Project
Inflammatory response time course, HUVEC (Wada, 2009)
Navigation
Downloads
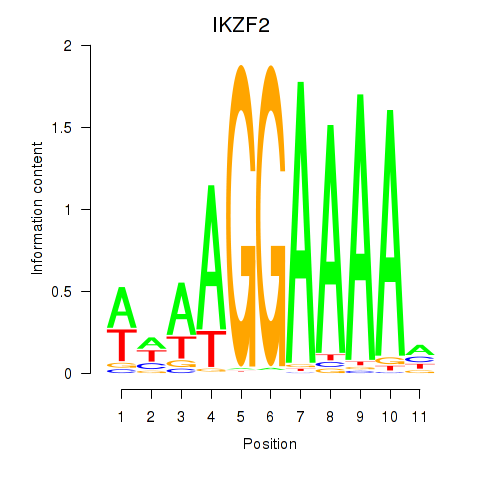
Results for IKZF2
Z-value: 0.96
Transcription factors associated with IKZF2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
IKZF2
|
ENSG00000030419.17 | IKZF2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| IKZF2 | hg38_v1_chr2_-_213151590_213151619 | -0.07 | 7.4e-01 | Click! |
Activity profile of IKZF2 motif
Sorted Z-values of IKZF2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of IKZF2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_+_100719734 | 4.12 |
ENST00000370119.8
ENST00000294728.7 ENST00000347652.6 ENST00000370115.1 |
VCAM1
|
vascular cell adhesion molecule 1 |
| chr7_+_94394886 | 2.10 |
ENST00000297268.11
ENST00000620463.1 |
COL1A2
|
collagen type I alpha 2 chain |
| chr17_-_69141878 | 2.08 |
ENST00000590645.1
ENST00000284425.7 |
ABCA6
|
ATP binding cassette subfamily A member 6 |
| chr8_+_96584920 | 2.03 |
ENST00000521590.5
|
SDC2
|
syndecan 2 |
| chr6_-_142945028 | 1.86 |
ENST00000012134.7
|
HIVEP2
|
HIVEP zinc finger 2 |
| chr6_-_142945160 | 1.84 |
ENST00000367603.8
|
HIVEP2
|
HIVEP zinc finger 2 |
| chr8_-_6563044 | 1.82 |
ENST00000338312.10
|
ANGPT2
|
angiopoietin 2 |
| chr17_+_28042839 | 1.62 |
ENST00000582037.2
|
NLK
|
nemo like kinase |
| chr8_-_6563238 | 1.62 |
ENST00000629816.3
ENST00000523120.2 |
ANGPT2
|
angiopoietin 2 |
| chr8_-_6563409 | 1.58 |
ENST00000325203.9
|
ANGPT2
|
angiopoietin 2 |
| chr6_-_10412367 | 1.55 |
ENST00000379608.9
|
TFAP2A
|
transcription factor AP-2 alpha |
| chr6_+_106098933 | 1.47 |
ENST00000369089.3
|
PRDM1
|
PR/SET domain 1 |
| chr1_-_173050931 | 1.43 |
ENST00000404377.5
|
TNFSF18
|
TNF superfamily member 18 |
| chr12_+_26195647 | 1.31 |
ENST00000535504.1
|
SSPN
|
sarcospan |
| chr10_+_113709261 | 1.27 |
ENST00000672138.1
ENST00000452490.3 |
CASP7
|
caspase 7 |
| chr1_-_56579555 | 1.24 |
ENST00000371250.4
|
PLPP3
|
phospholipid phosphatase 3 |
| chr2_+_102070360 | 1.22 |
ENST00000409929.5
ENST00000424272.5 |
IL1R1
|
interleukin 1 receptor type 1 |
| chr15_-_55249029 | 1.20 |
ENST00000566877.5
|
RAB27A
|
RAB27A, member RAS oncogene family |
| chr6_+_106086316 | 1.18 |
ENST00000369091.6
ENST00000369096.9 |
PRDM1
|
PR/SET domain 1 |
| chr14_+_63204859 | 1.15 |
ENST00000555125.1
|
RHOJ
|
ras homolog family member J |
| chr2_-_24328113 | 1.13 |
ENST00000622089.4
|
ITSN2
|
intersectin 2 |
| chr10_-_114684612 | 1.07 |
ENST00000533213.6
ENST00000369252.8 |
ABLIM1
|
actin binding LIM protein 1 |
| chr1_+_148748774 | 1.02 |
ENST00000322209.5
|
NUDT4B
|
nudix hydrolase 4B |
| chr15_-_59372863 | 1.01 |
ENST00000288235.9
|
MYO1E
|
myosin IE |
| chr17_+_41226648 | 0.96 |
ENST00000377721.3
|
KRTAP9-2
|
keratin associated protein 9-2 |
| chr2_+_188974364 | 0.95 |
ENST00000304636.9
ENST00000317840.9 |
COL3A1
|
collagen type III alpha 1 chain |
| chr3_-_149333407 | 0.93 |
ENST00000470080.5
|
TM4SF18
|
transmembrane 4 L six family member 18 |
| chr4_-_145938473 | 0.91 |
ENST00000513320.5
|
ZNF827
|
zinc finger protein 827 |
| chr12_-_104050112 | 0.85 |
ENST00000547583.1
ENST00000546851.1 ENST00000360814.9 |
GLT8D2
|
glycosyltransferase 8 domain containing 2 |
| chr3_-_71583683 | 0.84 |
ENST00000649631.1
ENST00000648718.1 |
FOXP1
|
forkhead box P1 |
| chr4_+_133149307 | 0.83 |
ENST00000618019.1
|
PCDH10
|
protocadherin 10 |
| chr11_-_75306688 | 0.82 |
ENST00000532525.1
|
ARRB1
|
arrestin beta 1 |
| chr7_+_30852273 | 0.81 |
ENST00000509504.2
|
ENSG00000250424.4
|
novel protein, MINDY4 and AQP1 readthrough |
| chr8_+_122781621 | 0.75 |
ENST00000314393.6
|
ZHX2
|
zinc fingers and homeoboxes 2 |
| chr2_+_27078598 | 0.75 |
ENST00000380320.9
|
EMILIN1
|
elastin microfibril interfacer 1 |
| chr15_-_82045998 | 0.75 |
ENST00000329713.5
|
MEX3B
|
mex-3 RNA binding family member B |
| chr15_-_82046119 | 0.74 |
ENST00000558133.1
|
MEX3B
|
mex-3 RNA binding family member B |
| chr3_-_71583592 | 0.74 |
ENST00000650156.1
ENST00000649596.1 |
FOXP1
|
forkhead box P1 |
| chr3_-_112610262 | 0.72 |
ENST00000479368.1
|
CCDC80
|
coiled-coil domain containing 80 |
| chr13_+_97222296 | 0.70 |
ENST00000343600.8
ENST00000376673.7 ENST00000679496.1 ENST00000345429.10 |
MBNL2
|
muscleblind like splicing regulator 2 |
| chr15_+_73683938 | 0.69 |
ENST00000567189.5
|
CD276
|
CD276 molecule |
| chr4_-_145938422 | 0.68 |
ENST00000656985.1
ENST00000652097.1 ENST00000503462.3 ENST00000379448.9 ENST00000513840.2 |
ZNF827
|
zinc finger protein 827 |
| chr2_-_222320124 | 0.66 |
ENST00000678139.1
|
ENSG00000288658.1
|
novel protein, ortholog of Gm2102 (M. musculus) |
| chr14_+_63204436 | 0.66 |
ENST00000316754.8
|
RHOJ
|
ras homolog family member J |
| chrX_+_9543103 | 0.66 |
ENST00000683056.1
|
TBL1X
|
transducin beta like 1 X-linked |
| chr14_+_22281097 | 0.65 |
ENST00000390465.2
|
TRAV38-2DV8
|
T cell receptor alpha variable 38-2/delta variable 8 |
| chr2_-_24360299 | 0.64 |
ENST00000361999.7
|
ITSN2
|
intersectin 2 |
| chr14_+_32076939 | 0.64 |
ENST00000556611.5
ENST00000539826.6 |
ARHGAP5
|
Rho GTPase activating protein 5 |
| chr17_-_676348 | 0.63 |
ENST00000681510.1
ENST00000679680.1 |
VPS53
|
VPS53 subunit of GARP complex |
| chr15_-_58014097 | 0.62 |
ENST00000559517.5
|
ALDH1A2
|
aldehyde dehydrogenase 1 family member A2 |
| chr6_+_29395631 | 0.61 |
ENST00000642051.1
|
OR12D2
|
olfactory receptor family 12 subfamily D member 2 |
| chr22_-_45212431 | 0.61 |
ENST00000496226.1
ENST00000251993.11 |
KIAA0930
|
KIAA0930 |
| chr2_-_159798043 | 0.60 |
ENST00000664982.1
ENST00000259053.6 |
ENSG00000287091.1
CD302
|
novel transcript, sense intronic to CD302and LY75-CD302 CD302 molecule |
| chr3_-_71583713 | 0.60 |
ENST00000649528.3
ENST00000471386.3 ENST00000493089.7 |
FOXP1
|
forkhead box P1 |
| chr9_-_16705062 | 0.60 |
ENST00000471301.3
|
BNC2
|
basonuclin 2 |
| chr18_-_55403682 | 0.59 |
ENST00000564228.5
ENST00000630828.2 |
TCF4
|
transcription factor 4 |
| chr9_+_89605004 | 0.59 |
ENST00000252506.11
ENST00000375769.1 |
GADD45G
|
growth arrest and DNA damage inducible gamma |
| chr9_+_2159672 | 0.59 |
ENST00000634343.1
|
SMARCA2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr19_+_11089446 | 0.57 |
ENST00000557933.5
ENST00000455727.6 ENST00000535915.5 ENST00000545707.5 ENST00000558518.6 ENST00000558013.5 |
LDLR
|
low density lipoprotein receptor |
| chrX_+_136148440 | 0.57 |
ENST00000627383.2
ENST00000630084.2 |
FHL1
|
four and a half LIM domains 1 |
| chr2_-_175005357 | 0.57 |
ENST00000409156.7
ENST00000444573.2 ENST00000409900.9 |
CHN1
|
chimerin 1 |
| chr2_-_101151253 | 0.57 |
ENST00000376840.8
ENST00000409318.2 |
TBC1D8
|
TBC1 domain family member 8 |
| chr16_-_20900319 | 0.56 |
ENST00000564349.5
ENST00000324344.9 |
ERI2
DCUN1D3
|
ERI1 exoribonuclease family member 2 defective in cullin neddylation 1 domain containing 3 |
| chr17_-_40782544 | 0.53 |
ENST00000301656.4
|
KRT27
|
keratin 27 |
| chr8_-_71356653 | 0.53 |
ENST00000388742.8
ENST00000388740.4 |
EYA1
|
EYA transcriptional coactivator and phosphatase 1 |
| chr3_+_179148341 | 0.53 |
ENST00000263967.4
|
PIK3CA
|
phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha |
| chr5_-_140633167 | 0.53 |
ENST00000302014.11
|
CD14
|
CD14 molecule |
| chr7_-_106284524 | 0.52 |
ENST00000681936.1
ENST00000680786.1 ENST00000681550.1 |
NAMPT
|
nicotinamide phosphoribosyltransferase |
| chr1_-_150269051 | 0.52 |
ENST00000414276.6
ENST00000360244.8 |
APH1A
|
aph-1 homolog A, gamma-secretase subunit |
| chr10_+_30434176 | 0.51 |
ENST00000263056.6
ENST00000375322.2 |
MAP3K8
|
mitogen-activated protein kinase kinase kinase 8 |
| chr12_+_121132869 | 0.51 |
ENST00000328963.10
|
P2RX7
|
purinergic receptor P2X 7 |
| chr10_+_30434021 | 0.50 |
ENST00000542547.5
|
MAP3K8
|
mitogen-activated protein kinase kinase kinase 8 |
| chr6_+_27138588 | 0.49 |
ENST00000615353.1
|
H4C9
|
H4 clustered histone 9 |
| chrX_+_28587411 | 0.49 |
ENST00000378993.6
|
IL1RAPL1
|
interleukin 1 receptor accessory protein like 1 |
| chr19_-_42765657 | 0.49 |
ENST00000406636.7
ENST00000404209.8 ENST00000306511.5 |
PSG8
|
pregnancy specific beta-1-glycoprotein 8 |
| chr10_+_113125536 | 0.48 |
ENST00000349937.7
|
TCF7L2
|
transcription factor 7 like 2 |
| chr9_+_2159850 | 0.48 |
ENST00000416751.2
|
SMARCA2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr8_-_71356511 | 0.48 |
ENST00000419131.6
ENST00000388743.6 |
EYA1
|
EYA transcriptional coactivator and phosphatase 1 |
| chr1_+_99646025 | 0.47 |
ENST00000263174.9
ENST00000605497.5 ENST00000615664.1 |
PALMD
|
palmdelphin |
| chr20_-_10420737 | 0.46 |
ENST00000649912.1
|
ENSG00000285723.1
|
novel protein |
| chr1_-_155978524 | 0.46 |
ENST00000361247.9
|
ARHGEF2
|
Rho/Rac guanine nucleotide exchange factor 2 |
| chr3_-_183099508 | 0.46 |
ENST00000476176.5
|
MCCC1
|
methylcrotonoyl-CoA carboxylase 1 |
| chr11_-_33869816 | 0.45 |
ENST00000395833.7
|
LMO2
|
LIM domain only 2 |
| chr19_-_57559833 | 0.45 |
ENST00000457177.5
|
ZNF550
|
zinc finger protein 550 |
| chr16_+_74999312 | 0.44 |
ENST00000566250.5
ENST00000567962.5 |
ZNRF1
|
zinc and ring finger 1 |
| chr17_+_28042660 | 0.44 |
ENST00000407008.8
|
NLK
|
nemo like kinase |
| chr12_-_123436664 | 0.44 |
ENST00000280571.10
|
RILPL2
|
Rab interacting lysosomal protein like 2 |
| chr9_-_72365198 | 0.44 |
ENST00000376962.10
ENST00000376960.8 |
ZFAND5
|
zinc finger AN1-type containing 5 |
| chr3_-_27722699 | 0.43 |
ENST00000461503.2
|
EOMES
|
eomesodermin |
| chr17_+_7884783 | 0.43 |
ENST00000380358.9
|
CHD3
|
chromodomain helicase DNA binding protein 3 |
| chrX_+_136169833 | 0.43 |
ENST00000628032.2
|
FHL1
|
four and a half LIM domains 1 |
| chr12_-_91179472 | 0.43 |
ENST00000550099.5
ENST00000546391.5 |
DCN
|
decorin |
| chr5_-_111756245 | 0.42 |
ENST00000447165.6
|
NREP
|
neuronal regeneration related protein |
| chr17_-_17836973 | 0.41 |
ENST00000261646.11
ENST00000355815.8 |
SREBF1
|
sterol regulatory element binding transcription factor 1 |
| chrX_+_136169624 | 0.41 |
ENST00000394153.6
|
FHL1
|
four and a half LIM domains 1 |
| chr12_-_55927756 | 0.41 |
ENST00000549939.1
|
PYM1
|
PYM homolog 1, exon junction complex associated factor |
| chr4_+_133149278 | 0.41 |
ENST00000264360.7
|
PCDH10
|
protocadherin 10 |
| chr5_-_176388563 | 0.40 |
ENST00000509257.1
ENST00000616685.1 ENST00000614830.5 |
NOP16
|
NOP16 nucleolar protein |
| chr13_-_40982880 | 0.40 |
ENST00000635415.1
|
ELF1
|
E74 like ETS transcription factor 1 |
| chr12_-_71157872 | 0.40 |
ENST00000546561.2
|
TSPAN8
|
tetraspanin 8 |
| chrX_-_136780925 | 0.40 |
ENST00000250617.7
|
ARHGEF6
|
Rac/Cdc42 guanine nucleotide exchange factor 6 |
| chr16_+_74999003 | 0.40 |
ENST00000335325.9
ENST00000320619.10 |
ZNRF1
|
zinc and ring finger 1 |
| chr1_-_155978427 | 0.40 |
ENST00000313667.8
|
ARHGEF2
|
Rho/Rac guanine nucleotide exchange factor 2 |
| chrX_+_136169664 | 0.39 |
ENST00000456445.5
|
FHL1
|
four and a half LIM domains 1 |
| chr12_-_71157992 | 0.39 |
ENST00000247829.8
|
TSPAN8
|
tetraspanin 8 |
| chr4_+_168832005 | 0.39 |
ENST00000393726.7
ENST00000507735.6 |
PALLD
|
palladin, cytoskeletal associated protein |
| chr5_-_176388629 | 0.39 |
ENST00000619979.4
ENST00000621444.4 |
NOP16
|
NOP16 nucleolar protein |
| chr9_-_70869076 | 0.39 |
ENST00000677594.1
|
TRPM3
|
transient receptor potential cation channel subfamily M member 3 |
| chr18_+_58196736 | 0.39 |
ENST00000675221.1
|
NEDD4L
|
NEDD4 like E3 ubiquitin protein ligase |
| chr3_+_160842143 | 0.38 |
ENST00000464260.5
ENST00000295839.9 |
PPM1L
|
protein phosphatase, Mg2+/Mn2+ dependent 1L |
| chr11_-_86068743 | 0.38 |
ENST00000356360.9
|
PICALM
|
phosphatidylinositol binding clathrin assembly protein |
| chr18_-_57586668 | 0.38 |
ENST00000592699.6
ENST00000382873.8 ENST00000262093.11 ENST00000652755.1 |
FECH
|
ferrochelatase |
| chr9_+_34989641 | 0.38 |
ENST00000453597.8
ENST00000312316.9 ENST00000458263.6 ENST00000537321.5 ENST00000682809.1 ENST00000684748.1 |
DNAJB5
|
DnaJ heat shock protein family (Hsp40) member B5 |
| chr14_-_44961889 | 0.38 |
ENST00000579157.1
ENST00000396128.9 ENST00000556500.1 |
KLHL28
|
kelch like family member 28 |
| chr18_-_55422306 | 0.37 |
ENST00000566777.5
ENST00000626584.2 |
TCF4
|
transcription factor 4 |
| chr5_+_154038953 | 0.37 |
ENST00000439768.2
ENST00000522782.6 ENST00000322602.9 ENST00000522177.1 ENST00000520899.1 |
MFAP3
|
microfibril associated protein 3 |
| chr22_-_35824373 | 0.36 |
ENST00000473487.6
|
RBFOX2
|
RNA binding fox-1 homolog 2 |
| chr6_+_31655888 | 0.36 |
ENST00000375916.4
|
APOM
|
apolipoprotein M |
| chr7_-_93219564 | 0.36 |
ENST00000341723.8
|
HEPACAM2
|
HEPACAM family member 2 |
| chr7_-_93219545 | 0.36 |
ENST00000440868.5
|
HEPACAM2
|
HEPACAM family member 2 |
| chr17_+_41249687 | 0.35 |
ENST00000334109.3
|
KRTAP9-4
|
keratin associated protein 9-4 |
| chr12_+_59664677 | 0.35 |
ENST00000548610.5
|
SLC16A7
|
solute carrier family 16 member 7 |
| chr9_+_5510492 | 0.35 |
ENST00000397747.5
|
PDCD1LG2
|
programmed cell death 1 ligand 2 |
| chr14_-_106235582 | 0.35 |
ENST00000390607.2
|
IGHV3-21
|
immunoglobulin heavy variable 3-21 |
| chr7_+_1086800 | 0.34 |
ENST00000413368.5
ENST00000397092.5 ENST00000297469.3 |
GPER1
|
G protein-coupled estrogen receptor 1 |
| chr17_+_34285734 | 0.34 |
ENST00000305869.4
|
CCL11
|
C-C motif chemokine ligand 11 |
| chr20_+_38805686 | 0.34 |
ENST00000299824.6
ENST00000373331.2 |
PPP1R16B
|
protein phosphatase 1 regulatory subunit 16B |
| chr3_+_41194741 | 0.33 |
ENST00000643541.1
ENST00000426215.5 ENST00000645210.1 ENST00000646381.1 ENST00000405570.6 ENST00000642248.1 ENST00000433400.6 |
CTNNB1
|
catenin beta 1 |
| chrX_+_81202066 | 0.33 |
ENST00000373212.6
|
SH3BGRL
|
SH3 domain binding glutamate rich protein like |
| chrX_-_101407893 | 0.33 |
ENST00000676156.1
ENST00000675592.1 ENST00000674634.2 ENST00000649178.1 ENST00000218516.4 |
GLA
|
galactosidase alpha |
| chr6_+_87344812 | 0.33 |
ENST00000388923.5
|
C6orf163
|
chromosome 6 open reading frame 163 |
| chr2_-_127675459 | 0.33 |
ENST00000355119.9
|
LIMS2
|
LIM zinc finger domain containing 2 |
| chr2_-_55334529 | 0.33 |
ENST00000645860.1
ENST00000642563.1 ENST00000647396.1 |
CCDC88A
|
coiled-coil domain containing 88A |
| chrX_+_41339931 | 0.32 |
ENST00000642424.1
|
DDX3X
|
DEAD-box helicase 3 X-linked |
| chr18_-_55422492 | 0.32 |
ENST00000561992.5
ENST00000630712.2 |
TCF4
|
transcription factor 4 |
| chr15_+_40439721 | 0.31 |
ENST00000561234.5
|
BAHD1
|
bromo adjacent homology domain containing 1 |
| chr17_-_7217206 | 0.31 |
ENST00000447163.6
ENST00000647975.1 |
DLG4
|
discs large MAGUK scaffold protein 4 |
| chr2_-_127675065 | 0.31 |
ENST00000545738.6
ENST00000409808.6 |
LIMS2
|
LIM zinc finger domain containing 2 |
| chr11_-_55936400 | 0.31 |
ENST00000301532.3
|
OR5I1
|
olfactory receptor family 5 subfamily I member 1 |
| chr5_+_173889337 | 0.31 |
ENST00000520867.5
ENST00000334035.9 |
CPEB4
|
cytoplasmic polyadenylation element binding protein 4 |
| chr2_-_208255055 | 0.31 |
ENST00000345146.7
|
IDH1
|
isocitrate dehydrogenase (NADP(+)) 1 |
| chr20_-_44504850 | 0.30 |
ENST00000411544.5
|
SERINC3
|
serine incorporator 3 |
| chr10_+_110919595 | 0.29 |
ENST00000369452.9
|
SHOC2
|
SHOC2 leucine rich repeat scaffold protein |
| chrX_-_41665766 | 0.29 |
ENST00000643043.2
ENST00000486402.1 ENST00000646087.2 |
CASK
|
calcium/calmodulin dependent serine protein kinase |
| chr10_-_92497727 | 0.29 |
ENST00000496903.5
ENST00000678824.1 ENST00000371581.9 |
IDE
|
insulin degrading enzyme |
| chr2_-_227379297 | 0.28 |
ENST00000304568.4
|
TM4SF20
|
transmembrane 4 L six family member 20 |
| chr12_-_55927865 | 0.27 |
ENST00000454792.2
ENST00000408946.7 |
PYM1
|
PYM homolog 1, exon junction complex associated factor |
| chr1_+_50105666 | 0.27 |
ENST00000651347.1
|
ELAVL4
|
ELAV like RNA binding protein 4 |
| chr17_-_41041447 | 0.26 |
ENST00000306271.5
|
KRTAP1-1
|
keratin associated protein 1-1 |
| chrX_+_136648214 | 0.26 |
ENST00000370628.2
|
CD40LG
|
CD40 ligand |
| chr11_-_72785932 | 0.26 |
ENST00000539138.1
ENST00000542989.5 |
STARD10
|
StAR related lipid transfer domain containing 10 |
| chr15_+_73684204 | 0.26 |
ENST00000537340.6
ENST00000318424.9 |
CD276
|
CD276 molecule |
| chrX_+_136648138 | 0.26 |
ENST00000370629.7
|
CD40LG
|
CD40 ligand |
| chr13_-_46182136 | 0.26 |
ENST00000323076.7
|
LCP1
|
lymphocyte cytosolic protein 1 |
| chr6_-_131063207 | 0.26 |
ENST00000530481.5
|
EPB41L2
|
erythrocyte membrane protein band 4.1 like 2 |
| chr11_-_65662780 | 0.26 |
ENST00000534283.1
ENST00000527749.5 ENST00000533187.5 ENST00000525693.5 ENST00000534558.5 ENST00000532879.5 ENST00000406246.8 ENST00000532999.5 |
RELA
|
RELA proto-oncogene, NF-kB subunit |
| chr1_+_171248471 | 0.25 |
ENST00000402921.6
ENST00000617670.6 ENST00000367750.7 |
FMO1
|
flavin containing dimethylaniline monoxygenase 1 |
| chr15_+_81000913 | 0.25 |
ENST00000267984.4
|
TLNRD1
|
talin rod domain containing 1 |
| chrX_+_103776493 | 0.25 |
ENST00000433491.5
ENST00000612423.4 ENST00000443502.5 |
PLP1
|
proteolipid protein 1 |
| chr19_+_30372364 | 0.25 |
ENST00000355537.4
|
ZNF536
|
zinc finger protein 536 |
| chr19_-_54941610 | 0.24 |
ENST00000328092.9
ENST00000590030.5 |
NLRP7
|
NLR family pyrin domain containing 7 |
| chr1_-_17045219 | 0.24 |
ENST00000491274.5
|
SDHB
|
succinate dehydrogenase complex iron sulfur subunit B |
| chr2_-_196926670 | 0.24 |
ENST00000354764.9
|
PGAP1
|
post-GPI attachment to proteins inositol deacylase 1 |
| chr6_+_108166015 | 0.24 |
ENST00000368986.9
|
NR2E1
|
nuclear receptor subfamily 2 group E member 1 |
| chrX_+_103776831 | 0.24 |
ENST00000621218.5
ENST00000619236.1 |
PLP1
|
proteolipid protein 1 |
| chr19_+_43716095 | 0.24 |
ENST00000596627.1
|
IRGC
|
immunity related GTPase cinema |
| chr6_+_31670167 | 0.24 |
ENST00000375864.4
|
LY6G5B
|
lymphocyte antigen 6 family member G5B |
| chr17_+_7888783 | 0.23 |
ENST00000330494.12
ENST00000358181.8 |
CHD3
|
chromodomain helicase DNA binding protein 3 |
| chrX_+_101550537 | 0.23 |
ENST00000372829.8
|
ARMCX1
|
armadillo repeat containing X-linked 1 |
| chr1_+_174964750 | 0.23 |
ENST00000367688.3
|
RABGAP1L
|
RAB GTPase activating protein 1 like |
| chr22_+_39901075 | 0.23 |
ENST00000344138.9
|
GRAP2
|
GRB2 related adaptor protein 2 |
| chr4_-_167234266 | 0.23 |
ENST00000511269.5
ENST00000506697.5 ENST00000512042.1 |
SPOCK3
|
SPARC (osteonectin), cwcv and kazal like domains proteoglycan 3 |
| chr19_-_43205551 | 0.23 |
ENST00000599391.1
ENST00000244295.13 ENST00000596907.5 ENST00000405312.8 ENST00000451895.1 ENST00000433626.6 |
PSG4
|
pregnancy specific beta-1-glycoprotein 4 |
| chr20_+_58891302 | 0.23 |
ENST00000371095.7
ENST00000265620.11 ENST00000354359.12 ENST00000371085.8 |
GNAS
|
GNAS complex locus |
| chr6_-_131063272 | 0.23 |
ENST00000445890.6
ENST00000368128.6 ENST00000628542.2 |
EPB41L2
|
erythrocyte membrane protein band 4.1 like 2 |
| chr18_-_55321640 | 0.23 |
ENST00000637169.2
|
TCF4
|
transcription factor 4 |
| chr20_+_45881218 | 0.23 |
ENST00000372523.1
|
ZSWIM1
|
zinc finger SWIM-type containing 1 |
| chr8_-_140921188 | 0.23 |
ENST00000517887.5
|
PTK2
|
protein tyrosine kinase 2 |
| chr2_+_233032672 | 0.22 |
ENST00000233840.3
|
NEU2
|
neuraminidase 2 |
| chr6_+_32976347 | 0.22 |
ENST00000606059.1
|
BRD2
|
bromodomain containing 2 |
| chr7_+_134986483 | 0.22 |
ENST00000436302.6
ENST00000435976.6 ENST00000455283.2 |
AGBL3
|
ATP/GTP binding protein like 3 |
| chr12_+_122021850 | 0.22 |
ENST00000261822.5
|
BCL7A
|
BAF chromatin remodeling complex subunit BCL7A |
| chr6_-_131063233 | 0.21 |
ENST00000392427.7
ENST00000337057.8 ENST00000525271.5 ENST00000527411.5 |
EPB41L2
|
erythrocyte membrane protein band 4.1 like 2 |
| chr19_-_12666695 | 0.21 |
ENST00000221363.8
ENST00000598876.1 ENST00000456935.7 ENST00000486847.2 |
MAN2B1
|
mannosidase alpha class 2B member 1 |
| chr1_-_159714581 | 0.21 |
ENST00000255030.9
ENST00000437342.1 ENST00000368112.5 ENST00000368111.5 ENST00000368110.1 |
CRP
|
C-reactive protein |
| chr12_-_86838867 | 0.21 |
ENST00000621808.4
|
MGAT4C
|
MGAT4 family member C |
| chr11_-_65662931 | 0.21 |
ENST00000615805.4
ENST00000612991.4 |
RELA
|
RELA proto-oncogene, NF-kB subunit |
| chr3_+_46577778 | 0.21 |
ENST00000296145.6
|
TDGF1
|
teratocarcinoma-derived growth factor 1 |
| chr1_+_161749762 | 0.21 |
ENST00000367943.5
|
DUSP12
|
dual specificity phosphatase 12 |
| chr17_-_7217178 | 0.20 |
ENST00000485100.5
|
DLG4
|
discs large MAGUK scaffold protein 4 |
| chr15_+_84603896 | 0.20 |
ENST00000541040.5
ENST00000538076.5 ENST00000485222.2 |
ZSCAN2
|
zinc finger and SCAN domain containing 2 |
| chr1_+_158831323 | 0.20 |
ENST00000368141.5
|
MNDA
|
myeloid cell nuclear differentiation antigen |
| chr1_-_34165219 | 0.20 |
ENST00000373381.9
|
CSMD2
|
CUB and Sushi multiple domains 2 |
| chr13_-_72781871 | 0.20 |
ENST00000377767.9
ENST00000377780.8 |
DIS3
|
DIS3 homolog, exosome endoribonuclease and 3'-5' exoribonuclease |
| chr6_-_26056460 | 0.20 |
ENST00000343677.4
|
H1-2
|
H1.2 linker histone, cluster member |
| chr1_-_68232514 | 0.19 |
ENST00000262348.9
ENST00000370973.2 ENST00000370971.1 |
WLS
|
Wnt ligand secretion mediator |
| chr1_+_209768597 | 0.19 |
ENST00000487271.5
ENST00000477431.1 |
TRAF3IP3
|
TRAF3 interacting protein 3 |
| chr9_-_127905313 | 0.19 |
ENST00000622357.5
|
ST6GALNAC6
|
ST6 N-acetylgalactosaminide alpha-2,6-sialyltransferase 6 |
| chr7_+_55365317 | 0.19 |
ENST00000254770.3
|
LANCL2
|
LanC like 2 |
| chr5_-_16508990 | 0.19 |
ENST00000399793.6
|
RETREG1
|
reticulophagy regulator 1 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 5.0 | GO:0050928 | negative regulation of positive chemotaxis(GO:0050928) |
| 0.9 | 2.6 | GO:0033082 | regulation of extrathymic T cell differentiation(GO:0033082) sebum secreting cell proliferation(GO:1990654) |
| 0.6 | 4.1 | GO:0022614 | membrane to membrane docking(GO:0022614) |
| 0.4 | 1.2 | GO:2000661 | positive regulation of interleukin-1-mediated signaling pathway(GO:2000661) |
| 0.4 | 1.6 | GO:0003409 | optic cup structural organization(GO:0003409) |
| 0.4 | 1.4 | GO:2000329 | negative regulation of T-helper 17 cell lineage commitment(GO:2000329) |
| 0.3 | 2.0 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.2 | 0.7 | GO:1903259 | exon-exon junction complex disassembly(GO:1903259) |
| 0.2 | 1.2 | GO:1903435 | positive regulation of constitutive secretory pathway(GO:1903435) |
| 0.2 | 0.9 | GO:0071802 | negative regulation of podosome assembly(GO:0071802) |
| 0.2 | 0.5 | GO:1904172 | positive regulation of interleukin-1 alpha secretion(GO:0050717) regulation of bleb assembly(GO:1904170) positive regulation of bleb assembly(GO:1904172) |
| 0.2 | 1.3 | GO:0072733 | response to staurosporine(GO:0072733) cellular response to staurosporine(GO:0072734) |
| 0.2 | 0.5 | GO:0006117 | acetaldehyde metabolic process(GO:0006117) |
| 0.1 | 0.7 | GO:1903978 | regulation of microglial cell activation(GO:1903978) |
| 0.1 | 0.4 | GO:0002302 | CD8-positive, alpha-beta T cell differentiation involved in immune response(GO:0002302) |
| 0.1 | 2.1 | GO:0043589 | skin morphogenesis(GO:0043589) |
| 0.1 | 0.9 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.1 | 2.2 | GO:0032625 | interleukin-21 production(GO:0032625) interleukin-21 secretion(GO:0072619) |
| 0.1 | 1.0 | GO:0035166 | post-embryonic hemopoiesis(GO:0035166) |
| 0.1 | 1.8 | GO:0061299 | retina vasculature morphogenesis in camera-type eye(GO:0061299) |
| 0.1 | 0.3 | GO:0031959 | mineralocorticoid receptor signaling pathway(GO:0031959) positive regulation of cardiac vascular smooth muscle cell differentiation(GO:2000724) |
| 0.1 | 0.3 | GO:1902309 | negative regulation of peptidyl-serine dephosphorylation(GO:1902309) |
| 0.1 | 1.0 | GO:0072513 | positive regulation of secondary heart field cardioblast proliferation(GO:0072513) |
| 0.1 | 0.4 | GO:0046909 | intermembrane transport(GO:0046909) protein transport from ciliary membrane to plasma membrane(GO:1903445) |
| 0.1 | 0.5 | GO:2000653 | regulation of genetic imprinting(GO:2000653) |
| 0.1 | 0.5 | GO:0038123 | toll-like receptor TLR1:TLR2 signaling pathway(GO:0038123) response to triacyl bacterial lipopeptide(GO:0071725) cellular response to triacyl bacterial lipopeptide(GO:0071727) |
| 0.1 | 1.4 | GO:0008354 | germ cell migration(GO:0008354) |
| 0.1 | 0.3 | GO:0006097 | glyoxylate cycle(GO:0006097) |
| 0.1 | 0.8 | GO:0042699 | follicle-stimulating hormone signaling pathway(GO:0042699) |
| 0.1 | 0.4 | GO:0034444 | regulation of plasma lipoprotein particle oxidation(GO:0034444) negative regulation of plasma lipoprotein particle oxidation(GO:0034445) |
| 0.1 | 0.4 | GO:0061357 | positive regulation of Wnt protein secretion(GO:0061357) |
| 0.1 | 0.2 | GO:0060164 | regulation of timing of neuron differentiation(GO:0060164) |
| 0.1 | 0.3 | GO:1901143 | insulin catabolic process(GO:1901143) |
| 0.1 | 0.6 | GO:0042904 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.1 | 0.3 | GO:0035962 | response to interleukin-13(GO:0035962) |
| 0.1 | 2.1 | GO:0042501 | serine phosphorylation of STAT protein(GO:0042501) |
| 0.1 | 0.2 | GO:1901421 | positive regulation of response to alcohol(GO:1901421) |
| 0.1 | 0.8 | GO:0044334 | canonical Wnt signaling pathway involved in positive regulation of epithelial to mesenchymal transition(GO:0044334) |
| 0.1 | 0.2 | GO:2000174 | regulation of pro-T cell differentiation(GO:2000174) positive regulation of pro-T cell differentiation(GO:2000176) |
| 0.1 | 1.0 | GO:0045078 | positive regulation of interferon-gamma biosynthetic process(GO:0045078) |
| 0.1 | 0.4 | GO:1902963 | regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902962) negative regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902963) |
| 0.1 | 0.3 | GO:1903566 | positive regulation of protein localization to cilium(GO:1903566) |
| 0.0 | 0.6 | GO:0097105 | presynaptic membrane assembly(GO:0097105) |
| 0.0 | 0.3 | GO:0016139 | glycoside catabolic process(GO:0016139) |
| 0.0 | 0.2 | GO:0018008 | N-terminal peptidyl-glycine N-myristoylation(GO:0018008) |
| 0.0 | 0.4 | GO:0006848 | pyruvate transport(GO:0006848) pyruvate transmembrane transport(GO:1901475) |
| 0.0 | 0.4 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.0 | 0.3 | GO:0009597 | detection of virus(GO:0009597) |
| 0.0 | 1.9 | GO:1903861 | positive regulation of dendrite extension(GO:1903861) |
| 0.0 | 0.1 | GO:1903015 | regulation of exo-alpha-sialidase activity(GO:1903015) |
| 0.0 | 0.2 | GO:0071043 | CUT catabolic process(GO:0071034) CUT metabolic process(GO:0071043) |
| 0.0 | 0.2 | GO:0035610 | protein side chain deglutamylation(GO:0035610) |
| 0.0 | 0.5 | GO:0097113 | AMPA glutamate receptor clustering(GO:0097113) glutamate receptor clustering(GO:0097688) |
| 0.0 | 0.9 | GO:0042789 | mRNA transcription from RNA polymerase II promoter(GO:0042789) |
| 0.0 | 0.4 | GO:0006552 | leucine catabolic process(GO:0006552) |
| 0.0 | 0.3 | GO:0071651 | positive regulation of chemokine (C-C motif) ligand 5 production(GO:0071651) |
| 0.0 | 0.2 | GO:0033133 | positive regulation of glucokinase activity(GO:0033133) |
| 0.0 | 0.3 | GO:2000346 | negative regulation of hepatocyte proliferation(GO:2000346) |
| 0.0 | 0.1 | GO:0036269 | swimming behavior(GO:0036269) |
| 0.0 | 0.6 | GO:0010225 | response to UV-C(GO:0010225) |
| 0.0 | 0.3 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.0 | 0.1 | GO:0070845 | misfolded protein transport(GO:0070843) polyubiquitinated protein transport(GO:0070844) polyubiquitinated misfolded protein transport(GO:0070845) Hsp90 deacetylation(GO:0070846) |
| 0.0 | 0.2 | GO:0033591 | response to L-ascorbic acid(GO:0033591) |
| 0.0 | 0.5 | GO:0007220 | Notch receptor processing(GO:0007220) |
| 0.0 | 0.1 | GO:1903926 | signal transduction involved in intra-S DNA damage checkpoint(GO:0072428) response to bisphenol A(GO:1903925) cellular response to bisphenol A(GO:1903926) |
| 0.0 | 0.1 | GO:0021965 | spinal cord ventral commissure morphogenesis(GO:0021965) |
| 0.0 | 0.1 | GO:0099558 | maintenance of synapse structure(GO:0099558) |
| 0.0 | 0.2 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.0 | 0.1 | GO:0044855 | plasma membrane raft distribution(GO:0044855) plasma membrane raft localization(GO:0044856) plasma membrane raft polarization(GO:0044858) regulation of plasma membrane raft polarization(GO:1903906) |
| 0.0 | 0.2 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.0 | 0.6 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.0 | 0.4 | GO:0017085 | response to insecticide(GO:0017085) |
| 0.0 | 0.2 | GO:0003199 | endocardial cushion to mesenchymal transition involved in heart valve formation(GO:0003199) positive regulation of blood vessel remodeling(GO:2000504) |
| 0.0 | 0.6 | GO:1900745 | positive regulation of p38MAPK cascade(GO:1900745) |
| 0.0 | 0.2 | GO:0090666 | scaRNA localization to Cajal body(GO:0090666) |
| 0.0 | 0.5 | GO:0042759 | long-chain fatty acid biosynthetic process(GO:0042759) |
| 0.0 | 0.5 | GO:0034356 | NAD biosynthesis via nicotinamide riboside salvage pathway(GO:0034356) |
| 0.0 | 0.5 | GO:2000353 | positive regulation of endothelial cell apoptotic process(GO:2000353) |
| 0.0 | 0.2 | GO:0006689 | ganglioside catabolic process(GO:0006689) |
| 0.0 | 0.0 | GO:1902725 | negative regulation of satellite cell differentiation(GO:1902725) |
| 0.0 | 0.1 | GO:1904381 | Golgi apparatus mannose trimming(GO:1904381) |
| 0.0 | 0.1 | GO:0045410 | positive regulation of interleukin-6 biosynthetic process(GO:0045410) |
| 0.0 | 0.3 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.0 | 0.3 | GO:1900364 | negative regulation of mRNA polyadenylation(GO:1900364) |
| 0.0 | 0.1 | GO:0033159 | negative regulation of protein import into nucleus, translocation(GO:0033159) |
| 0.0 | 0.1 | GO:0090427 | activation of meiosis(GO:0090427) |
| 0.0 | 0.3 | GO:0032782 | bile acid secretion(GO:0032782) |
| 0.0 | 0.2 | GO:0032534 | regulation of microvillus assembly(GO:0032534) |
| 0.0 | 0.4 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.0 | 0.1 | GO:0035910 | ascending aorta development(GO:0035905) ascending aorta morphogenesis(GO:0035910) |
| 0.0 | 0.5 | GO:0031069 | hair follicle morphogenesis(GO:0031069) |
| 0.0 | 0.0 | GO:0040030 | regulation of molecular function, epigenetic(GO:0040030) |
| 0.0 | 1.6 | GO:0043268 | positive regulation of potassium ion transport(GO:0043268) |
| 0.0 | 0.3 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.0 | 0.1 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.0 | 0.2 | GO:0019375 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.0 | 0.1 | GO:0010716 | negative regulation of extracellular matrix disassembly(GO:0010716) |
| 0.0 | 0.6 | GO:0003416 | endochondral bone growth(GO:0003416) |
| 0.0 | 0.1 | GO:0015798 | myo-inositol transport(GO:0015798) |
| 0.0 | 0.0 | GO:0061086 | negative regulation of histone H3-K27 methylation(GO:0061086) |
| 0.0 | 0.1 | GO:2000435 | regulation of protein neddylation(GO:2000434) negative regulation of protein neddylation(GO:2000435) |
| 0.0 | 0.2 | GO:0001887 | selenium compound metabolic process(GO:0001887) |
| 0.0 | 0.0 | GO:0010607 | negative regulation of cytoplasmic mRNA processing body assembly(GO:0010607) |
| 0.0 | 0.1 | GO:0070858 | negative regulation of bile acid biosynthetic process(GO:0070858) negative regulation of bile acid metabolic process(GO:1904252) |
| 0.0 | 0.2 | GO:0050882 | voluntary musculoskeletal movement(GO:0050882) |
| 0.0 | 0.4 | GO:0050860 | negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.0 | 0.2 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.0 | 0.3 | GO:0048745 | smooth muscle tissue development(GO:0048745) |
| 0.0 | 0.8 | GO:0070207 | protein homotrimerization(GO:0070207) |
| 0.0 | 0.1 | GO:0060440 | trachea formation(GO:0060440) |
| 0.0 | 0.6 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.0 | 0.2 | GO:0040032 | post-embryonic body morphogenesis(GO:0040032) |
| 0.0 | 0.1 | GO:0006600 | creatine metabolic process(GO:0006600) |
| 0.0 | 0.3 | GO:0031507 | heterochromatin assembly(GO:0031507) |
| 0.0 | 0.2 | GO:0032930 | positive regulation of superoxide anion generation(GO:0032930) |
| 0.0 | 0.2 | GO:0006105 | succinate metabolic process(GO:0006105) |
| 0.0 | 0.2 | GO:0038007 | netrin-activated signaling pathway(GO:0038007) negative regulation of synapse assembly(GO:0051964) |
| 0.0 | 0.1 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.0 | 0.1 | GO:0010751 | regulation of nitric oxide mediated signal transduction(GO:0010749) negative regulation of nitric oxide mediated signal transduction(GO:0010751) |
| 0.0 | 1.0 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.0 | 0.4 | GO:0016048 | detection of temperature stimulus(GO:0016048) |
| 0.0 | 0.2 | GO:0006677 | glycosylceramide metabolic process(GO:0006677) |
| 0.0 | 0.2 | GO:0035641 | locomotory exploration behavior(GO:0035641) |
| 0.0 | 1.4 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.0 | 0.1 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.0 | 0.2 | GO:0007351 | blastoderm segmentation(GO:0007350) tripartite regional subdivision(GO:0007351) anterior/posterior axis specification, embryo(GO:0008595) |
| 0.0 | 0.0 | GO:0070221 | sulfide oxidation(GO:0019418) sulfide oxidation, using sulfide:quinone oxidoreductase(GO:0070221) |
| 0.0 | 0.3 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 0.2 | GO:0030889 | negative regulation of B cell proliferation(GO:0030889) |
| 0.0 | 0.2 | GO:0032310 | prostaglandin secretion(GO:0032310) |
| 0.0 | 0.2 | GO:0019885 | antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 0.0 | 0.0 | GO:0010979 | regulation of vitamin D 24-hydroxylase activity(GO:0010979) positive regulation of vitamin D 24-hydroxylase activity(GO:0010980) vitamin D catabolic process(GO:0042369) |
| 0.0 | 0.2 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.0 | 0.0 | GO:1903031 | regulation of microtubule plus-end binding(GO:1903031) positive regulation of microtubule plus-end binding(GO:1903033) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 4.1 | GO:0071065 | alpha9-beta1 integrin-vascular cell adhesion molecule-1 complex(GO:0071065) |
| 0.2 | 2.8 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.2 | 0.6 | GO:1990666 | PCSK9-LDLR complex(GO:1990666) |
| 0.2 | 0.5 | GO:0035525 | NF-kappaB p50/p65 complex(GO:0035525) |
| 0.1 | 0.5 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.1 | 0.4 | GO:0034365 | discoidal high-density lipoprotein particle(GO:0034365) |
| 0.1 | 0.3 | GO:0098843 | postsynaptic endocytic zone(GO:0098843) |
| 0.1 | 1.2 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.1 | 0.4 | GO:0070381 | endosome to plasma membrane transport vesicle(GO:0070381) |
| 0.1 | 0.8 | GO:0070369 | beta-catenin-TCF7L2 complex(GO:0070369) |
| 0.1 | 0.5 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.1 | 0.5 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.0 | 0.4 | GO:0002169 | 3-methylcrotonyl-CoA carboxylase complex, mitochondrial(GO:0002169) methylcrotonoyl-CoA carboxylase complex(GO:1905202) |
| 0.0 | 0.2 | GO:0005749 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.0 | 0.2 | GO:0071149 | TEAD-2-YAP complex(GO:0071149) |
| 0.0 | 0.7 | GO:0008091 | spectrin(GO:0008091) |
| 0.0 | 0.3 | GO:1990745 | EARP complex(GO:1990745) |
| 0.0 | 0.3 | GO:0044327 | dendritic spine head(GO:0044327) |
| 0.0 | 0.5 | GO:0005677 | chromatin silencing complex(GO:0005677) |
| 0.0 | 0.5 | GO:0098839 | postsynaptic density membrane(GO:0098839) |
| 0.0 | 0.5 | GO:0032059 | bleb(GO:0032059) |
| 0.0 | 1.1 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.8 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.0 | 0.4 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 1.6 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.4 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 0.3 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.0 | 0.1 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.0 | 0.6 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.7 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 0.7 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 2.7 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.0 | 1.5 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 0.3 | GO:0031597 | cytosolic proteasome complex(GO:0031597) |
| 0.0 | 0.2 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.0 | 0.3 | GO:0005652 | nuclear lamina(GO:0005652) |
| 0.0 | 0.2 | GO:0002199 | zona pellucida receptor complex(GO:0002199) |
| 0.0 | 0.3 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.0 | 0.1 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.0 | 2.2 | GO:0032993 | protein-DNA complex(GO:0032993) |
| 0.0 | 0.4 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.0 | 1.1 | GO:0016459 | myosin complex(GO:0016459) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 4.1 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.3 | 1.2 | GO:0004909 | interleukin-1, Type I, activating receptor activity(GO:0004909) |
| 0.3 | 0.8 | GO:0031896 | V2 vasopressin receptor binding(GO:0031896) |
| 0.3 | 0.8 | GO:0098640 | integrin binding involved in cell-matrix adhesion(GO:0098640) |
| 0.2 | 3.0 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.2 | 1.2 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 0.2 | 0.5 | GO:0005174 | CD40 receptor binding(GO:0005174) |
| 0.1 | 0.4 | GO:0005477 | pyruvate secondary active transmembrane transporter activity(GO:0005477) |
| 0.1 | 0.5 | GO:0047280 | nicotinamide phosphoribosyltransferase activity(GO:0047280) |
| 0.1 | 0.3 | GO:0051990 | isocitrate dehydrogenase (NADP+) activity(GO:0004450) (R)-2-hydroxyglutarate dehydrogenase activity(GO:0051990) |
| 0.1 | 0.3 | GO:0031626 | beta-endorphin binding(GO:0031626) |
| 0.1 | 0.5 | GO:0016019 | peptidoglycan receptor activity(GO:0016019) |
| 0.1 | 0.4 | GO:0032810 | sterol response element binding(GO:0032810) |
| 0.1 | 0.3 | GO:0043273 | CTPase activity(GO:0043273) |
| 0.1 | 0.6 | GO:0030229 | very-low-density lipoprotein particle receptor activity(GO:0030229) |
| 0.1 | 1.5 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.1 | 0.3 | GO:0031728 | CCR3 chemokine receptor binding(GO:0031728) |
| 0.1 | 0.3 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.1 | 0.5 | GO:0031812 | G-protein coupled nucleotide receptor binding(GO:0031811) P2Y1 nucleotide receptor binding(GO:0031812) |
| 0.1 | 0.7 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.1 | 2.0 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.1 | 0.2 | GO:0051538 | succinate dehydrogenase (ubiquinone) activity(GO:0008177) 3 iron, 4 sulfur cluster binding(GO:0051538) |
| 0.1 | 2.6 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.1 | 0.5 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.1 | 0.2 | GO:0033265 | choline binding(GO:0033265) |
| 0.1 | 0.6 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.0 | 0.4 | GO:0004485 | methylcrotonoyl-CoA carboxylase activity(GO:0004485) |
| 0.0 | 5.1 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.0 | 0.5 | GO:0004931 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.0 | 0.2 | GO:0052795 | exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 0.0 | 0.2 | GO:0019107 | glycylpeptide N-tetradecanoyltransferase activity(GO:0004379) myristoyltransferase activity(GO:0019107) |
| 0.0 | 0.1 | GO:0033754 | indoleamine 2,3-dioxygenase activity(GO:0033754) |
| 0.0 | 1.2 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 0.1 | GO:0035663 | Toll-like receptor 2 binding(GO:0035663) |
| 0.0 | 0.3 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.0 | 0.1 | GO:0070506 | high-density lipoprotein particle receptor activity(GO:0070506) |
| 0.0 | 0.3 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.2 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.0 | 0.2 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.5 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.4 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.0 | 0.9 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.0 | 0.1 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.0 | 0.3 | GO:1990239 | steroid hormone binding(GO:1990239) |
| 0.0 | 2.4 | GO:0050681 | androgen receptor binding(GO:0050681) |
| 0.0 | 0.2 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 0.0 | 1.0 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 2.8 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 0.3 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.0 | 0.6 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 1.0 | GO:0000146 | microfilament motor activity(GO:0000146) |
| 0.0 | 0.9 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.1 | GO:0016639 | oxidoreductase activity, acting on the CH-NH2 group of donors, NAD or NADP as acceptor(GO:0016639) |
| 0.0 | 0.6 | GO:0031852 | mu-type opioid receptor binding(GO:0031852) |
| 0.0 | 0.2 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.0 | 1.5 | GO:0032813 | tumor necrosis factor receptor superfamily binding(GO:0032813) |
| 0.0 | 0.3 | GO:0019966 | interleukin-1 binding(GO:0019966) |
| 0.0 | 0.3 | GO:0004559 | alpha-mannosidase activity(GO:0004559) |
| 0.0 | 1.0 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.0 | 1.3 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.0 | 0.1 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.0 | 0.1 | GO:0010348 | lithium:proton antiporter activity(GO:0010348) |
| 0.0 | 0.8 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.0 | 0.1 | GO:0042903 | tubulin deacetylase activity(GO:0042903) |
| 0.0 | 0.7 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.0 | 0.4 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.5 | GO:0098811 | transcriptional activator activity, RNA polymerase II transcription factor binding(GO:0001190) transcriptional repressor activity, RNA polymerase II activating transcription factor binding(GO:0098811) |
| 0.0 | 0.4 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.0 | 0.1 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.1 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.0 | 2.0 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 0.2 | GO:0050694 | galactosylceramide sulfotransferase activity(GO:0001733) galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.0 | 0.1 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.0 | 0.1 | GO:0035614 | snRNA stem-loop binding(GO:0035614) |
| 0.0 | 0.1 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.0 | 0.4 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.0 | 0.2 | GO:0008430 | selenium binding(GO:0008430) |
| 0.0 | 0.7 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.0 | 0.2 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 0.4 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.0 | 1.0 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.0 | 0.3 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.0 | 0.0 | GO:0071987 | WD40-repeat domain binding(GO:0071987) |
| 0.0 | 0.5 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 1.1 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.0 | 0.3 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.3 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 1.4 | GO:0042626 | ATPase activity, coupled to transmembrane movement of substances(GO:0042626) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 4.1 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.1 | 6.1 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.1 | 1.1 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.1 | 2.6 | PID LYMPH ANGIOGENESIS PATHWAY | VEGFR3 signaling in lymphatic endothelium |
| 0.1 | 2.4 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.0 | 1.3 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 2.3 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 1.4 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 1.2 | ST P38 MAPK PATHWAY | p38 MAPK Pathway |
| 0.0 | 0.5 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 0.9 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.0 | 0.6 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 0.5 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.0 | 1.1 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.0 | 1.4 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 0.6 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 0.2 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 0.5 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.0 | 2.6 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 1.0 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 0.8 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 0.7 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.5 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.1 | ST G ALPHA S PATHWAY | G alpha s Pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 5.5 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.1 | 2.1 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.1 | 2.1 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.1 | 1.0 | REACTOME REGULATED PROTEOLYSIS OF P75NTR | Genes involved in Regulated proteolysis of p75NTR |
| 0.1 | 0.5 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.1 | 2.3 | REACTOME A TETRASACCHARIDE LINKER SEQUENCE IS REQUIRED FOR GAG SYNTHESIS | Genes involved in A tetrasaccharide linker sequence is required for GAG synthesis |
| 0.0 | 1.4 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 4.7 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 1.4 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 1.2 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 2.3 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.0 | 0.8 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.0 | 0.6 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 1.1 | REACTOME RORA ACTIVATES CIRCADIAN EXPRESSION | Genes involved in RORA Activates Circadian Expression |
| 0.0 | 0.5 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 1.8 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.0 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.0 | 0.4 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.2 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.0 | 1.4 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.2 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 0.4 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 0.2 | REACTOME PD1 SIGNALING | Genes involved in PD-1 signaling |
| 0.0 | 0.1 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 0.7 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 0.2 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |