GO:0031084 | BLOC-2 complex(GO:0031084) |
| 0.1 | 0.3 | GO:0008275 | gamma-tubulin small complex(GO:0008275) |
| 0.1 | 0.4 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.0 | 0.1 | GO:0005606 | laminin-1 complex(GO:0005606) |
| 0.0 | 0.8 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.0 | 0.7 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.1 | GO:1990667 | PCSK9-AnxA2 complex(GO:1990667) |
| 0.0 | 0.1 | GO:1990913 | sperm head plasma membrane(GO:1990913) ooplasm(GO:1990917) |
| 0.0 | 0.3 | GO:0032389 | MutLalpha complex(GO:0032389) |
| 0.0 | 0.1 | GO:0071062 | alphav-beta3 integrin-vitronectin complex(GO:0071062) |
| 0.0 | 0.2 | GO:1990589 | ATF4-CREB1 transcription factor complex(GO:1990589) |
| 0.0 | 0.2 | GO:0097209 | epidermal lamellar body(GO:0097209) |
| 0.0 | 1.2 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.0 | 0.1 | GO:0035363 | histone locus body(GO:0035363) |
| 0.0 | 1.8 | GO:0099738 | cell cortex region(GO:0099738) |
| 0.0 | 0.3 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 0.5 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 2.4 | GO:0097517 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 0.8 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 1.2 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 0.2 | GO:1990761 | growth cone lamellipodium(GO:1990761) |
| 0.0 | 0.1 | GO:0070436 | Grb2-EGFR complex(GO:0070436) |
| 0.0 | 0.4 | GO:0043220 | Schmidt-Lanterman incisure(GO:0043220) |
| 0.0 | 0.2 | GO:0070531 | BRCA1-A complex(GO:0070531) |
| 0.0 | 0.2 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.0 | 0.1 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 0.0 | 0.3 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.0 | 0.1 | GO:0030905 | retromer, tubulation complex(GO:0030905) |
| 0.0 | 0.1 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.0 | 0.2 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 0.4 | GO:0005639 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) |
| 0.0 | 0.1 | GO:0043190 | ATP-binding cassette (ABC) transporter complex(GO:0043190) |
| 0.0 | 0.2 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.1 | GO:0032009 | early phagosome(GO:0032009) |
| 0.0 | 0.2 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.2 | GO:0016600 | flotillin complex(GO:0016600) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0000247 | C-8 sterol isomerase activity(GO:0000247) cholestenol delta-isomerase activity(GO:0047750) |
| 0.2 | 0.9 | GO:0047179 | platelet-activating factor acetyltransferase activity(GO:0047179) |
| 0.2 | 0.9 | GO:0052845 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) inositol diphosphate tetrakisphosphate diphosphatase activity(GO:0052840) inositol bisdiphosphate tetrakisphosphate diphosphatase activity(GO:0052841) inositol diphosphate pentakisphosphate diphosphatase activity(GO:0052842) inositol-1-diphosphate-2,3,4,5,6-pentakisphosphate diphosphatase activity(GO:0052843) inositol-3-diphosphate-1,2,4,5,6-pentakisphosphate diphosphatase activity(GO:0052844) inositol-5-diphosphate-1,2,3,4,6-pentakisphosphate diphosphatase activity(GO:0052845) inositol-1,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 1-diphosphatase activity(GO:0052846) inositol-1,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 5-diphosphatase activity(GO:0052847) inositol-3,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 5-diphosphatase activity(GO:0052848) |
| 0.1 | 0.6 | GO:0042132 | fructose 1,6-bisphosphate 1-phosphatase activity(GO:0042132) |
| 0.1 | 1.1 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.1 | 0.4 | GO:0050613 | delta14-sterol reductase activity(GO:0050613) |
| 0.1 | 0.8 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 0.1 | 0.3 | GO:0016608 | growth hormone-releasing hormone activity(GO:0016608) |
| 0.1 | 0.4 | GO:0086020 | gap junction channel activity involved in SA node cell-atrial cardiac muscle cell electrical coupling(GO:0086020) |
| 0.1 | 0.2 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.1 | 0.4 | GO:0032558 | adenyl deoxyribonucleotide binding(GO:0032558) dATP binding(GO:0032564) |
| 0.1 | 0.3 | GO:0003858 | 3-hydroxybutyrate dehydrogenase activity(GO:0003858) |
| 0.1 | 0.3 | GO:0052594 | tryptamine:oxygen oxidoreductase (deaminating) activity(GO:0052593) aminoacetone:oxygen oxidoreductase(deaminating) activity(GO:0052594) aliphatic-amine oxidase activity(GO:0052595) phenethylamine:oxygen oxidoreductase (deaminating) activity(GO:0052596) |
| 0.1 | 0.9 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.1 | 0.3 | GO:0015111 | iodide transmembrane transporter activity(GO:0015111) |
| 0.1 | 0.4 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 0.1 | 0.2 | GO:0008476 | protein-tyrosine sulfotransferase activity(GO:0008476) |
| 0.1 | 0.3 | GO:1903135 | superoxide dismutase copper chaperone activity(GO:0016532) cupric ion binding(GO:1903135) |
| 0.1 | 0.2 | GO:0052739 | phosphatidylserine 1-acylhydrolase activity(GO:0052739) |
| 0.1 | 0.5 | GO:0019863 | IgE binding(GO:0019863) |
| 0.1 | 0.8 | GO:0032395 | MHC class II receptor activity(GO:0032395) |
| 0.0 | 0.1 | GO:0005477 | pyruvate secondary active transmembrane transporter activity(GO:0005477) |
| 0.0 | 0.8 | GO:0004716 | receptor signaling protein tyrosine kinase activity(GO:0004716) |
| 0.0 | 0.1 | GO:0004980 | melanocyte-stimulating hormone receptor activity(GO:0004980) |
| 0.0 | 0.5 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.0 | 0.1 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.0 | 0.2 | GO:0034186 | apolipoprotein A-I binding(GO:0034186) |
| 0.0 | 0.1 | GO:0097258 | tocopherol omega-hydroxylase activity(GO:0052870) alpha-tocopherol omega-hydroxylase activity(GO:0052871) 20-hydroxy-leukotriene B4 omega oxidase activity(GO:0097258) 20-aldehyde-leukotriene B4 20-monooxygenase activity(GO:0097259) |
| 0.0 | 0.4 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.0 | 0.3 | GO:0005127 | ciliary neurotrophic factor receptor binding(GO:0005127) |
| 0.0 | 0.3 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.0 | 0.7 | GO:1901567 | icosanoid binding(GO:0050542) fatty acid derivative binding(GO:1901567) |
| 0.0 | 0.2 | GO:0052836 | inositol 5-diphosphate pentakisphosphate 5-kinase activity(GO:0052836) inositol diphosphate tetrakisphosphate kinase activity(GO:0052839) |
| 0.0 | 0.3 | GO:0004962 | endothelin receptor activity(GO:0004962) |
| 0.0 | 0.1 | GO:0008426 | protein kinase C inhibitor activity(GO:0008426) |
| 0.0 | 0.6 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.0 | 0.8 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.0 | 0.2 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 0.0 | 0.1 | GO:0047391 | alkylglycerophosphoethanolamine phosphodiesterase activity(GO:0047391) |
| 0.0 | 0.1 | GO:0000309 | nicotinamide-nucleotide adenylyltransferase activity(GO:0000309) |
| 0.0 | 0.1 | GO:0004598 | peptidylglycine monooxygenase activity(GO:0004504) peptidylamidoglycolate lyase activity(GO:0004598) |
| 0.0 | 0.1 | GO:0032093 | SAM domain binding(GO:0032093) |
| 0.0 | 1.6 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.1 | GO:1990460 | leptin receptor binding(GO:1990460) |
| 0.0 | 0.1 | GO:0004878 | complement component C5a receptor activity(GO:0004878) |
| 0.0 | 0.7 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.2 | GO:1990763 | arrestin family protein binding(GO:1990763) |
| 0.0 | 0.1 | GO:0030549 | acetylcholine receptor activator activity(GO:0030549) |
| 0.0 | 0.1 | GO:0050253 | retinyl-palmitate esterase activity(GO:0050253) |
| 0.0 | 0.3 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.0 | 0.3 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.0 | 0.2 | GO:0015165 | pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
| 0.0 | 0.1 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.0 | 0.6 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.0 | 0.1 | GO:0008478 | pyridoxal kinase activity(GO:0008478) |
| 0.0 | 0.3 | GO:0046790 | virion binding(GO:0046790) |
| 0.0 | 0.1 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.0 | 1.1 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.0 | 0.4 | GO:0005537 | mannose binding(GO:0005537) |
| 0.0 | 0.1 | GO:0035514 | DNA demethylase activity(GO:0035514) |
| 0.0 | 0.1 | GO:0032767 | copper-dependent protein binding(GO:0032767) |
| 0.0 | 0.1 | GO:0004069 | L-aspartate:2-oxoglutarate aminotransferase activity(GO:0004069) |
| 0.0 | 0.7 | GO:0001530 | lipopolysaccharide binding(GO:0001530) |
| 0.0 | 0.4 | GO:0043225 | anion transmembrane-transporting ATPase activity(GO:0043225) |
| 0.0 | 0.3 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.6 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.2 | GO:0016433 | rRNA (adenine) methyltransferase activity(GO:0016433) |
| 0.0 | 0.2 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.0 | 0.2 | GO:0008430 | selenium binding(GO:0008430) |
| 0.0 | 0.1 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.0 | 0.2 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 0.1 | GO:0016402 | pristanoyl-CoA oxidase activity(GO:0016402) |
| 0.0 | 0.1 | GO:0008172 | S-methyltransferase activity(GO:0008172) DNA-methyltransferase activity(GO:0009008) |
| 0.0 | 0.2 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.0 | 1.1 | GO:0004180 | carboxypeptidase activity(GO:0004180) |
| 0.0 | 0.6 | GO:0019956 | chemokine binding(GO:0019956) |
| 0.0 | 0.2 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.0 | 0.6 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.3 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.0 | 0.1 | GO:0004996 | thyroid-stimulating hormone receptor activity(GO:0004996) |
| 0.0 | 0.1 | GO:0015184 | L-cystine transmembrane transporter activity(GO:0015184) |
| 0.0 | 0.2 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 0.1 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.0 | 0.3 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.0 | 0.3 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 0.1 | GO:0047066 | phospholipid-hydroperoxide glutathione peroxidase activity(GO:0047066) |
| 0.0 | 0.3 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.0 | 0.5 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.1 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 0.0 | 0.3 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.1 | GO:0051425 | PTB domain binding(GO:0051425) |
| 0.0 | 0.1 | GO:0034617 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced pteridine as one donor, and incorporation of one atom of oxygen(GO:0016714) tetrahydrobiopterin binding(GO:0034617) |
| 0.0 | 0.1 | GO:0023024 | MHC class I protein complex binding(GO:0023024) |
| 0.0 | 0.4 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.0 | 0.1 | GO:0072542 | protein phosphatase activator activity(GO:0072542) |
| 0.0 | 0.0 | GO:0004074 | biliverdin reductase activity(GO:0004074) |
| 0.0 | 0.1 | GO:0015166 | polyol transmembrane transporter activity(GO:0015166) |
| 0.0 | 0.1 | GO:0005384 | manganese ion transmembrane transporter activity(GO:0005384) |
| 0.0 | 0.3 | GO:0031852 | mu-type opioid receptor binding(GO:0031852) |
| 0.0 | 0.2 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 0.0 | GO:0090541 | MIT domain binding(GO:0090541) |
| 0.0 | 0.1 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.0 | 0.1 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) protein phosphatase 2B binding(GO:0030346) |
| 0.0 | 0.0 | GO:0032428 | sphingolipid activator protein activity(GO:0030290) beta-N-acetylgalactosaminidase activity(GO:0032428) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.6 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.0 | 1.0 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 0.9 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.0 | 1.0 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 0.4 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 0.9 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 0.3 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 0.4 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 1.0 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.0 | 0.8 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 0.6 | PID IL12 2PATHWAY | IL12-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.6 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.1 | 0.9 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.1 | 2.0 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 0.9 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.0 | 0.8 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.0 | 1.0 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 1.6 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.0 | 2.0 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 0.6 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.0 | 0.6 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 1.0 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF CAMKII | Genes involved in CREB phosphorylation through the activation of CaMKII |
| 0.0 | 0.1 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.0 | 0.6 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 0.5 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.0 | 0.3 | REACTOME SIGNALING BY NOTCH3 | Genes involved in Signaling by NOTCH3 |
| 0.0 | 0.9 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.5 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.3 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.0 | 0.3 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.0 | 0.5 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.4 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.0 | 0.8 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 0.4 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.0 | 0.3 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 0.0 | 0.3 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.0 | 1.6 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 0.1 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 0.2 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 0.2 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 0.1 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.0 | 0.4 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.3 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.3 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |