Project
Inflammatory response time course, HUVEC (Wada, 2009)
Navigation
Downloads
Results for IRF2_STAT2_IRF8_IRF1
Z-value: 10.71
Transcription factors associated with IRF2_STAT2_IRF8_IRF1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
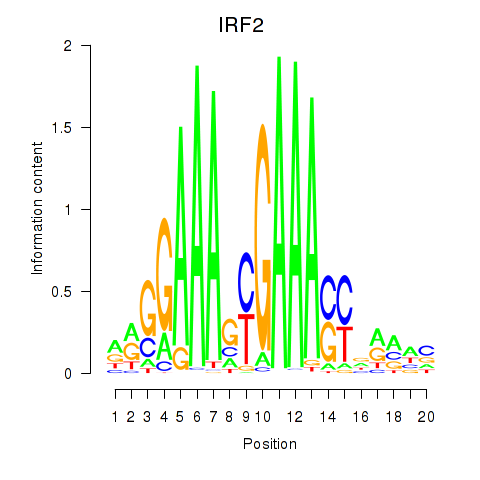
IRF2
|
ENSG00000168310.11 | IRF2 |
|
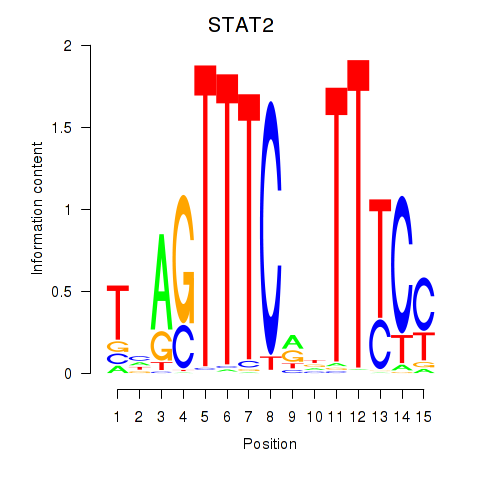
STAT2
|
ENSG00000170581.14 | STAT2 |
|
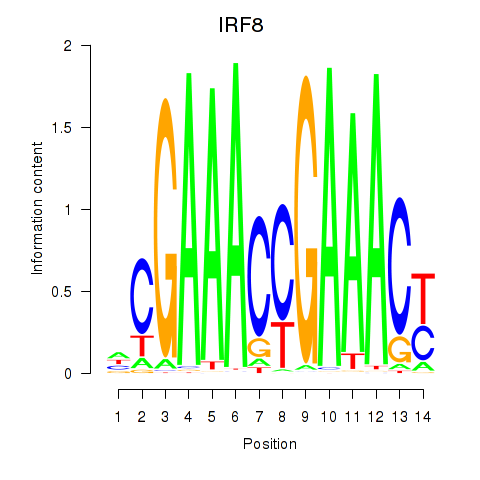
IRF8
|
ENSG00000140968.11 | IRF8 |
|
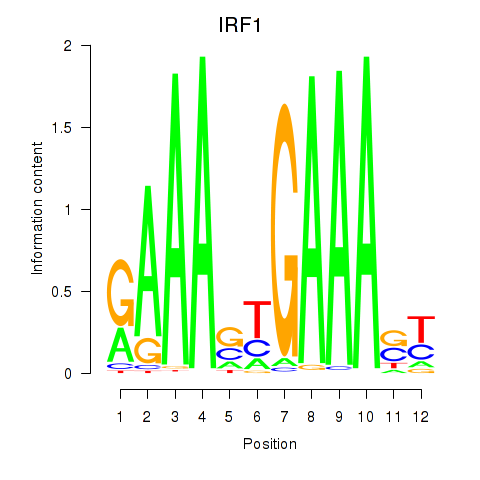
IRF1
|
ENSG00000125347.15 | IRF1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| IRF2 | hg38_v1_chr4_-_184474518_184474556 | 0.59 | 1.9e-03 | Click! |
| IRF8 | hg38_v1_chr16_+_85908988_85909033 | 0.56 | 3.6e-03 | Click! |
| IRF1 | hg38_v1_chr5_-_132490750_132490840 | 0.46 | 2.0e-02 | Click! |
| STAT2 | hg38_v1_chr12_-_56360084_56360146 | 0.37 | 7.2e-02 | Click! |
Activity profile of IRF2_STAT2_IRF8_IRF1 motif
Sorted Z-values of IRF2_STAT2_IRF8_IRF1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of IRF2_STAT2_IRF8_IRF1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 78.0 | 234.0 | GO:0046967 | cytosol to ER transport(GO:0046967) |
| 57.5 | 172.4 | GO:0009139 | dUDP biosynthetic process(GO:0006227) dTDP biosynthetic process(GO:0006233) pyrimidine nucleoside diphosphate biosynthetic process(GO:0009139) pyrimidine deoxyribonucleoside diphosphate metabolic process(GO:0009196) pyrimidine deoxyribonucleoside diphosphate biosynthetic process(GO:0009197) dTDP metabolic process(GO:0046072) dUDP metabolic process(GO:0046077) |
| 52.1 | 156.4 | GO:0045062 | extrathymic T cell selection(GO:0045062) |
| 39.5 | 118.6 | GO:0051097 | negative regulation of helicase activity(GO:0051097) |
| 38.8 | 194.2 | GO:1900245 | positive regulation of MDA-5 signaling pathway(GO:1900245) |
| 37.9 | 189.7 | GO:0034343 | type III interferon production(GO:0034343) regulation of type III interferon production(GO:0034344) |
| 34.2 | 136.9 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 29.2 | 350.3 | GO:0018377 | protein myristoylation(GO:0018377) |
| 26.4 | 396.7 | GO:0035457 | cellular response to interferon-alpha(GO:0035457) |
| 23.7 | 474.2 | GO:0003374 | dynamin polymerization involved in membrane fission(GO:0003373) dynamin polymerization involved in mitochondrial fission(GO:0003374) |
| 23.5 | 234.9 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 22.2 | 88.7 | GO:0050823 | peptide stabilization(GO:0050822) peptide antigen stabilization(GO:0050823) |
| 21.0 | 252.3 | GO:0060700 | regulation of ribonuclease activity(GO:0060700) |
| 20.7 | 124.3 | GO:0034165 | positive regulation of toll-like receptor 9 signaling pathway(GO:0034165) |
| 20.3 | 81.3 | GO:0042488 | positive regulation of odontogenesis of dentin-containing tooth(GO:0042488) mammary gland fat development(GO:0060611) positive regulation of macrophage colony-stimulating factor signaling pathway(GO:1902228) positive regulation of response to macrophage colony-stimulating factor(GO:1903971) positive regulation of cellular response to macrophage colony-stimulating factor stimulus(GO:1903974) positive regulation of microglial cell migration(GO:1904141) |
| 17.4 | 208.6 | GO:1901740 | negative regulation of myoblast fusion(GO:1901740) |
| 17.0 | 67.8 | GO:0090086 | negative regulation of protein deubiquitination(GO:0090086) |
| 16.3 | 49.0 | GO:0034769 | basement membrane disassembly(GO:0034769) |
| 14.7 | 103.2 | GO:2000110 | negative regulation of macrophage apoptotic process(GO:2000110) |
| 14.4 | 143.9 | GO:0045345 | positive regulation of MHC class I biosynthetic process(GO:0045345) |
| 14.3 | 42.9 | GO:0050717 | positive regulation of interleukin-1 alpha secretion(GO:0050717) |
| 13.5 | 40.4 | GO:0032829 | regulation of CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation(GO:0032829) positive regulation of CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation(GO:0032831) negative regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000562) |
| 12.9 | 12.9 | GO:0002590 | regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002589) negative regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002590) |
| 11.6 | 34.8 | GO:1902524 | positive regulation of protein K48-linked ubiquitination(GO:1902524) |
| 11.4 | 91.2 | GO:0070842 | aggresome assembly(GO:0070842) |
| 11.2 | 67.1 | GO:1900041 | negative regulation of interleukin-2 secretion(GO:1900041) |
| 9.8 | 78.1 | GO:0071351 | interleukin-18-mediated signaling pathway(GO:0035655) cellular response to interleukin-18(GO:0071351) |
| 9.5 | 57.1 | GO:0070383 | DNA cytosine deamination(GO:0070383) |
| 8.5 | 33.9 | GO:0006238 | CMP salvage(GO:0006238) CMP biosynthetic process(GO:0009224) CMP metabolic process(GO:0046035) |
| 8.2 | 98.2 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 7.9 | 31.8 | GO:2000329 | negative regulation of T-helper 17 cell lineage commitment(GO:2000329) |
| 7.8 | 78.0 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 6.6 | 52.8 | GO:0072734 | response to staurosporine(GO:0072733) cellular response to staurosporine(GO:0072734) |
| 6.5 | 13.0 | GO:0046642 | negative regulation of alpha-beta T cell proliferation(GO:0046642) |
| 6.5 | 25.8 | GO:1990108 | protein linear deubiquitination(GO:1990108) |
| 6.4 | 19.3 | GO:0009720 | detection of hormone stimulus(GO:0009720) |
| 6.1 | 30.4 | GO:2000834 | androgen secretion(GO:0035935) regulation of androgen secretion(GO:2000834) positive regulation of androgen secretion(GO:2000836) |
| 5.7 | 17.1 | GO:1901254 | modulation by host of viral RNA genome replication(GO:0044830) positive regulation of intracellular transport of viral material(GO:1901254) |
| 5.2 | 139.4 | GO:0072643 | interferon-gamma secretion(GO:0072643) |
| 5.2 | 36.1 | GO:0000255 | allantoin metabolic process(GO:0000255) |
| 4.5 | 49.5 | GO:0098795 | mRNA cleavage involved in gene silencing by miRNA(GO:0035279) mRNA cleavage involved in gene silencing(GO:0098795) |
| 4.1 | 33.2 | GO:0097039 | protein linear polyubiquitination(GO:0097039) |
| 4.1 | 500.4 | GO:0034340 | response to type I interferon(GO:0034340) |
| 4.0 | 12.0 | GO:2000523 | regulation of T cell costimulation(GO:2000523) positive regulation of T cell costimulation(GO:2000525) |
| 3.9 | 11.6 | GO:0051821 | negative regulation of transcription from RNA polymerase II promoter in response to UV-induced DNA damage(GO:0010768) transmission of virus(GO:0019089) dissemination or transmission of symbiont from host(GO:0044007) dissemination or transmission of organism from other organism involved in symbiotic interaction(GO:0051821) |
| 3.8 | 102.3 | GO:1900116 | extracellular regulation of signal transduction(GO:1900115) extracellular negative regulation of signal transduction(GO:1900116) |
| 3.8 | 7.6 | GO:0044333 | Wnt signaling pathway involved in digestive tract morphogenesis(GO:0044333) |
| 3.7 | 33.1 | GO:0030578 | PML body organization(GO:0030578) |
| 3.6 | 25.5 | GO:0019885 | antigen processing and presentation of endogenous peptide antigen(GO:0002483) antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 3.5 | 24.4 | GO:0033590 | response to cobalamin(GO:0033590) cellular response to erythropoietin(GO:0036018) |
| 3.2 | 9.6 | GO:0006147 | guanine catabolic process(GO:0006147) |
| 3.2 | 57.1 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 3.2 | 15.8 | GO:0010513 | positive regulation of phosphatidylinositol biosynthetic process(GO:0010513) |
| 3.2 | 12.6 | GO:0035455 | response to interferon-alpha(GO:0035455) |
| 3.1 | 299.3 | GO:0006521 | regulation of cellular amino acid metabolic process(GO:0006521) |
| 3.1 | 9.2 | GO:0000964 | mitochondrial RNA 5'-end processing(GO:0000964) rRNA import into mitochondrion(GO:0035928) |
| 3.1 | 6.1 | GO:1903660 | negative regulation of complement-dependent cytotoxicity(GO:1903660) |
| 3.0 | 29.7 | GO:0034983 | peptidyl-lysine deacetylation(GO:0034983) |
| 2.9 | 8.8 | GO:0035915 | pore formation in membrane of other organism(GO:0035915) |
| 2.9 | 45.9 | GO:1902221 | L-phenylalanine metabolic process(GO:0006558) L-phenylalanine catabolic process(GO:0006559) erythrose 4-phosphate/phosphoenolpyruvate family amino acid metabolic process(GO:1902221) erythrose 4-phosphate/phosphoenolpyruvate family amino acid catabolic process(GO:1902222) |
| 2.8 | 11.3 | GO:0003409 | optic cup structural organization(GO:0003409) |
| 2.8 | 11.2 | GO:0090290 | positive regulation of osteoclast proliferation(GO:0090290) |
| 2.8 | 19.5 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 2.6 | 36.8 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 2.6 | 7.9 | GO:0003308 | negative regulation of Wnt signaling pathway involved in heart development(GO:0003308) regulation of calcium-independent cell-cell adhesion(GO:0051040) corticotropin hormone secreting cell differentiation(GO:0060128) |
| 2.5 | 7.6 | GO:1905006 | negative regulation of epithelial to mesenchymal transition involved in endocardial cushion formation(GO:1905006) |
| 2.5 | 7.6 | GO:2000661 | positive regulation of interleukin-1-mediated signaling pathway(GO:2000661) |
| 2.3 | 7.0 | GO:0061394 | regulation of transcription from RNA polymerase II promoter in response to arsenic-containing substance(GO:0061394) |
| 2.3 | 28.1 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 2.3 | 6.9 | GO:0097272 | ammonia homeostasis(GO:0097272) |
| 2.3 | 9.2 | GO:0009443 | pyridoxal 5'-phosphate salvage(GO:0009443) |
| 2.3 | 34.2 | GO:1902004 | positive regulation of beta-amyloid formation(GO:1902004) |
| 2.3 | 2.3 | GO:1904933 | regulation of cell proliferation in midbrain(GO:1904933) |
| 2.2 | 6.7 | GO:0018395 | peptidyl-lysine hydroxylation to 5-hydroxy-L-lysine(GO:0018395) histone arginine demethylation(GO:0070077) histone H3-R2 demethylation(GO:0070078) histone H4-R3 demethylation(GO:0070079) |
| 2.2 | 6.6 | GO:0036451 | cap mRNA methylation(GO:0036451) |
| 2.2 | 6.5 | GO:0033634 | positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
| 2.1 | 4.2 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 2.1 | 8.3 | GO:0030037 | actin filament reorganization involved in cell cycle(GO:0030037) |
| 2.1 | 16.5 | GO:0015760 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 2.0 | 8.1 | GO:2000638 | regulation of SREBP signaling pathway(GO:2000638) negative regulation of SREBP signaling pathway(GO:2000639) |
| 1.9 | 5.6 | GO:2000755 | regulation of phospholipid scramblase activity(GO:1900161) positive regulation of phospholipid scramblase activity(GO:1900163) regulation of glucosylceramide catabolic process(GO:2000752) positive regulation of glucosylceramide catabolic process(GO:2000753) regulation of sphingomyelin catabolic process(GO:2000754) positive regulation of sphingomyelin catabolic process(GO:2000755) |
| 1.8 | 9.0 | GO:1903413 | cellular response to bile acid(GO:1903413) |
| 1.8 | 10.8 | GO:0003431 | growth plate cartilage chondrocyte development(GO:0003431) |
| 1.8 | 8.9 | GO:1903899 | positive regulation of PERK-mediated unfolded protein response(GO:1903899) |
| 1.8 | 10.5 | GO:0061767 | negative regulation of lung blood pressure(GO:0061767) |
| 1.7 | 13.8 | GO:0043402 | glucocorticoid mediated signaling pathway(GO:0043402) |
| 1.7 | 5.2 | GO:1904582 | regulation of intracellular mRNA localization(GO:1904580) positive regulation of intracellular mRNA localization(GO:1904582) |
| 1.7 | 323.4 | GO:0015992 | proton transport(GO:0015992) |
| 1.7 | 8.5 | GO:2000051 | negative regulation of non-canonical Wnt signaling pathway(GO:2000051) |
| 1.7 | 5.0 | GO:0006566 | threonine metabolic process(GO:0006566) |
| 1.6 | 19.5 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 1.5 | 57.4 | GO:0010390 | histone monoubiquitination(GO:0010390) |
| 1.3 | 9.3 | GO:0006154 | adenosine catabolic process(GO:0006154) inosine biosynthetic process(GO:0046103) |
| 1.3 | 10.6 | GO:2001137 | positive regulation of endocytic recycling(GO:2001137) |
| 1.3 | 10.5 | GO:0070164 | negative regulation of adiponectin secretion(GO:0070164) |
| 1.3 | 10.3 | GO:0032962 | positive regulation of inositol trisphosphate biosynthetic process(GO:0032962) |
| 1.3 | 38.0 | GO:0032897 | negative regulation of viral transcription(GO:0032897) |
| 1.2 | 4.9 | GO:0009236 | cobalamin biosynthetic process(GO:0009236) |
| 1.2 | 325.5 | GO:0051607 | defense response to virus(GO:0051607) |
| 1.2 | 10.7 | GO:0042905 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 1.2 | 8.3 | GO:0046449 | creatinine metabolic process(GO:0046449) |
| 1.2 | 8.2 | GO:0035616 | histone H2B conserved C-terminal lysine deubiquitination(GO:0035616) |
| 1.2 | 34.9 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 1.2 | 24.4 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 1.2 | 1.2 | GO:0051595 | response to methylglyoxal(GO:0051595) |
| 1.1 | 6.9 | GO:1903588 | negative regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903588) |
| 1.1 | 6.6 | GO:1904627 | response to phorbol 13-acetate 12-myristate(GO:1904627) cellular response to phorbol 13-acetate 12-myristate(GO:1904628) |
| 1.1 | 6.4 | GO:0090070 | positive regulation of ribosome biogenesis(GO:0090070) positive regulation of rRNA processing(GO:2000234) |
| 1.1 | 6.3 | GO:1904274 | tricellular tight junction assembly(GO:1904274) |
| 1.0 | 15.7 | GO:0031953 | negative regulation of protein autophosphorylation(GO:0031953) |
| 1.0 | 5.2 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 1.0 | 4.2 | GO:1900244 | retrograde trans-synaptic signaling by soluble gas(GO:0098923) trans-synaptic signaling by soluble gas(GO:0099543) positive regulation of synaptic vesicle endocytosis(GO:1900244) positive regulation of synaptic vesicle recycling(GO:1903423) |
| 1.0 | 5.2 | GO:0045204 | MAPK export from nucleus(GO:0045204) |
| 1.0 | 4.1 | GO:1905244 | regulation of modification of synaptic structure(GO:1905244) |
| 1.0 | 2.0 | GO:0021524 | visceral motor neuron differentiation(GO:0021524) |
| 1.0 | 4.1 | GO:0098886 | modification of dendritic spine(GO:0098886) |
| 1.0 | 5.8 | GO:0000430 | regulation of transcription from RNA polymerase II promoter by glucose(GO:0000430) positive regulation of transcription from RNA polymerase II promoter by glucose(GO:0000432) |
| 1.0 | 6.8 | GO:0090074 | negative regulation of protein homodimerization activity(GO:0090074) |
| 1.0 | 3.8 | GO:0044837 | assembly of actomyosin apparatus involved in cytokinesis(GO:0000912) actomyosin contractile ring assembly(GO:0000915) actomyosin contractile ring organization(GO:0044837) |
| 1.0 | 13.5 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 1.0 | 6.7 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.9 | 35.3 | GO:0019835 | cytolysis(GO:0019835) |
| 0.9 | 2.6 | GO:0090031 | regulation of growth plate cartilage chondrocyte proliferation(GO:0003420) positive regulation of steroid hormone biosynthetic process(GO:0090031) |
| 0.9 | 7.0 | GO:0002353 | kinin cascade(GO:0002254) plasma kallikrein-kinin cascade(GO:0002353) |
| 0.9 | 2.6 | GO:0090234 | regulation of kinetochore assembly(GO:0090234) |
| 0.8 | 3.4 | GO:2000152 | regulation of ubiquitin-specific protease activity(GO:2000152) positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 0.8 | 2.5 | GO:0033078 | extrathymic T cell differentiation(GO:0033078) |
| 0.8 | 10.7 | GO:0035095 | behavioral response to nicotine(GO:0035095) |
| 0.8 | 4.8 | GO:0034141 | positive regulation of toll-like receptor 3 signaling pathway(GO:0034141) |
| 0.8 | 4.8 | GO:0038172 | interleukin-33-mediated signaling pathway(GO:0038172) |
| 0.8 | 3.1 | GO:0006617 | SRP-dependent cotranslational protein targeting to membrane, signal sequence recognition(GO:0006617) |
| 0.7 | 11.2 | GO:0046597 | negative regulation of viral entry into host cell(GO:0046597) |
| 0.7 | 6.0 | GO:0045657 | positive regulation of monocyte differentiation(GO:0045657) |
| 0.7 | 9.6 | GO:0021840 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) ERBB3 signaling pathway(GO:0038129) |
| 0.7 | 2.9 | GO:0010609 | mRNA localization resulting in posttranscriptional regulation of gene expression(GO:0010609) platelet alpha granule organization(GO:0070889) regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902162) positive regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902164) |
| 0.7 | 4.9 | GO:1902261 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) |
| 0.7 | 2.1 | GO:0007518 | myoblast fate determination(GO:0007518) |
| 0.7 | 0.7 | GO:0010847 | regulation of chromatin assembly(GO:0010847) |
| 0.7 | 2.1 | GO:0003221 | right ventricular cardiac muscle tissue morphogenesis(GO:0003221) |
| 0.7 | 8.8 | GO:0006307 | DNA dealkylation involved in DNA repair(GO:0006307) |
| 0.7 | 5.3 | GO:0031584 | activation of phospholipase D activity(GO:0031584) |
| 0.7 | 3.3 | GO:1903265 | positive regulation of tumor necrosis factor-mediated signaling pathway(GO:1903265) |
| 0.7 | 17.0 | GO:0043011 | myeloid dendritic cell differentiation(GO:0043011) |
| 0.6 | 5.8 | GO:1904879 | positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1904879) |
| 0.6 | 1.9 | GO:1904468 | negative regulation of synaptic vesicle recycling(GO:1903422) negative regulation of tumor necrosis factor secretion(GO:1904468) negative regulation of membrane invagination(GO:1905154) calcium ion regulated lysosome exocytosis(GO:1990927) |
| 0.6 | 2.5 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.6 | 4.8 | GO:0061737 | leukotriene signaling pathway(GO:0061737) |
| 0.6 | 9.6 | GO:0038003 | opioid receptor signaling pathway(GO:0038003) |
| 0.6 | 12.5 | GO:0006516 | glycoprotein catabolic process(GO:0006516) |
| 0.6 | 2.4 | GO:0061484 | hematopoietic stem cell homeostasis(GO:0061484) |
| 0.6 | 4.7 | GO:1901098 | positive regulation of autophagosome maturation(GO:1901098) |
| 0.6 | 8.8 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.6 | 1.8 | GO:1903862 | positive regulation of oxidative phosphorylation(GO:1903862) |
| 0.6 | 49.5 | GO:0002456 | T cell mediated immunity(GO:0002456) |
| 0.5 | 2.7 | GO:0060022 | hard palate development(GO:0060022) |
| 0.5 | 8.7 | GO:0010818 | T cell chemotaxis(GO:0010818) |
| 0.5 | 2.1 | GO:0098957 | anterograde axonal transport of mitochondrion(GO:0098957) |
| 0.5 | 1.5 | GO:1902871 | regulation of transcription from RNA polymerase II promoter involved in forebrain neuron fate commitment(GO:0021882) cerebral cortex GABAergic interneuron fate commitment(GO:0021893) positive regulation of neural retina development(GO:0061075) positive regulation of retina development in camera-type eye(GO:1902868) positive regulation of amacrine cell differentiation(GO:1902871) |
| 0.5 | 3.0 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.5 | 8.0 | GO:0021891 | olfactory bulb interneuron development(GO:0021891) |
| 0.5 | 1.0 | GO:0002860 | positive regulation of response to tumor cell(GO:0002836) positive regulation of immune response to tumor cell(GO:0002839) positive regulation of natural killer cell mediated immune response to tumor cell(GO:0002857) positive regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002860) |
| 0.5 | 2.0 | GO:2000653 | regulation of genetic imprinting(GO:2000653) |
| 0.5 | 1.9 | GO:1902775 | mitochondrial large ribosomal subunit assembly(GO:1902775) |
| 0.5 | 37.9 | GO:0060333 | interferon-gamma-mediated signaling pathway(GO:0060333) |
| 0.5 | 8.6 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.5 | 7.6 | GO:0060315 | negative regulation of ryanodine-sensitive calcium-release channel activity(GO:0060315) |
| 0.5 | 4.7 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.5 | 1.9 | GO:0097368 | establishment of Sertoli cell barrier(GO:0097368) |
| 0.5 | 1.4 | GO:0046292 | formaldehyde metabolic process(GO:0046292) |
| 0.5 | 2.3 | GO:0061304 | retinal blood vessel morphogenesis(GO:0061304) |
| 0.5 | 6.9 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.5 | 6.4 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.5 | 4.1 | GO:0089700 | protein kinase D signaling(GO:0089700) |
| 0.4 | 1.3 | GO:0071492 | cellular response to UV-A(GO:0071492) |
| 0.4 | 0.9 | GO:1900020 | regulation of protein kinase C activity(GO:1900019) positive regulation of protein kinase C activity(GO:1900020) |
| 0.4 | 0.8 | GO:0070318 | positive regulation of G0 to G1 transition(GO:0070318) |
| 0.4 | 1.3 | GO:0003366 | cell-matrix adhesion involved in ameboidal cell migration(GO:0003366) |
| 0.4 | 19.0 | GO:0045668 | negative regulation of osteoblast differentiation(GO:0045668) |
| 0.4 | 2.4 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.4 | 8.9 | GO:0071294 | cellular response to zinc ion(GO:0071294) |
| 0.4 | 4.0 | GO:0042256 | mature ribosome assembly(GO:0042256) |
| 0.4 | 2.8 | GO:0035791 | platelet-derived growth factor receptor-beta signaling pathway(GO:0035791) |
| 0.4 | 1.6 | GO:0072086 | specification of loop of Henle identity(GO:0072086) |
| 0.4 | 6.7 | GO:0036066 | protein O-linked fucosylation(GO:0036066) |
| 0.4 | 1.2 | GO:0044854 | plasma membrane raft assembly(GO:0044854) plasma membrane raft organization(GO:0044857) caveola assembly(GO:0070836) |
| 0.4 | 4.9 | GO:0046149 | porphyrin-containing compound catabolic process(GO:0006787) tetrapyrrole catabolic process(GO:0033015) heme catabolic process(GO:0042167) pigment catabolic process(GO:0046149) |
| 0.4 | 4.5 | GO:0002441 | histamine production involved in inflammatory response(GO:0002349) histamine secretion involved in inflammatory response(GO:0002441) histamine secretion by mast cell(GO:0002553) |
| 0.4 | 1.5 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.4 | 2.2 | GO:0060040 | retinal bipolar neuron differentiation(GO:0060040) |
| 0.4 | 7.2 | GO:0042531 | positive regulation of tyrosine phosphorylation of STAT protein(GO:0042531) |
| 0.4 | 5.4 | GO:0070935 | 3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 0.3 | 10.8 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.3 | 1.6 | GO:0007352 | zygotic specification of dorsal/ventral axis(GO:0007352) |
| 0.3 | 3.3 | GO:0008343 | adult feeding behavior(GO:0008343) |
| 0.3 | 1.9 | GO:0035105 | sterol regulatory element binding protein import into nucleus(GO:0035105) |
| 0.3 | 1.0 | GO:0017198 | N-terminal peptidyl-serine acetylation(GO:0017198) N-terminal peptidyl-glutamic acid acetylation(GO:0018002) peptidyl-serine acetylation(GO:0030920) |
| 0.3 | 2.5 | GO:1903237 | negative regulation of leukocyte tethering or rolling(GO:1903237) |
| 0.3 | 0.6 | GO:1903371 | regulation of endoplasmic reticulum tubular network organization(GO:1903371) |
| 0.3 | 1.5 | GO:0032439 | endosome localization(GO:0032439) |
| 0.3 | 7.6 | GO:0042359 | vitamin D metabolic process(GO:0042359) |
| 0.3 | 4.2 | GO:0060155 | platelet dense granule organization(GO:0060155) |
| 0.3 | 2.9 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.3 | 2.3 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.3 | 2.0 | GO:0007000 | nucleolus organization(GO:0007000) |
| 0.3 | 76.4 | GO:0042157 | lipoprotein metabolic process(GO:0042157) |
| 0.3 | 2.3 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 0.3 | 1.4 | GO:0002154 | thyroid hormone mediated signaling pathway(GO:0002154) regulation of thyroid hormone mediated signaling pathway(GO:0002155) |
| 0.3 | 12.8 | GO:0046834 | lipid phosphorylation(GO:0046834) |
| 0.3 | 11.6 | GO:0070207 | protein homotrimerization(GO:0070207) |
| 0.3 | 11.3 | GO:0051497 | negative regulation of stress fiber assembly(GO:0051497) |
| 0.3 | 1.1 | GO:0002439 | chronic inflammatory response to antigenic stimulus(GO:0002439) |
| 0.3 | 7.7 | GO:0006750 | glutathione biosynthetic process(GO:0006750) |
| 0.3 | 0.3 | GO:0086092 | regulation of the force of heart contraction by cardiac conduction(GO:0086092) |
| 0.3 | 1.5 | GO:0060631 | regulation of meiosis I(GO:0060631) |
| 0.3 | 4.6 | GO:0089711 | L-glutamate transmembrane transport(GO:0089711) |
| 0.3 | 0.3 | GO:1902869 | regulation of amacrine cell differentiation(GO:1902869) |
| 0.3 | 2.3 | GO:0006828 | manganese ion transport(GO:0006828) |
| 0.3 | 3.5 | GO:0010867 | positive regulation of triglyceride biosynthetic process(GO:0010867) |
| 0.2 | 1.0 | GO:0090080 | positive regulation of MAPKKK cascade by fibroblast growth factor receptor signaling pathway(GO:0090080) |
| 0.2 | 2.7 | GO:0061088 | regulation of sequestering of zinc ion(GO:0061088) |
| 0.2 | 4.1 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.2 | 1.2 | GO:0006850 | mitochondrial pyruvate transport(GO:0006850) mitochondrial pyruvate transmembrane transport(GO:1902361) |
| 0.2 | 1.8 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.2 | 3.2 | GO:0031274 | positive regulation of pseudopodium assembly(GO:0031274) |
| 0.2 | 0.7 | GO:0071626 | mastication(GO:0071626) learned vocalization behavior(GO:0098583) |
| 0.2 | 2.9 | GO:0090179 | regulation of establishment of planar polarity involved in neural tube closure(GO:0090178) planar cell polarity pathway involved in neural tube closure(GO:0090179) |
| 0.2 | 2.7 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.2 | 7.3 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.2 | 0.9 | GO:1903251 | multi-ciliated epithelial cell differentiation(GO:1903251) |
| 0.2 | 2.2 | GO:0031086 | nuclear-transcribed mRNA catabolic process, deadenylation-independent decay(GO:0031086) |
| 0.2 | 25.4 | GO:0009615 | response to virus(GO:0009615) |
| 0.2 | 1.1 | GO:0061743 | motor learning(GO:0061743) |
| 0.2 | 1.3 | GO:0032488 | Cdc42 protein signal transduction(GO:0032488) |
| 0.2 | 0.6 | GO:0046968 | peptide antigen transport(GO:0046968) |
| 0.2 | 3.2 | GO:0006297 | nucleotide-excision repair, DNA gap filling(GO:0006297) |
| 0.2 | 2.7 | GO:2000543 | positive regulation of gastrulation(GO:2000543) |
| 0.2 | 4.0 | GO:0002360 | T cell lineage commitment(GO:0002360) |
| 0.2 | 7.7 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.2 | 1.6 | GO:1902748 | melanocyte migration(GO:0097324) positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.2 | 11.3 | GO:0006298 | mismatch repair(GO:0006298) |
| 0.2 | 1.6 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.2 | 2.6 | GO:0006590 | thyroid hormone generation(GO:0006590) |
| 0.2 | 1.0 | GO:0021764 | amygdala development(GO:0021764) |
| 0.2 | 0.6 | GO:0033385 | geranylgeranyl diphosphate metabolic process(GO:0033385) geranylgeranyl diphosphate biosynthetic process(GO:0033386) |
| 0.2 | 9.6 | GO:0006513 | protein monoubiquitination(GO:0006513) |
| 0.2 | 10.8 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.2 | 4.9 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.2 | 0.4 | GO:0036496 | regulation of translational initiation by eIF2 alpha dephosphorylation(GO:0036496) |
| 0.2 | 1.8 | GO:0006657 | CDP-choline pathway(GO:0006657) |
| 0.2 | 101.7 | GO:0019221 | cytokine-mediated signaling pathway(GO:0019221) |
| 0.2 | 3.1 | GO:0072610 | interleukin-12 secretion(GO:0072610) regulation of interleukin-12 secretion(GO:2001182) |
| 0.2 | 1.3 | GO:0001705 | ectoderm formation(GO:0001705) ectodermal cell fate commitment(GO:0001712) |
| 0.2 | 0.7 | GO:0051758 | homologous chromosome movement towards spindle pole involved in homologous chromosome segregation(GO:0051758) |
| 0.2 | 3.2 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.2 | 3.8 | GO:0090200 | positive regulation of release of cytochrome c from mitochondria(GO:0090200) |
| 0.2 | 4.5 | GO:0007567 | parturition(GO:0007567) |
| 0.2 | 4.6 | GO:0045109 | intermediate filament organization(GO:0045109) |
| 0.2 | 27.4 | GO:0006661 | phosphatidylinositol biosynthetic process(GO:0006661) |
| 0.2 | 3.6 | GO:0051131 | chaperone-mediated protein complex assembly(GO:0051131) |
| 0.2 | 1.5 | GO:0009249 | protein lipoylation(GO:0009249) |
| 0.2 | 1.0 | GO:0010667 | negative regulation of cardiac muscle cell apoptotic process(GO:0010667) |
| 0.2 | 1.8 | GO:0060019 | radial glial cell differentiation(GO:0060019) |
| 0.2 | 1.0 | GO:0050916 | sensory perception of sweet taste(GO:0050916) sensory perception of umami taste(GO:0050917) |
| 0.2 | 3.4 | GO:0045943 | positive regulation of transcription from RNA polymerase I promoter(GO:0045943) |
| 0.2 | 6.2 | GO:0043392 | negative regulation of DNA binding(GO:0043392) |
| 0.2 | 1.6 | GO:0010571 | positive regulation of nuclear cell cycle DNA replication(GO:0010571) |
| 0.2 | 1.9 | GO:0017085 | response to insecticide(GO:0017085) |
| 0.2 | 2.5 | GO:0002087 | regulation of respiratory gaseous exchange by neurological system process(GO:0002087) |
| 0.2 | 1.2 | GO:0042403 | thyroid hormone metabolic process(GO:0042403) |
| 0.2 | 2.6 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.1 | 0.9 | GO:0045078 | positive regulation of interferon-gamma biosynthetic process(GO:0045078) |
| 0.1 | 0.4 | GO:0061073 | ciliary body morphogenesis(GO:0061073) |
| 0.1 | 1.7 | GO:1904424 | regulation of GTP binding(GO:1904424) |
| 0.1 | 3.3 | GO:0006044 | N-acetylglucosamine metabolic process(GO:0006044) |
| 0.1 | 1.2 | GO:0045905 | translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 0.1 | 2.1 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.1 | 0.3 | GO:1902003 | regulation of beta-amyloid formation(GO:1902003) |
| 0.1 | 0.8 | GO:1904261 | regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904259) positive regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904261) basement membrane assembly involved in embryonic body morphogenesis(GO:2001197) |
| 0.1 | 4.0 | GO:0036150 | phosphatidylserine acyl-chain remodeling(GO:0036150) |
| 0.1 | 5.2 | GO:0035338 | long-chain fatty-acyl-CoA biosynthetic process(GO:0035338) |
| 0.1 | 2.4 | GO:0042759 | long-chain fatty acid biosynthetic process(GO:0042759) |
| 0.1 | 2.2 | GO:0006450 | regulation of translational fidelity(GO:0006450) |
| 0.1 | 1.8 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.1 | 0.9 | GO:0032971 | regulation of muscle filament sliding(GO:0032971) |
| 0.1 | 0.3 | GO:0090383 | phagosome acidification(GO:0090383) |
| 0.1 | 2.7 | GO:0016445 | somatic diversification of immunoglobulins(GO:0016445) |
| 0.1 | 3.7 | GO:1904659 | hexose transmembrane transport(GO:0035428) glucose transmembrane transport(GO:1904659) |
| 0.1 | 9.6 | GO:0006400 | tRNA modification(GO:0006400) |
| 0.1 | 0.2 | GO:0048852 | diencephalon morphogenesis(GO:0048852) |
| 0.1 | 1.4 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 0.1 | 0.7 | GO:0033227 | dsRNA transport(GO:0033227) |
| 0.1 | 0.2 | GO:0042335 | cuticle development(GO:0042335) |
| 0.1 | 2.1 | GO:1902259 | regulation of delayed rectifier potassium channel activity(GO:1902259) |
| 0.1 | 5.4 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.1 | 1.5 | GO:1904321 | response to forskolin(GO:1904321) cellular response to forskolin(GO:1904322) |
| 0.1 | 6.1 | GO:0031929 | TOR signaling(GO:0031929) |
| 0.1 | 1.0 | GO:0072321 | chaperone-mediated protein transport(GO:0072321) |
| 0.1 | 1.8 | GO:0032717 | negative regulation of interleukin-8 production(GO:0032717) |
| 0.1 | 1.2 | GO:0008298 | intracellular mRNA localization(GO:0008298) |
| 0.1 | 5.2 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.1 | 0.7 | GO:0035989 | tendon development(GO:0035989) |
| 0.1 | 0.7 | GO:0043697 | dedifferentiation(GO:0043696) cell dedifferentiation(GO:0043697) |
| 0.1 | 2.0 | GO:0033344 | cholesterol efflux(GO:0033344) |
| 0.1 | 3.1 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.1 | 7.9 | GO:0043154 | negative regulation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0043154) |
| 0.1 | 2.5 | GO:0010259 | multicellular organism aging(GO:0010259) |
| 0.1 | 0.4 | GO:1900454 | positive regulation of norepinephrine secretion(GO:0010701) positive regulation of long term synaptic depression(GO:1900454) |
| 0.1 | 0.7 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.1 | 0.3 | GO:0033037 | polysaccharide localization(GO:0033037) |
| 0.1 | 1.8 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.1 | 0.8 | GO:0043923 | positive regulation by host of viral transcription(GO:0043923) |
| 0.1 | 1.3 | GO:0032836 | glomerular basement membrane development(GO:0032836) |
| 0.1 | 0.9 | GO:0006032 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) glucosamine-containing compound catabolic process(GO:1901072) |
| 0.1 | 0.4 | GO:1990928 | response to amino acid starvation(GO:1990928) |
| 0.1 | 0.6 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.1 | 0.6 | GO:0021943 | formation of radial glial scaffolds(GO:0021943) |
| 0.1 | 1.7 | GO:0006853 | carnitine shuttle(GO:0006853) |
| 0.1 | 2.7 | GO:0000045 | autophagosome assembly(GO:0000045) |
| 0.1 | 1.1 | GO:0021794 | thalamus development(GO:0021794) |
| 0.1 | 0.5 | GO:0090336 | positive regulation of brown fat cell differentiation(GO:0090336) |
| 0.1 | 0.3 | GO:1902410 | mitotic cytokinetic process(GO:1902410) |
| 0.1 | 0.9 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.1 | 0.7 | GO:0009134 | nucleoside diphosphate catabolic process(GO:0009134) |
| 0.1 | 1.3 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.1 | 5.1 | GO:0030574 | collagen catabolic process(GO:0030574) |
| 0.1 | 1.3 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.1 | 1.1 | GO:0034501 | protein localization to kinetochore(GO:0034501) |
| 0.1 | 0.2 | GO:0090309 | positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.1 | 0.5 | GO:0050862 | positive regulation of T cell receptor signaling pathway(GO:0050862) |
| 0.1 | 0.2 | GO:1901079 | negative regulation of lipoprotein particle clearance(GO:0010985) negative regulation of low-density lipoprotein particle clearance(GO:0010989) positive regulation of relaxation of muscle(GO:1901079) occluding junction disassembly(GO:1905071) regulation of occluding junction disassembly(GO:1905073) positive regulation of occluding junction disassembly(GO:1905075) |
| 0.1 | 0.5 | GO:0007144 | female meiosis I(GO:0007144) |
| 0.1 | 4.9 | GO:0042273 | ribosomal large subunit biogenesis(GO:0042273) |
| 0.1 | 1.0 | GO:0046827 | positive regulation of protein export from nucleus(GO:0046827) |
| 0.1 | 0.8 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.1 | 15.0 | GO:0010506 | regulation of autophagy(GO:0010506) |
| 0.1 | 1.1 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.1 | 1.3 | GO:0048147 | negative regulation of fibroblast proliferation(GO:0048147) |
| 0.1 | 0.2 | GO:0060272 | embryonic skeletal joint morphogenesis(GO:0060272) |
| 0.1 | 3.9 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.1 | 0.3 | GO:0006987 | activation of signaling protein activity involved in unfolded protein response(GO:0006987) |
| 0.0 | 2.8 | GO:0060395 | SMAD protein signal transduction(GO:0060395) |
| 0.0 | 0.8 | GO:0015812 | gamma-aminobutyric acid transport(GO:0015812) |
| 0.0 | 1.7 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.0 | 4.7 | GO:0016575 | histone deacetylation(GO:0016575) |
| 0.0 | 0.9 | GO:0060765 | regulation of androgen receptor signaling pathway(GO:0060765) |
| 0.0 | 4.3 | GO:1902017 | regulation of cilium assembly(GO:1902017) |
| 0.0 | 1.7 | GO:1901385 | regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.0 | 0.8 | GO:0000722 | telomere maintenance via recombination(GO:0000722) |
| 0.0 | 0.4 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.0 | 1.9 | GO:0006370 | 7-methylguanosine mRNA capping(GO:0006370) |
| 0.0 | 3.6 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.0 | 1.4 | GO:0050919 | negative chemotaxis(GO:0050919) |
| 0.0 | 0.2 | GO:0090394 | negative regulation of excitatory postsynaptic potential(GO:0090394) |
| 0.0 | 1.6 | GO:0006458 | 'de novo' protein folding(GO:0006458) |
| 0.0 | 1.1 | GO:0030033 | microvillus assembly(GO:0030033) |
| 0.0 | 1.9 | GO:0032210 | regulation of telomere maintenance via telomerase(GO:0032210) |
| 0.0 | 0.6 | GO:0031284 | positive regulation of guanylate cyclase activity(GO:0031284) |
| 0.0 | 0.5 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.0 | 1.4 | GO:0009948 | anterior/posterior axis specification(GO:0009948) |
| 0.0 | 0.6 | GO:0007196 | adenylate cyclase-inhibiting G-protein coupled glutamate receptor signaling pathway(GO:0007196) |
| 0.0 | 1.5 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.0 | 0.6 | GO:0098743 | cell aggregation(GO:0098743) |
| 0.0 | 1.5 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.0 | 0.1 | GO:0014055 | acetylcholine secretion, neurotransmission(GO:0014055) regulation of acetylcholine secretion, neurotransmission(GO:0014056) positive regulation of acetylcholine secretion, neurotransmission(GO:0014057) negative regulation of gonadotropin secretion(GO:0032277) negative regulation of luteinizing hormone secretion(GO:0033685) |
| 0.0 | 1.6 | GO:0048661 | positive regulation of smooth muscle cell proliferation(GO:0048661) |
| 0.0 | 0.4 | GO:2000172 | regulation of branching morphogenesis of a nerve(GO:2000172) |
| 0.0 | 1.4 | GO:0021762 | substantia nigra development(GO:0021762) |
| 0.0 | 3.7 | GO:0048706 | embryonic skeletal system development(GO:0048706) |
| 0.0 | 0.3 | GO:0042738 | exogenous drug catabolic process(GO:0042738) |
| 0.0 | 0.4 | GO:0048935 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.0 | 0.3 | GO:1901660 | calcium ion export(GO:1901660) calcium ion export from cell(GO:1990034) |
| 0.0 | 0.1 | GO:0051612 | negative regulation of neurotransmitter uptake(GO:0051581) serotonin uptake(GO:0051610) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.0 | 3.2 | GO:0007030 | Golgi organization(GO:0007030) |
| 0.0 | 0.7 | GO:0006654 | phosphatidic acid biosynthetic process(GO:0006654) |
| 0.0 | 0.2 | GO:0019720 | Mo-molybdopterin cofactor biosynthetic process(GO:0006777) Mo-molybdopterin cofactor metabolic process(GO:0019720) |
| 0.0 | 1.0 | GO:0007340 | acrosome reaction(GO:0007340) |
| 0.0 | 1.4 | GO:0036498 | IRE1-mediated unfolded protein response(GO:0036498) |
| 0.0 | 0.9 | GO:0050918 | positive chemotaxis(GO:0050918) |
| 0.0 | 1.5 | GO:0018279 | protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.0 | 2.0 | GO:0000724 | double-strand break repair via homologous recombination(GO:0000724) recombinational repair(GO:0000725) |
| 0.0 | 0.5 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.0 | 0.8 | GO:0006693 | prostanoid metabolic process(GO:0006692) prostaglandin metabolic process(GO:0006693) |
| 0.0 | 0.5 | GO:1901663 | ubiquinone metabolic process(GO:0006743) ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.0 | 0.1 | GO:0010578 | regulation of adenylate cyclase activity involved in G-protein coupled receptor signaling pathway(GO:0010578) positive regulation of adenylate cyclase activity involved in G-protein coupled receptor signaling pathway(GO:0010579) |
| 0.0 | 0.5 | GO:0016048 | detection of temperature stimulus(GO:0016048) |
| 0.0 | 0.6 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.0 | 0.5 | GO:0044364 | killing of cells of other organism(GO:0031640) disruption of cells of other organism(GO:0044364) |
| 0.0 | 0.3 | GO:0055003 | cardiac myofibril assembly(GO:0055003) |
| 0.0 | 0.0 | GO:1903336 | negative regulation of vacuolar transport(GO:1903336) |
| 0.0 | 0.1 | GO:0086028 | bundle of His cell to Purkinje myocyte signaling(GO:0086028) bundle of His cell action potential(GO:0086043) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 46.1 | 322.7 | GO:0042825 | TAP complex(GO:0042825) |
| 35.8 | 286.3 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 14.8 | 59.1 | GO:0030895 | apolipoprotein B mRNA editing enzyme complex(GO:0030895) |
| 13.5 | 81.3 | GO:1990682 | CSF1-CSF1R complex(GO:1990682) |
| 9.2 | 82.8 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 8.7 | 34.9 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 7.5 | 52.7 | GO:0036021 | endolysosome lumen(GO:0036021) |
| 6.5 | 25.9 | GO:0097179 | protease inhibitor complex(GO:0097179) |
| 5.7 | 17.1 | GO:1902737 | dendritic filopodium(GO:1902737) |
| 4.7 | 33.2 | GO:0071797 | LUBAC complex(GO:0071797) |
| 4.3 | 64.9 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 2.8 | 41.9 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 2.6 | 15.8 | GO:0020018 | ciliary pocket(GO:0020016) ciliary pocket membrane(GO:0020018) |
| 2.6 | 18.3 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 2.3 | 7.0 | GO:1990622 | CHOP-ATF3 complex(GO:1990622) |
| 2.1 | 46.5 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) |
| 2.0 | 165.2 | GO:0015030 | Cajal body(GO:0015030) |
| 1.7 | 5.2 | GO:0005600 | collagen type XIII trimer(GO:0005600) transmembrane collagen trimer(GO:0030936) |
| 1.7 | 10.1 | GO:0043527 | tRNA methyltransferase complex(GO:0043527) |
| 1.5 | 94.5 | GO:0016235 | aggresome(GO:0016235) |
| 1.4 | 18.8 | GO:0030870 | Mre11 complex(GO:0030870) |
| 1.4 | 789.8 | GO:0031965 | nuclear membrane(GO:0031965) |
| 1.4 | 4.2 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 1.4 | 4.1 | GO:0097444 | spine apparatus(GO:0097444) |
| 1.4 | 8.1 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 1.3 | 86.0 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 1.2 | 14.6 | GO:0005614 | interstitial matrix(GO:0005614) |
| 1.2 | 14.2 | GO:0044352 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 1.2 | 9.2 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 1.2 | 13.8 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 1.1 | 10.0 | GO:0072487 | MSL complex(GO:0072487) |
| 1.1 | 42.2 | GO:0034385 | very-low-density lipoprotein particle(GO:0034361) triglyceride-rich lipoprotein particle(GO:0034385) |
| 1.1 | 17.0 | GO:0032059 | bleb(GO:0032059) |
| 1.0 | 11.5 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 1.0 | 106.8 | GO:0005811 | lipid particle(GO:0005811) |
| 1.0 | 15.8 | GO:0033391 | chromatoid body(GO:0033391) |
| 1.0 | 14.6 | GO:0030478 | actin cap(GO:0030478) |
| 0.9 | 1.9 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.9 | 63.3 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.8 | 41.5 | GO:0043657 | host(GO:0018995) host cell(GO:0043657) |
| 0.7 | 8.9 | GO:0071439 | clathrin complex(GO:0071439) |
| 0.7 | 17.1 | GO:0036020 | endolysosome membrane(GO:0036020) |
| 0.6 | 7.8 | GO:0071556 | integral component of lumenal side of endoplasmic reticulum membrane(GO:0071556) lumenal side of endoplasmic reticulum membrane(GO:0098553) |
| 0.6 | 3.6 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.6 | 4.1 | GO:0098837 | postsynaptic recycling endosome(GO:0098837) |
| 0.6 | 5.2 | GO:0019907 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) |
| 0.6 | 5.1 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.6 | 1.7 | GO:0034455 | t-UTP complex(GO:0034455) |
| 0.6 | 36.1 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.6 | 42.8 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.5 | 3.3 | GO:0097149 | centralspindlin complex(GO:0097149) |
| 0.5 | 2.6 | GO:0097454 | Schwann cell microvillus(GO:0097454) |
| 0.5 | 1.5 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.5 | 1262.0 | GO:0005739 | mitochondrion(GO:0005739) |
| 0.5 | 222.3 | GO:0098852 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.5 | 84.2 | GO:0005769 | early endosome(GO:0005769) |
| 0.5 | 18.8 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.4 | 11.6 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.4 | 3.7 | GO:0005587 | collagen type IV trimer(GO:0005587) |
| 0.4 | 6.4 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.4 | 755.2 | GO:0005615 | extracellular space(GO:0005615) |
| 0.4 | 2.0 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.4 | 3.5 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.4 | 6.6 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.4 | 35.5 | GO:0035097 | histone methyltransferase complex(GO:0035097) |
| 0.4 | 5.0 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.4 | 6.0 | GO:0030119 | AP-type membrane coat adaptor complex(GO:0030119) |
| 0.3 | 13.6 | GO:0035580 | specific granule lumen(GO:0035580) |
| 0.3 | 4.2 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.3 | 1.3 | GO:0034669 | integrin alpha4-beta7 complex(GO:0034669) |
| 0.3 | 58.8 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.3 | 15.5 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.3 | 5.4 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.3 | 5.8 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.3 | 4.9 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.3 | 3.1 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) |
| 0.3 | 9.6 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.3 | 7.5 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.3 | 8.2 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.3 | 90.6 | GO:0010008 | endosome membrane(GO:0010008) |
| 0.3 | 12.5 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.2 | 1.7 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.2 | 6.4 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.2 | 4.3 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.2 | 1.9 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.2 | 4.8 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.2 | 48.5 | GO:0045111 | intermediate filament cytoskeleton(GO:0045111) |
| 0.2 | 0.9 | GO:0005863 | striated muscle myosin thick filament(GO:0005863) |
| 0.2 | 1.8 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.2 | 6.5 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.2 | 142.2 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.2 | 22.1 | GO:0031967 | organelle envelope(GO:0031967) envelope(GO:0031975) |
| 0.2 | 0.9 | GO:0030669 | clathrin-coated endocytic vesicle membrane(GO:0030669) |
| 0.2 | 0.8 | GO:0045180 | basal cortex(GO:0045180) |
| 0.2 | 32.1 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.2 | 0.8 | GO:0035061 | interchromatin granule(GO:0035061) |
| 0.2 | 6.6 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.1 | 14.8 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.1 | 2.4 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.1 | 0.4 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.1 | 0.9 | GO:0098536 | deuterosome(GO:0098536) |
| 0.1 | 4.1 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.1 | 1.0 | GO:0031415 | NatA complex(GO:0031415) |
| 0.1 | 1.6 | GO:0001520 | outer dense fiber(GO:0001520) |
| 0.1 | 10.0 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.1 | 0.5 | GO:0016471 | vacuolar proton-transporting V-type ATPase complex(GO:0016471) |
| 0.1 | 1.5 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.1 | 0.6 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.1 | 2.1 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.1 | 3.0 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.1 | 9.4 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.1 | 1.3 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.1 | 1.7 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.1 | 0.6 | GO:0005865 | striated muscle thin filament(GO:0005865) myofilament(GO:0036379) |
| 0.1 | 2.0 | GO:0030686 | 90S preribosome(GO:0030686) |
| 0.1 | 9.4 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.1 | 0.9 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.1 | 1.0 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.1 | 11.1 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.1 | 375.6 | GO:0005829 | cytosol(GO:0005829) |
| 0.1 | 0.7 | GO:0001673 | male germ cell nucleus(GO:0001673) |
| 0.1 | 1.3 | GO:0005844 | polysome(GO:0005844) |
| 0.1 | 7.0 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.8 | GO:0035267 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.0 | 1.4 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 1.5 | GO:0000795 | synaptonemal complex(GO:0000795) |
| 0.0 | 2.8 | GO:0005930 | axoneme(GO:0005930) ciliary plasm(GO:0097014) |
| 0.0 | 1.4 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 0.3 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.0 | 0.6 | GO:0097381 | photoreceptor disc membrane(GO:0097381) |
| 0.0 | 0.3 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 3.0 | GO:0005874 | microtubule(GO:0005874) |
| 0.0 | 0.6 | GO:0042383 | sarcolemma(GO:0042383) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 57.5 | 172.4 | GO:0004798 | thymidylate kinase activity(GO:0004798) |
| 47.7 | 716.0 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 46.0 | 322.3 | GO:0022897 | peptide:proton symporter activity(GO:0015333) proton-dependent peptide secondary active transmembrane transporter activity(GO:0022897) |
| 45.6 | 136.9 | GO:0008859 | exoribonuclease II activity(GO:0008859) |
| 40.3 | 322.7 | GO:0015440 | peptide-transporting ATPase activity(GO:0015440) |
| 34.0 | 101.9 | GO:0042007 | interleukin-18 binding(GO:0042007) |
| 31.0 | 155.2 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 26.0 | 78.1 | GO:0042008 | interleukin-18 receptor activity(GO:0042008) |
| 15.3 | 45.9 | GO:0001716 | L-amino-acid oxidase activity(GO:0001716) |
| 12.2 | 85.2 | GO:0005157 | macrophage colony-stimulating factor receptor binding(GO:0005157) |
| 10.4 | 51.8 | GO:0047844 | deoxycytidine deaminase activity(GO:0047844) |
| 10.2 | 295.9 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 8.2 | 32.8 | GO:0005151 | interleukin-1, Type II receptor binding(GO:0005151) |
| 8.1 | 357.0 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 7.4 | 52.1 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 6.7 | 40.4 | GO:0048030 | disaccharide binding(GO:0048030) |
| 6.1 | 30.4 | GO:0004882 | androgen receptor activity(GO:0004882) |
| 4.8 | 19.3 | GO:0034041 | sterol-transporting ATPase activity(GO:0034041) |
| 4.5 | 27.3 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 4.4 | 34.9 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 4.3 | 98.8 | GO:0032183 | SUMO binding(GO:0032183) |
| 4.1 | 128.6 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 3.8 | 91.2 | GO:0070628 | proteasome binding(GO:0070628) |
| 3.8 | 26.3 | GO:0034046 | poly(G) binding(GO:0034046) |
| 3.6 | 17.8 | GO:0030298 | receptor signaling protein tyrosine kinase activator activity(GO:0030298) CCR1 chemokine receptor binding(GO:0031726) |
| 3.4 | 17.0 | GO:0055077 | gap junction hemi-channel activity(GO:0055077) |
| 3.2 | 461.7 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 3.2 | 9.6 | GO:0008892 | guanine deaminase activity(GO:0008892) |
| 3.1 | 15.7 | GO:0061513 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 3.1 | 33.9 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 3.0 | 75.3 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 3.0 | 41.9 | GO:0031386 | protein tag(GO:0031386) |
| 2.7 | 24.4 | GO:0032407 | MutSalpha complex binding(GO:0032407) |
| 2.6 | 7.7 | GO:0102008 | cytosolic dipeptidase activity(GO:0102008) |
| 2.5 | 10.1 | GO:0098770 | FBXO family protein binding(GO:0098770) |
| 2.5 | 12.5 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 2.5 | 19.7 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 2.4 | 21.3 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 2.3 | 9.2 | GO:0008478 | pyridoxal kinase activity(GO:0008478) |
| 2.2 | 6.6 | GO:0004483 | mRNA (nucleoside-2'-O-)-methyltransferase activity(GO:0004483) |
| 2.1 | 122.7 | GO:0051539 | 4 iron, 4 sulfur cluster binding(GO:0051539) |
| 2.1 | 12.6 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 1.9 | 7.6 | GO:0004909 | interleukin-1, Type I, activating receptor activity(GO:0004909) |
| 1.8 | 10.8 | GO:0061649 | ubiquitinated histone binding(GO:0061649) |
| 1.8 | 10.5 | GO:0031962 | mineralocorticoid receptor binding(GO:0031962) |
| 1.7 | 35.6 | GO:0051400 | BH domain binding(GO:0051400) |
| 1.6 | 19.5 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 1.6 | 4.8 | GO:0052739 | phosphatidylserine 1-acylhydrolase activity(GO:0052739) |
| 1.5 | 6.1 | GO:0005136 | interleukin-4 receptor binding(GO:0005136) |
| 1.5 | 1.5 | GO:0016979 | lipoate-protein ligase activity(GO:0016979) |
| 1.5 | 7.5 | GO:0031735 | CCR10 chemokine receptor binding(GO:0031735) |
| 1.5 | 29.7 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 1.4 | 25.8 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 1.4 | 28.1 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 1.4 | 67.0 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 1.3 | 7.6 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 1.3 | 15.1 | GO:0043176 | amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 1.2 | 677.7 | GO:0005525 | GTP binding(GO:0005525) |
| 1.2 | 4.9 | GO:0004074 | biliverdin reductase activity(GO:0004074) |
| 1.2 | 54.8 | GO:0043394 | proteoglycan binding(GO:0043394) |
| 1.1 | 100.9 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
| 1.1 | 3.2 | GO:0005519 | cytoskeletal regulatory protein binding(GO:0005519) |
| 1.1 | 4.2 | GO:0045569 | TRAIL binding(GO:0045569) |
| 1.0 | 176.6 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 1.0 | 10.3 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 1.0 | 5.1 | GO:0008281 | sulfonylurea receptor activity(GO:0008281) |
| 1.0 | 30.5 | GO:0005521 | lamin binding(GO:0005521) |
| 1.0 | 14.0 | GO:0031996 | thioesterase binding(GO:0031996) |
| 1.0 | 6.8 | GO:0043140 | ATP-dependent 3'-5' DNA helicase activity(GO:0043140) |
| 0.9 | 5.6 | GO:0004699 | calcium-independent protein kinase C activity(GO:0004699) |
| 0.9 | 82.7 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.8 | 8.4 | GO:0043813 | phosphatidylinositol-3,5-bisphosphate 5-phosphatase activity(GO:0043813) |
| 0.8 | 5.0 | GO:0030060 | L-malate dehydrogenase activity(GO:0030060) |
| 0.8 | 4.8 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.7 | 8.8 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.7 | 63.1 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.7 | 2.1 | GO:0004060 | arylamine N-acetyltransferase activity(GO:0004060) |
| 0.7 | 4.9 | GO:0060698 | endoribonuclease inhibitor activity(GO:0060698) |
| 0.7 | 4.8 | GO:0004974 | leukotriene receptor activity(GO:0004974) |
| 0.7 | 10.7 | GO:0004028 | 3-chloroallyl aldehyde dehydrogenase activity(GO:0004028) |
| 0.6 | 24.4 | GO:0001848 | complement binding(GO:0001848) |
| 0.6 | 15.2 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.6 | 3.2 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.6 | 1.2 | GO:0016639 | oxidoreductase activity, acting on the CH-NH2 group of donors, NAD or NADP as acceptor(GO:0016639) |
| 0.6 | 9.6 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.6 | 3.5 | GO:0004977 | melanocortin receptor activity(GO:0004977) |
| 0.6 | 6.2 | GO:0000099 | sulfur amino acid transmembrane transporter activity(GO:0000099) |
| 0.6 | 10.1 | GO:0016423 | tRNA (guanine) methyltransferase activity(GO:0016423) |
| 0.5 | 16.9 | GO:0005035 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.5 | 2.7 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.5 | 7.0 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.5 | 20.4 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.5 | 6.9 | GO:0042301 | phosphate ion binding(GO:0042301) |
| 0.5 | 9.6 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.5 | 2.6 | GO:0008941 | nitric oxide dioxygenase activity(GO:0008941) oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of two atoms of oxygen into one donor(GO:0016708) |
| 0.5 | 6.8 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.5 | 2.6 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.5 | 132.6 | GO:0005125 | cytokine activity(GO:0005125) |
| 0.5 | 9.9 | GO:0008035 | high-density lipoprotein particle binding(GO:0008035) |
| 0.5 | 66.0 | GO:0004386 | helicase activity(GO:0004386) |
| 0.5 | 1.5 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 0.5 | 3.3 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 0.5 | 1.4 | GO:0060001 | minus-end directed microfilament motor activity(GO:0060001) |
| 0.5 | 25.7 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.4 | 4.5 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.4 | 8.3 | GO:0031702 | type 1 angiotensin receptor binding(GO:0031702) |
| 0.4 | 10.8 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.4 | 2.4 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.4 | 7.6 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.4 | 1.6 | GO:0004793 | glycine hydroxymethyltransferase activity(GO:0004372) threonine aldolase activity(GO:0004793) L-allo-threonine aldolase activity(GO:0008732) |
| 0.4 | 5.8 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.4 | 28.5 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.4 | 2.6 | GO:0005280 | hydrogen:amino acid symporter activity(GO:0005280) |
| 0.4 | 19.6 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.4 | 0.7 | GO:0051033 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.4 | 1.4 | GO:0031531 | thyrotropin-releasing hormone receptor binding(GO:0031531) |
| 0.3 | 7.2 | GO:0042974 | retinoic acid receptor binding(GO:0042974) |
| 0.3 | 11.4 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.3 | 3.9 | GO:0016713 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced iron-sulfur protein as one donor, and incorporation of one atom of oxygen(GO:0016713) |
| 0.3 | 1.0 | GO:1990190 | peptide-glutamate-N-acetyltransferase activity(GO:1990190) |
| 0.3 | 8.2 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.3 | 46.1 | GO:0005254 | chloride channel activity(GO:0005254) |
| 0.3 | 2.2 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.3 | 3.0 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.3 | 1.5 | GO:0047374 | methylumbelliferyl-acetate deacetylase activity(GO:0047374) |
| 0.3 | 6.7 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.3 | 6.4 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.3 | 5.7 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.3 | 4.7 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.3 | 4.9 | GO:0043495 | adenylate cyclase binding(GO:0008179) protein anchor(GO:0043495) |
| 0.3 | 5.2 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.3 | 299.6 | GO:0008270 | zinc ion binding(GO:0008270) |
| 0.3 | 2.9 | GO:0031698 | beta-2 adrenergic receptor binding(GO:0031698) |
| 0.3 | 3.5 | GO:0000182 | rDNA binding(GO:0000182) |
| 0.3 | 2.3 | GO:0030023 | extracellular matrix constituent conferring elasticity(GO:0030023) |
| 0.2 | 1.0 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) |
| 0.2 | 1.2 | GO:0005502 | 11-cis retinal binding(GO:0005502) |
| 0.2 | 10.5 | GO:0050681 | androgen receptor binding(GO:0050681) |
| 0.2 | 2.4 | GO:0046703 | natural killer cell lectin-like receptor binding(GO:0046703) |
| 0.2 | 6.3 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.2 | 6.0 | GO:0036459 | thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) |
| 0.2 | 1.6 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.2 | 2.2 | GO:0002161 | aminoacyl-tRNA editing activity(GO:0002161) |
| 0.2 | 0.4 | GO:0048019 | receptor antagonist activity(GO:0048019) |
| 0.2 | 3.7 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.2 | 3.1 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.2 | 1.1 | GO:0015087 | cadmium ion transmembrane transporter activity(GO:0015086) cobalt ion transmembrane transporter activity(GO:0015087) lead ion transmembrane transporter activity(GO:0015094) ferrous iron uptake transmembrane transporter activity(GO:0015639) |
| 0.2 | 5.2 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.2 | 13.6 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.2 | 8.8 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.2 | 1.0 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.2 | 4.8 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.2 | 0.6 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.2 | 0.9 | GO:0003756 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.2 | 9.2 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.2 | 31.2 | GO:0005319 | lipid transporter activity(GO:0005319) |
| 0.2 | 1.1 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.2 | 2.0 | GO:0043560 | insulin receptor substrate binding(GO:0043560) |
| 0.2 | 2.6 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.2 | 0.7 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 0.2 | 11.6 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.2 | 2.4 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.2 | 0.5 | GO:0017077 | oxidative phosphorylation uncoupler activity(GO:0017077) |
| 0.2 | 1.3 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.2 | 15.7 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.2 | 1.4 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.2 | 0.8 | GO:0015562 | efflux transmembrane transporter activity(GO:0015562) |
| 0.2 | 3.3 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.1 | 5.5 | GO:0031369 | translation initiation factor binding(GO:0031369) |
| 0.1 | 1.2 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.1 | 2.4 | GO:0008198 | ferrous iron binding(GO:0008198) |
| 0.1 | 0.6 | GO:0031545 | peptidyl-proline dioxygenase activity(GO:0031543) peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.1 | 2.0 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.1 | 1.9 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.1 | 3.2 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.1 | 1.3 | GO:0015288 | porin activity(GO:0015288) |
| 0.1 | 3.0 | GO:0031005 | filamin binding(GO:0031005) |
| 0.1 | 1.8 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.1 | 0.5 | GO:0031208 | POZ domain binding(GO:0031208) |
| 0.1 | 0.5 | GO:0008310 | single-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008310) |
| 0.1 | 9.3 | GO:0016667 | oxidoreductase activity, acting on a sulfur group of donors(GO:0016667) |
| 0.1 | 5.4 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.1 | 224.6 | GO:0042802 | identical protein binding(GO:0042802) |
| 0.1 | 45.8 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.1 | 2.1 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.1 | 6.6 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.1 | 0.5 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.1 | 21.4 | GO:0042393 | histone binding(GO:0042393) |
| 0.1 | 1.7 | GO:0019864 | IgG binding(GO:0019864) |
| 0.1 | 4.2 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.1 | 0.5 | GO:0030171 | voltage-gated proton channel activity(GO:0030171) |
| 0.1 | 3.9 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.1 | 0.8 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.1 | 17.8 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.1 | 6.4 | GO:0016279 | protein-lysine N-methyltransferase activity(GO:0016279) |
| 0.1 | 0.8 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) gamma-aminobutyric acid transmembrane transporter activity(GO:0015185) |
| 0.1 | 3.4 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.1 | 3.2 | GO:0005549 | odorant binding(GO:0005549) |
| 0.1 | 2.5 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.1 | 3.0 | GO:0036442 | hydrogen-exporting ATPase activity(GO:0036442) |
| 0.1 | 0.9 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.1 | 10.0 | GO:0017048 | Rho GTPase binding(GO:0017048) |
| 0.1 | 3.1 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.1 | 1.4 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.1 | 1.7 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.1 | 9.4 | GO:0047485 | protein N-terminus binding(GO:0047485) |
| 0.1 | 2.7 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.1 | 2.5 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.1 | 4.1 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.1 | 0.9 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.1 | 0.4 | GO:0019776 | Atg8 ligase activity(GO:0019776) |
| 0.1 | 0.6 | GO:0008048 | calcium sensitive guanylate cyclase activator activity(GO:0008048) |
| 0.1 | 1.1 | GO:0008235 | metalloexopeptidase activity(GO:0008235) |
| 0.1 | 2.0 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.1 | 14.6 | GO:0004713 | protein tyrosine kinase activity(GO:0004713) |
| 0.1 | 0.3 | GO:0008665 | 2'-phosphotransferase activity(GO:0008665) |
| 0.1 | 8.3 | GO:0005179 | hormone activity(GO:0005179) |
| 0.1 | 2.1 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.1 | 4.5 | GO:0019212 | protein phosphatase inhibitor activity(GO:0004864) phosphatase inhibitor activity(GO:0019212) |
| 0.1 | 1.5 | GO:0070034 | telomerase RNA binding(GO:0070034) |
| 0.1 | 4.2 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.1 | 1.1 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.1 | 1.2 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.1 | 1.7 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.1 | 1.9 | GO:0033558 | histone deacetylase activity(GO:0004407) protein deacetylase activity(GO:0033558) |
| 0.1 | 0.7 | GO:0003951 | NAD+ kinase activity(GO:0003951) |
| 0.1 | 1.4 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.1 | 2.9 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.1 | 2.6 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.1 | 1.2 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.1 | 1.7 | GO:0030553 | cGMP binding(GO:0030553) |
| 0.0 | 0.3 | GO:0050659 | N-acetylgalactosamine 4-sulfate 6-O-sulfotransferase activity(GO:0050659) |
| 0.0 | 0.5 | GO:0005227 | calcium activated cation channel activity(GO:0005227) |
| 0.0 | 0.4 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 3.5 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.0 | 0.8 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.0 | 0.4 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.0 | 1.4 | GO:0030145 | manganese ion binding(GO:0030145) |
| 0.0 | 1.2 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.0 | 1.5 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.0 | 3.1 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 0.3 | GO:0099580 | ion antiporter activity involved in regulation of postsynaptic membrane potential(GO:0099580) |
| 0.0 | 0.1 | GO:0016497 | substance K receptor activity(GO:0016497) |
| 0.0 | 2.1 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.0 | 1.1 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 0.3 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.0 | 0.1 | GO:0005041 | low-density lipoprotein receptor activity(GO:0005041) |
| 0.0 | 0.4 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.0 | 0.6 | GO:0008066 | glutamate receptor activity(GO:0008066) |
| 0.0 | 1.0 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.0 | 0.4 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.0 | 0.1 | GO:0015143 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.0 | 0.3 | GO:0019825 | oxygen binding(GO:0019825) |
| 0.0 | 0.6 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.5 | 27.3 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 1.8 | 146.5 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 1.8 | 88.3 | PID MYC PATHWAY | C-MYC pathway |
| 1.7 | 36.4 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 1.7 | 52.8 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 1.7 | 63.7 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 1.6 | 103.1 | SIG PIP3 SIGNALING IN B LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
| 1.5 | 78.1 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 1.4 | 49.0 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 1.4 | 25.9 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 1.1 | 98.1 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 1.0 | 26.1 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.9 | 193.9 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.8 | 22.4 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.8 | 101.5 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.8 | 36.6 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.7 | 57.1 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.6 | 29.1 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.6 | 5.3 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.6 | 35.6 | SIG PIP3 SIGNALING IN CARDIAC MYOCTES | Genes related to PIP3 signaling in cardiac myocytes |
| 0.5 | 16.9 | SIG CHEMOTAXIS | Genes related to chemotaxis |
| 0.5 | 12.6 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.5 | 4.0 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.5 | 17.3 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.5 | 196.5 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.5 | 7.8 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.5 | 28.6 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.4 | 6.6 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.4 | 26.0 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.4 | 12.3 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.4 | 10.6 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.4 | 24.5 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.4 | 16.5 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.4 | 3.4 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.3 | 81.1 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.3 | 7.5 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.3 | 14.1 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.3 | 3.7 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.3 | 5.6 | PID IGF1 PATHWAY | IGF1 pathway |
| 0.3 | 8.0 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.3 | 16.0 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.2 | 6.7 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.2 | 17.4 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.2 | 54.2 | NABA MATRISOME ASSOCIATED | Ensemble of genes encoding ECM-associated proteins including ECM-affilaited proteins, ECM regulators and secreted factors |
| 0.2 | 3.1 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.1 | 1.6 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.1 | 3.1 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.1 | 4.9 | PID ENDOTHELIN PATHWAY | Endothelins |
| 0.1 | 0.4 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.1 | 3.6 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.1 | 4.8 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.1 | 6.8 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.1 | 6.7 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.1 | 1.4 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.1 | 1.3 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.1 | 6.9 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.1 | 3.4 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.1 | 2.2 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.1 | 1.5 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.1 | 2.3 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.1 | 16.5 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.1 | 1.0 | PID FGF PATHWAY | FGF signaling pathway |
| 0.1 | 0.8 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.1 | 0.6 | PID IL2 1PATHWAY | IL2-mediated signaling events |
| 0.0 | 0.5 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 2.0 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 0.7 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.0 | 0.6 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 0.8 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.0 | 0.6 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 20.2 | 2287.1 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 12.3 | 49.0 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 11.0 | 253.0 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 10.2 | 172.7 | REACTOME TRAF3 DEPENDENT IRF ACTIVATION PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |
| 5.1 | 349.8 | REACTOME VIF MEDIATED DEGRADATION OF APOBEC3G | Genes involved in Vif-mediated degradation of APOBEC3G |
| 3.4 | 322.6 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 3.1 | 49.6 | REACTOME PD1 SIGNALING | Genes involved in PD-1 signaling |
| 2.8 | 132.7 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 2.7 | 183.8 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 2.6 | 56.6 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 2.5 | 172.9 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 2.2 | 40.1 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 1.7 | 75.1 | REACTOME RIG I MDA5 MEDIATED INDUCTION OF IFN ALPHA BETA PATHWAYS | Genes involved in RIG-I/MDA5 mediated induction of IFN-alpha/beta pathways |
| 1.7 | 52.8 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 1.3 | 15.4 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 1.3 | 47.3 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 1.1 | 15.9 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 1.1 | 15.1 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 1.0 | 22.0 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 1.0 | 7.8 | REACTOME ACTIVATION OF IRF3 IRF7 MEDIATED BY TBK1 IKK EPSILON | Genes involved in Activation of IRF3/IRF7 mediated by TBK1/IKK epsilon |
| 0.8 | 19.3 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.8 | 33.1 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.7 | 9.6 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.6 | 134.1 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.4 | 26.7 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.4 | 12.3 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.4 | 9.0 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.4 | 2.6 | REACTOME INFLUENZA VIRAL RNA TRANSCRIPTION AND REPLICATION | Genes involved in Influenza Viral RNA Transcription and Replication |
| 0.4 | 3.3 | REACTOME NUCLEOTIDE BINDING DOMAIN LEUCINE RICH REPEAT CONTAINING RECEPTOR NLR SIGNALING PATHWAYS | Genes involved in Nucleotide-binding domain, leucine rich repeat containing receptor (NLR) signaling pathways |
| 0.4 | 10.5 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.4 | 13.8 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.3 | 26.9 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.3 | 11.2 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.3 | 3.3 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.3 | 10.3 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.3 | 7.3 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.3 | 7.0 | REACTOME ACTIVATION OF GENES BY ATF4 | Genes involved in Activation of Genes by ATF4 |
| 0.3 | 26.5 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.3 | 7.6 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.3 | 10.7 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.3 | 4.7 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.3 | 2.5 | REACTOME G PROTEIN ACTIVATION | Genes involved in G-protein activation |
| 0.2 | 11.7 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.2 | 5.2 | REACTOME DESTABILIZATION OF MRNA BY BRF1 | Genes involved in Destabilization of mRNA by Butyrate Response Factor 1 (BRF1) |
| 0.2 | 5.8 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.2 | 15.0 | REACTOME COSTIMULATION BY THE CD28 FAMILY | Genes involved in Costimulation by the CD28 family |
| 0.2 | 6.8 | REACTOME THROMBOXANE SIGNALLING THROUGH TP RECEPTOR | Genes involved in Thromboxane signalling through TP receptor |
| 0.2 | 4.8 | REACTOME EICOSANOID LIGAND BINDING RECEPTORS | Genes involved in Eicosanoid ligand-binding receptors |
| 0.2 | 5.0 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.2 | 6.2 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.2 | 5.2 | REACTOME CYCLIN E ASSOCIATED EVENTS DURING G1 S TRANSITION | Genes involved in Cyclin E associated events during G1/S transition |
| 0.2 | 1.4 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.2 | 4.9 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.2 | 3.1 | REACTOME ACYL CHAIN REMODELLING OF PI | Genes involved in Acyl chain remodelling of PI |
| 0.2 | 5.7 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.1 | 5.2 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.1 | 5.1 | REACTOME INWARDLY RECTIFYING K CHANNELS | Genes involved in Inwardly rectifying K+ channels |
| 0.1 | 7.4 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.1 | 5.8 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.1 | 3.7 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.1 | 10.2 | REACTOME INTERFERON SIGNALING | Genes involved in Interferon Signaling |
| 0.1 | 12.3 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.1 | 2.9 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.1 | 7.5 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.1 | 2.2 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.1 | 1.5 | REACTOME GABA B RECEPTOR ACTIVATION | Genes involved in GABA B receptor activation |
| 0.1 | 5.6 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.1 | 3.0 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.1 | 11.2 | REACTOME POTASSIUM CHANNELS | Genes involved in Potassium Channels |
| 0.1 | 3.6 | REACTOME PREFOLDIN MEDIATED TRANSFER OF SUBSTRATE TO CCT TRIC | Genes involved in Prefoldin mediated transfer of substrate to CCT/TriC |
| 0.1 | 1.5 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.1 | 2.9 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.1 | 3.1 | REACTOME SRP DEPENDENT COTRANSLATIONAL PROTEIN TARGETING TO MEMBRANE | Genes involved in SRP-dependent cotranslational protein targeting to membrane |
| 0.1 | 9.0 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.1 | 6.5 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.1 | 1.7 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.1 | 1.8 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.1 | 2.0 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.1 | 1.8 | REACTOME SIGNALING BY EGFR IN CANCER | Genes involved in Signaling by EGFR in Cancer |
| 0.1 | 2.0 | REACTOME KERATAN SULFATE BIOSYNTHESIS | Genes involved in Keratan sulfate biosynthesis |
| 0.1 | 1.6 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.1 | 0.5 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.1 | 1.8 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.1 | 0.1 | REACTOME NEF MEDIATES DOWN MODULATION OF CELL SURFACE RECEPTORS BY RECRUITING THEM TO CLATHRIN ADAPTERS | Genes involved in Nef-mediates down modulation of cell surface receptors by recruiting them to clathrin adapters |
| 0.0 | 0.6 | REACTOME PROTEIN FOLDING | Genes involved in Protein folding |
| 0.0 | 1.3 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.0 | 0.9 | REACTOME PIP3 ACTIVATES AKT SIGNALING | Genes involved in PIP3 activates AKT signaling |
| 0.0 | 0.8 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 0.8 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 1.4 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 0.6 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 1.4 | REACTOME ION CHANNEL TRANSPORT | Genes involved in Ion channel transport |
| 0.0 | 0.9 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 0.4 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.0 | 4.0 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 1.2 | REACTOME PTM GAMMA CARBOXYLATION HYPUSINE FORMATION AND ARYLSULFATASE ACTIVATION | Genes involved in PTM: gamma carboxylation, hypusine formation and arylsulfatase activation |
| 0.0 | 1.0 | REACTOME FGFR LIGAND BINDING AND ACTIVATION | Genes involved in FGFR ligand binding and activation |
| 0.0 | 0.8 | REACTOME HOST INTERACTIONS OF HIV FACTORS | Genes involved in Host Interactions of HIV factors |
| 0.0 | 0.3 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.0 | 0.4 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 2.8 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 0.4 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.0 | 1.0 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 2.1 | REACTOME NONSENSE MEDIATED DECAY ENHANCED BY THE EXON JUNCTION COMPLEX | Genes involved in Nonsense Mediated Decay Enhanced by the Exon Junction Complex |
| 0.0 | 0.3 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 1.7 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.0 | 0.8 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |