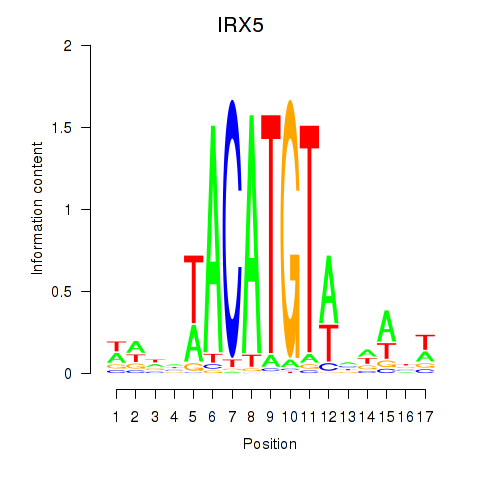
|
chr4_+_41359599
Show fit
|
2.10 |
ENST00000513024.5
|
LIMCH1
|
LIM and calponin homology domains 1
|
|
chr9_-_41908681
Show fit
|
1.68 |
ENST00000476961.5
|
CNTNAP3B
|
contactin associated protein family member 3B
|
|
chr1_+_61203496
Show fit
|
1.60 |
ENST00000663597.1
|
NFIA
|
nuclear factor I A
|
|
chr15_+_66293217
Show fit
|
1.41 |
ENST00000319194.9
ENST00000525134.6
|
DIS3L
|
DIS3 like exosome 3'-5' exoribonuclease
|
|
chr1_-_145910031
Show fit
|
1.30 |
ENST00000369304.8
|
ITGA10
|
integrin subunit alpha 10
|
|
chr1_+_61081728
Show fit
|
1.23 |
ENST00000371189.8
|
NFIA
|
nuclear factor I A
|
|
chr1_+_61082398
Show fit
|
1.23 |
ENST00000664149.1
|
NFIA
|
nuclear factor I A
|
|
chr9_-_90642855
Show fit
|
1.18 |
ENST00000637905.1
|
DIRAS2
|
DIRAS family GTPase 2
|
|
chr1_-_145910066
Show fit
|
1.14 |
ENST00000539363.2
|
ITGA10
|
integrin subunit alpha 10
|
|
chr2_-_182242031
Show fit
|
1.00 |
ENST00000358139.6
|
PDE1A
|
phosphodiesterase 1A
|
|
chr12_+_32502114
Show fit
|
0.86 |
ENST00000682739.1
ENST00000427716.7
ENST00000583694.2
|
FGD4
|
FYVE, RhoGEF and PH domain containing 4
|
|
chr7_-_120858066
Show fit
|
0.80 |
ENST00000222747.8
|
TSPAN12
|
tetraspanin 12
|
|
chr8_-_17697654
Show fit
|
0.80 |
ENST00000297488.10
|
MTUS1
|
microtubule associated scaffold protein 1
|
|
chr15_+_66293541
Show fit
|
0.78 |
ENST00000319212.9
ENST00000525109.1
|
DIS3L
|
DIS3 like exosome 3'-5' exoribonuclease
|
|
chr3_-_169147734
Show fit
|
0.75 |
ENST00000464456.5
|
MECOM
|
MDS1 and EVI1 complex locus
|
|
chr11_+_31509744
Show fit
|
0.73 |
ENST00000639878.1
ENST00000379163.10
ENST00000638347.1
ENST00000350638.10
ENST00000638764.1
ENST00000639570.1
ENST00000640533.1
ENST00000638482.1
ENST00000640961.2
ENST00000640342.1
ENST00000640231.1
ENST00000640954.1
ENST00000395934.2
|
ELP4
|
elongator acetyltransferase complex subunit 4
|
|
chr2_+_172821575
Show fit
|
0.73 |
ENST00000397087.7
|
RAPGEF4
|
Rap guanine nucleotide exchange factor 4
|
|
chr1_+_196819731
Show fit
|
0.72 |
ENST00000320493.10
ENST00000367424.4
|
CFHR1
|
complement factor H related 1
|
|
chr6_+_34889228
Show fit
|
0.71 |
ENST00000360359.5
ENST00000649117.1
ENST00000650178.1
|
ANKS1A
|
ankyrin repeat and sterile alpha motif domain containing 1A
|
|
chr11_-_102874974
Show fit
|
0.69 |
ENST00000571244.3
|
MMP12
|
matrix metallopeptidase 12
|
|
chr6_+_130421086
Show fit
|
0.67 |
ENST00000545622.5
|
TMEM200A
|
transmembrane protein 200A
|
|
chr11_+_31509819
Show fit
|
0.64 |
ENST00000638184.1
|
ELP4
|
elongator acetyltransferase complex subunit 4
|
|
chr4_+_140343443
Show fit
|
0.63 |
ENST00000338517.8
ENST00000394203.7
ENST00000506322.5
|
SCOC
|
short coiled-coil protein
|
|
chr2_+_190927649
Show fit
|
0.57 |
ENST00000409428.5
ENST00000409215.5
|
GLS
|
glutaminase
|
|
chr1_+_96722628
Show fit
|
0.53 |
ENST00000675401.1
|
PTBP2
|
polypyrimidine tract binding protein 2
|
|
chr19_-_12881460
Show fit
|
0.49 |
ENST00000592506.1
|
DNASE2
|
deoxyribonuclease 2, lysosomal
|
|
chr20_+_56358938
Show fit
|
0.47 |
ENST00000371384.4
ENST00000437418.1
|
FAM210B
|
family with sequence similarity 210 member B
|
|
chr6_-_111483700
Show fit
|
0.47 |
ENST00000435970.5
ENST00000358835.7
|
REV3L
|
REV3 like, DNA directed polymerase zeta catalytic subunit
|
|
chr1_+_53014926
Show fit
|
0.45 |
ENST00000430330.6
ENST00000408941.7
ENST00000478274.6
ENST00000484100.5
ENST00000435345.6
ENST00000488965.1
|
SCP2
|
sterol carrier protein 2
|
|
chr17_-_31314066
Show fit
|
0.45 |
ENST00000577894.1
|
EVI2B
|
ecotropic viral integration site 2B
|
|
chr12_+_10212867
Show fit
|
0.44 |
ENST00000545047.5
ENST00000266458.10
ENST00000629504.1
ENST00000543602.5
ENST00000545887.1
|
GABARAPL1
|
GABA type A receptor associated protein like 1
|
|
chr20_+_10218808
Show fit
|
0.44 |
ENST00000254976.7
ENST00000304886.6
|
SNAP25
|
synaptosome associated protein 25
|
|
chr17_-_49646581
Show fit
|
0.43 |
ENST00000510476.5
ENST00000503676.5
|
SPOP
|
speckle type BTB/POZ protein
|
|
chr12_-_21604840
Show fit
|
0.42 |
ENST00000261195.3
|
GYS2
|
glycogen synthase 2
|
|
chr3_+_159069252
Show fit
|
0.39 |
ENST00000640015.1
ENST00000476809.7
ENST00000485419.7
|
IQCJ-SCHIP1
|
IQCJ-SCHIP1 readthrough
|
|
chr10_+_125973373
Show fit
|
0.39 |
ENST00000417114.5
ENST00000445510.5
ENST00000368691.5
|
FANK1
|
fibronectin type III and ankyrin repeat domains 1
|
|
chrX_+_100644183
Show fit
|
0.38 |
ENST00000640889.1
ENST00000373004.5
|
SRPX2
|
sushi repeat containing protein X-linked 2
|
|
chr19_+_50188180
Show fit
|
0.38 |
ENST00000598205.5
|
MYH14
|
myosin heavy chain 14
|
|
chr12_+_15322257
Show fit
|
0.38 |
ENST00000674316.1
|
PTPRO
|
protein tyrosine phosphatase receptor type O
|
|
chr1_-_61725121
Show fit
|
0.37 |
ENST00000371177.2
ENST00000606498.5
|
TM2D1
|
TM2 domain containing 1
|
|
chr5_-_94111627
Show fit
|
0.37 |
ENST00000505869.5
ENST00000395965.8
ENST00000509163.5
|
FAM172A
|
family with sequence similarity 172 member A
|
|
chr5_+_141192330
Show fit
|
0.35 |
ENST00000239446.6
|
PCDHB10
|
protocadherin beta 10
|
|
chr9_-_20382461
Show fit
|
0.35 |
ENST00000380321.5
ENST00000629733.3
|
MLLT3
|
MLLT3 super elongation complex subunit
|
|
chr11_-_60243103
Show fit
|
0.35 |
ENST00000651255.1
|
MS4A4E
|
membrane spanning 4-domains A4E
|
|
chr5_-_156963222
Show fit
|
0.35 |
ENST00000407087.4
ENST00000274532.7
|
TIMD4
|
T cell immunoglobulin and mucin domain containing 4
|
|
chr17_-_7205116
Show fit
|
0.34 |
ENST00000649520.1
ENST00000649186.1
|
DLG4
|
discs large MAGUK scaffold protein 4
|
|
chr4_+_89894846
Show fit
|
0.34 |
ENST00000264790.7
|
MMRN1
|
multimerin 1
|
|
chrX_-_33211540
Show fit
|
0.34 |
ENST00000357033.9
|
DMD
|
dystrophin
|
|
chr14_-_106025628
Show fit
|
0.33 |
ENST00000631943.1
|
IGHV7-4-1
|
immunoglobulin heavy variable 7-4-1
|
|
chr4_-_103198331
Show fit
|
0.33 |
ENST00000265148.9
ENST00000514974.1
|
CENPE
|
centromere protein E
|
|
chr15_+_40765620
Show fit
|
0.32 |
ENST00000558467.1
|
GCHFR
|
GTP cyclohydrolase I feedback regulator
|
|
chr7_-_64982021
Show fit
|
0.32 |
ENST00000610793.1
ENST00000620222.4
|
ZNF117
|
zinc finger protein 117
|
|
chr11_-_60183191
Show fit
|
0.32 |
ENST00000412309.6
|
MS4A6A
|
membrane spanning 4-domains A6A
|
|
chr9_-_110337808
Show fit
|
0.32 |
ENST00000374510.8
ENST00000374507.4
ENST00000423740.7
ENST00000374511.7
|
TXNDC8
|
thioredoxin domain containing 8
|
|
chr1_+_63523490
Show fit
|
0.32 |
ENST00000371088.5
|
EFCAB7
|
EF-hand calcium binding domain 7
|
|
chr4_-_103198371
Show fit
|
0.31 |
ENST00000611174.4
ENST00000380026.8
|
CENPE
|
centromere protein E
|
|
chr3_+_180868748
Show fit
|
0.30 |
ENST00000465551.5
|
FXR1
|
FMR1 autosomal homolog 1
|
|
chr1_+_158999963
Show fit
|
0.30 |
ENST00000566111.5
|
IFI16
|
interferon gamma inducible protein 16
|
|
chr14_+_55661272
Show fit
|
0.30 |
ENST00000555573.5
|
KTN1
|
kinectin 1
|
|
chr14_-_106154113
Show fit
|
0.28 |
ENST00000390603.2
|
IGHV3-15
|
immunoglobulin heavy variable 3-15
|
|
chr1_+_196943738
Show fit
|
0.27 |
ENST00000367415.8
ENST00000367421.5
ENST00000649283.1
ENST00000476712.6
ENST00000496448.6
ENST00000473386.1
ENST00000649960.1
|
CFHR2
|
complement factor H related 2
|
|
chr9_-_122905340
Show fit
|
0.27 |
ENST00000423239.6
ENST00000357244.7
|
RC3H2
|
ring finger and CCCH-type domains 2
|
|
chr15_+_24954912
Show fit
|
0.26 |
ENST00000584968.5
ENST00000346403.10
ENST00000554227.6
ENST00000390687.9
ENST00000579070.5
ENST00000577565.1
ENST00000577949.5
ENST00000338327.4
|
SNRPN
SNURF
|
small nuclear ribonucleoprotein polypeptide N
SNRPN upstream reading frame
|
|
chr1_-_21050952
Show fit
|
0.25 |
ENST00000264211.12
|
EIF4G3
|
eukaryotic translation initiation factor 4 gamma 3
|
|
chr5_+_65563239
Show fit
|
0.25 |
ENST00000535264.5
ENST00000538977.5
ENST00000261308.10
|
PPWD1
|
peptidylprolyl isomerase domain and WD repeat containing 1
|
|
chr11_+_22666604
Show fit
|
0.24 |
ENST00000454584.6
|
GAS2
|
growth arrest specific 2
|
|
chr3_-_197260722
Show fit
|
0.22 |
ENST00000654733.1
ENST00000661808.1
|
DLG1
|
discs large MAGUK scaffold protein 1
|
|
chr11_-_35526024
Show fit
|
0.22 |
ENST00000615849.4
|
PAMR1
|
peptidase domain containing associated with muscle regeneration 1
|
|
chr11_-_60183011
Show fit
|
0.22 |
ENST00000533023.5
ENST00000420732.6
ENST00000528851.6
|
MS4A6A
|
membrane spanning 4-domains A6A
|
|
chr9_-_41189310
Show fit
|
0.22 |
ENST00000456520.5
ENST00000377391.8
ENST00000613716.4
ENST00000617933.1
|
CBWD6
|
COBW domain containing 6
|
|
chr11_+_103109522
Show fit
|
0.22 |
ENST00000334267.11
|
DYNC2H1
|
dynein cytoplasmic 2 heavy chain 1
|
|
chr12_+_57941499
Show fit
|
0.21 |
ENST00000300145.4
|
ATP23
|
ATP23 metallopeptidase and ATP synthase assembly factor homolog
|
|
chr9_-_146140
Show fit
|
0.21 |
ENST00000475990.5
|
CBWD1
|
COBW domain containing 1
|
|
chr3_-_3179674
Show fit
|
0.21 |
ENST00000424814.5
ENST00000450014.1
ENST00000231948.9
ENST00000432408.6
ENST00000639284.1
|
CRBN
|
cereblon
|
|
chr1_-_85404494
Show fit
|
0.21 |
ENST00000633113.1
|
DDAH1
|
dimethylarginine dimethylaminohydrolase 1
|
|
chr9_+_68241854
Show fit
|
0.20 |
ENST00000616550.4
ENST00000618217.4
ENST00000377342.9
ENST00000478048.5
ENST00000360171.11
|
CBWD3
|
COBW domain containing 3
|
|
chr14_-_23154369
Show fit
|
0.20 |
ENST00000453702.5
|
SLC7A8
|
solute carrier family 7 member 8
|
|
chr3_-_9952337
Show fit
|
0.18 |
ENST00000411976.2
ENST00000412055.6
|
PRRT3
|
proline rich transmembrane protein 3
|
|
chr14_-_23155302
Show fit
|
0.18 |
ENST00000529705.6
|
SLC7A8
|
solute carrier family 7 member 8
|
|
chr4_+_74445126
Show fit
|
0.18 |
ENST00000395748.8
|
AREG
|
amphiregulin
|
|
chr1_-_212847649
Show fit
|
0.18 |
ENST00000332912.3
|
SPATA45
|
spermatogenesis associated 45
|
|
chr1_-_158554405
Show fit
|
0.18 |
ENST00000641282.1
ENST00000641622.1
|
OR6Y1
|
olfactory receptor family 6 subfamily Y member 1
|
|
chr19_-_40090860
Show fit
|
0.17 |
ENST00000599972.1
ENST00000450241.6
ENST00000595687.6
ENST00000340963.9
|
ZNF780A
|
zinc finger protein 780A
|
|
chr13_-_41194485
Show fit
|
0.17 |
ENST00000379483.4
|
KBTBD7
|
kelch repeat and BTB domain containing 7
|
|
chr1_+_235367360
Show fit
|
0.17 |
ENST00000651186.1
ENST00000406207.5
ENST00000645899.1
ENST00000644578.1
ENST00000645372.1
ENST00000647407.1
ENST00000642610.2
ENST00000543662.4
ENST00000366601.8
|
TBCE
|
tubulin folding cofactor E
|
|
chr9_+_65675834
Show fit
|
0.17 |
ENST00000377392.9
ENST00000377384.5
ENST00000430059.6
ENST00000429800.6
ENST00000382405.8
ENST00000377395.8
|
CBWD5
|
COBW domain containing 5
|
|
chr2_+_159733958
Show fit
|
0.17 |
ENST00000409591.5
|
MARCHF7
|
membrane associated ring-CH-type finger 7
|
|
chr19_-_12881429
Show fit
|
0.17 |
ENST00000222219.8
|
DNASE2
|
deoxyribonuclease 2, lysosomal
|
|
chr1_+_220094086
Show fit
|
0.16 |
ENST00000366922.3
|
IARS2
|
isoleucyl-tRNA synthetase 2, mitochondrial
|
|
chr10_+_116427839
Show fit
|
0.16 |
ENST00000369230.4
|
PNLIPRP3
|
pancreatic lipase related protein 3
|
|
chr8_-_107498041
Show fit
|
0.16 |
ENST00000297450.7
|
ANGPT1
|
angiopoietin 1
|
|
chr19_-_51723968
Show fit
|
0.16 |
ENST00000222115.5
ENST00000540069.7
|
HAS1
|
hyaluronan synthase 1
|
|
chr1_-_243843164
Show fit
|
0.16 |
ENST00000491219.6
ENST00000680056.1
ENST00000492957.2
|
AKT3
|
AKT serine/threonine kinase 3
|
|
chr15_-_63156774
Show fit
|
0.15 |
ENST00000462430.5
|
RPS27L
|
ribosomal protein S27 like
|
|
chr14_-_95157890
Show fit
|
0.15 |
ENST00000526495.6
|
DICER1
|
dicer 1, ribonuclease III
|
|
chrX_-_19970298
Show fit
|
0.14 |
ENST00000379687.7
ENST00000379682.8
|
BCLAF3
|
BCLAF1 and THRAP3 family member 3
|
|
chr1_+_116754422
Show fit
|
0.14 |
ENST00000369478.4
ENST00000369477.1
|
CD2
|
CD2 molecule
|
|
chr19_-_23274194
Show fit
|
0.14 |
ENST00000640920.1
ENST00000639997.1
ENST00000639327.1
ENST00000640517.1
ENST00000639752.1
ENST00000640354.1
ENST00000638919.1
ENST00000638822.1
ENST00000640838.1
ENST00000611392.5
ENST00000594653.1
|
ENSG00000284428.1
ENSG00000283201.1
ENSG00000269107.1
|
novel transcript
zinc finger protein 724
novel transcript
|
|
chr4_-_142560669
Show fit
|
0.14 |
ENST00000510812.5
|
INPP4B
|
inositol polyphosphate-4-phosphatase type II B
|
|
chr6_-_52840843
Show fit
|
0.14 |
ENST00000370989.6
|
GSTA5
|
glutathione S-transferase alpha 5
|
|
chr14_+_71586261
Show fit
|
0.14 |
ENST00000358550.6
|
SIPA1L1
|
signal induced proliferation associated 1 like 1
|
|
chr10_-_114144599
Show fit
|
0.14 |
ENST00000428953.1
|
CCDC186
|
coiled-coil domain containing 186
|
|
chr13_+_75788838
Show fit
|
0.14 |
ENST00000497947.6
|
LMO7
|
LIM domain 7
|
|
chr3_-_197260369
Show fit
|
0.13 |
ENST00000658155.1
ENST00000453607.5
|
DLG1
|
discs large MAGUK scaffold protein 1
|
|
chr8_-_107497909
Show fit
|
0.13 |
ENST00000517746.6
|
ANGPT1
|
angiopoietin 1
|
|
chr4_-_173399102
Show fit
|
0.13 |
ENST00000296506.8
|
SCRG1
|
stimulator of chondrogenesis 1
|
|
chr9_-_6008469
Show fit
|
0.13 |
ENST00000399933.8
|
KIAA2026
|
KIAA2026
|
|
chr3_+_111911604
Show fit
|
0.13 |
ENST00000495180.1
|
PHLDB2
|
pleckstrin homology like domain family B member 2
|
|
chr4_+_129096144
Show fit
|
0.13 |
ENST00000425929.6
ENST00000508622.1
|
C4orf33
|
chromosome 4 open reading frame 33
|
|
chr19_+_16829387
Show fit
|
0.12 |
ENST00000248054.10
ENST00000596802.5
ENST00000379803.5
|
SIN3B
|
SIN3 transcription regulator family member B
|
|
chr19_-_40090921
Show fit
|
0.12 |
ENST00000595508.1
ENST00000414720.6
ENST00000455521.5
ENST00000595773.5
ENST00000683561.1
|
ENSG00000269749.1
ZNF780A
|
novel transcript
zinc finger protein 780A
|
|
chr19_-_54463762
Show fit
|
0.12 |
ENST00000611161.2
|
LENG9
|
leukocyte receptor cluster member 9
|
|
chr12_-_121669646
Show fit
|
0.12 |
ENST00000355329.7
|
MORN3
|
MORN repeat containing 3
|
|
chr5_-_36001006
Show fit
|
0.12 |
ENST00000625798.2
|
UGT3A1
|
UDP glycosyltransferase family 3 member A1
|
|
chr14_+_96256194
Show fit
|
0.12 |
ENST00000216629.11
ENST00000553356.1
|
BDKRB1
|
bradykinin receptor B1
|
|
chr12_-_9208388
Show fit
|
0.11 |
ENST00000261336.7
|
PZP
|
PZP alpha-2-macroglobulin like
|
|
chr2_-_40430257
Show fit
|
0.11 |
ENST00000408028.6
ENST00000332839.8
ENST00000406391.2
ENST00000405901.7
|
SLC8A1
|
solute carrier family 8 member A1
|
|
chr5_+_148063971
Show fit
|
0.11 |
ENST00000398454.5
ENST00000359874.7
ENST00000508733.5
ENST00000256084.8
|
SPINK5
|
serine peptidase inhibitor Kazal type 5
|
|
chr6_+_116369837
Show fit
|
0.11 |
ENST00000645988.1
|
DSE
|
dermatan sulfate epimerase
|
|
chr1_+_67207614
Show fit
|
0.11 |
ENST00000425614.3
|
IL23R
|
interleukin 23 receptor
|
|
chr5_+_55160161
Show fit
|
0.11 |
ENST00000296734.6
ENST00000515370.1
ENST00000503787.6
|
GPX8
|
glutathione peroxidase 8 (putative)
|
|
chr8_+_67064316
Show fit
|
0.11 |
ENST00000675306.2
ENST00000678017.1
ENST00000262210.11
ENST00000675869.1
ENST00000677009.1
ENST00000676847.1
ENST00000676471.1
ENST00000678542.1
ENST00000677619.1
ENST00000676605.1
ENST00000678553.1
ENST00000674993.1
ENST00000678318.1
ENST00000676573.1
ENST00000676317.1
ENST00000677592.1
ENST00000679226.1
ENST00000675955.1
ENST00000676882.1
ENST00000678616.1
ENST00000678645.1
ENST00000678747.1
|
CSPP1
|
centrosome and spindle pole associated protein 1
|
|
chr12_-_52926459
Show fit
|
0.11 |
ENST00000552150.5
|
KRT8
|
keratin 8
|
|
chr1_+_108712903
Show fit
|
0.11 |
ENST00000370017.9
|
FNDC7
|
fibronectin type III domain containing 7
|
|
chr5_-_64768619
Show fit
|
0.11 |
ENST00000513458.9
|
SREK1IP1
|
SREK1 interacting protein 1
|
|
chr3_-_132684685
Show fit
|
0.11 |
ENST00000512094.5
ENST00000632629.1
|
NPHP3
NPHP3-ACAD11
|
nephrocystin 3
NPHP3-ACAD11 readthrough (NMD candidate)
|
|
chr12_+_133181409
Show fit
|
0.10 |
ENST00000416488.5
ENST00000228289.9
ENST00000541211.6
ENST00000536435.7
ENST00000500625.7
ENST00000539248.6
ENST00000542711.6
ENST00000536899.6
ENST00000542986.6
ENST00000611984.4
ENST00000541975.2
|
ZNF268
|
zinc finger protein 268
|
|
chr19_+_57746799
Show fit
|
0.10 |
ENST00000317178.10
ENST00000431353.1
|
ZNF776
|
zinc finger protein 776
|
|
chr1_-_38881587
Show fit
|
0.10 |
ENST00000357771.5
ENST00000621281.1
|
GJA9
ENSG00000274944.4
|
gap junction protein alpha 9
novel protein
|
|
chr7_+_77840122
Show fit
|
0.09 |
ENST00000450574.5
ENST00000248550.7
|
PHTF2
|
putative homeodomain transcription factor 2
|
|
chr3_+_12351470
Show fit
|
0.09 |
ENST00000287820.10
|
PPARG
|
peroxisome proliferator activated receptor gamma
|
|
chr7_+_143620925
Show fit
|
0.09 |
ENST00000444908.7
|
TCAF2
|
TRPM8 channel associated factor 2
|
|
chr6_-_154356735
Show fit
|
0.09 |
ENST00000367220.8
ENST00000265198.8
ENST00000520261.1
|
IPCEF1
|
interaction protein for cytohesin exchange factors 1
|
|
chr11_+_57805541
Show fit
|
0.09 |
ENST00000683201.1
ENST00000683769.1
|
CTNND1
|
catenin delta 1
|
|
chr1_+_201780490
Show fit
|
0.09 |
ENST00000430015.5
|
NAV1
|
neuron navigator 1
|
|
chr2_-_55917699
Show fit
|
0.08 |
ENST00000634374.1
|
EFEMP1
|
EGF containing fibulin extracellular matrix protein 1
|
|
chr21_+_37420299
Show fit
|
0.08 |
ENST00000455097.6
ENST00000643854.1
ENST00000645424.1
ENST00000642309.1
ENST00000645774.1
ENST00000398956.2
|
DYRK1A
|
dual specificity tyrosine phosphorylation regulated kinase 1A
|
|
chr4_-_176269213
Show fit
|
0.08 |
ENST00000296525.7
|
ASB5
|
ankyrin repeat and SOCS box containing 5
|
|
chr16_+_19523811
Show fit
|
0.08 |
ENST00000396212.6
ENST00000381396.9
|
CCP110
|
centriolar coiled-coil protein 110
|
|
chr12_-_100092890
Show fit
|
0.08 |
ENST00000550544.5
ENST00000551980.1
ENST00000548045.5
ENST00000545232.6
ENST00000551973.5
|
UHRF1BP1L
|
UHRF1 binding protein 1 like
|
|
chr17_+_3475959
Show fit
|
0.08 |
ENST00000263080.3
|
ASPA
|
aspartoacylase
|
|
chrX_+_100666854
Show fit
|
0.08 |
ENST00000640282.1
|
SRPX2
|
sushi repeat containing protein X-linked 2
|
|
chr13_+_111186773
Show fit
|
0.07 |
ENST00000426073.6
|
ARHGEF7
|
Rho guanine nucleotide exchange factor 7
|
|
chr2_-_69961624
Show fit
|
0.07 |
ENST00000320256.6
|
ASPRV1
|
aspartic peptidase retroviral like 1
|
|
chrX_-_100874351
Show fit
|
0.07 |
ENST00000372966.8
|
NOX1
|
NADPH oxidase 1
|
|
chr12_+_101475319
Show fit
|
0.07 |
ENST00000551346.2
|
SPIC
|
Spi-C transcription factor
|
|
chr2_+_225399684
Show fit
|
0.07 |
ENST00000636099.1
|
NYAP2
|
neuronal tyrosine-phosphorylated phosphoinositide-3-kinase adaptor 2
|
|
chr11_+_57763820
Show fit
|
0.07 |
ENST00000674106.1
|
CTNND1
|
catenin delta 1
|
|
chr18_+_36129888
Show fit
|
0.07 |
ENST00000351393.10
ENST00000442325.6
ENST00000423854.6
ENST00000358232.11
ENST00000350494.10
ENST00000542824.5
|
ELP2
|
elongator acetyltransferase complex subunit 2
|
|
chr3_-_52679713
Show fit
|
0.07 |
ENST00000296302.11
ENST00000356770.8
ENST00000337303.8
ENST00000409057.5
ENST00000410007.5
ENST00000409114.7
ENST00000409767.5
ENST00000423351.5
|
PBRM1
|
polybromo 1
|
|
chr1_-_114581589
Show fit
|
0.07 |
ENST00000369541.4
|
BCAS2
|
BCAS2 pre-mRNA processing factor
|
|
chrX_-_100874332
Show fit
|
0.06 |
ENST00000372960.8
|
NOX1
|
NADPH oxidase 1
|
|
chr11_-_75096876
Show fit
|
0.06 |
ENST00000641541.1
ENST00000641593.1
ENST00000641504.1
ENST00000641931.1
ENST00000647690.1
|
OR2AT4
ENSG00000284722.2
|
olfactory receptor family 2 subfamily AT member 4
novel transcript
|
|
chr12_+_55549602
Show fit
|
0.06 |
ENST00000641569.1
ENST00000641851.1
|
OR6C4
|
olfactory receptor family 6 subfamily C member 4
|
|
chr19_-_38253238
Show fit
|
0.06 |
ENST00000587515.5
|
PPP1R14A
|
protein phosphatase 1 regulatory inhibitor subunit 14A
|
|
chr3_+_138347648
Show fit
|
0.05 |
ENST00000614350.4
ENST00000289104.8
|
MRAS
|
muscle RAS oncogene homolog
|
|
chr8_-_11201799
Show fit
|
0.05 |
ENST00000416569.3
|
XKR6
|
XK related 6
|
|
chr2_+_233729042
Show fit
|
0.05 |
ENST00000482026.6
|
UGT1A3
|
UDP glucuronosyltransferase family 1 member A3
|
|
chr11_-_30992665
Show fit
|
0.05 |
ENST00000406071.6
|
DCDC1
|
doublecortin domain containing 1
|
|
chrY_-_6874027
Show fit
|
0.05 |
ENST00000215479.10
|
AMELY
|
amelogenin Y-linked
|
|
chr7_+_143620967
Show fit
|
0.05 |
ENST00000684770.1
|
TCAF2
|
TRPM8 channel associated factor 2
|
|
chr2_+_37950432
Show fit
|
0.05 |
ENST00000407257.5
ENST00000417700.6
ENST00000234195.7
ENST00000442857.5
|
RMDN2
|
regulator of microtubule dynamics 2
|
|
chr5_-_56233287
Show fit
|
0.04 |
ENST00000513241.2
ENST00000341048.9
|
ANKRD55
|
ankyrin repeat domain 55
|
|
chr1_-_111563934
Show fit
|
0.04 |
ENST00000443498.5
|
TMIGD3
|
transmembrane and immunoglobulin domain containing 3
|
|
chr4_+_76074701
Show fit
|
0.04 |
ENST00000355810.9
ENST00000349321.7
|
ART3
|
ADP-ribosyltransferase 3 (inactive)
|
|
chr12_+_5432101
Show fit
|
0.04 |
ENST00000423158.4
|
NTF3
|
neurotrophin 3
|
|
chr1_+_196888014
Show fit
|
0.04 |
ENST00000367416.6
ENST00000608469.6
ENST00000251424.8
ENST00000367418.2
|
CFHR4
|
complement factor H related 4
|
|
chr3_+_98166696
Show fit
|
0.04 |
ENST00000641450.1
|
OR5H15
|
olfactory receptor family 5 subfamily H member 15
|
|
chr6_+_26216928
Show fit
|
0.04 |
ENST00000303910.4
|
H2AC8
|
H2A clustered histone 8
|
|
chr19_-_20661563
Show fit
|
0.04 |
ENST00000601440.6
ENST00000291750.6
ENST00000595094.1
|
ZNF626
ENSG00000269110.1
|
zinc finger protein 626
novel transcript
|
|
chr20_-_3781440
Show fit
|
0.04 |
ENST00000379756.3
|
SPEF1
|
sperm flagellar 1
|
|
chr16_-_55833085
Show fit
|
0.04 |
ENST00000360526.8
|
CES1
|
carboxylesterase 1
|
|
chr6_+_149566356
Show fit
|
0.04 |
ENST00000367419.10
|
GINM1
|
glycoprotein integral membrane 1
|
|
chr8_+_232137
Show fit
|
0.04 |
ENST00000521145.5
ENST00000320552.6
ENST00000308811.8
ENST00000640035.1
ENST00000522866.5
|
ZNF596
|
zinc finger protein 596
|
|
chr6_-_26216673
Show fit
|
0.04 |
ENST00000541790.3
|
H2BC8
|
H2B clustered histone 8
|
|
chr12_-_10130241
Show fit
|
0.04 |
ENST00000353231.9
ENST00000525605.1
|
CLEC7A
|
C-type lectin domain containing 7A
|
|
chr14_+_73097027
Show fit
|
0.03 |
ENST00000532192.1
|
RBM25
|
RNA binding motif protein 25
|
|
chr12_-_10130143
Show fit
|
0.03 |
ENST00000298523.9
ENST00000396484.6
ENST00000310002.4
ENST00000304084.13
|
CLEC7A
|
C-type lectin domain containing 7A
|
|
chr17_-_15619512
Show fit
|
0.03 |
ENST00000395667.7
|
CDRT1
|
CMT1A duplicated region transcript 1
|
|
chr6_+_29461440
Show fit
|
0.03 |
ENST00000396792.2
|
OR2H1
|
olfactory receptor family 2 subfamily H member 1
|
|
chr1_+_10450004
Show fit
|
0.03 |
ENST00000377049.4
|
CORT
|
cortistatin
|
|
chr8_+_28090229
Show fit
|
0.03 |
ENST00000521015.5
ENST00000521570.5
|
ELP3
|
elongator acetyltransferase complex subunit 3
|
|
chr4_-_154550376
Show fit
|
0.03 |
ENST00000302078.9
ENST00000499023.7
|
PLRG1
|
pleiotropic regulator 1
|
|
chr6_-_33711684
Show fit
|
0.03 |
ENST00000374231.8
ENST00000607484.6
|
UQCC2
|
ubiquinol-cytochrome c reductase complex assembly factor 2
|
|
chr12_-_52473798
Show fit
|
0.02 |
ENST00000252250.7
|
KRT6C
|
keratin 6C
|
|
chr7_-_80512041
Show fit
|
0.02 |
ENST00000398291.4
|
GNAT3
|
G protein subunit alpha transducin 3
|
|
chr12_-_7503744
Show fit
|
0.02 |
ENST00000396620.7
ENST00000432237.3
|
CD163
|
CD163 molecule
|
|
chr4_-_185471744
Show fit
|
0.02 |
ENST00000510617.5
|
CCDC110
|
coiled-coil domain containing 110
|
|
chr11_-_4288083
Show fit
|
0.02 |
ENST00000638166.1
|
SSU72P4
|
SSU72 pseudogene 4
|
|
chr17_+_7884783
Show fit
|
0.02 |
ENST00000380358.9
|
CHD3
|
chromodomain helicase DNA binding protein 3
|
|
chr18_+_24426633
Show fit
|
0.02 |
ENST00000648078.1
ENST00000284202.9
|
IMPACT
|
impact RWD domain protein
|
|
chr10_+_93566659
Show fit
|
0.02 |
ENST00000371481.9
ENST00000371483.8
ENST00000604414.1
|
FFAR4
|
free fatty acid receptor 4
|
|
chr3_-_20012250
Show fit
|
0.02 |
ENST00000389050.5
|
PP2D1
|
protein phosphatase 2C like domain containing 1
|
|
chr3_-_38816217
Show fit
|
0.02 |
ENST00000449082.3
ENST00000655275.1
|
SCN10A
|
sodium voltage-gated channel alpha subunit 10
|
|
chr10_-_89643870
Show fit
|
0.02 |
ENST00000322191.10
ENST00000342512.3
|
PANK1
|
pantothenate kinase 1
|
|
chr13_-_35855758
Show fit
|
0.02 |
ENST00000615680.4
|
DCLK1
|
doublecortin like kinase 1
|
|
chr9_-_21187671
Show fit
|
0.02 |
ENST00000421715.2
|
IFNA4
|
interferon alpha 4
|
|
chr6_-_127918540
Show fit
|
0.02 |
ENST00000368250.5
|
THEMIS
|
thymocyte selection associated
|
|
chr14_+_19743571
Show fit
|
0.01 |
ENST00000642117.2
|
OR4Q3
|
olfactory receptor family 4 subfamily Q member 3
|
|
chr16_-_55833186
Show fit
|
0.01 |
ENST00000361503.8
ENST00000422046.6
|
CES1
|
carboxylesterase 1
|