Project
Inflammatory response time course, HUVEC (Wada, 2009)
Navigation
Downloads
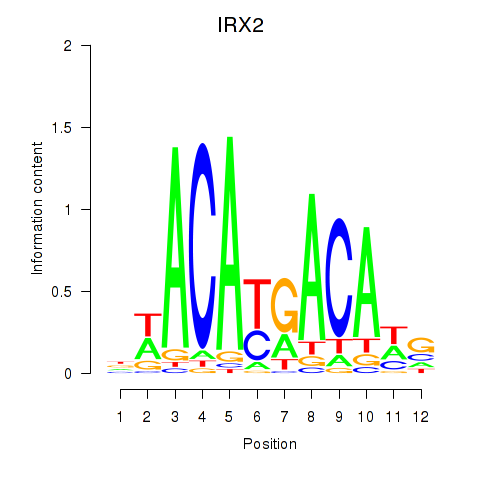
Results for IRX2
Z-value: 0.55
Transcription factors associated with IRX2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
IRX2
|
ENSG00000170561.13 | IRX2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| IRX2 | hg38_v1_chr5_-_2751670_2751694 | -0.28 | 1.8e-01 | Click! |
Activity profile of IRX2 motif
Sorted Z-values of IRX2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of IRX2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr9_+_76459152 | 1.74 |
ENST00000444201.6
ENST00000376730.5 |
GCNT1
|
glucosaminyl (N-acetyl) transferase 1 |
| chr7_+_150514851 | 1.17 |
ENST00000313543.5
|
GIMAP7
|
GTPase, IMAP family member 7 |
| chr17_-_3691887 | 0.90 |
ENST00000552050.5
|
P2RX5
|
purinergic receptor P2X 5 |
| chr13_+_102656933 | 0.87 |
ENST00000650757.1
|
TPP2
|
tripeptidyl peptidase 2 |
| chr18_-_55403682 | 0.79 |
ENST00000564228.5
ENST00000630828.2 |
TCF4
|
transcription factor 4 |
| chr9_-_137302264 | 0.78 |
ENST00000356628.4
|
NRARP
|
NOTCH regulated ankyrin repeat protein |
| chr7_+_107044689 | 0.64 |
ENST00000265717.5
|
PRKAR2B
|
protein kinase cAMP-dependent type II regulatory subunit beta |
| chr17_-_27800524 | 0.57 |
ENST00000313735.11
|
NOS2
|
nitric oxide synthase 2 |
| chr8_-_85341659 | 0.57 |
ENST00000522389.5
|
CA1
|
carbonic anhydrase 1 |
| chr9_-_35812238 | 0.56 |
ENST00000396638.7
ENST00000340291.6 |
SPAG8
|
sperm associated antigen 8 |
| chr7_-_21945866 | 0.55 |
ENST00000356195.9
ENST00000447180.5 ENST00000373934.4 ENST00000406877.8 ENST00000457951.5 |
CDCA7L
|
cell division cycle associated 7 like |
| chr5_-_88731827 | 0.53 |
ENST00000627170.2
|
MEF2C
|
myocyte enhancer factor 2C |
| chr12_+_8992029 | 0.51 |
ENST00000543895.1
|
KLRG1
|
killer cell lectin like receptor G1 |
| chr12_+_109098118 | 0.49 |
ENST00000336865.6
|
UNG
|
uracil DNA glycosylase |
| chr2_-_163735989 | 0.48 |
ENST00000333129.4
ENST00000409634.5 |
FIGN
|
fidgetin, microtubule severing factor |
| chr12_+_32502114 | 0.46 |
ENST00000682739.1
ENST00000427716.7 ENST00000583694.2 |
FGD4
|
FYVE, RhoGEF and PH domain containing 4 |
| chr1_+_74235377 | 0.43 |
ENST00000326637.8
|
TNNI3K
|
TNNI3 interacting kinase |
| chr15_-_19988117 | 0.43 |
ENST00000558565.2
|
IGHV3OR15-7
|
immunoglobulin heavy variable 3/OR15-7 (pseudogene) |
| chr2_+_137964446 | 0.40 |
ENST00000280096.5
ENST00000280097.5 |
HNMT
|
histamine N-methyltransferase |
| chr2_+_190927649 | 0.38 |
ENST00000409428.5
ENST00000409215.5 |
GLS
|
glutaminase |
| chr6_+_24774925 | 0.38 |
ENST00000356509.7
ENST00000230056.8 |
GMNN
|
geminin DNA replication inhibitor |
| chr6_-_159045010 | 0.36 |
ENST00000338313.5
|
TAGAP
|
T cell activation RhoGTPase activating protein |
| chr8_-_85341705 | 0.35 |
ENST00000517618.5
|
CA1
|
carbonic anhydrase 1 |
| chr15_+_37934626 | 0.35 |
ENST00000559502.5
ENST00000558148.5 ENST00000319669.5 ENST00000558158.5 |
TMCO5A
|
transmembrane and coiled-coil domains 5A |
| chr8_+_97887903 | 0.35 |
ENST00000520016.5
|
MATN2
|
matrilin 2 |
| chr6_-_111605859 | 0.34 |
ENST00000651359.1
ENST00000650859.1 ENST00000359831.8 ENST00000368761.11 |
TRAF3IP2
|
TRAF3 interacting protein 2 |
| chr7_-_105691637 | 0.34 |
ENST00000472195.1
|
ATXN7L1
|
ataxin 7 like 1 |
| chr3_+_98168700 | 0.32 |
ENST00000383696.4
|
OR5H15
|
olfactory receptor family 5 subfamily H member 15 |
| chr17_+_7857987 | 0.32 |
ENST00000332439.5
|
CYB5D1
|
cytochrome b5 domain containing 1 |
| chr9_-_13175824 | 0.32 |
ENST00000545857.5
|
MPDZ
|
multiple PDZ domain crumbs cell polarity complex component |
| chr14_-_100375333 | 0.32 |
ENST00000557297.5
ENST00000555813.5 ENST00000392882.7 ENST00000557135.5 ENST00000556698.5 ENST00000554509.5 ENST00000555410.5 |
WARS1
|
tryptophanyl-tRNA synthetase 1 |
| chr14_-_24508251 | 0.30 |
ENST00000250378.7
ENST00000206446.4 |
CMA1
|
chymase 1 |
| chr22_-_28306645 | 0.28 |
ENST00000612946.4
|
TTC28
|
tetratricopeptide repeat domain 28 |
| chr1_+_162381703 | 0.28 |
ENST00000458626.4
|
C1orf226
|
chromosome 1 open reading frame 226 |
| chr17_+_35844077 | 0.28 |
ENST00000604694.1
|
TAF15
|
TATA-box binding protein associated factor 15 |
| chr6_-_27873525 | 0.27 |
ENST00000618305.2
|
H4C13
|
H4 clustered histone 13 |
| chr1_+_15756659 | 0.27 |
ENST00000375771.5
|
FBLIM1
|
filamin binding LIM protein 1 |
| chr22_-_35617321 | 0.27 |
ENST00000397326.7
ENST00000442617.1 |
MB
|
myoglobin |
| chr7_-_5959083 | 0.27 |
ENST00000539903.5
|
RSPH10B
|
radial spoke head 10 homolog B |
| chr1_-_158554405 | 0.27 |
ENST00000641282.1
ENST00000641622.1 |
OR6Y1
|
olfactory receptor family 6 subfamily Y member 1 |
| chr12_-_7503744 | 0.26 |
ENST00000396620.7
ENST00000432237.3 |
CD163
|
CD163 molecule |
| chr16_+_82035245 | 0.26 |
ENST00000199936.9
|
HSD17B2
|
hydroxysteroid 17-beta dehydrogenase 2 |
| chr21_-_30492008 | 0.26 |
ENST00000334063.6
|
KRTAP19-3
|
keratin associated protein 19-3 |
| chr1_+_152719522 | 0.26 |
ENST00000368775.3
|
C1orf68
|
chromosome 1 open reading frame 68 |
| chr2_+_48617798 | 0.25 |
ENST00000448460.5
ENST00000437125.5 ENST00000430487.6 |
GTF2A1L
|
general transcription factor IIA subunit 1 like |
| chr2_+_137964279 | 0.25 |
ENST00000329366.8
|
HNMT
|
histamine N-methyltransferase |
| chr2_+_108588286 | 0.25 |
ENST00000332345.10
|
LIMS1
|
LIM zinc finger domain containing 1 |
| chr14_-_75069478 | 0.25 |
ENST00000555463.1
|
ACYP1
|
acylphosphatase 1 |
| chr4_-_149815826 | 0.24 |
ENST00000636793.2
ENST00000636414.1 |
IQCM
|
IQ motif containing M |
| chr16_-_74607088 | 0.24 |
ENST00000565260.1
ENST00000447066.6 ENST00000205061.9 ENST00000422840.7 ENST00000627032.2 |
GLG1
|
golgi glycoprotein 1 |
| chr7_+_73667824 | 0.24 |
ENST00000324941.5
ENST00000451519.1 |
VPS37D
|
VPS37D subunit of ESCRT-I |
| chr17_-_7857854 | 0.23 |
ENST00000333775.9
|
NAA38
|
N-alpha-acetyltransferase 38, NatC auxiliary subunit |
| chr17_+_39700046 | 0.23 |
ENST00000269571.10
|
ERBB2
|
erb-b2 receptor tyrosine kinase 2 |
| chr17_-_7857938 | 0.23 |
ENST00000575208.2
|
NAA38
|
N-alpha-acetyltransferase 38, NatC auxiliary subunit |
| chrX_-_149595314 | 0.23 |
ENST00000598963.3
|
HSFX2
|
heat shock transcription factor family, X-linked 2 |
| chr11_+_33016106 | 0.23 |
ENST00000311388.7
|
DEPDC7
|
DEP domain containing 7 |
| chr19_-_35228699 | 0.23 |
ENST00000324675.3
|
FAM187B
|
family with sequence similarity 187 member B |
| chr15_-_44194407 | 0.22 |
ENST00000484674.5
|
FRMD5
|
FERM domain containing 5 |
| chr5_+_161850597 | 0.21 |
ENST00000634335.1
ENST00000635880.1 |
GABRA1
|
gamma-aminobutyric acid type A receptor subunit alpha1 |
| chr16_+_67570741 | 0.21 |
ENST00000644753.1
ENST00000642819.1 ENST00000645306.1 |
CTCF
|
CCCTC-binding factor |
| chr9_+_137086857 | 0.21 |
ENST00000682881.1
ENST00000683324.1 ENST00000542372.2 ENST00000683355.1 ENST00000682117.1 ENST00000682212.1 ENST00000684144.1 ENST00000683987.1 ENST00000371589.9 ENST00000535144.6 ENST00000475449.7 ENST00000684759.1 |
MAN1B1
|
mannosidase alpha class 1B member 1 |
| chr11_+_104036624 | 0.21 |
ENST00000302259.5
|
DDI1
|
DNA damage inducible 1 homolog 1 |
| chr9_-_35079923 | 0.20 |
ENST00000378643.8
ENST00000448890.1 |
FANCG
|
FA complementation group G |
| chr3_-_58433810 | 0.20 |
ENST00000474765.1
ENST00000485460.5 ENST00000302746.11 ENST00000383714.8 |
PDHB
|
pyruvate dehydrogenase E1 subunit beta |
| chr3_-_185821092 | 0.20 |
ENST00000421047.3
|
IGF2BP2
|
insulin like growth factor 2 mRNA binding protein 2 |
| chr5_+_147878703 | 0.20 |
ENST00000296694.5
|
SCGB3A2
|
secretoglobin family 3A member 2 |
| chr14_+_22462932 | 0.19 |
ENST00000390477.2
|
TRDC
|
T cell receptor delta constant |
| chr9_-_110337808 | 0.19 |
ENST00000374510.8
ENST00000374507.4 ENST00000423740.7 ENST00000374511.7 |
TXNDC8
|
thioredoxin domain containing 8 |
| chr2_+_233729042 | 0.19 |
ENST00000482026.6
|
UGT1A3
|
UDP glucuronosyltransferase family 1 member A3 |
| chrX_-_100874351 | 0.19 |
ENST00000372966.8
|
NOX1
|
NADPH oxidase 1 |
| chr13_+_34942263 | 0.18 |
ENST00000379939.7
ENST00000400445.7 |
NBEA
|
neurobeachin |
| chr3_-_123980727 | 0.17 |
ENST00000620893.4
|
ROPN1
|
rhophilin associated tail protein 1 |
| chr4_-_69639642 | 0.17 |
ENST00000604629.6
ENST00000604021.1 |
UGT2A2
|
UDP glucuronosyltransferase family 2 member A2 |
| chr2_-_46617020 | 0.17 |
ENST00000474980.1
ENST00000281382.11 ENST00000306465.8 |
PIGF
|
phosphatidylinositol glycan anchor biosynthesis class F |
| chr10_+_99782628 | 0.16 |
ENST00000648689.1
ENST00000647814.1 |
ABCC2
|
ATP binding cassette subfamily C member 2 |
| chr17_+_36064265 | 0.16 |
ENST00000616054.2
|
CCL18
|
C-C motif chemokine ligand 18 |
| chr16_+_33459650 | 0.16 |
ENST00000617705.1
|
TP53TG3F
|
TP53 target 3 family member F |
| chr19_-_46654657 | 0.16 |
ENST00000300875.4
|
DACT3
|
dishevelled binding antagonist of beta catenin 3 |
| chr14_-_105845677 | 0.16 |
ENST00000390556.6
|
IGHD
|
immunoglobulin heavy constant delta |
| chr5_-_72507354 | 0.16 |
ENST00000414109.2
ENST00000318442.6 |
ZNF366
|
zinc finger protein 366 |
| chr2_+_46617180 | 0.15 |
ENST00000238892.4
|
CRIPT
|
CXXC repeat containing interactor of PDZ3 domain |
| chr3_-_114624921 | 0.15 |
ENST00000393785.6
|
ZBTB20
|
zinc finger and BTB domain containing 20 |
| chr5_+_73813518 | 0.15 |
ENST00000296799.8
|
ARHGEF28
|
Rho guanine nucleotide exchange factor 28 |
| chr12_-_123272234 | 0.15 |
ENST00000544658.5
|
CDK2AP1
|
cyclin dependent kinase 2 associated protein 1 |
| chr4_+_82900679 | 0.15 |
ENST00000302236.10
|
THAP9
|
THAP domain containing 9 |
| chr18_+_22933819 | 0.15 |
ENST00000399722.6
ENST00000399725.6 ENST00000399721.6 |
RBBP8
|
RB binding protein 8, endonuclease |
| chr14_-_64823148 | 0.14 |
ENST00000389722.7
|
SPTB
|
spectrin beta, erythrocytic |
| chr7_+_143959927 | 0.14 |
ENST00000624504.1
|
OR2F1
|
olfactory receptor family 2 subfamily F member 1 |
| chr10_-_95069489 | 0.14 |
ENST00000371270.6
ENST00000535898.5 ENST00000623108.3 |
CYP2C8
|
cytochrome P450 family 2 subfamily C member 8 |
| chr4_+_113292838 | 0.14 |
ENST00000672411.1
ENST00000673231.1 |
ANK2
|
ankyrin 2 |
| chrX_-_24027186 | 0.14 |
ENST00000328046.8
|
KLHL15
|
kelch like family member 15 |
| chr5_-_138875290 | 0.13 |
ENST00000521094.2
ENST00000274711.7 |
LRRTM2
|
leucine rich repeat transmembrane neuronal 2 |
| chr6_+_46693835 | 0.13 |
ENST00000450697.1
|
TDRD6
|
tudor domain containing 6 |
| chr21_+_42403856 | 0.13 |
ENST00000291535.11
|
UBASH3A
|
ubiquitin associated and SH3 domain containing A |
| chr17_+_997101 | 0.13 |
ENST00000327158.5
|
TIMM22
|
translocase of inner mitochondrial membrane 22 |
| chr17_-_58353069 | 0.13 |
ENST00000580947.1
|
SUPT4H1
|
SPT4 homolog, DSIF elongation factor subunit |
| chr3_+_51975026 | 0.12 |
ENST00000458031.6
ENST00000273596.8 ENST00000463937.1 ENST00000635952.1 |
ABHD14A
ABHD14A-ACY1
|
abhydrolase domain containing 14A ABHD14A-ACY1 readthrough |
| chr21_+_42403874 | 0.12 |
ENST00000319294.11
ENST00000398367.1 |
UBASH3A
|
ubiquitin associated and SH3 domain containing A |
| chr4_+_113292925 | 0.12 |
ENST00000673353.1
ENST00000505342.6 ENST00000672915.1 ENST00000509550.5 |
ANK2
|
ankyrin 2 |
| chr4_+_48805137 | 0.12 |
ENST00000504654.5
|
OCIAD1
|
OCIA domain containing 1 |
| chr21_-_30166782 | 0.12 |
ENST00000286808.5
|
CLDN17
|
claudin 17 |
| chr3_-_114624979 | 0.12 |
ENST00000676079.1
|
ZBTB20
|
zinc finger and BTB domain containing 20 |
| chr16_-_69339493 | 0.12 |
ENST00000562595.5
ENST00000615447.1 ENST00000306875.10 ENST00000562081.2 |
COG8
|
component of oligomeric golgi complex 8 |
| chr7_-_150323489 | 0.12 |
ENST00000683684.1
ENST00000478393.5 |
ACTR3C
|
actin related protein 3C |
| chr8_+_12104389 | 0.11 |
ENST00000400085.7
|
ZNF705D
|
zinc finger protein 705D |
| chr16_+_81238682 | 0.11 |
ENST00000258168.7
ENST00000564552.1 |
BCO1
|
beta-carotene oxygenase 1 |
| chr20_+_45207025 | 0.11 |
ENST00000372781.4
|
SEMG1
|
semenogelin 1 |
| chr6_-_159044980 | 0.10 |
ENST00000367066.8
|
TAGAP
|
T cell activation RhoGTPase activating protein |
| chr7_-_83649097 | 0.10 |
ENST00000643230.2
|
SEMA3E
|
semaphorin 3E |
| chr7_+_100088957 | 0.10 |
ENST00000303904.8
ENST00000419210.1 ENST00000418625.5 |
COPS6
|
COP9 signalosome subunit 6 |
| chr13_-_21459253 | 0.10 |
ENST00000400590.8
ENST00000382466.7 ENST00000415724.2 ENST00000542645.5 |
ZDHHC20
|
zinc finger DHHC-type palmitoyltransferase 20 |
| chr17_-_3433841 | 0.09 |
ENST00000248384.1
|
OR1E2
|
olfactory receptor family 1 subfamily E member 2 |
| chr16_-_71289609 | 0.09 |
ENST00000338099.9
ENST00000563876.1 |
CMTR2
|
cap methyltransferase 2 |
| chr4_-_127965930 | 0.08 |
ENST00000641447.1
ENST00000296468.8 ENST00000641134.1 ENST00000641147.1 ENST00000641178.1 |
MFSD8
|
major facilitator superfamily domain containing 8 |
| chr3_-_194351290 | 0.08 |
ENST00000429275.1
ENST00000323830.4 |
CPN2
|
carboxypeptidase N subunit 2 |
| chr20_+_45221362 | 0.08 |
ENST00000372769.4
|
SEMG2
|
semenogelin 2 |
| chr10_-_73358709 | 0.08 |
ENST00000340329.7
|
CFAP70
|
cilia and flagella associated protein 70 |
| chr1_-_112618204 | 0.07 |
ENST00000369664.1
|
ST7L
|
suppression of tumorigenicity 7 like |
| chr6_-_159045104 | 0.07 |
ENST00000326965.7
|
TAGAP
|
T cell activation RhoGTPase activating protein |
| chr6_+_29396555 | 0.07 |
ENST00000623183.1
|
OR12D2
|
olfactory receptor family 12 subfamily D member 2 |
| chr1_+_67207614 | 0.06 |
ENST00000425614.3
|
IL23R
|
interleukin 23 receptor |
| chr4_-_167234266 | 0.05 |
ENST00000511269.5
ENST00000506697.5 ENST00000512042.1 |
SPOCK3
|
SPARC (osteonectin), cwcv and kazal like domains proteoglycan 3 |
| chr5_-_95081482 | 0.05 |
ENST00000312216.12
ENST00000512425.5 ENST00000505208.5 ENST00000429576.6 ENST00000508509.5 ENST00000510732.5 |
MCTP1
|
multiple C2 and transmembrane domain containing 1 |
| chr1_+_166989089 | 0.05 |
ENST00000367870.6
|
MAEL
|
maelstrom spermatogenic transposon silencer |
| chr12_-_91178520 | 0.05 |
ENST00000425043.5
ENST00000420120.6 ENST00000441303.6 ENST00000456569.2 |
DCN
|
decorin |
| chr7_-_108130349 | 0.05 |
ENST00000205386.8
ENST00000418464.1 ENST00000388781.8 |
LAMB4
|
laminin subunit beta 4 |
| chrX_-_21658324 | 0.05 |
ENST00000379499.3
|
KLHL34
|
kelch like family member 34 |
| chrX_-_49186328 | 0.05 |
ENST00000599218.6
ENST00000376317.4 |
PRICKLE3
|
prickle planar cell polarity protein 3 |
| chr7_+_72925075 | 0.05 |
ENST00000434423.5
|
POM121
|
POM121 transmembrane nucleoporin |
| chr16_+_58392391 | 0.05 |
ENST00000426538.6
ENST00000328514.11 |
GINS3
|
GINS complex subunit 3 |
| chr5_-_83720813 | 0.05 |
ENST00000515590.1
ENST00000274341.9 |
HAPLN1
|
hyaluronan and proteoglycan link protein 1 |
| chr4_-_22443110 | 0.05 |
ENST00000508133.5
|
ADGRA3
|
adhesion G protein-coupled receptor A3 |
| chr18_+_52752032 | 0.04 |
ENST00000412726.5
ENST00000578080.1 ENST00000582875.1 |
DCC
|
DCC netrin 1 receptor |
| chr4_+_186266183 | 0.04 |
ENST00000403665.7
ENST00000492972.6 ENST00000264692.8 |
F11
|
coagulation factor XI |
| chr4_+_188139438 | 0.04 |
ENST00000332517.4
|
TRIML1
|
tripartite motif family like 1 |
| chr11_+_111255982 | 0.04 |
ENST00000637637.1
|
C11orf53
|
chromosome 11 open reading frame 53 |
| chr6_-_22302826 | 0.04 |
ENST00000651245.1
|
PRL
|
prolactin |
| chrX_-_100874209 | 0.04 |
ENST00000372964.5
ENST00000217885.5 |
NOX1
|
NADPH oxidase 1 |
| chr6_-_31652678 | 0.04 |
ENST00000437771.5
ENST00000362049.10 ENST00000439687.6 ENST00000211379.9 ENST00000375964.11 ENST00000676571.1 ENST00000676615.1 |
BAG6
|
BAG cochaperone 6 |
| chr16_+_58392462 | 0.04 |
ENST00000318129.6
|
GINS3
|
GINS complex subunit 3 |
| chr21_-_14658812 | 0.04 |
ENST00000647101.1
|
SAMSN1
|
SAM domain, SH3 domain and nuclear localization signals 1 |
| chrX_+_155997706 | 0.04 |
ENST00000369423.7
|
IL9R
|
interleukin 9 receptor |
| chr6_+_30940970 | 0.04 |
ENST00000462446.6
ENST00000304311.3 |
MUCL3
|
mucin like 3 |
| chr13_-_40982880 | 0.04 |
ENST00000635415.1
|
ELF1
|
E74 like ETS transcription factor 1 |
| chr7_+_69967464 | 0.03 |
ENST00000664521.1
|
AUTS2
|
activator of transcription and developmental regulator AUTS2 |
| chr1_-_206921987 | 0.03 |
ENST00000530505.1
ENST00000442471.4 |
FCMR
|
Fc fragment of IgM receptor |
| chr21_-_44801769 | 0.03 |
ENST00000330942.9
|
UBE2G2
|
ubiquitin conjugating enzyme E2 G2 |
| chr19_-_39846329 | 0.03 |
ENST00000599134.1
ENST00000597634.5 ENST00000598417.5 ENST00000601274.5 ENST00000594309.5 ENST00000221801.8 |
FBL
|
fibrillarin |
| chr9_-_21217311 | 0.03 |
ENST00000380216.1
|
IFNA16
|
interferon alpha 16 |
| chr1_+_166989254 | 0.03 |
ENST00000367872.9
ENST00000447624.1 |
MAEL
|
maelstrom spermatogenic transposon silencer |
| chr2_-_233013228 | 0.03 |
ENST00000264051.8
|
NGEF
|
neuronal guanine nucleotide exchange factor |
| chrX_+_155997581 | 0.03 |
ENST00000244174.10
|
IL9R
|
interleukin 9 receptor |
| chr17_+_76079182 | 0.02 |
ENST00000334586.10
|
ZACN
|
zinc activated ion channel |
| chr2_+_112721486 | 0.02 |
ENST00000327581.4
|
NT5DC4
|
5'-nucleotidase domain containing 4 |
| chr19_-_41959266 | 0.02 |
ENST00000600292.5
ENST00000601078.5 ENST00000601891.5 ENST00000222008.11 |
RABAC1
|
Rab acceptor 1 |
| chr20_-_31390483 | 0.02 |
ENST00000376315.2
|
DEFB119
|
defensin beta 119 |
| chr7_-_130687432 | 0.02 |
ENST00000456951.5
|
TSGA13
|
testis specific 13 |
| chr7_-_151080787 | 0.02 |
ENST00000540185.5
ENST00000297532.11 |
FASTK
|
Fas activated serine/threonine kinase |
| chr2_+_170178136 | 0.01 |
ENST00000409044.7
ENST00000408978.9 |
MYO3B
|
myosin IIIB |
| chr1_-_149812359 | 0.01 |
ENST00000369167.2
ENST00000545683.1 |
H2BC18
|
H2B clustered histone 18 |
| chr6_+_27957241 | 0.01 |
ENST00000244623.1
|
OR2B6
|
olfactory receptor family 2 subfamily B member 6 |
| chr7_-_151080833 | 0.01 |
ENST00000353841.6
ENST00000482571.2 |
FASTK
|
Fas activated serine/threonine kinase |
| chr10_+_125973373 | 0.01 |
ENST00000417114.5
ENST00000445510.5 ENST00000368691.5 |
FANK1
|
fibronectin type III and ankyrin repeat domains 1 |
| chr4_+_56505782 | 0.00 |
ENST00000640821.3
|
ARL9
|
ADP ribosylation factor like GTPase 9 |
| chr2_-_88861258 | 0.00 |
ENST00000390240.2
|
IGKJ3
|
immunoglobulin kappa joining 3 |
| chr7_+_95485934 | 0.00 |
ENST00000325885.6
|
ASB4
|
ankyrin repeat and SOCS box containing 4 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.7 | GO:0060352 | cell adhesion molecule production(GO:0060352) |
| 0.1 | 0.6 | GO:0001692 | histamine metabolic process(GO:0001692) |
| 0.1 | 0.3 | GO:0050720 | interleukin-1 beta biosynthetic process(GO:0050720) |
| 0.1 | 0.6 | GO:0097338 | response to clozapine(GO:0097338) |
| 0.1 | 0.8 | GO:0014807 | regulation of somitogenesis(GO:0014807) |
| 0.1 | 0.4 | GO:0071163 | DNA replication preinitiation complex assembly(GO:0071163) |
| 0.1 | 0.2 | GO:1990451 | cellular stress response to acidic pH(GO:1990451) |
| 0.1 | 0.2 | GO:0036510 | trimming of terminal mannose on C branch(GO:0036510) |
| 0.1 | 0.5 | GO:0003172 | primary heart field specification(GO:0003138) sinoatrial valve development(GO:0003172) sinoatrial valve morphogenesis(GO:0003185) |
| 0.1 | 0.4 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 0.1 | 0.2 | GO:0016999 | antibiotic metabolic process(GO:0016999) detoxification of mercury ion(GO:0050787) |
| 0.0 | 0.2 | GO:0040030 | regulation of molecular function, epigenetic(GO:0040030) |
| 0.0 | 0.2 | GO:1900005 | positive regulation of serine-type endopeptidase activity(GO:1900005) positive regulation of serine-type peptidase activity(GO:1902573) |
| 0.0 | 0.3 | GO:0031444 | slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 0.0 | 0.6 | GO:0032310 | innate immune response in mucosa(GO:0002227) positive regulation of guanylate cyclase activity(GO:0031284) prostaglandin secretion(GO:0032310) |
| 0.0 | 0.1 | GO:0036451 | cap mRNA methylation(GO:0036451) |
| 0.0 | 0.5 | GO:0045008 | depyrimidination(GO:0045008) |
| 0.0 | 0.1 | GO:0071630 | nucleus-associated proteasomal ubiquitin-dependent protein catabolic process(GO:0071630) |
| 0.0 | 0.1 | GO:0016119 | carotene metabolic process(GO:0016119) |
| 0.0 | 0.2 | GO:0033088 | negative regulation of immature T cell proliferation in thymus(GO:0033088) |
| 0.0 | 0.3 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 0.0 | 0.8 | GO:0035590 | purinergic nucleotide receptor signaling pathway(GO:0035590) |
| 0.0 | 0.4 | GO:0086069 | bundle of His cell to Purkinje myocyte communication(GO:0086069) |
| 0.0 | 0.3 | GO:0033623 | regulation of integrin activation(GO:0033623) |
| 0.0 | 0.1 | GO:0002933 | lipid hydroxylation(GO:0002933) |
| 0.0 | 1.2 | GO:0046039 | GTP metabolic process(GO:0046039) |
| 0.0 | 0.1 | GO:0010792 | DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) double-strand break repair via single-strand annealing(GO:0045002) |
| 0.0 | 0.3 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.0 | 0.2 | GO:1902897 | regulation of postsynaptic density protein 95 clustering(GO:1902897) |
| 0.0 | 0.1 | GO:0038155 | interleukin-23-mediated signaling pathway(GO:0038155) |
| 0.0 | 0.5 | GO:0010569 | regulation of double-strand break repair via homologous recombination(GO:0010569) |
| 0.0 | 0.2 | GO:0052697 | flavonoid glucuronidation(GO:0052696) xenobiotic glucuronidation(GO:0052697) |
| 0.0 | 0.2 | GO:0006662 | glycerol ether metabolic process(GO:0006662) |
| 0.0 | 0.9 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.0 | 0.1 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.0 | 0.3 | GO:0002230 | positive regulation of defense response to virus by host(GO:0002230) |
| 0.0 | 0.0 | GO:0021965 | spinal cord ventral commissure morphogenesis(GO:0021965) |
| 0.0 | 0.0 | GO:0051919 | positive regulation of fibrinolysis(GO:0051919) |
| 0.0 | 0.8 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.0 | 0.0 | GO:1990258 | box C/D snoRNA 3'-end processing(GO:0000494) box C/D snoRNA metabolic process(GO:0033967) box C/D snoRNA processing(GO:0034963) histone glutamine methylation(GO:1990258) |
| 0.0 | 0.0 | GO:1904379 | tail-anchored membrane protein insertion into ER membrane(GO:0071816) protein localization to cytosolic proteasome complex(GO:1904327) protein localization to cytosolic proteasome complex involved in ERAD pathway(GO:1904379) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0031417 | NatC complex(GO:0031417) |
| 0.0 | 0.9 | GO:0031229 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) |
| 0.0 | 0.6 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.2 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.0 | 0.1 | GO:0030849 | autosome(GO:0030849) |
| 0.0 | 0.1 | GO:0032044 | DSIF complex(GO:0032044) |
| 0.0 | 0.1 | GO:0042721 | mitochondrial inner membrane protein insertion complex(GO:0042721) |
| 0.0 | 0.8 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 0.1 | GO:0072536 | interleukin-23 receptor complex(GO:0072536) |
| 0.0 | 0.2 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.0 | 0.2 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 0.2 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 0.3 | GO:0043220 | Schmidt-Lanterman incisure(GO:0043220) |
| 0.0 | 0.2 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 1.7 | GO:0031985 | Golgi cisterna(GO:0031985) |
| 0.0 | 1.2 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.0 | GO:0072379 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.7 | GO:0003829 | beta-1,3-galactosyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity(GO:0003829) |
| 0.2 | 0.9 | GO:0008240 | tripeptidyl-peptidase activity(GO:0008240) |
| 0.1 | 0.6 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 0.1 | 0.9 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.1 | 0.3 | GO:0047006 | 17-alpha,20-alpha-dihydroxypregn-4-en-3-one dehydrogenase activity(GO:0047006) |
| 0.1 | 0.9 | GO:0004931 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.1 | 0.2 | GO:0043035 | chromatin insulator sequence binding(GO:0043035) |
| 0.1 | 0.4 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.0 | 0.5 | GO:0097506 | uracil DNA N-glycosylase activity(GO:0004844) deaminated base DNA N-glycosylase activity(GO:0097506) |
| 0.0 | 0.2 | GO:0030171 | voltage-gated proton channel activity(GO:0030171) |
| 0.0 | 0.4 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.0 | 0.2 | GO:0004307 | ethanolaminephosphotransferase activity(GO:0004307) |
| 0.0 | 0.2 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.0 | 0.8 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.0 | 0.6 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.0 | 0.1 | GO:0004483 | mRNA (nucleoside-2'-O-)-methyltransferase activity(GO:0004483) |
| 0.0 | 0.1 | GO:0004803 | transposase activity(GO:0004803) |
| 0.0 | 0.5 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 0.1 | GO:0034875 | oxidoreductase activity, acting on CH or CH2 groups, quinone or similar compound as acceptor(GO:0033695) caffeine oxidase activity(GO:0034875) |
| 0.0 | 0.2 | GO:0099529 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.0 | 0.2 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 0.0 | 0.2 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.0 | 0.1 | GO:0004739 | pyruvate dehydrogenase (acetyl-transferring) activity(GO:0004739) |
| 0.0 | 0.2 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.0 | 0.2 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 0.0 | 0.1 | GO:0042019 | interleukin-23 binding(GO:0042019) interleukin-23 receptor activity(GO:0042020) |
| 0.0 | 0.1 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.0 | 0.8 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 0.3 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.0 | 0.0 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.0 | 0.0 | GO:1990259 | protein-glutamine N-methyltransferase activity(GO:0036009) histone-glutamine methyltransferase activity(GO:1990259) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.1 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.0 | 1.7 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 0.2 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.0 | 0.4 | REACTOME ASSOCIATION OF LICENSING FACTORS WITH THE PRE REPLICATIVE COMPLEX | Genes involved in Association of licensing factors with the pre-replicative complex |
| 0.0 | 0.6 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.0 | 1.3 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.5 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.2 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 0.4 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 0.6 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |