Project
Inflammatory response time course, HUVEC (Wada, 2009)
Navigation
Downloads
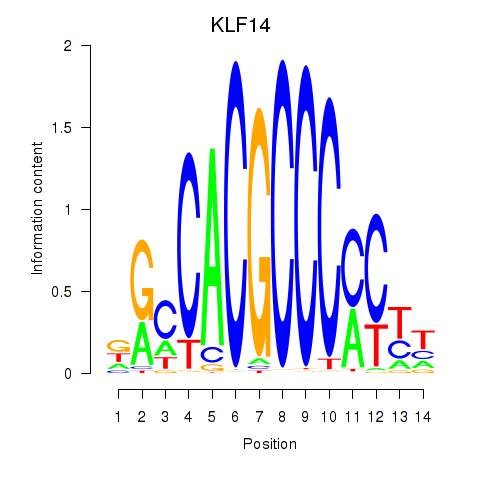
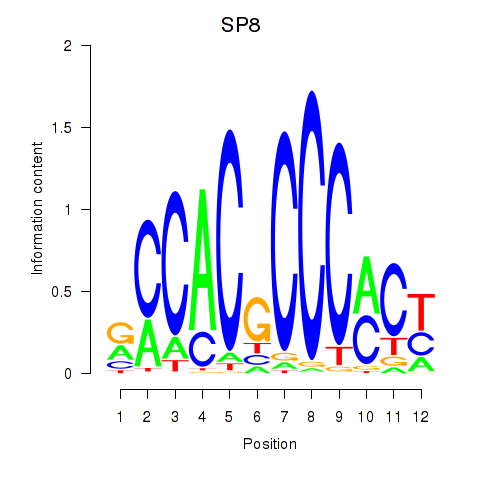
Results for KLF14_SP8
Z-value: 2.03
Transcription factors associated with KLF14_SP8
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
KLF14
|
ENSG00000266265.4 | KLF14 |
|
SP8
|
ENSG00000164651.17 | SP8 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| SP8 | hg38_v1_chr7_-_20786879_20786891 | 0.84 | 1.2e-07 | Click! |
| KLF14 | hg38_v1_chr7_-_130734207_130734207 | -0.38 | 6.3e-02 | Click! |
Activity profile of KLF14_SP8 motif
Sorted Z-values of KLF14_SP8 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of KLF14_SP8
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.3 | 9.9 | GO:0055048 | regulation of spindle elongation(GO:0032887) regulation of mitotic spindle elongation(GO:0032888) anastral spindle assembly(GO:0055048) protein localization to spindle pole body(GO:0071988) regulation of protein localization to spindle pole body(GO:1902363) positive regulation of protein localization to spindle pole body(GO:1902365) positive regulation of mitotic spindle elongation(GO:1902846) |
| 2.4 | 7.2 | GO:1903892 | negative regulation of ATF6-mediated unfolded protein response(GO:1903892) |
| 2.0 | 2.0 | GO:1903384 | neuron intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:0036482) positive regulation of mitochondrial electron transport, NADH to ubiquinone(GO:1902958) regulation of hydrogen peroxide-induced neuron intrinsic apoptotic signaling pathway(GO:1903383) negative regulation of hydrogen peroxide-induced neuron intrinsic apoptotic signaling pathway(GO:1903384) |
| 1.7 | 6.9 | GO:1904980 | positive regulation of endosome organization(GO:1904980) |
| 1.7 | 5.0 | GO:0038162 | erythropoietin-mediated signaling pathway(GO:0038162) |
| 1.6 | 4.7 | GO:1901420 | negative regulation of response to alcohol(GO:1901420) |
| 1.4 | 5.6 | GO:1904428 | negative regulation of tubulin deacetylation(GO:1904428) |
| 1.2 | 4.8 | GO:1905205 | positive regulation of connective tissue replacement(GO:1905205) |
| 1.2 | 3.6 | GO:0060152 | peroxisome localization(GO:0060151) microtubule-based peroxisome localization(GO:0060152) |
| 1.2 | 7.1 | GO:0061687 | detoxification of inorganic compound(GO:0061687) |
| 1.2 | 3.5 | GO:0032877 | positive regulation of DNA endoreduplication(GO:0032877) |
| 1.2 | 3.5 | GO:0016476 | calcium-dependent cell-matrix adhesion(GO:0016340) regulation of embryonic cell shape(GO:0016476) |
| 1.1 | 4.2 | GO:1903296 | regulation of glutamate secretion, neurotransmission(GO:1903294) positive regulation of glutamate secretion, neurotransmission(GO:1903296) |
| 1.0 | 9.4 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 1.0 | 4.9 | GO:0043988 | histone H3-S28 phosphorylation(GO:0043988) |
| 1.0 | 2.9 | GO:0051793 | medium-chain fatty acid catabolic process(GO:0051793) |
| 0.9 | 3.8 | GO:0006208 | pyrimidine nucleobase catabolic process(GO:0006208) thymine catabolic process(GO:0006210) thymine metabolic process(GO:0019859) |
| 0.9 | 3.6 | GO:1904381 | Golgi apparatus mannose trimming(GO:1904381) |
| 0.8 | 5.0 | GO:1903285 | negative regulation of dopamine uptake involved in synaptic transmission(GO:0051585) norepinephrine uptake(GO:0051620) regulation of norepinephrine uptake(GO:0051621) negative regulation of norepinephrine uptake(GO:0051622) negative regulation of catecholamine uptake involved in synaptic transmission(GO:0051945) regulation of glutathione peroxidase activity(GO:1903282) positive regulation of glutathione peroxidase activity(GO:1903284) positive regulation of hydrogen peroxide catabolic process(GO:1903285) positive regulation of peroxidase activity(GO:2000470) |
| 0.8 | 4.1 | GO:0090118 | receptor-mediated endocytosis of low-density lipoprotein particle involved in cholesterol transport(GO:0090118) |
| 0.8 | 2.3 | GO:2000562 | negative regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000562) |
| 0.8 | 3.0 | GO:0006663 | platelet activating factor biosynthetic process(GO:0006663) |
| 0.7 | 10.3 | GO:0021853 | cerebral cortex GABAergic interneuron migration(GO:0021853) interneuron migration(GO:1904936) |
| 0.7 | 4.3 | GO:0038016 | insulin receptor internalization(GO:0038016) |
| 0.7 | 7.8 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.7 | 6.9 | GO:1901525 | negative regulation of macromitophagy(GO:1901525) |
| 0.7 | 2.1 | GO:0036451 | cap mRNA methylation(GO:0036451) |
| 0.7 | 6.8 | GO:2000601 | positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.7 | 2.0 | GO:0060032 | notochord regression(GO:0060032) |
| 0.7 | 2.7 | GO:0046909 | intermembrane transport(GO:0046909) protein transport from ciliary membrane to plasma membrane(GO:1903445) |
| 0.6 | 1.9 | GO:0051228 | mitotic spindle disassembly(GO:0051228) spindle disassembly(GO:0051230) |
| 0.6 | 2.6 | GO:0097032 | respiratory chain complex II assembly(GO:0034552) mitochondrial respiratory chain complex II assembly(GO:0034553) mitochondrial respiratory chain complex II biogenesis(GO:0097032) |
| 0.6 | 3.8 | GO:0006021 | inositol biosynthetic process(GO:0006021) |
| 0.6 | 2.5 | GO:0044878 | mitotic cytokinesis checkpoint(GO:0044878) |
| 0.6 | 1.9 | GO:0002415 | immunoglobulin transcytosis in epithelial cells mediated by polymeric immunoglobulin receptor(GO:0002415) |
| 0.6 | 1.2 | GO:0071284 | cellular response to lead ion(GO:0071284) |
| 0.6 | 10.3 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.6 | 1.8 | GO:0097156 | fasciculation of motor neuron axon(GO:0097156) |
| 0.6 | 0.6 | GO:0060995 | cell-cell signaling involved in kidney development(GO:0060995) Wnt signaling pathway involved in kidney development(GO:0061289) canonical Wnt signaling pathway involved in metanephric kidney development(GO:0061290) cell-cell signaling involved in metanephros development(GO:0072204) |
| 0.6 | 1.7 | GO:0043973 | histone H3-K4 acetylation(GO:0043973) |
| 0.6 | 2.2 | GO:0061086 | negative regulation of histone H3-K27 methylation(GO:0061086) |
| 0.6 | 1.7 | GO:0035470 | positive regulation of vascular wound healing(GO:0035470) |
| 0.6 | 3.9 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.5 | 1.6 | GO:0002416 | IgG immunoglobulin transcytosis in epithelial cells mediated by FcRn immunoglobulin receptor(GO:0002416) |
| 0.5 | 6.5 | GO:0042492 | gamma-delta T cell differentiation(GO:0042492) |
| 0.5 | 2.7 | GO:0010760 | negative regulation of macrophage chemotaxis(GO:0010760) |
| 0.5 | 4.3 | GO:0060398 | regulation of growth hormone receptor signaling pathway(GO:0060398) |
| 0.5 | 1.1 | GO:1902310 | positive regulation of peptidyl-serine dephosphorylation(GO:1902310) |
| 0.5 | 2.1 | GO:0006669 | sphinganine-1-phosphate biosynthetic process(GO:0006669) |
| 0.5 | 13.2 | GO:1902894 | negative regulation of pri-miRNA transcription from RNA polymerase II promoter(GO:1902894) |
| 0.5 | 4.2 | GO:0038108 | negative regulation of appetite by leptin-mediated signaling pathway(GO:0038108) |
| 0.5 | 2.0 | GO:0021816 | extension of a leading process involved in cell motility in cerebral cortex radial glia guided migration(GO:0021816) |
| 0.5 | 2.0 | GO:0001927 | exocyst assembly(GO:0001927) |
| 0.5 | 1.5 | GO:0006097 | glyoxylate cycle(GO:0006097) |
| 0.5 | 1.4 | GO:0036071 | N-glycan fucosylation(GO:0036071) |
| 0.5 | 1.4 | GO:2001151 | regulation of renal water transport(GO:2001151) positive regulation of renal water transport(GO:2001153) |
| 0.5 | 1.9 | GO:0006272 | leading strand elongation(GO:0006272) |
| 0.5 | 3.7 | GO:1903300 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.5 | 1.4 | GO:1900169 | regulation of glucocorticoid mediated signaling pathway(GO:1900169) |
| 0.4 | 1.8 | GO:0060992 | response to fungicide(GO:0060992) |
| 0.4 | 1.8 | GO:0015770 | disaccharide transport(GO:0015766) sucrose transport(GO:0015770) oligosaccharide transport(GO:0015772) |
| 0.4 | 0.4 | GO:0002086 | diaphragm contraction(GO:0002086) |
| 0.4 | 6.0 | GO:0043983 | histone H4-K12 acetylation(GO:0043983) |
| 0.4 | 1.7 | GO:0061511 | centriole elongation(GO:0061511) |
| 0.4 | 5.5 | GO:0044387 | negative regulation of protein kinase activity by regulation of protein phosphorylation(GO:0044387) |
| 0.4 | 2.5 | GO:0033489 | cholesterol biosynthetic process via desmosterol(GO:0033489) cholesterol biosynthetic process via lathosterol(GO:0033490) |
| 0.4 | 2.1 | GO:1901093 | regulation of protein tetramerization(GO:1901090) negative regulation of protein tetramerization(GO:1901091) regulation of protein homotetramerization(GO:1901093) negative regulation of protein homotetramerization(GO:1901094) |
| 0.4 | 9.1 | GO:0002115 | store-operated calcium entry(GO:0002115) |
| 0.4 | 1.2 | GO:0000103 | sulfate assimilation(GO:0000103) |
| 0.4 | 5.3 | GO:0031580 | membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) |
| 0.4 | 2.8 | GO:0090206 | negative regulation of cholesterol biosynthetic process(GO:0045541) negative regulation of cholesterol metabolic process(GO:0090206) |
| 0.4 | 0.4 | GO:0060689 | cell differentiation involved in salivary gland development(GO:0060689) |
| 0.4 | 2.0 | GO:0000379 | tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 0.4 | 3.5 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.4 | 1.2 | GO:2000777 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process involved in cellular response to hypoxia(GO:2000777) |
| 0.4 | 0.4 | GO:2000417 | negative regulation of eosinophil migration(GO:2000417) |
| 0.4 | 2.3 | GO:0044339 | canonical Wnt signaling pathway involved in osteoblast differentiation(GO:0044339) |
| 0.4 | 1.1 | GO:0031247 | actin rod assembly(GO:0031247) |
| 0.4 | 1.1 | GO:0021718 | superior olivary nucleus development(GO:0021718) superior olivary nucleus maturation(GO:0021722) |
| 0.4 | 1.1 | GO:0009085 | lysine biosynthetic process(GO:0009085) lysine biosynthetic process via aminoadipic acid(GO:0019878) |
| 0.4 | 1.8 | GO:0090245 | axis elongation involved in somitogenesis(GO:0090245) |
| 0.4 | 1.5 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.4 | 1.8 | GO:0093001 | glycolysis from storage polysaccharide through glucose-1-phosphate(GO:0093001) |
| 0.4 | 2.5 | GO:0097338 | response to clozapine(GO:0097338) |
| 0.4 | 1.1 | GO:0035674 | tricarboxylic acid transmembrane transport(GO:0035674) |
| 0.3 | 2.1 | GO:0016128 | ergosterol biosynthetic process(GO:0006696) ergosterol metabolic process(GO:0008204) phytosteroid metabolic process(GO:0016128) phytosteroid biosynthetic process(GO:0016129) |
| 0.3 | 1.0 | GO:0007314 | oocyte construction(GO:0007308) oocyte axis specification(GO:0007309) oocyte anterior/posterior axis specification(GO:0007314) pole plasm assembly(GO:0007315) maternal determination of anterior/posterior axis, embryo(GO:0008358) P granule organization(GO:0030719) |
| 0.3 | 3.7 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.3 | 0.3 | GO:1902954 | regulation of early endosome to recycling endosome transport(GO:1902954) |
| 0.3 | 2.0 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.3 | 3.3 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.3 | 1.0 | GO:1901301 | regulation of cargo loading into COPII-coated vesicle(GO:1901301) |
| 0.3 | 1.0 | GO:0019427 | acetate biosynthetic process(GO:0019413) acetyl-CoA biosynthetic process from acetate(GO:0019427) propionate biosynthetic process(GO:0019542) |
| 0.3 | 3.6 | GO:0046826 | negative regulation of protein export from nucleus(GO:0046826) |
| 0.3 | 2.9 | GO:0071376 | response to corticotropin-releasing hormone(GO:0043435) cellular response to corticotropin-releasing hormone stimulus(GO:0071376) |
| 0.3 | 1.0 | GO:1905051 | regulation of base-excision repair(GO:1905051) positive regulation of base-excision repair(GO:1905053) |
| 0.3 | 2.6 | GO:0038028 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) |
| 0.3 | 1.3 | GO:2000314 | regulation of neural crest formation(GO:0090299) negative regulation of neural crest formation(GO:0090301) negative regulation of fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:2000314) |
| 0.3 | 0.9 | GO:0098758 | regulation of interleukin-1 biosynthetic process(GO:0045360) positive regulation of interleukin-1 biosynthetic process(GO:0045362) interleukin-1 beta biosynthetic process(GO:0050720) response to interleukin-8(GO:0098758) cellular response to interleukin-8(GO:0098759) regulation of progesterone biosynthetic process(GO:2000182) |
| 0.3 | 1.2 | GO:0070221 | sulfide oxidation(GO:0019418) sulfide oxidation, using sulfide:quinone oxidoreductase(GO:0070221) |
| 0.3 | 1.9 | GO:0010748 | regulation of plasma membrane long-chain fatty acid transport(GO:0010746) negative regulation of plasma membrane long-chain fatty acid transport(GO:0010748) |
| 0.3 | 3.7 | GO:1904896 | ESCRT complex disassembly(GO:1904896) ESCRT III complex disassembly(GO:1904903) |
| 0.3 | 2.1 | GO:0015798 | myo-inositol transport(GO:0015798) |
| 0.3 | 0.9 | GO:0090234 | regulation of kinetochore assembly(GO:0090234) |
| 0.3 | 0.9 | GO:0030327 | prenylated protein catabolic process(GO:0030327) |
| 0.3 | 0.9 | GO:0070124 | mitochondrial translational initiation(GO:0070124) |
| 0.3 | 1.2 | GO:0021592 | fourth ventricle development(GO:0021592) |
| 0.3 | 2.0 | GO:0018406 | protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 0.3 | 6.2 | GO:0070734 | histone H3-K27 methylation(GO:0070734) |
| 0.3 | 2.3 | GO:0003138 | primary heart field specification(GO:0003138) sinoatrial valve development(GO:0003172) sinoatrial valve morphogenesis(GO:0003185) |
| 0.3 | 4.8 | GO:0036149 | phosphatidylinositol acyl-chain remodeling(GO:0036149) |
| 0.3 | 1.7 | GO:0033277 | abortive mitotic cell cycle(GO:0033277) |
| 0.3 | 8.8 | GO:0060065 | uterus development(GO:0060065) |
| 0.3 | 7.9 | GO:0045898 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) |
| 0.3 | 2.2 | GO:0038033 | positive regulation of endothelial cell chemotaxis by VEGF-activated vascular endothelial growth factor receptor signaling pathway(GO:0038033) |
| 0.3 | 0.8 | GO:0035494 | SNARE complex disassembly(GO:0035494) |
| 0.3 | 1.3 | GO:0060355 | positive regulation of cell adhesion molecule production(GO:0060355) |
| 0.3 | 1.1 | GO:0061083 | regulation of protein refolding(GO:0061083) negative regulation of protein refolding(GO:0061084) |
| 0.3 | 1.3 | GO:0046512 | diol biosynthetic process(GO:0034312) sphingosine biosynthetic process(GO:0046512) |
| 0.3 | 1.8 | GO:1902075 | cellular response to salt(GO:1902075) |
| 0.3 | 0.8 | GO:0010808 | positive regulation of synaptic vesicle priming(GO:0010808) |
| 0.3 | 2.1 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 0.3 | 0.8 | GO:0002528 | regulation of vascular permeability involved in acute inflammatory response(GO:0002528) |
| 0.3 | 0.8 | GO:0046416 | D-amino acid metabolic process(GO:0046416) |
| 0.3 | 2.3 | GO:1904684 | negative regulation of metalloendopeptidase activity(GO:1904684) |
| 0.3 | 2.5 | GO:0046600 | negative regulation of centriole replication(GO:0046600) |
| 0.2 | 1.5 | GO:0097114 | striatal medium spiny neuron differentiation(GO:0021773) NMDA glutamate receptor clustering(GO:0097114) |
| 0.2 | 0.7 | GO:0034729 | histone H3-K79 methylation(GO:0034729) |
| 0.2 | 2.5 | GO:0032525 | somite rostral/caudal axis specification(GO:0032525) |
| 0.2 | 2.0 | GO:0032803 | regulation of low-density lipoprotein particle receptor catabolic process(GO:0032803) |
| 0.2 | 7.5 | GO:0051412 | response to corticosterone(GO:0051412) |
| 0.2 | 1.0 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.2 | 1.2 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.2 | 0.5 | GO:0072720 | response to dithiothreitol(GO:0072720) |
| 0.2 | 0.7 | GO:0061763 | multivesicular body-lysosome fusion(GO:0061763) |
| 0.2 | 0.5 | GO:0042636 | negative regulation of hair cycle(GO:0042636) |
| 0.2 | 0.7 | GO:1904908 | negative regulation of maintenance of sister chromatid cohesion(GO:0034092) negative regulation of maintenance of mitotic sister chromatid cohesion(GO:0034183) maintenance of mitotic sister chromatid cohesion, telomeric(GO:0099403) mitotic sister chromatid cohesion, telomeric(GO:0099404) regulation of maintenance of mitotic sister chromatid cohesion, telomeric(GO:1904907) negative regulation of maintenance of mitotic sister chromatid cohesion, telomeric(GO:1904908) |
| 0.2 | 1.1 | GO:2001295 | malonyl-CoA biosynthetic process(GO:2001295) |
| 0.2 | 1.6 | GO:0030421 | defecation(GO:0030421) |
| 0.2 | 2.2 | GO:0016139 | glycoside catabolic process(GO:0016139) |
| 0.2 | 1.7 | GO:0035021 | negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.2 | 1.9 | GO:0060754 | positive regulation of mast cell chemotaxis(GO:0060754) |
| 0.2 | 0.6 | GO:1902683 | regulation of receptor localization to synapse(GO:1902683) |
| 0.2 | 0.6 | GO:0018312 | peptidyl-serine ADP-ribosylation(GO:0018312) |
| 0.2 | 1.0 | GO:0016480 | negative regulation of transcription from RNA polymerase III promoter(GO:0016480) |
| 0.2 | 0.6 | GO:0048633 | positive regulation of skeletal muscle tissue growth(GO:0048633) |
| 0.2 | 0.8 | GO:0051257 | meiotic metaphase I plate congression(GO:0043060) meiotic spindle midzone assembly(GO:0051257) meiotic metaphase plate congression(GO:0051311) |
| 0.2 | 1.0 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.2 | 1.0 | GO:0014886 | transition between slow and fast fiber(GO:0014886) |
| 0.2 | 0.6 | GO:0007113 | endomitotic cell cycle(GO:0007113) |
| 0.2 | 0.4 | GO:0060431 | primary lung bud formation(GO:0060431) |
| 0.2 | 1.6 | GO:1901187 | regulation of ephrin receptor signaling pathway(GO:1901187) |
| 0.2 | 0.8 | GO:0018076 | N-terminal peptidyl-lysine acetylation(GO:0018076) |
| 0.2 | 1.8 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.2 | 1.8 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.2 | 2.9 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.2 | 0.6 | GO:0015917 | aminophospholipid transport(GO:0015917) |
| 0.2 | 4.8 | GO:0014733 | regulation of skeletal muscle adaptation(GO:0014733) |
| 0.2 | 0.8 | GO:0043585 | negative regulation of histone deacetylation(GO:0031064) nose morphogenesis(GO:0043585) |
| 0.2 | 0.6 | GO:1902961 | regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902959) positive regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902961) regulation of aspartic-type peptidase activity(GO:1905245) positive regulation of aspartic-type peptidase activity(GO:1905247) |
| 0.2 | 0.6 | GO:0097274 | urea homeostasis(GO:0097274) |
| 0.2 | 0.9 | GO:0035645 | enteric smooth muscle cell differentiation(GO:0035645) |
| 0.2 | 0.9 | GO:2001181 | positive regulation of interleukin-10 secretion(GO:2001181) |
| 0.2 | 0.4 | GO:1903061 | positive regulation of lipoprotein metabolic process(GO:0050747) positive regulation of protein lipidation(GO:1903061) |
| 0.2 | 1.9 | GO:0050861 | positive regulation of B cell receptor signaling pathway(GO:0050861) |
| 0.2 | 0.6 | GO:0050760 | negative regulation of thymidylate synthase biosynthetic process(GO:0050760) |
| 0.2 | 2.9 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.2 | 0.5 | GO:0021503 | neural fold bending(GO:0021503) |
| 0.2 | 0.2 | GO:1904868 | telomerase catalytic core complex assembly(GO:1904868) regulation of telomerase catalytic core complex assembly(GO:1904882) positive regulation of telomerase catalytic core complex assembly(GO:1904884) |
| 0.2 | 2.6 | GO:0045003 | DNA recombinase assembly(GO:0000730) double-strand break repair via synthesis-dependent strand annealing(GO:0045003) |
| 0.2 | 0.7 | GO:0044313 | protein K6-linked deubiquitination(GO:0044313) |
| 0.2 | 0.7 | GO:1990166 | protein localization to site of double-strand break(GO:1990166) |
| 0.2 | 1.5 | GO:0007172 | signal complex assembly(GO:0007172) |
| 0.2 | 1.0 | GO:0070829 | response to vitamin B2(GO:0033274) heterochromatin maintenance(GO:0070829) |
| 0.2 | 1.2 | GO:1902231 | positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902231) |
| 0.2 | 1.0 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.2 | 0.7 | GO:0031455 | glycine betaine biosynthetic process from choline(GO:0019285) glycine betaine metabolic process(GO:0031455) glycine betaine biosynthetic process(GO:0031456) |
| 0.2 | 1.8 | GO:0030050 | vesicle transport along actin filament(GO:0030050) |
| 0.2 | 0.3 | GO:0090526 | regulation of gluconeogenesis involved in cellular glucose homeostasis(GO:0090526) |
| 0.2 | 0.3 | GO:0045653 | negative regulation of megakaryocyte differentiation(GO:0045653) |
| 0.2 | 2.4 | GO:0021891 | olfactory bulb interneuron development(GO:0021891) |
| 0.2 | 1.4 | GO:0060179 | male mating behavior(GO:0060179) |
| 0.2 | 1.4 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.2 | 1.4 | GO:0070673 | response to interleukin-18(GO:0070673) |
| 0.2 | 1.9 | GO:0044829 | positive regulation by host of viral genome replication(GO:0044829) |
| 0.2 | 0.9 | GO:1903553 | positive regulation of extracellular exosome assembly(GO:1903553) |
| 0.2 | 1.7 | GO:0010533 | regulation of activation of Janus kinase activity(GO:0010533) |
| 0.1 | 0.4 | GO:0038193 | thromboxane A2 signaling pathway(GO:0038193) |
| 0.1 | 0.4 | GO:1900113 | negative regulation of histone H3-K9 trimethylation(GO:1900113) |
| 0.1 | 0.1 | GO:2000301 | negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.1 | 0.6 | GO:0048073 | regulation of eye pigmentation(GO:0048073) |
| 0.1 | 0.4 | GO:0009786 | regulation of asymmetric cell division(GO:0009786) |
| 0.1 | 0.4 | GO:0042412 | taurine biosynthetic process(GO:0042412) |
| 0.1 | 0.6 | GO:0006550 | isoleucine catabolic process(GO:0006550) |
| 0.1 | 1.0 | GO:0061343 | cell adhesion involved in heart morphogenesis(GO:0061343) |
| 0.1 | 0.1 | GO:1900369 | negative regulation of RNA interference(GO:1900369) |
| 0.1 | 1.1 | GO:0035583 | sequestering of TGFbeta in extracellular matrix(GO:0035583) |
| 0.1 | 1.0 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.1 | 0.6 | GO:0006288 | base-excision repair, DNA ligation(GO:0006288) |
| 0.1 | 3.1 | GO:0016254 | preassembly of GPI anchor in ER membrane(GO:0016254) |
| 0.1 | 1.4 | GO:0043555 | regulation of translation in response to stress(GO:0043555) |
| 0.1 | 3.6 | GO:0030208 | dermatan sulfate biosynthetic process(GO:0030208) |
| 0.1 | 0.5 | GO:0097069 | cellular response to thyroxine stimulus(GO:0097069) cellular response to L-phenylalanine derivative(GO:1904387) |
| 0.1 | 0.7 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.1 | 0.9 | GO:0075071 | autophagy of host cells involved in interaction with symbiont(GO:0075044) autophagy involved in symbiotic interaction(GO:0075071) |
| 0.1 | 0.4 | GO:0030221 | basophil differentiation(GO:0030221) |
| 0.1 | 2.9 | GO:0000052 | citrulline metabolic process(GO:0000052) |
| 0.1 | 0.8 | GO:0010796 | regulation of multivesicular body size(GO:0010796) |
| 0.1 | 3.8 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.1 | 0.4 | GO:0090235 | regulation of metaphase plate congression(GO:0090235) |
| 0.1 | 0.6 | GO:0015862 | uridine transport(GO:0015862) |
| 0.1 | 0.5 | GO:0006701 | progesterone biosynthetic process(GO:0006701) |
| 0.1 | 5.8 | GO:0043928 | exonucleolytic nuclear-transcribed mRNA catabolic process involved in deadenylation-dependent decay(GO:0043928) |
| 0.1 | 0.6 | GO:0042264 | peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 0.1 | 4.0 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.1 | 0.6 | GO:0048861 | leukemia inhibitory factor signaling pathway(GO:0048861) |
| 0.1 | 0.2 | GO:0086100 | endothelin receptor signaling pathway(GO:0086100) |
| 0.1 | 0.4 | GO:0007225 | patched ligand maturation(GO:0007225) signal maturation(GO:0035638) |
| 0.1 | 0.4 | GO:0035871 | protein K11-linked deubiquitination(GO:0035871) |
| 0.1 | 0.7 | GO:0070940 | dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 0.1 | 1.3 | GO:0031547 | brain-derived neurotrophic factor receptor signaling pathway(GO:0031547) |
| 0.1 | 1.3 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.1 | 0.5 | GO:0090308 | regulation of methylation-dependent chromatin silencing(GO:0090308) |
| 0.1 | 0.7 | GO:0035965 | cardiolipin acyl-chain remodeling(GO:0035965) |
| 0.1 | 0.3 | GO:0090071 | negative regulation of ribosome biogenesis(GO:0090071) |
| 0.1 | 0.6 | GO:0006574 | valine catabolic process(GO:0006574) |
| 0.1 | 0.2 | GO:0015911 | plasma membrane long-chain fatty acid transport(GO:0015911) |
| 0.1 | 0.8 | GO:0072383 | plus-end-directed vesicle transport along microtubule(GO:0072383) |
| 0.1 | 1.3 | GO:0031936 | negative regulation of chromatin silencing(GO:0031936) |
| 0.1 | 0.3 | GO:0006421 | asparaginyl-tRNA aminoacylation(GO:0006421) |
| 0.1 | 0.7 | GO:0014005 | microglia differentiation(GO:0014004) microglia development(GO:0014005) |
| 0.1 | 0.2 | GO:1904049 | negative regulation of spontaneous neurotransmitter secretion(GO:1904049) |
| 0.1 | 0.1 | GO:1900110 | negative regulation of histone H3-K9 dimethylation(GO:1900110) |
| 0.1 | 0.8 | GO:0070120 | ciliary neurotrophic factor-mediated signaling pathway(GO:0070120) |
| 0.1 | 0.4 | GO:0018199 | peptidyl-glutamine modification(GO:0018199) |
| 0.1 | 2.2 | GO:0035563 | positive regulation of chromatin binding(GO:0035563) |
| 0.1 | 2.3 | GO:0016024 | CDP-diacylglycerol biosynthetic process(GO:0016024) |
| 0.1 | 0.3 | GO:0098507 | polynucleotide 5' dephosphorylation(GO:0098507) |
| 0.1 | 1.4 | GO:1902414 | protein localization to cell junction(GO:1902414) |
| 0.1 | 0.3 | GO:1903674 | regulation of cap-dependent translational initiation(GO:1903674) positive regulation of cap-dependent translational initiation(GO:1903676) |
| 0.1 | 0.8 | GO:0042415 | norepinephrine metabolic process(GO:0042415) |
| 0.1 | 1.4 | GO:0030903 | notochord development(GO:0030903) |
| 0.1 | 2.7 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.1 | 1.2 | GO:0033299 | secretion of lysosomal enzymes(GO:0033299) |
| 0.1 | 2.1 | GO:0010839 | negative regulation of keratinocyte proliferation(GO:0010839) |
| 0.1 | 3.0 | GO:0043153 | entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.1 | 0.2 | GO:0010979 | regulation of vitamin D 24-hydroxylase activity(GO:0010979) positive regulation of vitamin D 24-hydroxylase activity(GO:0010980) |
| 0.1 | 0.8 | GO:0036091 | positive regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0036091) |
| 0.1 | 0.9 | GO:1904179 | regulation of adipose tissue development(GO:1904177) positive regulation of adipose tissue development(GO:1904179) |
| 0.1 | 1.3 | GO:1900028 | negative regulation of ruffle assembly(GO:1900028) |
| 0.1 | 1.0 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.1 | 0.3 | GO:0070370 | heat acclimation(GO:0010286) cellular heat acclimation(GO:0070370) |
| 0.1 | 0.5 | GO:0023016 | signal transduction by trans-phosphorylation(GO:0023016) |
| 0.1 | 0.5 | GO:0003228 | atrial cardiac muscle tissue development(GO:0003228) atrial cardiac muscle tissue morphogenesis(GO:0055009) |
| 0.1 | 0.4 | GO:0070245 | positive regulation of thymocyte apoptotic process(GO:0070245) |
| 0.1 | 1.4 | GO:0042373 | vitamin K metabolic process(GO:0042373) |
| 0.1 | 1.3 | GO:0098535 | de novo centriole assembly(GO:0098535) |
| 0.1 | 1.0 | GO:0000733 | DNA strand renaturation(GO:0000733) |
| 0.1 | 1.5 | GO:0033127 | regulation of histone phosphorylation(GO:0033127) |
| 0.1 | 0.2 | GO:0031133 | regulation of axon diameter(GO:0031133) |
| 0.1 | 0.7 | GO:0045629 | positive regulation of T-helper 1 cell differentiation(GO:0045627) negative regulation of T-helper 2 cell differentiation(GO:0045629) negative regulation of monocyte chemotactic protein-1 production(GO:0071638) |
| 0.1 | 0.1 | GO:0002238 | response to molecule of fungal origin(GO:0002238) cellular response to molecule of fungal origin(GO:0071226) |
| 0.1 | 0.3 | GO:0046292 | formaldehyde metabolic process(GO:0046292) |
| 0.1 | 0.2 | GO:0003408 | optic cup formation involved in camera-type eye development(GO:0003408) |
| 0.1 | 0.6 | GO:2000504 | positive regulation of blood vessel remodeling(GO:2000504) |
| 0.1 | 0.7 | GO:1990253 | cellular response to leucine starvation(GO:1990253) |
| 0.1 | 0.1 | GO:1990785 | response to water-immersion restraint stress(GO:1990785) |
| 0.1 | 0.5 | GO:0045048 | protein insertion into ER membrane(GO:0045048) |
| 0.1 | 3.1 | GO:0032094 | response to food(GO:0032094) |
| 0.1 | 1.1 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.1 | 0.2 | GO:1902595 | regulation of DNA replication origin binding(GO:1902595) |
| 0.1 | 4.1 | GO:0045737 | positive regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045737) |
| 0.1 | 0.6 | GO:1901909 | diadenosine polyphosphate catabolic process(GO:0015961) diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 0.1 | 0.1 | GO:0051877 | pigment granule aggregation in cell center(GO:0051877) |
| 0.1 | 0.2 | GO:0022615 | protein to membrane docking(GO:0022615) |
| 0.1 | 0.4 | GO:0035330 | regulation of hippo signaling(GO:0035330) |
| 0.1 | 0.3 | GO:0061355 | Wnt protein secretion(GO:0061355) regulation of Wnt protein secretion(GO:0061356) positive regulation of Wnt protein secretion(GO:0061357) |
| 0.1 | 0.1 | GO:0001827 | inner cell mass cell fate commitment(GO:0001827) |
| 0.1 | 0.9 | GO:0046951 | ketone body biosynthetic process(GO:0046951) |
| 0.1 | 0.5 | GO:1902268 | negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.1 | 0.6 | GO:0070272 | proton-transporting ATP synthase complex assembly(GO:0043461) proton-transporting ATP synthase complex biogenesis(GO:0070272) |
| 0.1 | 1.5 | GO:0006853 | carnitine shuttle(GO:0006853) |
| 0.1 | 0.6 | GO:0014057 | positive regulation of acetylcholine secretion, neurotransmission(GO:0014057) |
| 0.1 | 2.8 | GO:0006817 | phosphate ion transport(GO:0006817) |
| 0.1 | 2.4 | GO:2000811 | negative regulation of anoikis(GO:2000811) |
| 0.1 | 0.3 | GO:0030576 | Cajal body organization(GO:0030576) |
| 0.1 | 0.3 | GO:0007195 | adenylate cyclase-inhibiting dopamine receptor signaling pathway(GO:0007195) |
| 0.1 | 3.2 | GO:1904030 | negative regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045736) negative regulation of cyclin-dependent protein kinase activity(GO:1904030) |
| 0.1 | 0.4 | GO:0090169 | regulation of spindle assembly(GO:0090169) regulation of mitotic spindle assembly(GO:1901673) |
| 0.1 | 0.4 | GO:0021966 | corticospinal neuron axon guidance(GO:0021966) |
| 0.1 | 0.5 | GO:0010792 | DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) |
| 0.1 | 1.6 | GO:0070986 | left/right axis specification(GO:0070986) |
| 0.1 | 2.7 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.1 | 0.9 | GO:0045176 | apical protein localization(GO:0045176) |
| 0.1 | 2.1 | GO:0035162 | embryonic hemopoiesis(GO:0035162) |
| 0.1 | 1.8 | GO:0060213 | regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060211) positive regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060213) |
| 0.1 | 0.6 | GO:2000860 | positive regulation of mineralocorticoid secretion(GO:2000857) positive regulation of aldosterone secretion(GO:2000860) |
| 0.1 | 0.4 | GO:0015742 | alpha-ketoglutarate transport(GO:0015742) |
| 0.1 | 0.6 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.1 | 0.3 | GO:0015910 | peroxisomal long-chain fatty acid import(GO:0015910) |
| 0.1 | 0.3 | GO:1902445 | regulation of mitochondrial membrane permeability involved in programmed necrotic cell death(GO:1902445) |
| 0.1 | 2.7 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) |
| 0.1 | 0.6 | GO:1900262 | regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.1 | 0.1 | GO:0030047 | actin modification(GO:0030047) |
| 0.1 | 1.3 | GO:0030422 | production of siRNA involved in RNA interference(GO:0030422) |
| 0.1 | 0.4 | GO:0033088 | negative regulation of immature T cell proliferation in thymus(GO:0033088) |
| 0.1 | 0.7 | GO:0003150 | muscular septum morphogenesis(GO:0003150) |
| 0.1 | 0.5 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.1 | 2.5 | GO:0043616 | keratinocyte proliferation(GO:0043616) |
| 0.1 | 0.6 | GO:0006689 | ganglioside catabolic process(GO:0006689) |
| 0.1 | 0.3 | GO:1903232 | melanosome assembly(GO:1903232) |
| 0.1 | 0.6 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.1 | 1.1 | GO:0002087 | regulation of respiratory gaseous exchange by neurological system process(GO:0002087) |
| 0.1 | 1.1 | GO:0061179 | negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 0.1 | 0.2 | GO:0021776 | smoothened signaling pathway involved in ventral spinal cord interneuron specification(GO:0021775) smoothened signaling pathway involved in spinal cord motor neuron cell fate specification(GO:0021776) |
| 0.1 | 0.3 | GO:0021798 | forebrain dorsal/ventral pattern formation(GO:0021798) |
| 0.1 | 1.3 | GO:0036119 | response to platelet-derived growth factor(GO:0036119) cellular response to platelet-derived growth factor stimulus(GO:0036120) |
| 0.1 | 0.2 | GO:1904395 | positive regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904395) |
| 0.1 | 0.6 | GO:1901029 | negative regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway(GO:1901029) |
| 0.1 | 1.0 | GO:0034453 | microtubule anchoring(GO:0034453) |
| 0.1 | 0.5 | GO:2000394 | positive regulation of lamellipodium morphogenesis(GO:2000394) |
| 0.1 | 1.3 | GO:0070072 | proton-transporting V-type ATPase complex assembly(GO:0070070) vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.1 | 0.6 | GO:0040034 | regulation of development, heterochronic(GO:0040034) regulation of timing of cell differentiation(GO:0048505) |
| 0.1 | 0.2 | GO:0032431 | activation of phospholipase A2 activity(GO:0032431) |
| 0.1 | 0.3 | GO:0018125 | peptidyl-cysteine methylation(GO:0018125) |
| 0.1 | 1.0 | GO:1904424 | regulation of GTP binding(GO:1904424) |
| 0.1 | 0.1 | GO:0010764 | negative regulation of fibroblast migration(GO:0010764) |
| 0.1 | 0.4 | GO:0000720 | pyrimidine dimer repair by nucleotide-excision repair(GO:0000720) positive regulation of t-circle formation(GO:1904431) |
| 0.1 | 0.6 | GO:0061198 | fungiform papilla formation(GO:0061198) |
| 0.1 | 0.3 | GO:1904569 | regulation of selenocysteine incorporation(GO:1904569) |
| 0.1 | 0.9 | GO:0014051 | gamma-aminobutyric acid secretion(GO:0014051) |
| 0.1 | 2.0 | GO:0032228 | regulation of synaptic transmission, GABAergic(GO:0032228) |
| 0.1 | 0.6 | GO:0046498 | S-adenosylhomocysteine metabolic process(GO:0046498) |
| 0.1 | 1.2 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.1 | 0.7 | GO:1901621 | negative regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901621) |
| 0.1 | 0.9 | GO:0051593 | response to folic acid(GO:0051593) |
| 0.1 | 0.9 | GO:0002052 | positive regulation of neuroblast proliferation(GO:0002052) |
| 0.1 | 0.3 | GO:0070885 | negative regulation of calcineurin-NFAT signaling cascade(GO:0070885) |
| 0.1 | 0.6 | GO:0007512 | adult heart development(GO:0007512) |
| 0.1 | 0.7 | GO:0001955 | blood vessel maturation(GO:0001955) |
| 0.1 | 0.6 | GO:2000270 | negative regulation of fibroblast apoptotic process(GO:2000270) |
| 0.1 | 0.5 | GO:0070537 | histone H2A K63-linked deubiquitination(GO:0070537) |
| 0.1 | 0.5 | GO:0043380 | regulation of memory T cell differentiation(GO:0043380) |
| 0.1 | 1.4 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.1 | 0.2 | GO:0098971 | anterograde dendritic transport of neurotransmitter receptor complex(GO:0098971) axo-dendritic protein transport(GO:0099640) |
| 0.1 | 1.3 | GO:0006105 | succinate metabolic process(GO:0006105) |
| 0.1 | 0.3 | GO:0021678 | third ventricle development(GO:0021678) |
| 0.1 | 0.2 | GO:2001287 | negative regulation of caveolin-mediated endocytosis(GO:2001287) |
| 0.1 | 1.2 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.1 | 0.8 | GO:0051085 | chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.1 | 1.5 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.1 | 0.5 | GO:0006203 | dGTP catabolic process(GO:0006203) dATP catabolic process(GO:0046061) |
| 0.1 | 0.3 | GO:0010968 | regulation of microtubule nucleation(GO:0010968) |
| 0.1 | 0.3 | GO:1901350 | cell-cell signaling involved in cell-cell junction organization(GO:1901350) |
| 0.1 | 2.5 | GO:0007143 | female meiotic division(GO:0007143) |
| 0.1 | 1.0 | GO:0032463 | negative regulation of protein homooligomerization(GO:0032463) |
| 0.1 | 3.1 | GO:0014003 | oligodendrocyte development(GO:0014003) |
| 0.1 | 0.7 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.1 | 1.6 | GO:1904707 | positive regulation of vascular smooth muscle cell proliferation(GO:1904707) |
| 0.1 | 0.3 | GO:0006169 | adenosine salvage(GO:0006169) dATP biosynthetic process(GO:0006175) |
| 0.1 | 0.2 | GO:0070269 | pyroptosis(GO:0070269) |
| 0.1 | 0.6 | GO:0035999 | tetrahydrofolate interconversion(GO:0035999) |
| 0.1 | 8.3 | GO:0022904 | respiratory electron transport chain(GO:0022904) |
| 0.1 | 0.5 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.1 | 0.9 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.1 | 0.7 | GO:0036089 | cleavage furrow formation(GO:0036089) |
| 0.1 | 0.2 | GO:0033591 | response to L-ascorbic acid(GO:0033591) |
| 0.1 | 0.4 | GO:0034499 | late endosome to Golgi transport(GO:0034499) |
| 0.1 | 0.7 | GO:0035608 | protein deglutamylation(GO:0035608) |
| 0.1 | 0.2 | GO:0060455 | negative regulation of gastric acid secretion(GO:0060455) |
| 0.1 | 1.2 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.1 | 0.3 | GO:0070086 | ubiquitin-dependent endocytosis(GO:0070086) |
| 0.1 | 0.5 | GO:0006578 | amino-acid betaine biosynthetic process(GO:0006578) carnitine biosynthetic process(GO:0045329) |
| 0.1 | 0.6 | GO:2000580 | regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.1 | 0.3 | GO:0045204 | MAPK export from nucleus(GO:0045204) |
| 0.1 | 0.3 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.1 | 0.2 | GO:0030950 | establishment or maintenance of actin cytoskeleton polarity(GO:0030950) |
| 0.1 | 0.2 | GO:0018008 | N-terminal peptidyl-glycine N-myristoylation(GO:0018008) |
| 0.1 | 0.7 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.1 | 0.2 | GO:0061739 | protein lipidation involved in autophagosome assembly(GO:0061739) |
| 0.1 | 0.3 | GO:0006931 | substrate-dependent cell migration, cell attachment to substrate(GO:0006931) |
| 0.1 | 0.5 | GO:0022417 | protein maturation by protein folding(GO:0022417) |
| 0.1 | 0.2 | GO:1904058 | positive regulation of sensory perception of pain(GO:1904058) |
| 0.1 | 0.4 | GO:0072718 | response to cisplatin(GO:0072718) |
| 0.1 | 2.4 | GO:1900026 | positive regulation of substrate adhesion-dependent cell spreading(GO:1900026) |
| 0.1 | 0.3 | GO:0007549 | dosage compensation(GO:0007549) dosage compensation by inactivation of X chromosome(GO:0009048) |
| 0.1 | 0.3 | GO:0048820 | hair follicle maturation(GO:0048820) |
| 0.1 | 2.5 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.1 | 2.9 | GO:0050982 | detection of mechanical stimulus(GO:0050982) |
| 0.1 | 1.7 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.0 | 0.3 | GO:0051958 | methotrexate transport(GO:0051958) |
| 0.0 | 0.6 | GO:0036152 | phosphatidylethanolamine acyl-chain remodeling(GO:0036152) |
| 0.0 | 0.2 | GO:0080154 | regulation of fertilization(GO:0080154) |
| 0.0 | 0.2 | GO:0031161 | phosphatidylinositol catabolic process(GO:0031161) |
| 0.0 | 0.7 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.0 | 0.8 | GO:0003016 | respiratory system process(GO:0003016) |
| 0.0 | 0.2 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.0 | 0.9 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.0 | 1.3 | GO:0050718 | positive regulation of interleukin-1 beta secretion(GO:0050718) |
| 0.0 | 0.5 | GO:2000774 | positive regulation of cellular senescence(GO:2000774) |
| 0.0 | 0.3 | GO:0007567 | parturition(GO:0007567) |
| 0.0 | 0.6 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 0.0 | 0.5 | GO:0048227 | plasma membrane to endosome transport(GO:0048227) |
| 0.0 | 0.2 | GO:0016557 | peroxisome membrane biogenesis(GO:0016557) |
| 0.0 | 0.4 | GO:0042074 | cell migration involved in gastrulation(GO:0042074) |
| 0.0 | 0.7 | GO:1902187 | negative regulation of viral release from host cell(GO:1902187) |
| 0.0 | 1.6 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.0 | 4.6 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.0 | 0.2 | GO:1990637 | response to prolactin(GO:1990637) |
| 0.0 | 0.3 | GO:1904354 | negative regulation of telomere capping(GO:1904354) |
| 0.0 | 0.4 | GO:0051026 | chiasma assembly(GO:0051026) |
| 0.0 | 1.5 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.0 | 0.5 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.0 | 0.3 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.0 | 0.5 | GO:2001212 | regulation of vasculogenesis(GO:2001212) |
| 0.0 | 0.2 | GO:0044806 | G-quadruplex DNA unwinding(GO:0044806) |
| 0.0 | 0.6 | GO:0039532 | negative regulation of viral-induced cytoplasmic pattern recognition receptor signaling pathway(GO:0039532) |
| 0.0 | 0.6 | GO:1903504 | regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090266) regulation of mitotic spindle checkpoint(GO:1903504) |
| 0.0 | 0.8 | GO:1900273 | positive regulation of long-term synaptic potentiation(GO:1900273) |
| 0.0 | 0.7 | GO:0035988 | chondrocyte proliferation(GO:0035988) |
| 0.0 | 0.1 | GO:0007538 | primary sex determination(GO:0007538) |
| 0.0 | 0.2 | GO:0046726 | positive regulation by virus of viral protein levels in host cell(GO:0046726) |
| 0.0 | 0.4 | GO:0038180 | nerve growth factor signaling pathway(GO:0038180) |
| 0.0 | 0.2 | GO:0031086 | nuclear-transcribed mRNA catabolic process, deadenylation-independent decay(GO:0031086) deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.0 | 0.3 | GO:2000615 | regulation of histone H3-K9 acetylation(GO:2000615) |
| 0.0 | 0.5 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.0 | 0.0 | GO:0005997 | xylulose metabolic process(GO:0005997) D-xylose metabolic process(GO:0042732) |
| 0.0 | 1.7 | GO:0006699 | bile acid biosynthetic process(GO:0006699) |
| 0.0 | 0.6 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 1.5 | GO:0006362 | transcription elongation from RNA polymerase I promoter(GO:0006362) |
| 0.0 | 0.6 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.0 | 1.1 | GO:0000460 | maturation of 5.8S rRNA(GO:0000460) |
| 0.0 | 0.2 | GO:2000535 | regulation of entry of bacterium into host cell(GO:2000535) |
| 0.0 | 0.5 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.0 | 0.5 | GO:1904714 | regulation of chaperone-mediated autophagy(GO:1904714) |
| 0.0 | 8.0 | GO:0006367 | transcription initiation from RNA polymerase II promoter(GO:0006367) |
| 0.0 | 1.3 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 0.6 | GO:0044030 | regulation of DNA methylation(GO:0044030) |
| 0.0 | 0.4 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.0 | 0.1 | GO:0043335 | protein unfolding(GO:0043335) |
| 0.0 | 0.2 | GO:0032484 | Ral protein signal transduction(GO:0032484) regulation of Ral protein signal transduction(GO:0032485) |
| 0.0 | 0.5 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.0 | 0.1 | GO:0090521 | glomerular visceral epithelial cell migration(GO:0090521) |
| 0.0 | 0.3 | GO:0030202 | heparin metabolic process(GO:0030202) heparin biosynthetic process(GO:0030210) |
| 0.0 | 0.1 | GO:0044240 | multicellular organism lipid catabolic process(GO:0044240) |
| 0.0 | 0.3 | GO:0072663 | protein targeting to peroxisome(GO:0006625) protein localization to peroxisome(GO:0072662) establishment of protein localization to peroxisome(GO:0072663) |
| 0.0 | 0.1 | GO:0006499 | N-terminal protein myristoylation(GO:0006499) |
| 0.0 | 0.6 | GO:0006004 | fucose metabolic process(GO:0006004) |
| 0.0 | 0.6 | GO:0061462 | protein localization to lysosome(GO:0061462) |
| 0.0 | 0.3 | GO:0051415 | interphase microtubule nucleation by interphase microtubule organizing center(GO:0051415) microtubule nucleation by microtubule organizing center(GO:0051418) |
| 0.0 | 0.6 | GO:0035246 | peptidyl-arginine N-methylation(GO:0035246) |
| 0.0 | 0.2 | GO:1904217 | regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904217) positive regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904219) positive regulation of serine C-palmitoyltransferase activity(GO:1904222) |
| 0.0 | 0.5 | GO:0048266 | behavioral response to pain(GO:0048266) |
| 0.0 | 0.3 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.0 | 1.2 | GO:0030262 | apoptotic nuclear changes(GO:0030262) |
| 0.0 | 0.3 | GO:0046618 | drug export(GO:0046618) |
| 0.0 | 0.1 | GO:0034444 | regulation of lipoprotein oxidation(GO:0034442) negative regulation of lipoprotein oxidation(GO:0034443) regulation of plasma lipoprotein particle oxidation(GO:0034444) negative regulation of plasma lipoprotein particle oxidation(GO:0034445) |
| 0.0 | 0.1 | GO:0035519 | protein K29-linked ubiquitination(GO:0035519) |
| 0.0 | 0.2 | GO:0033131 | regulation of glucokinase activity(GO:0033131) regulation of hexokinase activity(GO:1903299) |
| 0.0 | 0.3 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.0 | 0.7 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.0 | 0.3 | GO:0043536 | positive regulation of blood vessel endothelial cell migration(GO:0043536) |
| 0.0 | 0.4 | GO:0060263 | regulation of respiratory burst(GO:0060263) |
| 0.0 | 0.0 | GO:0007500 | mesodermal cell fate determination(GO:0007500) |
| 0.0 | 0.4 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.0 | 0.7 | GO:1901663 | ubiquinone metabolic process(GO:0006743) ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.0 | 0.1 | GO:2000969 | positive regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000969) |
| 0.0 | 0.1 | GO:2000381 | negative regulation of mesoderm development(GO:2000381) |
| 0.0 | 0.1 | GO:1901874 | regulation of post-translational protein modification(GO:1901873) negative regulation of post-translational protein modification(GO:1901874) |
| 0.0 | 0.5 | GO:0043570 | maintenance of DNA repeat elements(GO:0043570) |
| 0.0 | 0.2 | GO:0035093 | spermatogenesis, exchange of chromosomal proteins(GO:0035093) |
| 0.0 | 0.3 | GO:0001561 | fatty acid alpha-oxidation(GO:0001561) |
| 0.0 | 0.3 | GO:1900227 | positive regulation of NLRP3 inflammasome complex assembly(GO:1900227) |
| 0.0 | 0.2 | GO:0070172 | positive regulation of tooth mineralization(GO:0070172) |
| 0.0 | 1.0 | GO:0060976 | coronary vasculature development(GO:0060976) |
| 0.0 | 0.3 | GO:0045603 | positive regulation of endothelial cell differentiation(GO:0045603) |
| 0.0 | 0.1 | GO:0031291 | Ran protein signal transduction(GO:0031291) |
| 0.0 | 1.5 | GO:0036297 | interstrand cross-link repair(GO:0036297) |
| 0.0 | 0.3 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) netrin-activated signaling pathway(GO:0038007) |
| 0.0 | 0.5 | GO:0030497 | fatty acid elongation(GO:0030497) |
| 0.0 | 0.1 | GO:0099525 | presynaptic dense core granule exocytosis(GO:0099525) |
| 0.0 | 1.7 | GO:0006809 | nitric oxide biosynthetic process(GO:0006809) |
| 0.0 | 0.2 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.0 | 0.4 | GO:0016558 | protein import into peroxisome matrix(GO:0016558) |
| 0.0 | 0.5 | GO:0090083 | regulation of inclusion body assembly(GO:0090083) |
| 0.0 | 0.2 | GO:0006616 | SRP-dependent cotranslational protein targeting to membrane, translocation(GO:0006616) |
| 0.0 | 0.5 | GO:2000780 | negative regulation of DNA repair(GO:0045738) negative regulation of double-strand break repair(GO:2000780) |
| 0.0 | 0.9 | GO:0002042 | cell migration involved in sprouting angiogenesis(GO:0002042) |
| 0.0 | 0.3 | GO:0060252 | positive regulation of glial cell proliferation(GO:0060252) |
| 0.0 | 0.1 | GO:0009414 | response to water deprivation(GO:0009414) |
| 0.0 | 0.0 | GO:0048320 | commitment of multipotent stem cells to neuronal lineage in forebrain(GO:0021898) axial mesoderm morphogenesis(GO:0048319) axial mesoderm formation(GO:0048320) |
| 0.0 | 0.3 | GO:0042998 | positive regulation of Golgi to plasma membrane protein transport(GO:0042998) |
| 0.0 | 0.3 | GO:0021942 | radial glia guided migration of Purkinje cell(GO:0021942) |
| 0.0 | 0.1 | GO:0001869 | regulation of complement activation, lectin pathway(GO:0001868) negative regulation of complement activation, lectin pathway(GO:0001869) |
| 0.0 | 1.0 | GO:0045103 | intermediate filament-based process(GO:0045103) |
| 0.0 | 1.7 | GO:0006027 | glycosaminoglycan catabolic process(GO:0006027) |
| 0.0 | 0.2 | GO:0071712 | ER-associated misfolded protein catabolic process(GO:0071712) |
| 0.0 | 1.1 | GO:0008088 | axo-dendritic transport(GO:0008088) |
| 0.0 | 0.9 | GO:0007566 | embryo implantation(GO:0007566) |
| 0.0 | 0.4 | GO:0006506 | GPI anchor metabolic process(GO:0006505) GPI anchor biosynthetic process(GO:0006506) |
| 0.0 | 0.4 | GO:0045070 | positive regulation of viral genome replication(GO:0045070) |
| 0.0 | 1.3 | GO:0000266 | mitochondrial fission(GO:0000266) |
| 0.0 | 0.2 | GO:0045054 | constitutive secretory pathway(GO:0045054) |
| 0.0 | 0.3 | GO:0002138 | retinoic acid biosynthetic process(GO:0002138) |
| 0.0 | 0.3 | GO:1903963 | arachidonic acid secretion(GO:0050482) arachidonate transport(GO:1903963) |
| 0.0 | 0.4 | GO:1901748 | leukotriene D4 metabolic process(GO:1901748) leukotriene D4 biosynthetic process(GO:1901750) |
| 0.0 | 0.2 | GO:0015871 | choline transport(GO:0015871) |
| 0.0 | 1.2 | GO:0030835 | negative regulation of actin filament depolymerization(GO:0030835) negative regulation of protein depolymerization(GO:1901880) |
| 0.0 | 0.3 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.0 | 0.4 | GO:0006228 | UTP biosynthetic process(GO:0006228) |
| 0.0 | 0.2 | GO:0035020 | regulation of Rac protein signal transduction(GO:0035020) |
| 0.0 | 0.0 | GO:0044340 | canonical Wnt signaling pathway involved in regulation of cell proliferation(GO:0044340) |
| 0.0 | 0.1 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.0 | 0.6 | GO:0043304 | regulation of mast cell activation involved in immune response(GO:0033006) regulation of mast cell degranulation(GO:0043304) |
| 0.0 | 0.3 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.0 | 0.2 | GO:0006600 | creatine metabolic process(GO:0006600) |
| 0.0 | 0.3 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 0.4 | GO:0008209 | androgen metabolic process(GO:0008209) |
| 0.0 | 0.3 | GO:0060539 | diaphragm development(GO:0060539) |
| 0.0 | 0.3 | GO:1904776 | regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.0 | 0.2 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.0 | 0.1 | GO:0009305 | protein biotinylation(GO:0009305) response to biotin(GO:0070781) histone biotinylation(GO:0071110) |
| 0.0 | 0.1 | GO:0008215 | spermine metabolic process(GO:0008215) |
| 0.0 | 0.2 | GO:0042670 | retinal cone cell differentiation(GO:0042670) retinal cone cell development(GO:0046549) |
| 0.0 | 0.2 | GO:2000427 | positive regulation of apoptotic cell clearance(GO:2000427) |
| 0.0 | 0.2 | GO:0030091 | protein repair(GO:0030091) |
| 0.0 | 0.1 | GO:0016137 | glycoside metabolic process(GO:0016137) |
| 0.0 | 0.2 | GO:0070932 | histone H3 deacetylation(GO:0070932) |
| 0.0 | 0.6 | GO:0007140 | male meiosis(GO:0007140) |
| 0.0 | 0.3 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.0 | 0.0 | GO:0050651 | dermatan sulfate proteoglycan biosynthetic process(GO:0050651) dermatan sulfate proteoglycan metabolic process(GO:0050655) |
| 0.0 | 0.3 | GO:0045879 | negative regulation of smoothened signaling pathway(GO:0045879) |
| 0.0 | 0.0 | GO:1900126 | negative regulation of hyaluronan biosynthetic process(GO:1900126) |
| 0.0 | 0.3 | GO:0006450 | regulation of translational fidelity(GO:0006450) |
| 0.0 | 0.0 | GO:0030259 | lipid glycosylation(GO:0030259) |
| 0.0 | 2.1 | GO:0007286 | spermatid development(GO:0007286) |
| 0.0 | 0.3 | GO:0048384 | retinoic acid receptor signaling pathway(GO:0048384) |
| 0.0 | 0.3 | GO:0002076 | osteoblast development(GO:0002076) |
| 0.0 | 0.5 | GO:0042044 | fluid transport(GO:0042044) |
| 0.0 | 0.6 | GO:0001662 | behavioral fear response(GO:0001662) behavioral defense response(GO:0002209) |
| 0.0 | 0.1 | GO:0038003 | opioid receptor signaling pathway(GO:0038003) |
| 0.0 | 0.2 | GO:0018206 | peptidyl-methionine modification(GO:0018206) |
| 0.0 | 0.2 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.0 | 0.1 | GO:0010265 | SCF complex assembly(GO:0010265) |
| 0.0 | 0.2 | GO:0002097 | tRNA wobble base modification(GO:0002097) |
| 0.0 | 0.2 | GO:0031034 | myosin filament assembly(GO:0031034) |
| 0.0 | 0.1 | GO:0051697 | protein delipidation(GO:0051697) |
| 0.0 | 0.1 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.0 | 0.2 | GO:0010510 | regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) |
| 0.0 | 0.7 | GO:0030433 | ER-associated ubiquitin-dependent protein catabolic process(GO:0030433) |
| 0.0 | 0.1 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.0 | 0.1 | GO:0090043 | regulation of tubulin deacetylation(GO:0090043) |
| 0.0 | 0.1 | GO:0019323 | pentose-phosphate shunt, non-oxidative branch(GO:0009052) pentose catabolic process(GO:0019323) |
| 0.0 | 0.1 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.0 | 0.0 | GO:0070895 | transposon integration(GO:0070893) regulation of transposon integration(GO:0070894) negative regulation of transposon integration(GO:0070895) |
| 0.0 | 0.6 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.0 | 0.2 | GO:0044804 | nucleophagy(GO:0044804) |
| 0.0 | 0.3 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.0 | 0.0 | GO:0009644 | response to high light intensity(GO:0009644) |
| 0.0 | 0.1 | GO:0034244 | negative regulation of transcription elongation from RNA polymerase II promoter(GO:0034244) |
| 0.0 | 0.1 | GO:0070166 | enamel mineralization(GO:0070166) |
| 0.0 | 0.4 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.0 | 0.3 | GO:0061154 | endothelial tube morphogenesis(GO:0061154) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.3 | 9.9 | GO:0055028 | cortical microtubule(GO:0055028) |
| 0.7 | 3.7 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.7 | 2.8 | GO:0032798 | Swi5-Sfr1 complex(GO:0032798) |
| 0.6 | 8.5 | GO:0034464 | BBSome(GO:0034464) |
| 0.6 | 0.6 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.6 | 2.9 | GO:0032449 | CBM complex(GO:0032449) |
| 0.6 | 3.4 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.5 | 2.7 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.5 | 2.3 | GO:0072558 | NLRP1 inflammasome complex(GO:0072558) |
| 0.5 | 1.4 | GO:0000939 | condensed chromosome inner kinetochore(GO:0000939) |
| 0.4 | 3.6 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.4 | 4.8 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.4 | 0.4 | GO:0044614 | nuclear pore cytoplasmic filaments(GO:0044614) |
| 0.4 | 6.1 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.4 | 2.0 | GO:0031436 | BRCA1-BARD1 complex(GO:0031436) |
| 0.4 | 8.7 | GO:0000786 | nucleosome(GO:0000786) |
| 0.4 | 2.2 | GO:1990393 | 3M complex(GO:1990393) |
| 0.3 | 2.0 | GO:0031501 | mannosyltransferase complex(GO:0031501) |
| 0.3 | 0.9 | GO:0097543 | ciliary inversin compartment(GO:0097543) |
| 0.3 | 0.9 | GO:0000805 | X chromosome(GO:0000805) |
| 0.3 | 0.9 | GO:0097224 | sperm connecting piece(GO:0097224) |
| 0.3 | 3.5 | GO:0016011 | dystroglycan complex(GO:0016011) |
| 0.3 | 0.6 | GO:0090498 | extrinsic component of Golgi membrane(GO:0090498) |
| 0.3 | 1.8 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.3 | 1.8 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.3 | 2.0 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.3 | 2.8 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.3 | 1.4 | GO:0097513 | myosin II filament(GO:0097513) |
| 0.3 | 1.3 | GO:0043291 | RAVE complex(GO:0043291) |
| 0.3 | 1.3 | GO:0045283 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.3 | 6.6 | GO:0070822 | Sin3-type complex(GO:0070822) |
| 0.2 | 1.0 | GO:0071149 | TEAD-2-YAP complex(GO:0071149) |
| 0.2 | 3.9 | GO:0001520 | outer dense fiber(GO:0001520) |
| 0.2 | 2.3 | GO:0016589 | NURF complex(GO:0016589) |
| 0.2 | 0.9 | GO:0097629 | extrinsic component of omegasome membrane(GO:0097629) |
| 0.2 | 2.0 | GO:0070531 | BRCA1-A complex(GO:0070531) |
| 0.2 | 0.9 | GO:0012510 | trans-Golgi network transport vesicle membrane(GO:0012510) |
| 0.2 | 0.7 | GO:0071062 | alphav-beta3 integrin-vitronectin complex(GO:0071062) |
| 0.2 | 6.4 | GO:0000145 | exocyst(GO:0000145) |
| 0.2 | 5.5 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.2 | 2.2 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.2 | 1.0 | GO:0098837 | postsynaptic recycling endosome(GO:0098837) |
| 0.2 | 2.3 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.2 | 0.8 | GO:0005715 | late recombination nodule(GO:0005715) |
| 0.2 | 1.2 | GO:0014802 | terminal cisterna(GO:0014802) |
| 0.2 | 3.4 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.2 | 0.8 | GO:0097059 | CNTFR-CLCF1 complex(GO:0097059) |
| 0.2 | 4.1 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.2 | 3.4 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.2 | 7.3 | GO:0030132 | clathrin coat of coated pit(GO:0030132) |
| 0.2 | 3.7 | GO:0000177 | cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.2 | 0.4 | GO:0061673 | mitotic spindle astral microtubule(GO:0061673) |
| 0.2 | 1.0 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.2 | 1.9 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.2 | 2.7 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.2 | 0.8 | GO:0044308 | axonal spine(GO:0044308) |
| 0.2 | 1.8 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.2 | 6.4 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.2 | 1.7 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.2 | 2.7 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.2 | 0.8 | GO:1990761 | growth cone lamellipodium(GO:1990761) |
| 0.2 | 1.7 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.1 | 3.1 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.1 | 1.2 | GO:0043196 | varicosity(GO:0043196) |
| 0.1 | 0.9 | GO:0071458 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) |
| 0.1 | 1.1 | GO:0044815 | DNA packaging complex(GO:0044815) |
| 0.1 | 0.6 | GO:0034365 | discoidal high-density lipoprotein particle(GO:0034365) |
| 0.1 | 0.6 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 0.1 | 0.4 | GO:0000333 | telomerase catalytic core complex(GO:0000333) |
| 0.1 | 0.5 | GO:0017109 | glutamate-cysteine ligase complex(GO:0017109) |
| 0.1 | 0.3 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.1 | 3.4 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.1 | 1.9 | GO:0097470 | ribbon synapse(GO:0097470) |
| 0.1 | 1.1 | GO:0017059 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.1 | 2.6 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.1 | 5.6 | GO:0016592 | mediator complex(GO:0016592) |
| 0.1 | 0.7 | GO:0042272 | nuclear RNA export factor complex(GO:0042272) |
| 0.1 | 2.5 | GO:0097546 | ciliary base(GO:0097546) |
| 0.1 | 0.1 | GO:0005816 | spindle pole body(GO:0005816) |
| 0.1 | 2.9 | GO:0010369 | chromocenter(GO:0010369) |
| 0.1 | 0.6 | GO:0005958 | DNA-dependent protein kinase-DNA ligase 4 complex(GO:0005958) |
| 0.1 | 0.6 | GO:0070695 | FHF complex(GO:0070695) |
| 0.1 | 1.2 | GO:0001673 | male germ cell nucleus(GO:0001673) |
| 0.1 | 0.3 | GO:1990723 | cytoplasmic periphery of the nuclear pore complex(GO:1990723) |
| 0.1 | 5.9 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.1 | 0.6 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.1 | 5.9 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.1 | 1.8 | GO:0016461 | unconventional myosin complex(GO:0016461) |
| 0.1 | 0.5 | GO:0032302 | MutSbeta complex(GO:0032302) |
| 0.1 | 2.0 | GO:0033276 | transcription factor TFTC complex(GO:0033276) |
| 0.1 | 1.1 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.1 | 0.7 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.1 | 2.3 | GO:0044232 | organelle membrane contact site(GO:0044232) |
| 0.1 | 0.3 | GO:0032002 | interleukin-28 receptor complex(GO:0032002) |
| 0.1 | 3.1 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.1 | 0.5 | GO:1990131 | Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.1 | 0.8 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.1 | 2.6 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.1 | 1.5 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.1 | 5.4 | GO:0045334 | clathrin-coated endocytic vesicle(GO:0045334) |
| 0.1 | 0.5 | GO:0000835 | ER ubiquitin ligase complex(GO:0000835) Hrd1p ubiquitin ligase complex(GO:0000836) |
| 0.1 | 1.0 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.1 | 1.4 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.1 | 1.2 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.1 | 5.3 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.1 | 1.2 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.1 | 1.0 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.1 | 0.6 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.1 | 0.2 | GO:1990075 | periciliary membrane compartment(GO:1990075) |
| 0.1 | 2.5 | GO:0000314 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.1 | 1.0 | GO:0070938 | contractile ring(GO:0070938) |
| 0.1 | 1.8 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.1 | 0.8 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.1 | 0.6 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.1 | 0.4 | GO:0071012 | catalytic step 1 spliceosome(GO:0071012) |
| 0.1 | 0.1 | GO:0098592 | cytoplasmic side of apical plasma membrane(GO:0098592) |
| 0.1 | 0.5 | GO:0033165 | interphotoreceptor matrix(GO:0033165) |
| 0.1 | 0.6 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.1 | 0.4 | GO:0000110 | nucleotide-excision repair factor 1 complex(GO:0000110) |
| 0.1 | 6.2 | GO:0030315 | T-tubule(GO:0030315) |
| 0.1 | 0.5 | GO:0005851 | eukaryotic translation initiation factor 2B complex(GO:0005851) |
| 0.1 | 0.6 | GO:0031390 | Ctf18 RFC-like complex(GO:0031390) |
| 0.1 | 6.2 | GO:0031903 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.1 | 0.2 | GO:1990667 | PCSK9-AnxA2 complex(GO:1990667) |
| 0.1 | 0.5 | GO:1990909 | Wnt signalosome(GO:1990909) |
| 0.1 | 0.4 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.1 | 2.0 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.1 | 6.1 | GO:0042579 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.1 | 0.2 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.1 | 0.3 | GO:0008275 | gamma-tubulin small complex(GO:0008275) |
| 0.1 | 0.4 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.1 | 0.4 | GO:0030868 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 0.1 | 0.5 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.1 | 0.9 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.1 | 1.0 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.1 | 0.9 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 0.1 | 0.2 | GO:0034667 | integrin alpha3-beta1 complex(GO:0034667) |
| 0.1 | 8.8 | GO:0031227 | intrinsic component of endoplasmic reticulum membrane(GO:0031227) |
| 0.0 | 5.3 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.0 | 7.7 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.0 | 1.1 | GO:0043205 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.0 | 0.1 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.0 | 0.7 | GO:0030478 | actin cap(GO:0030478) |
| 0.0 | 0.4 | GO:0070775 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 0.2 | GO:0005854 | nascent polypeptide-associated complex(GO:0005854) |
| 0.0 | 0.3 | GO:0031673 | H zone(GO:0031673) |
| 0.0 | 2.7 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 0.6 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 0.1 | GO:0035189 | Rb-E2F complex(GO:0035189) |
| 0.0 | 0.6 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.7 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.4 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.0 | 0.2 | GO:0002079 | inner acrosomal membrane(GO:0002079) |
| 0.0 | 0.2 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.0 | 2.2 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.0 | 0.3 | GO:0005683 | U7 snRNP(GO:0005683) |
| 0.0 | 0.4 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.0 | 0.2 | GO:0036513 | Derlin-1 retrotranslocation complex(GO:0036513) |
| 0.0 | 1.8 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.0 | 0.4 | GO:0032300 | mismatch repair complex(GO:0032300) |
| 0.0 | 0.6 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.0 | 1.5 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.0 | 0.2 | GO:0071014 | post-mRNA release spliceosomal complex(GO:0071014) |
| 0.0 | 5.3 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 0.5 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.0 | 0.6 | GO:0070578 | RISC-loading complex(GO:0070578) |
| 0.0 | 0.6 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.0 | 0.6 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.0 | 0.4 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.0 | 1.1 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.0 | 0.7 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.0 | 0.3 | GO:0071203 | WASH complex(GO:0071203) |
| 0.0 | 0.5 | GO:0008091 | spectrin(GO:0008091) |
| 0.0 | 2.0 | GO:0031672 | A band(GO:0031672) |
| 0.0 | 0.5 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.0 | 0.4 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.0 | 0.5 | GO:0005652 | nuclear lamina(GO:0005652) |
| 0.0 | 0.0 | GO:0034448 | EGO complex(GO:0034448) |
| 0.0 | 0.5 | GO:0071011 | precatalytic spliceosome(GO:0071011) |
| 0.0 | 0.6 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.0 | 0.3 | GO:0044327 | dendritic spine head(GO:0044327) |
| 0.0 | 21.9 | GO:0016604 | nuclear body(GO:0016604) |
| 0.0 | 3.4 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 0.4 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.0 | 0.0 | GO:0005944 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) |
| 0.0 | 0.3 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.0 | 3.3 | GO:0048770 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 1.3 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.9 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.2 | GO:0031091 | platelet alpha granule(GO:0031091) |
| 0.0 | 0.2 | GO:0044194 | cytolytic granule(GO:0044194) |
| 0.0 | 1.5 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 0.1 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.0 | 0.3 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 0.0 | GO:0070435 | Shc-EGFR complex(GO:0070435) |
| 0.0 | 0.2 | GO:0031254 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.0 | 3.3 | GO:0043296 | apical junction complex(GO:0043296) |
| 0.0 | 0.1 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.0 | 0.4 | GO:0090665 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 0.1 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.0 | 0.2 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.0 | 0.1 | GO:0005971 | ribonucleoside-diphosphate reductase complex(GO:0005971) |
| 0.0 | 0.3 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.0 | 16.9 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
| 0.0 | 1.4 | GO:0101003 | ficolin-1-rich granule membrane(GO:0101003) |
| 0.0 | 0.5 | GO:0030686 | 90S preribosome(GO:0030686) |
| 0.0 | 0.1 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.0 | 0.1 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.2 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 0.3 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.0 | 1.5 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 0.5 | GO:0016235 | aggresome(GO:0016235) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.3 | 6.9 | GO:0005135 | interleukin-3 receptor binding(GO:0005135) |
| 1.0 | 5.0 | GO:0047179 | platelet-activating factor acetyltransferase activity(GO:0047179) |
| 1.0 | 6.9 | GO:0043812 | phosphatidylinositol-4-phosphate phosphatase activity(GO:0043812) |
| 0.8 | 10.8 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.8 | 5.0 | GO:0060961 | phospholipase D inhibitor activity(GO:0060961) |
| 0.8 | 10.2 | GO:0004723 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) |
| 0.8 | 0.8 | GO:0004741 | [pyruvate dehydrogenase (lipoamide)] phosphatase activity(GO:0004741) |
| 0.8 | 3.0 | GO:0047192 | 1-alkylglycerophosphocholine O-acetyltransferase activity(GO:0047192) |
| 0.7 | 2.9 | GO:0070991 | medium-chain-acyl-CoA dehydrogenase activity(GO:0070991) |
| 0.7 | 8.7 | GO:0030976 | thiamine pyrophosphate binding(GO:0030976) |
| 0.7 | 5.0 | GO:0071535 | RING-like zinc finger domain binding(GO:0071535) |
| 0.7 | 2.8 | GO:0047493 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.7 | 3.4 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.7 | 2.1 | GO:0004483 | mRNA (nucleoside-2'-O-)-methyltransferase activity(GO:0004483) |
| 0.7 | 2.0 | GO:0004584 | dolichyl-phosphate-mannose-glycolipid alpha-mannosyltransferase activity(GO:0004584) |
| 0.6 | 6.1 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.6 | 3.5 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.6 | 2.2 | GO:0047708 | biotinidase activity(GO:0047708) |
| 0.5 | 1.6 | GO:0030549 | acetylcholine receptor activator activity(GO:0030549) |
| 0.5 | 4.2 | GO:0015235 | cobalamin transporter activity(GO:0015235) |
| 0.5 | 4.7 | GO:0047144 | 2-acylglycerol-3-phosphate O-acyltransferase activity(GO:0047144) |
| 0.5 | 1.6 | GO:0051990 | (R)-2-hydroxyglutarate dehydrogenase activity(GO:0051990) |
| 0.5 | 4.0 | GO:0052834 | inositol monophosphate 1-phosphatase activity(GO:0008934) inositol monophosphate 3-phosphatase activity(GO:0052832) inositol monophosphate 4-phosphatase activity(GO:0052833) inositol monophosphate phosphatase activity(GO:0052834) |
| 0.5 | 1.4 | GO:0046921 | glycoprotein 6-alpha-L-fucosyltransferase activity(GO:0008424) alpha-(1->6)-fucosyltransferase activity(GO:0046921) |
| 0.5 | 1.9 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.4 | 1.3 | GO:0016434 | rRNA (cytosine) methyltransferase activity(GO:0016434) |
| 0.4 | 1.8 | GO:0015154 | sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.4 | 1.2 | GO:0090541 | MIT domain binding(GO:0090541) |
| 0.4 | 1.6 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.4 | 1.2 | GO:0032093 | SAM domain binding(GO:0032093) |
| 0.4 | 3.2 | GO:0016403 | dimethylargininase activity(GO:0016403) |
| 0.4 | 3.6 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.4 | 1.6 | GO:0001512 | dihydronicotinamide riboside quinone reductase activity(GO:0001512) melatonin binding(GO:1904408) |
| 0.4 | 4.6 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.4 | 3.5 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.4 | 1.1 | GO:0016534 | cyclin-dependent protein kinase 5 activator activity(GO:0016534) |
| 0.4 | 1.1 | GO:0004174 | electron-transferring-flavoprotein dehydrogenase activity(GO:0004174) oxidoreductase activity, acting on the CH-NH group of donors, quinone or similar compound as acceptor(GO:0016649) |
| 0.4 | 1.8 | GO:0008240 | tripeptidyl-peptidase activity(GO:0008240) |
| 0.3 | 1.0 | GO:0072590 | N-acetyl-L-aspartate-L-glutamate ligase activity(GO:0072590) |
| 0.3 | 2.1 | GO:0051996 | farnesyl-diphosphate farnesyltransferase activity(GO:0004310) squalene synthase activity(GO:0051996) |
| 0.3 | 7.9 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.3 | 5.3 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.3 | 5.7 | GO:0004716 | receptor signaling protein tyrosine kinase activity(GO:0004716) |
| 0.3 | 1.3 | GO:0008177 | succinate dehydrogenase (ubiquinone) activity(GO:0008177) |
| 0.3 | 6.4 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 0.3 | 4.8 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.3 | 0.9 | GO:0043035 | chromatin insulator sequence binding(GO:0043035) |
| 0.3 | 2.2 | GO:0005052 | peroxisome matrix targeting signal-1 binding(GO:0005052) |
| 0.3 | 2.8 | GO:0035252 | UDP-xylosyltransferase activity(GO:0035252) xylosyltransferase activity(GO:0042285) |
| 0.3 | 1.2 | GO:0050510 | N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase activity(GO:0050510) |
| 0.3 | 6.7 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.3 | 0.6 | GO:0047184 | 1-acylglycerophosphocholine O-acyltransferase activity(GO:0047184) |
| 0.3 | 2.1 | GO:0017050 | sphinganine kinase activity(GO:0008481) D-erythro-sphingosine kinase activity(GO:0017050) |
| 0.3 | 3.0 | GO:0001162 | RNA polymerase II intronic transcription regulatory region sequence-specific DNA binding(GO:0001162) |
| 0.3 | 1.2 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.3 | 1.4 | GO:0070679 | inositol 1,4,5 trisphosphate binding(GO:0070679) |
| 0.3 | 1.1 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) |
| 0.3 | 1.4 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.3 | 0.8 | GO:0070704 | C-5 sterol desaturase activity(GO:0000248) sterol desaturase activity(GO:0070704) |
| 0.3 | 1.9 | GO:0016900 | oxidoreductase activity, acting on the CH-OH group of donors, disulfide as acceptor(GO:0016900) vitamin-K-epoxide reductase (warfarin-sensitive) activity(GO:0047057) |
| 0.3 | 2.1 | GO:0047756 | chondroitin 4-sulfotransferase activity(GO:0047756) |
| 0.3 | 1.9 | GO:0030267 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 0.3 | 2.1 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.3 | 1.1 | GO:0047291 | neolactotetraosylceramide alpha-2,3-sialyltransferase activity(GO:0004513) lactosylceramide alpha-2,3-sialyltransferase activity(GO:0047291) |
| 0.3 | 1.8 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.2 | 1.0 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.2 | 1.5 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.2 | 1.7 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.2 | 2.9 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.2 | 1.0 | GO:0035033 | histone deacetylase regulator activity(GO:0035033) |
| 0.2 | 1.9 | GO:0005534 | galactose binding(GO:0005534) |
| 0.2 | 0.9 | GO:0004419 | hydroxymethylglutaryl-CoA lyase activity(GO:0004419) |
| 0.2 | 0.5 | GO:0017099 | very-long-chain-acyl-CoA dehydrogenase activity(GO:0017099) |
| 0.2 | 4.7 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.2 | 1.6 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) |
| 0.2 | 0.7 | GO:0003941 | L-serine ammonia-lyase activity(GO:0003941) |
| 0.2 | 1.1 | GO:0003989 | acetyl-CoA carboxylase activity(GO:0003989) |
| 0.2 | 1.1 | GO:0004647 | phosphoserine phosphatase activity(GO:0004647) |
| 0.2 | 1.8 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.2 | 2.0 | GO:0000213 | tRNA-intron endonuclease activity(GO:0000213) |
| 0.2 | 2.0 | GO:0016670 | oxidoreductase activity, acting on a sulfur group of donors, oxygen as acceptor(GO:0016670) |
| 0.2 | 1.1 | GO:0016454 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.2 | 0.6 | GO:0030272 | 5-formyltetrahydrofolate cyclo-ligase activity(GO:0030272) |
| 0.2 | 1.9 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.2 | 1.3 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.2 | 1.7 | GO:0001025 | RNA polymerase III transcription factor binding(GO:0001025) |
| 0.2 | 0.6 | GO:0034739 | histone deacetylase activity (H4-K16 specific)(GO:0034739) |
| 0.2 | 0.6 | GO:0097363 | protein O-GlcNAc transferase activity(GO:0097363) |
| 0.2 | 0.6 | GO:0047389 | glycerophosphocholine phosphodiesterase activity(GO:0047389) |
| 0.2 | 2.3 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.2 | 0.6 | GO:0003860 | 3-hydroxyisobutyryl-CoA hydrolase activity(GO:0003860) |
| 0.2 | 2.9 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.2 | 1.9 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.2 | 3.3 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.2 | 0.5 | GO:0044714 | GTP diphosphatase activity(GO:0036219) 2-hydroxy-adenosine triphosphate pyrophosphatase activity(GO:0044713) 2-hydroxy-(deoxy)adenosine-triphosphate pyrophosphatase activity(GO:0044714) ATP diphosphatase activity(GO:0047693) |
| 0.2 | 1.3 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.2 | 0.5 | GO:0004605 | phosphatidate cytidylyltransferase activity(GO:0004605) |
| 0.2 | 0.7 | GO:0004991 | parathyroid hormone receptor activity(GO:0004991) |
| 0.2 | 1.1 | GO:0015142 | citrate transmembrane transporter activity(GO:0015137) tricarboxylic acid transmembrane transporter activity(GO:0015142) |
| 0.2 | 3.8 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.2 | 1.2 | GO:0004962 | endothelin receptor activity(GO:0004962) |
| 0.2 | 1.0 | GO:0004489 | methylenetetrahydrofolate reductase (NAD(P)H) activity(GO:0004489) |
| 0.2 | 1.5 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.2 | 0.7 | GO:0001626 | nociceptin receptor activity(GO:0001626) |
| 0.2 | 1.5 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.2 | 1.4 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 0.2 | 0.9 | GO:0070568 | guanylyltransferase activity(GO:0070568) |
| 0.2 | 1.1 | GO:0004075 | biotin carboxylase activity(GO:0004075) |
| 0.2 | 0.9 | GO:0044729 | hemi-methylated DNA-binding(GO:0044729) |
| 0.2 | 0.8 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) |
| 0.2 | 0.5 | GO:0001537 | N-acetylgalactosamine 4-O-sulfotransferase activity(GO:0001537) |
| 0.2 | 0.8 | GO:0015199 | amino-acid betaine transmembrane transporter activity(GO:0015199) carnitine transmembrane transporter activity(GO:0015226) |
| 0.2 | 1.1 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 0.2 | 0.6 | GO:0052836 | inositol 5-diphosphate pentakisphosphate 5-kinase activity(GO:0052836) inositol diphosphate tetrakisphosphate kinase activity(GO:0052839) |
| 0.1 | 0.4 | GO:0004961 | thromboxane receptor activity(GO:0004960) thromboxane A2 receptor activity(GO:0004961) |
| 0.1 | 0.4 | GO:0015140 | malate transmembrane transporter activity(GO:0015140) |
| 0.1 | 1.0 | GO:0004305 | ethanolamine kinase activity(GO:0004305) |
| 0.1 | 7.5 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.1 | 3.5 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.1 | 1.0 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) oxidoreductase activity, acting on NAD(P)H, heme protein as acceptor(GO:0016653) |
| 0.1 | 0.9 | GO:0042301 | phosphate ion binding(GO:0042301) |
| 0.1 | 0.4 | GO:0004782 | sulfinoalanine decarboxylase activity(GO:0004782) |
| 0.1 | 1.3 | GO:0000403 | Y-form DNA binding(GO:0000403) |
| 0.1 | 0.5 | GO:0004357 | glutamate-cysteine ligase activity(GO:0004357) |
| 0.1 | 0.6 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.1 | 1.7 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.1 | 4.5 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.1 | 0.6 | GO:0048408 | epidermal growth factor binding(GO:0048408) |
| 0.1 | 0.5 | GO:0004739 | pyruvate dehydrogenase (acetyl-transferring) activity(GO:0004739) |
| 0.1 | 1.9 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.1 | 1.4 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.1 | 0.8 | GO:0032137 | guanine/thymine mispair binding(GO:0032137) |
| 0.1 | 0.7 | GO:0070004 | cysteine-type carboxypeptidase activity(GO:0016807) cysteine-type exopeptidase activity(GO:0070004) |
| 0.1 | 2.3 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.1 | 0.6 | GO:0001888 | glucuronyl-galactosyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0001888) |
| 0.1 | 2.0 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.1 | 3.8 | GO:0050750 | low-density lipoprotein particle receptor binding(GO:0050750) |
| 0.1 | 0.6 | GO:0046624 | sphingolipid transporter activity(GO:0046624) |
| 0.1 | 0.6 | GO:0050436 | microfibril binding(GO:0050436) |
| 0.1 | 0.5 | GO:0047391 | alkylglycerophosphoethanolamine phosphodiesterase activity(GO:0047391) |
| 0.1 | 0.6 | GO:0052656 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.1 | 0.6 | GO:0052795 | exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 0.1 | 0.8 | GO:0001224 | RNA polymerase II transcription cofactor binding(GO:0001224) RNA polymerase II transcription coactivator binding(GO:0001225) |
| 0.1 | 0.3 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.1 | 0.9 | GO:0009008 | DNA-methyltransferase activity(GO:0009008) |
| 0.1 | 0.4 | GO:0032450 | maltose alpha-glucosidase activity(GO:0032450) |
| 0.1 | 2.8 | GO:0071949 | FAD binding(GO:0071949) |
| 0.1 | 0.9 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.1 | 0.5 | GO:0000828 | inositol-1,3,4,5,6-pentakisphosphate kinase activity(GO:0000827) inositol hexakisphosphate kinase activity(GO:0000828) inositol heptakisphosphate kinase activity(GO:0000829) inositol hexakisphosphate 5-kinase activity(GO:0000832) inositol hexakisphosphate 1-kinase activity(GO:0052723) inositol hexakisphosphate 3-kinase activity(GO:0052724) |
| 0.1 | 1.1 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.1 | 1.0 | GO:0036310 | annealing helicase activity(GO:0036310) |
| 0.1 | 0.3 | GO:0071209 | U7 snRNA binding(GO:0071209) |
| 0.1 | 0.4 | GO:0033897 | ribonuclease T2 activity(GO:0033897) |
| 0.1 | 0.3 | GO:0047750 | cholestenol delta-isomerase activity(GO:0047750) |
| 0.1 | 1.1 | GO:0072542 | protein phosphatase activator activity(GO:0072542) |
| 0.1 | 0.6 | GO:0052844 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) inositol diphosphate tetrakisphosphate diphosphatase activity(GO:0052840) inositol bisdiphosphate tetrakisphosphate diphosphatase activity(GO:0052841) inositol diphosphate pentakisphosphate diphosphatase activity(GO:0052842) inositol-1-diphosphate-2,3,4,5,6-pentakisphosphate diphosphatase activity(GO:0052843) inositol-3-diphosphate-1,2,4,5,6-pentakisphosphate diphosphatase activity(GO:0052844) inositol-5-diphosphate-1,2,3,4,6-pentakisphosphate diphosphatase activity(GO:0052845) inositol-1,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 1-diphosphatase activity(GO:0052846) inositol-1,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 5-diphosphatase activity(GO:0052847) inositol-3,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 5-diphosphatase activity(GO:0052848) |
| 0.1 | 0.6 | GO:0090554 | phosphatidylcholine-translocating ATPase activity(GO:0090554) |
| 0.1 | 0.3 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.1 | 1.5 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.1 | 0.8 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.1 | 0.8 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.1 | 1.6 | GO:0016755 | transferase activity, transferring amino-acyl groups(GO:0016755) |
| 0.1 | 0.5 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.1 | 0.4 | GO:0010385 | double-stranded methylated DNA binding(GO:0010385) |
| 0.1 | 1.8 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.1 | 0.6 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.1 | 0.5 | GO:1902444 | riboflavin binding(GO:1902444) |
| 0.1 | 0.5 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.1 | 0.8 | GO:0043426 | MRF binding(GO:0043426) |
| 0.1 | 1.5 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.1 | 0.4 | GO:0004597 | peptide-aspartate beta-dioxygenase activity(GO:0004597) |
| 0.1 | 5.3 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.1 | 1.8 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.1 | 0.9 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.1 | 1.8 | GO:0000030 | mannosyltransferase activity(GO:0000030) |
| 0.1 | 0.3 | GO:0015056 | corticotrophin-releasing factor receptor activity(GO:0015056) |
| 0.1 | 0.1 | GO:0033142 | progesterone receptor binding(GO:0033142) |
| 0.1 | 1.0 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.1 | 0.5 | GO:0004522 | ribonuclease A activity(GO:0004522) |
| 0.1 | 2.7 | GO:0019198 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.1 | 0.9 | GO:0001134 | transcription factor activity, transcription factor recruiting(GO:0001134) |
| 0.1 | 8.2 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.1 | 1.7 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.1 | 0.6 | GO:0004677 | DNA-dependent protein kinase activity(GO:0004677) |
| 0.1 | 4.3 | GO:0016620 | oxidoreductase activity, acting on the aldehyde or oxo group of donors, NAD or NADP as acceptor(GO:0016620) |
| 0.1 | 1.5 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.1 | 0.9 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.1 | 0.7 | GO:0019788 | NEDD8 transferase activity(GO:0019788) |
| 0.1 | 2.3 | GO:0008656 | cysteine-type endopeptidase activator activity involved in apoptotic process(GO:0008656) |
| 0.1 | 0.8 | GO:0004897 | ciliary neurotrophic factor receptor activity(GO:0004897) |
| 0.1 | 1.5 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.1 | 2.5 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.1 | 0.4 | GO:0030346 | protein phosphatase 2B binding(GO:0030346) |
| 0.1 | 0.3 | GO:0051500 | D-aminoacyl-tRNA deacylase activity(GO:0051499) D-tyrosyl-tRNA(Tyr) deacylase activity(GO:0051500) |
| 0.1 | 0.4 | GO:0086062 | voltage-gated sodium channel activity involved in Purkinje myocyte action potential(GO:0086062) |
| 0.1 | 0.3 | GO:0004087 | carbamoyl-phosphate synthase (ammonia) activity(GO:0004087) carbamoyl-phosphate synthase (glutamine-hydrolyzing) activity(GO:0004088) |
| 0.1 | 1.8 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.1 | 1.1 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.1 | 0.5 | GO:0015252 | hydrogen ion channel activity(GO:0015252) |
| 0.1 | 8.9 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.1 | 1.0 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.1 | 2.2 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.1 | 2.5 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.1 | 0.7 | GO:0033592 | RNA strand annealing activity(GO:0033592) |
| 0.1 | 0.3 | GO:0004667 | prostaglandin-D synthase activity(GO:0004667) |
| 0.1 | 0.3 | GO:0061513 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 0.1 | 0.6 | GO:0033170 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.1 | 0.2 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.1 | 7.6 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.1 | 3.9 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.1 | 0.6 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.1 | 0.3 | GO:0004001 | adenosine kinase activity(GO:0004001) |
| 0.1 | 0.6 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.1 | 5.7 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.1 | 2.2 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.1 | 1.2 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.1 | 0.2 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.1 | 1.1 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.1 | 26.0 | GO:0001077 | transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) |
| 0.1 | 2.7 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.1 | 0.6 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.1 | 0.6 | GO:0030023 | extracellular matrix constituent conferring elasticity(GO:0030023) |
| 0.1 | 0.6 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.1 | 1.7 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.1 | 0.9 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.1 | 1.0 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.1 | 2.3 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.1 | 0.2 | GO:0052812 | phosphatidylinositol-3,4-bisphosphate 5-kinase activity(GO:0052812) |
| 0.1 | 0.2 | GO:0004379 | glycylpeptide N-tetradecanoyltransferase activity(GO:0004379) myristoyltransferase activity(GO:0019107) |
| 0.1 | 2.1 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.1 | 0.7 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) |
| 0.1 | 0.3 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.1 | 1.5 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.1 | 0.4 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.1 | 0.5 | GO:0003988 | acetyl-CoA C-acyltransferase activity(GO:0003988) |
| 0.1 | 0.3 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.1 | 4.8 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.1 | 0.4 | GO:0030345 | extracellular matrix structural constituent conferring compression resistance(GO:0030021) structural constituent of tooth enamel(GO:0030345) |
| 0.1 | 0.2 | GO:0016497 | substance K receptor activity(GO:0016497) |
| 0.1 | 1.6 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.1 | 0.2 | GO:1990189 | peptide-serine-N-acetyltransferase activity(GO:1990189) |
| 0.0 | 0.3 | GO:0015350 | methotrexate transporter activity(GO:0015350) |
| 0.0 | 0.3 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.0 | 0.2 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 1.9 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.0 | 0.5 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.0 | 0.4 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 0.0 | 0.6 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.0 | 0.6 | GO:0052744 | phosphatidylinositol-3-phosphatase activity(GO:0004438) phosphatidylinositol monophosphate phosphatase activity(GO:0052744) |
| 0.0 | 0.3 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.0 | 0.4 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.0 | 0.3 | GO:0015307 | drug:proton antiporter activity(GO:0015307) |
| 0.0 | 0.7 | GO:0016918 | retinal binding(GO:0016918) |
| 0.0 | 0.2 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.0 | 1.7 | GO:0016709 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen(GO:0016709) |
| 0.0 | 0.7 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.3 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.0 | 0.4 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.3 | GO:1990599 | 3' overhang single-stranded DNA endodeoxyribonuclease activity(GO:1990599) |
| 0.0 | 0.5 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.0 | 0.4 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.0 | 0.5 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 1.4 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.5 | GO:0022820 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.0 | 0.4 | GO:0097157 | pre-mRNA intronic binding(GO:0097157) |
| 0.0 | 1.5 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.4 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.0 | 0.4 | GO:0019534 | toxin transporter activity(GO:0019534) |
| 0.0 | 0.3 | GO:0035368 | selenocysteine insertion sequence binding(GO:0035368) |
| 0.0 | 0.4 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 0.9 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 1.3 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.2 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.0 | 0.1 | GO:0004750 | ribulose-phosphate 3-epimerase activity(GO:0004750) |
| 0.0 | 2.3 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 1.3 | GO:0008376 | acetylgalactosaminyltransferase activity(GO:0008376) |
| 0.0 | 1.3 | GO:0034062 | RNA polymerase activity(GO:0034062) |
| 0.0 | 0.2 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.0 | 2.1 | GO:0019213 | deacetylase activity(GO:0019213) |
| 0.0 | 0.3 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.1 | GO:0008106 | alcohol dehydrogenase (NADP+) activity(GO:0008106) |
| 0.0 | 0.8 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 0.5 | GO:0017151 | DEAD/H-box RNA helicase binding(GO:0017151) |
| 0.0 | 0.8 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 0.7 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.0 | 0.3 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.0 | 1.0 | GO:0070566 | adenylyltransferase activity(GO:0070566) |
| 0.0 | 0.3 | GO:0071253 | connexin binding(GO:0071253) |
| 0.0 | 0.5 | GO:0016273 | arginine N-methyltransferase activity(GO:0016273) protein-arginine N-methyltransferase activity(GO:0016274) |
| 0.0 | 0.2 | GO:0102345 | 3-hydroxy-behenoyl-CoA dehydratase activity(GO:0102344) 3-hydroxy-lignoceroyl-CoA dehydratase activity(GO:0102345) |
| 0.0 | 0.6 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.4 | GO:0017081 | chloride channel regulator activity(GO:0017081) |
| 0.0 | 0.1 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.0 | 1.1 | GO:0015149 | hexose transmembrane transporter activity(GO:0015149) |
| 0.0 | 0.9 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.3 | GO:0050694 | galactosylceramide sulfotransferase activity(GO:0001733) galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.0 | 1.2 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 0.7 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.2 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.0 | 0.3 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.0 | 1.0 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.1 | GO:0032428 | sphingolipid activator protein activity(GO:0030290) beta-N-acetylgalactosaminidase activity(GO:0032428) |
| 0.0 | 0.4 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.0 | 0.3 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.0 | 1.3 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 0.3 | GO:0005347 | ATP transmembrane transporter activity(GO:0005347) ADP transmembrane transporter activity(GO:0015217) |
| 0.0 | 0.7 | GO:0016881 | acid-amino acid ligase activity(GO:0016881) |
| 0.0 | 0.2 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.0 | 0.2 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.0 | 2.4 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.0 | 0.2 | GO:0031386 | protein tag(GO:0031386) |
| 0.0 | 1.1 | GO:0050660 | flavin adenine dinucleotide binding(GO:0050660) |
| 0.0 | 0.4 | GO:0005337 | nucleoside transmembrane transporter activity(GO:0005337) |
| 0.0 | 0.4 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.9 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.3 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.0 | 0.1 | GO:0019797 | procollagen-proline 3-dioxygenase activity(GO:0019797) |
| 0.0 | 0.6 | GO:0001221 | transcription cofactor binding(GO:0001221) |
| 0.0 | 0.4 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.0 | 0.8 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.0 | 0.3 | GO:0031802 | type 5 metabotropic glutamate receptor binding(GO:0031802) |
| 0.0 | 1.3 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.0 | 0.2 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.0 | 1.2 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 0.1 | GO:0008174 | mRNA methyltransferase activity(GO:0008174) |
| 0.0 | 0.3 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.0 | 0.3 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.0 | 0.1 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.0 | 3.2 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.0 | 2.0 | GO:0004553 | hydrolase activity, hydrolyzing O-glycosyl compounds(GO:0004553) |
| 0.0 | 0.4 | GO:0015197 | peptide transporter activity(GO:0015197) |
| 0.0 | 2.3 | GO:0004721 | phosphoprotein phosphatase activity(GO:0004721) |
| 0.0 | 0.3 | GO:0016780 | phosphotransferase activity, for other substituted phosphate groups(GO:0016780) |
| 0.0 | 0.1 | GO:0004078 | biotin-[acetyl-CoA-carboxylase] ligase activity(GO:0004077) biotin-[methylcrotonoyl-CoA-carboxylase] ligase activity(GO:0004078) biotin-[methylmalonyl-CoA-carboxytransferase] ligase activity(GO:0004079) biotin-[propionyl-CoA-carboxylase (ATP-hydrolyzing)] ligase activity(GO:0004080) biotin-protein ligase activity(GO:0018271) |
| 0.0 | 0.3 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.8 | GO:0030371 | translation repressor activity(GO:0030371) |
| 0.0 | 0.1 | GO:0003945 | N-acetyllactosamine synthase activity(GO:0003945) |
| 0.0 | 1.8 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
| 0.0 | 0.6 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 0.1 | GO:0003983 | UTP:glucose-1-phosphate uridylyltransferase activity(GO:0003983) pyrimidine ribonucleotide binding(GO:0032557) UTP-monosaccharide-1-phosphate uridylyltransferase activity(GO:0051748) |
| 0.0 | 0.2 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.0 | 0.1 | GO:0016428 | tRNA (cytosine-5-)-methyltransferase activity(GO:0016428) |
| 0.0 | 0.6 | GO:0005035 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.0 | 0.2 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.0 | 0.2 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.0 | 0.1 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) |
| 0.0 | 0.0 | GO:0008113 | peptide-methionine (S)-S-oxide reductase activity(GO:0008113) |
| 0.0 | 0.4 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.0 | 0.1 | GO:0016854 | racemase and epimerase activity(GO:0016854) |
| 0.0 | 0.1 | GO:0016728 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.0 | 0.2 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.0 | 0.1 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.0 | 0.2 | GO:0035381 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.0 | 1.2 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.1 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.0 | 0.5 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.0 | 0.1 | GO:0004439 | phosphatidylinositol-4,5-bisphosphate 5-phosphatase activity(GO:0004439) |
| 0.0 | 0.3 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.0 | 0.7 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.2 | GO:0016888 | endodeoxyribonuclease activity, producing 5'-phosphomonoesters(GO:0016888) |
| 0.0 | 0.1 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.0 | 0.2 | GO:0022889 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.0 | 0.1 | GO:0035197 | siRNA binding(GO:0035197) |
| 0.0 | 0.1 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.0 | 0.9 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 0.1 | GO:0042799 | histone methyltransferase activity (H4-K20 specific)(GO:0042799) |
| 0.0 | 0.1 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 4.7 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.3 | 1.9 | PID TXA2PATHWAY | Thromboxane A2 receptor signaling |
| 0.3 | 10.8 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.2 | 4.1 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.2 | 1.8 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.2 | 13.7 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.2 | 9.2 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.2 | 4.8 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.2 | 6.8 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.1 | 0.4 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.1 | 5.2 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.1 | 5.8 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.1 | 1.1 | PID LYMPH ANGIOGENESIS PATHWAY | VEGFR3 signaling in lymphatic endothelium |
| 0.1 | 4.7 | PID EPO PATHWAY | EPO signaling pathway |
| 0.1 | 12.2 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.1 | 7.2 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.1 | 11.5 | PID E2F PATHWAY | E2F transcription factor network |
| 0.1 | 2.1 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.1 | 0.7 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.1 | 10.6 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.1 | 4.6 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.1 | 2.2 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.1 | 4.0 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.1 | 5.4 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.1 | 12.2 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.1 | 0.5 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.1 | 4.6 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.1 | 4.0 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.1 | 1.1 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.1 | 0.4 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.1 | 1.6 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.1 | 2.5 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.1 | 1.0 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
| 0.1 | 2.2 | SIG PIP3 SIGNALING IN B LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
| 0.1 | 1.9 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.1 | 1.0 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.1 | 0.4 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.1 | 1.7 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.1 | 2.2 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.0 | 3.2 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 1.2 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.0 | 0.2 | PID ERBB1 RECEPTOR PROXIMAL PATHWAY | EGF receptor (ErbB1) signaling pathway |
| 0.0 | 1.6 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.0 | 0.6 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 0.2 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 1.0 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 1.1 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.0 | 0.9 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.0 | 0.5 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 1.4 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 1.2 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 0.3 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 1.3 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 0.7 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 2.1 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 0.3 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.0 | 0.3 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.0 | 1.8 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 0.6 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.0 | 0.6 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.0 | 0.4 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 1.3 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.0 | 0.8 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 1.3 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 1.0 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.4 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 0.2 | PID ENDOTHELIN PATHWAY | Endothelins |
| 0.0 | 0.9 | PID CD8 TCR PATHWAY | TCR signaling in naïve CD8+ T cells |
| 0.0 | 0.1 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 0.4 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.0 | 0.5 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 0.7 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 0.2 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 0.2 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 10.2 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.4 | 0.4 | REACTOME ARMS MEDIATED ACTIVATION | Genes involved in ARMS-mediated activation |
| 0.3 | 9.4 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.3 | 6.6 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.3 | 7.1 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.3 | 0.8 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.3 | 2.6 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.3 | 4.8 | REACTOME ACYL CHAIN REMODELLING OF PI | Genes involved in Acyl chain remodelling of PI |
| 0.2 | 4.0 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.2 | 1.9 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.2 | 2.2 | REACTOME ACYL CHAIN REMODELLING OF PC | Genes involved in Acyl chain remodelling of PC |
| 0.2 | 3.0 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.2 | 4.0 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.2 | 7.6 | REACTOME RNA POL I TRANSCRIPTION INITIATION | Genes involved in RNA Polymerase I Transcription Initiation |
| 0.2 | 0.4 | REACTOME RNA POL I TRANSCRIPTION | Genes involved in RNA Polymerase I Transcription |
| 0.2 | 6.5 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.2 | 8.5 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.2 | 7.0 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.2 | 2.4 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.1 | 2.7 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.1 | 1.9 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.1 | 3.7 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.1 | 5.3 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.1 | 5.1 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.1 | 5.6 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.1 | 5.2 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.1 | 2.1 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.1 | 6.1 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.1 | 2.9 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.1 | 2.5 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.1 | 0.2 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.1 | 1.3 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.1 | 2.7 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.1 | 1.3 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.1 | 1.9 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.1 | 4.7 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.1 | 1.7 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.1 | 3.7 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.1 | 2.7 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.1 | 0.2 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.1 | 1.8 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.1 | 1.8 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.1 | 1.1 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.1 | 4.6 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.1 | 8.0 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.1 | 0.9 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.1 | 1.0 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.1 | 1.7 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.1 | 2.0 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.1 | 0.7 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |
| 0.1 | 6.2 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.1 | 2.3 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.1 | 2.3 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.1 | 0.9 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.1 | 1.7 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.1 | 2.8 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.1 | 1.2 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.1 | 0.7 | REACTOME ASSOCIATION OF LICENSING FACTORS WITH THE PRE REPLICATIVE COMPLEX | Genes involved in Association of licensing factors with the pre-replicative complex |
| 0.1 | 0.2 | REACTOME SIGNALLING TO P38 VIA RIT AND RIN | Genes involved in Signalling to p38 via RIT and RIN |
| 0.1 | 2.4 | REACTOME FATTY ACYL COA BIOSYNTHESIS | Genes involved in Fatty Acyl-CoA Biosynthesis |
| 0.0 | 3.4 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 3.8 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.3 | REACTOME DOWNSTREAM TCR SIGNALING | Genes involved in Downstream TCR signaling |
| 0.0 | 1.1 | REACTOME GROWTH HORMONE RECEPTOR SIGNALING | Genes involved in Growth hormone receptor signaling |
| 0.0 | 1.3 | REACTOME INFLAMMASOMES | Genes involved in Inflammasomes |
| 0.0 | 4.8 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 3.9 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 1.1 | REACTOME N GLYCAN ANTENNAE ELONGATION IN THE MEDIAL TRANS GOLGI | Genes involved in N-glycan antennae elongation in the medial/trans-Golgi |
| 0.0 | 1.0 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.9 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 3.3 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 0.7 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.0 | 4.7 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 0.1 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.0 | 5.6 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.7 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 0.0 | 0.4 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.0 | 0.8 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 2.7 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 1.3 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.0 | 2.4 | REACTOME TRANSPORT OF MATURE TRANSCRIPT TO CYTOPLASM | Genes involved in Transport of Mature Transcript to Cytoplasm |
| 0.0 | 0.3 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.0 | 1.4 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 0.4 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 0.6 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.0 | 0.1 | REACTOME FORMATION OF THE HIV1 EARLY ELONGATION COMPLEX | Genes involved in Formation of the HIV-1 Early Elongation Complex |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.0 | 0.1 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.0 | 0.6 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 0.6 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.5 | REACTOME CD28 CO STIMULATION | Genes involved in CD28 co-stimulation |
| 0.0 | 0.3 | REACTOME OPSINS | Genes involved in Opsins |
| 0.0 | 0.5 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.0 | 0.4 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 0.4 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.5 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 0.1 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.0 | 1.9 | REACTOME GLYCEROPHOSPHOLIPID BIOSYNTHESIS | Genes involved in Glycerophospholipid biosynthesis |
| 0.0 | 0.6 | REACTOME PYRUVATE METABOLISM | Genes involved in Pyruvate metabolism |
| 0.0 | 0.2 | REACTOME MRNA CAPPING | Genes involved in mRNA Capping |
| 0.0 | 0.4 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.0 | 0.2 | REACTOME G ALPHA1213 SIGNALLING EVENTS | Genes involved in G alpha (12/13) signalling events |
| 0.0 | 0.3 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 0.4 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 0.5 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.0 | 0.4 | REACTOME FATTY ACID TRIACYLGLYCEROL AND KETONE BODY METABOLISM | Genes involved in Fatty acid, triacylglycerol, and ketone body metabolism |
| 0.0 | 0.6 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 0.6 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.3 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.0 | 0.3 | REACTOME FORMATION OF TRANSCRIPTION COUPLED NER TC NER REPAIR COMPLEX | Genes involved in Formation of transcription-coupled NER (TC-NER) repair complex |
| 0.0 | 0.3 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.0 | 0.4 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 0.3 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 0.2 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |