Project
Inflammatory response time course, HUVEC (Wada, 2009)
Navigation
Downloads
Results for KLF6
Z-value: 0.90
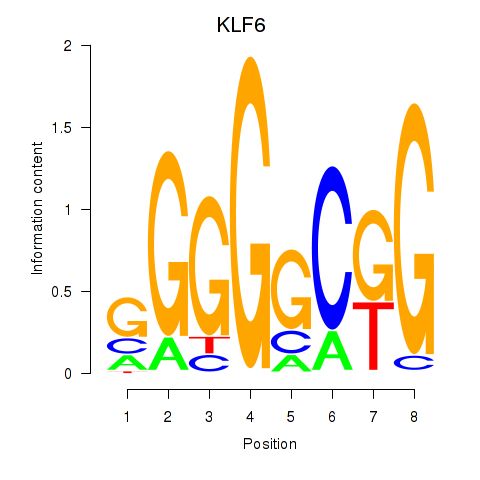
Transcription factors associated with KLF6
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
KLF6
|
ENSG00000067082.15 | KLF6 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| KLF6 | hg38_v1_chr10_-_3785225_3785281 | 0.51 | 9.5e-03 | Click! |
Activity profile of KLF6 motif
Sorted Z-values of KLF6 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of KLF6
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_+_123135938 | 4.91 |
ENST00000357878.7
|
HMX3
|
H6 family homeobox 3 |
| chr11_+_71527267 | 2.24 |
ENST00000398536.6
|
KRTAP5-7
|
keratin associated protein 5-7 |
| chr10_-_73874461 | 1.94 |
ENST00000305762.11
|
CAMK2G
|
calcium/calmodulin dependent protein kinase II gamma |
| chr10_-_129964240 | 1.79 |
ENST00000440978.2
ENST00000355311.10 |
EBF3
|
EBF transcription factor 3 |
| chr10_-_73874568 | 1.65 |
ENST00000322635.7
ENST00000322680.7 ENST00000394762.7 ENST00000680035.1 |
CAMK2G
|
calcium/calmodulin dependent protein kinase II gamma |
| chr4_+_4859658 | 1.52 |
ENST00000382723.5
|
MSX1
|
msh homeobox 1 |
| chr13_+_31846713 | 1.42 |
ENST00000645780.1
|
FRY
|
FRY microtubule binding protein |
| chr17_-_49764123 | 1.39 |
ENST00000240364.7
ENST00000506156.1 |
FAM117A
|
family with sequence similarity 117 member A |
| chr1_+_37474572 | 1.32 |
ENST00000373087.7
|
ZC3H12A
|
zinc finger CCCH-type containing 12A |
| chr20_+_43914801 | 1.31 |
ENST00000341197.9
|
TOX2
|
TOX high mobility group box family member 2 |
| chr4_+_41360759 | 1.23 |
ENST00000508501.5
ENST00000512946.5 ENST00000313860.11 ENST00000512632.5 ENST00000512820.5 |
LIMCH1
|
LIM and calponin homology domains 1 |
| chr1_+_113979460 | 1.14 |
ENST00000320334.5
|
OLFML3
|
olfactomedin like 3 |
| chr1_+_113979391 | 1.14 |
ENST00000393300.6
ENST00000369551.5 |
OLFML3
|
olfactomedin like 3 |
| chrX_+_151694967 | 1.14 |
ENST00000448726.5
ENST00000538575.5 |
PRRG3
|
proline rich and Gla domain 3 |
| chr2_-_85888958 | 1.12 |
ENST00000377332.8
ENST00000639119.1 ENST00000638572.2 |
ST3GAL5
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 5 |
| chr2_-_43226594 | 1.06 |
ENST00000282388.4
|
ZFP36L2
|
ZFP36 ring finger protein like 2 |
| chr2_-_85888685 | 1.05 |
ENST00000638178.1
ENST00000640982.1 ENST00000640992.1 |
ST3GAL5
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 5 |
| chr10_+_110871789 | 1.03 |
ENST00000393104.6
|
PDCD4
|
programmed cell death 4 |
| chr17_+_63477052 | 0.96 |
ENST00000290866.10
ENST00000428043.5 |
ACE
|
angiotensin I converting enzyme |
| chr5_-_108381109 | 0.93 |
ENST00000619412.4
|
FBXL17
|
F-box and leucine rich repeat protein 17 |
| chr16_+_29806519 | 0.92 |
ENST00000322945.11
ENST00000562337.5 ENST00000566906.6 ENST00000563402.1 ENST00000219782.10 |
MAZ
|
MYC associated zinc finger protein |
| chr7_-_27174274 | 0.91 |
ENST00000283921.5
|
HOXA10
|
homeobox A10 |
| chr7_-_27174253 | 0.90 |
ENST00000613671.1
|
HOXA10
|
homeobox A10 |
| chr17_+_7834200 | 0.89 |
ENST00000448097.7
|
KDM6B
|
lysine demethylase 6B |
| chr16_+_30395400 | 0.89 |
ENST00000320159.2
ENST00000613509.2 |
ZNF48
|
zinc finger protein 48 |
| chr2_-_85888897 | 0.89 |
ENST00000639305.1
ENST00000638986.1 |
ST3GAL5
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 5 |
| chr22_-_37519528 | 0.85 |
ENST00000403299.5
|
CARD10
|
caspase recruitment domain family member 10 |
| chr1_-_221742074 | 0.84 |
ENST00000366899.4
|
DUSP10
|
dual specificity phosphatase 10 |
| chr1_+_33256479 | 0.84 |
ENST00000539719.6
ENST00000483388.5 |
ZNF362
|
zinc finger protein 362 |
| chr22_-_37519349 | 0.81 |
ENST00000251973.10
|
CARD10
|
caspase recruitment domain family member 10 |
| chr1_-_114153863 | 0.80 |
ENST00000610222.3
ENST00000369547.6 ENST00000641643.2 |
SYT6
|
synaptotagmin 6 |
| chr19_+_42284020 | 0.77 |
ENST00000160740.7
|
CIC
|
capicua transcriptional repressor |
| chr8_-_79767843 | 0.77 |
ENST00000337919.9
|
HEY1
|
hes related family bHLH transcription factor with YRPW motif 1 |
| chr19_-_17245889 | 0.77 |
ENST00000291442.4
|
NR2F6
|
nuclear receptor subfamily 2 group F member 6 |
| chr10_-_73874502 | 0.77 |
ENST00000372765.5
ENST00000351293.7 ENST00000441192.2 ENST00000423381.6 |
CAMK2G
|
calcium/calmodulin dependent protein kinase II gamma |
| chr1_-_211579064 | 0.77 |
ENST00000367001.5
|
SLC30A1
|
solute carrier family 30 member 1 |
| chr12_+_57089094 | 0.76 |
ENST00000342556.6
ENST00000300131.8 |
NAB2
|
NGFI-A binding protein 2 |
| chr17_+_67825494 | 0.74 |
ENST00000306378.11
ENST00000544778.6 |
BPTF
|
bromodomain PHD finger transcription factor |
| chr5_+_138465472 | 0.73 |
ENST00000239938.5
|
EGR1
|
early growth response 1 |
| chr14_-_29927473 | 0.73 |
ENST00000616995.4
|
PRKD1
|
protein kinase D1 |
| chr14_-_29927801 | 0.73 |
ENST00000331968.11
|
PRKD1
|
protein kinase D1 |
| chr6_+_11537738 | 0.72 |
ENST00000379426.2
|
TMEM170B
|
transmembrane protein 170B |
| chr1_-_94541746 | 0.72 |
ENST00000334047.12
|
F3
|
coagulation factor III, tissue factor |
| chr12_-_49828394 | 0.72 |
ENST00000335999.7
|
NCKAP5L
|
NCK associated protein 5 like |
| chr11_+_44726811 | 0.71 |
ENST00000533202.5
ENST00000520358.7 ENST00000533080.5 ENST00000520999.6 |
TSPAN18
|
tetraspanin 18 |
| chr11_-_2885728 | 0.71 |
ENST00000647251.1
ENST00000380725.2 ENST00000430149.3 ENST00000414822.8 ENST00000440480.8 |
CDKN1C
|
cyclin dependent kinase inhibitor 1C |
| chr6_-_31897200 | 0.70 |
ENST00000395728.7
ENST00000375528.8 |
EHMT2
|
euchromatic histone lysine methyltransferase 2 |
| chr17_+_67825664 | 0.69 |
ENST00000321892.8
|
BPTF
|
bromodomain PHD finger transcription factor |
| chr14_-_29927596 | 0.69 |
ENST00000415220.6
|
PRKD1
|
protein kinase D1 |
| chr8_-_79767462 | 0.68 |
ENST00000674295.1
ENST00000518733.1 ENST00000674418.1 ENST00000674358.1 ENST00000354724.8 |
HEY1
|
hes related family bHLH transcription factor with YRPW motif 1 |
| chr14_-_105168753 | 0.67 |
ENST00000331782.8
ENST00000347004.2 |
JAG2
|
jagged canonical Notch ligand 2 |
| chr7_-_140478975 | 0.67 |
ENST00000474576.5
ENST00000473444.1 ENST00000471104.5 |
MKRN1
|
makorin ring finger protein 1 |
| chr16_+_31459950 | 0.64 |
ENST00000564900.1
|
ARMC5
|
armadillo repeat containing 5 |
| chr21_-_26845402 | 0.64 |
ENST00000284984.8
ENST00000676955.1 |
ADAMTS1
|
ADAM metallopeptidase with thrombospondin type 1 motif 1 |
| chr2_+_45651650 | 0.62 |
ENST00000306156.8
|
PRKCE
|
protein kinase C epsilon |
| chr12_+_19129779 | 0.61 |
ENST00000539256.5
ENST00000299275.10 ENST00000538714.5 |
PLEKHA5
|
pleckstrin homology domain containing A5 |
| chr6_-_110179995 | 0.61 |
ENST00000392586.5
ENST00000419252.1 ENST00000359451.6 ENST00000392588.5 |
WASF1
|
WASP family member 1 |
| chr17_-_42121330 | 0.61 |
ENST00000225916.10
|
KAT2A
|
lysine acetyltransferase 2A |
| chr12_-_94650506 | 0.61 |
ENST00000261226.9
|
TMCC3
|
transmembrane and coiled-coil domain family 3 |
| chr15_+_96330691 | 0.60 |
ENST00000394166.8
|
NR2F2
|
nuclear receptor subfamily 2 group F member 2 |
| chr16_+_68085344 | 0.59 |
ENST00000575270.5
ENST00000346183.8 |
NFATC3
|
nuclear factor of activated T cells 3 |
| chr11_-_63768762 | 0.59 |
ENST00000433688.2
|
C11orf95
|
chromosome 11 open reading frame 95 |
| chr10_+_110871903 | 0.58 |
ENST00000280154.12
|
PDCD4
|
programmed cell death 4 |
| chr10_-_102114935 | 0.58 |
ENST00000361198.9
|
LDB1
|
LIM domain binding 1 |
| chr4_-_13484313 | 0.58 |
ENST00000330852.10
ENST00000288723.9 ENST00000338176.8 ENST00000630951.1 |
RAB28
|
RAB28, member RAS oncogene family |
| chr1_+_87331668 | 0.58 |
ENST00000370542.1
|
LMO4
|
LIM domain only 4 |
| chr6_-_110179702 | 0.58 |
ENST00000392587.6
|
WASF1
|
WASP family member 1 |
| chr1_-_21937300 | 0.56 |
ENST00000374695.8
|
HSPG2
|
heparan sulfate proteoglycan 2 |
| chr3_-_125055987 | 0.56 |
ENST00000311127.9
|
HEG1
|
heart development protein with EGF like domains 1 |
| chr11_-_119364166 | 0.56 |
ENST00000525735.1
|
USP2
|
ubiquitin specific peptidase 2 |
| chr2_-_219543793 | 0.56 |
ENST00000243776.11
|
CHPF
|
chondroitin polymerizing factor |
| chr2_+_20667136 | 0.55 |
ENST00000272224.5
|
GDF7
|
growth differentiation factor 7 |
| chr3_+_124584625 | 0.55 |
ENST00000291478.9
ENST00000682363.1 ENST00000454902.1 |
KALRN
|
kalirin RhoGEF kinase |
| chr16_+_68085420 | 0.55 |
ENST00000349223.9
|
NFATC3
|
nuclear factor of activated T cells 3 |
| chr19_+_34254543 | 0.55 |
ENST00000588470.5
ENST00000299505.8 ENST00000589583.5 |
GARRE1
|
granule associated Rac and RHOG effector 1 |
| chr9_+_112750722 | 0.55 |
ENST00000374232.8
|
SNX30
|
sorting nexin family member 30 |
| chr15_-_55743086 | 0.54 |
ENST00000561292.1
ENST00000389286.9 |
PRTG
|
protogenin |
| chr3_+_50155305 | 0.53 |
ENST00000002829.8
ENST00000426511.5 |
SEMA3F
|
semaphorin 3F |
| chr5_-_81751022 | 0.53 |
ENST00000509013.2
ENST00000505980.5 ENST00000509053.5 |
SSBP2
|
single stranded DNA binding protein 2 |
| chr3_-_15859771 | 0.52 |
ENST00000399451.6
|
ANKRD28
|
ankyrin repeat domain 28 |
| chr11_+_64234569 | 0.52 |
ENST00000309422.7
ENST00000426086.3 |
VEGFB
|
vascular endothelial growth factor B |
| chr9_+_127611760 | 0.52 |
ENST00000625363.2
ENST00000626539.3 |
STXBP1
|
syntaxin binding protein 1 |
| chr6_-_110179623 | 0.52 |
ENST00000265601.7
ENST00000447287.5 ENST00000392589.6 ENST00000444391.5 |
WASF1
|
WASP family member 1 |
| chr16_+_68085552 | 0.52 |
ENST00000329524.8
|
NFATC3
|
nuclear factor of activated T cells 3 |
| chr4_-_25862979 | 0.52 |
ENST00000399878.8
|
SEL1L3
|
SEL1L family member 3 |
| chr17_-_21253398 | 0.51 |
ENST00000611551.1
|
NATD1
|
N-acetyltransferase domain containing 1 |
| chr19_+_1249870 | 0.51 |
ENST00000591446.6
|
MIDN
|
midnolin |
| chr16_+_31459479 | 0.51 |
ENST00000268314.9
|
ARMC5
|
armadillo repeat containing 5 |
| chr1_+_86914616 | 0.51 |
ENST00000370550.10
ENST00000370551.8 |
HS2ST1
|
heparan sulfate 2-O-sulfotransferase 1 |
| chr6_+_34889228 | 0.51 |
ENST00000360359.5
ENST00000649117.1 ENST00000650178.1 |
ANKS1A
|
ankyrin repeat and sterile alpha motif domain containing 1A |
| chr1_-_35557378 | 0.50 |
ENST00000325722.8
ENST00000469892.5 |
KIAA0319L
|
KIAA0319 like |
| chr9_+_2017383 | 0.50 |
ENST00000382194.6
|
SMARCA2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr3_+_50155024 | 0.50 |
ENST00000414301.5
ENST00000450338.5 ENST00000413852.5 |
SEMA3F
|
semaphorin 3F |
| chr10_+_74825445 | 0.49 |
ENST00000649006.1
ENST00000649463.1 ENST00000372725.6 |
KAT6B
|
lysine acetyltransferase 6B |
| chr6_+_107490103 | 0.49 |
ENST00000317357.10
|
SOBP
|
sine oculis binding protein homolog |
| chr18_+_62523002 | 0.48 |
ENST00000269499.10
|
ZCCHC2
|
zinc finger CCHC-type containing 2 |
| chr12_-_57430956 | 0.47 |
ENST00000347140.7
ENST00000402412.5 |
R3HDM2
|
R3H domain containing 2 |
| chr15_-_64775574 | 0.47 |
ENST00000300069.5
|
RBPMS2
|
RNA binding protein, mRNA processing factor 2 |
| chr7_-_28958321 | 0.47 |
ENST00000539664.3
|
TRIL
|
TLR4 interactor with leucine rich repeats |
| chr2_-_73233206 | 0.47 |
ENST00000258083.3
|
PRADC1
|
protease associated domain containing 1 |
| chrX_-_129654946 | 0.46 |
ENST00000429967.3
|
APLN
|
apelin |
| chr3_+_111071773 | 0.46 |
ENST00000485303.6
|
NECTIN3
|
nectin cell adhesion molecule 3 |
| chr12_-_57430778 | 0.46 |
ENST00000448732.1
ENST00000634871.1 |
R3HDM2
|
R3H domain containing 2 |
| chr20_+_34876603 | 0.46 |
ENST00000360596.7
ENST00000253382.5 |
ACSS2
|
acyl-CoA synthetase short chain family member 2 |
| chr6_-_31897675 | 0.46 |
ENST00000375530.8
ENST00000375537.8 |
EHMT2
|
euchromatic histone lysine methyltransferase 2 |
| chr11_+_68312542 | 0.45 |
ENST00000294304.12
|
LRP5
|
LDL receptor related protein 5 |
| chr13_-_36920227 | 0.45 |
ENST00000379826.5
ENST00000350148.10 |
SMAD9
|
SMAD family member 9 |
| chr7_-_87059639 | 0.45 |
ENST00000450689.7
|
ELAPOR2
|
endosome-lysosome associated apoptosis and autophagy regulator family member 2 |
| chr1_+_26696348 | 0.45 |
ENST00000457599.6
|
ARID1A
|
AT-rich interaction domain 1A |
| chr4_-_101347492 | 0.45 |
ENST00000394854.8
|
PPP3CA
|
protein phosphatase 3 catalytic subunit alpha |
| chr9_-_107489754 | 0.44 |
ENST00000610832.1
ENST00000374672.5 |
KLF4
|
Kruppel like factor 4 |
| chr20_-_31870216 | 0.44 |
ENST00000486996.5
ENST00000398084.6 |
DUSP15
|
dual specificity phosphatase 15 |
| chr17_+_28744034 | 0.43 |
ENST00000444415.7
ENST00000262396.10 |
TRAF4
|
TNF receptor associated factor 4 |
| chr9_+_126326809 | 0.43 |
ENST00000361171.8
ENST00000489637.3 |
MVB12B
|
multivesicular body subunit 12B |
| chr1_+_32072919 | 0.43 |
ENST00000438825.5
ENST00000336294.10 |
TMEM39B
|
transmembrane protein 39B |
| chr4_-_101347471 | 0.43 |
ENST00000323055.10
ENST00000512215.5 |
PPP3CA
|
protein phosphatase 3 catalytic subunit alpha |
| chr3_-_171460368 | 0.43 |
ENST00000436636.7
ENST00000465393.1 ENST00000341852.10 |
TNIK
|
TRAF2 and NCK interacting kinase |
| chr10_+_74826406 | 0.43 |
ENST00000648892.1
ENST00000647890.1 ENST00000372724.6 |
KAT6B
|
lysine acetyltransferase 6B |
| chr12_+_121626493 | 0.43 |
ENST00000617316.2
|
ORAI1
|
ORAI calcium release-activated calcium modulator 1 |
| chr17_+_83079595 | 0.42 |
ENST00000320095.12
|
METRNL
|
meteorin like, glial cell differentiation regulator |
| chr13_-_44436801 | 0.42 |
ENST00000261489.6
|
TSC22D1
|
TSC22 domain family member 1 |
| chr19_+_42284483 | 0.42 |
ENST00000575354.6
|
CIC
|
capicua transcriptional repressor |
| chr13_-_44437214 | 0.42 |
ENST00000622051.1
|
TSC22D1
|
TSC22 domain family member 1 |
| chr22_-_20437785 | 0.42 |
ENST00000622235.5
ENST00000623402.1 |
SCARF2
|
scavenger receptor class F member 2 |
| chr7_-_87059624 | 0.42 |
ENST00000444627.5
|
ELAPOR2
|
endosome-lysosome associated apoptosis and autophagy regulator family member 2 |
| chr10_+_97498881 | 0.41 |
ENST00000370664.4
|
UBTD1
|
ubiquitin domain containing 1 |
| chr11_-_66677748 | 0.41 |
ENST00000525754.5
ENST00000531969.5 ENST00000524637.1 ENST00000531036.2 ENST00000310046.9 |
RBM4B
|
RNA binding motif protein 4B |
| chr21_+_46324081 | 0.41 |
ENST00000359568.10
|
PCNT
|
pericentrin |
| chr16_+_53130921 | 0.41 |
ENST00000564845.5
|
CHD9
|
chromodomain helicase DNA binding protein 9 |
| chr12_+_8914525 | 0.41 |
ENST00000543824.5
|
PHC1
|
polyhomeotic homolog 1 |
| chr5_+_93584916 | 0.41 |
ENST00000647447.1
ENST00000615873.1 |
NR2F1
|
nuclear receptor subfamily 2 group F member 1 |
| chr7_-_122886706 | 0.41 |
ENST00000313070.11
ENST00000334010.11 ENST00000615869.4 |
CADPS2
|
calcium dependent secretion activator 2 |
| chr19_-_36032799 | 0.41 |
ENST00000592017.5
ENST00000360535.9 |
CLIP3
|
CAP-Gly domain containing linker protein 3 |
| chr16_-_49856105 | 0.40 |
ENST00000563137.7
|
ZNF423
|
zinc finger protein 423 |
| chr19_+_12938598 | 0.40 |
ENST00000586760.2
ENST00000316448.10 ENST00000588454.6 |
CALR
|
calreticulin |
| chr13_+_34942263 | 0.40 |
ENST00000379939.7
ENST00000400445.7 |
NBEA
|
neurobeachin |
| chr3_+_112562059 | 0.40 |
ENST00000261034.6
|
SLC35A5
|
solute carrier family 35 member A5 |
| chr2_-_96116565 | 0.39 |
ENST00000620793.2
|
ADRA2B
|
adrenoceptor alpha 2B |
| chr16_+_29812230 | 0.39 |
ENST00000300797.7
ENST00000637403.1 ENST00000572820.2 ENST00000637064.1 ENST00000636246.1 |
PRRT2
|
proline rich transmembrane protein 2 |
| chr6_+_71886900 | 0.39 |
ENST00000517960.5
ENST00000518273.5 ENST00000522291.5 ENST00000521978.5 ENST00000520567.5 ENST00000264839.11 |
RIMS1
|
regulating synaptic membrane exocytosis 1 |
| chr16_-_28211476 | 0.39 |
ENST00000569951.1
ENST00000565698.5 |
XPO6
|
exportin 6 |
| chr1_-_45206594 | 0.39 |
ENST00000359600.6
|
ZSWIM5
|
zinc finger SWIM-type containing 5 |
| chr2_+_219442023 | 0.39 |
ENST00000431523.5
ENST00000396698.5 |
SPEG
|
striated muscle enriched protein kinase |
| chr1_-_205680486 | 0.38 |
ENST00000367145.4
|
SLC45A3
|
solute carrier family 45 member 3 |
| chr3_+_9933805 | 0.38 |
ENST00000684493.1
ENST00000673935.2 ENST00000684181.1 ENST00000683189.1 ENST00000383811.8 ENST00000452070.6 ENST00000682642.1 ENST00000684659.1 ENST00000491527.2 ENST00000326434.9 ENST00000682783.1 ENST00000683835.1 ENST00000682570.1 |
CRELD1
|
cysteine rich with EGF like domains 1 |
| chr8_-_8386429 | 0.38 |
ENST00000615670.5
|
PRAG1
|
PEAK1 related, kinase-activating pseudokinase 1 |
| chr3_+_112562030 | 0.38 |
ENST00000468642.5
ENST00000492406.6 |
SLC35A5
|
solute carrier family 35 member A5 |
| chrX_-_120561424 | 0.37 |
ENST00000681206.1
ENST00000679927.1 ENST00000336592.11 |
CUL4B
|
cullin 4B |
| chr14_+_74763308 | 0.37 |
ENST00000325680.12
ENST00000552421.5 |
YLPM1
|
YLP motif containing 1 |
| chr5_+_79236092 | 0.37 |
ENST00000396137.5
|
JMY
|
junction mediating and regulatory protein, p53 cofactor |
| chr16_-_28211908 | 0.37 |
ENST00000566073.1
ENST00000304658.10 |
XPO6
|
exportin 6 |
| chr12_-_57006476 | 0.37 |
ENST00000300101.3
|
ZBTB39
|
zinc finger and BTB domain containing 39 |
| chr18_+_48539017 | 0.37 |
ENST00000256413.8
|
CTIF
|
cap binding complex dependent translation initiation factor |
| chr11_-_64810678 | 0.37 |
ENST00000312049.11
|
MEN1
|
menin 1 |
| chr1_-_114152915 | 0.36 |
ENST00000609117.5
ENST00000608879.1 ENST00000608203.1 ENST00000609577.1 |
SYT6
|
synaptotagmin 6 |
| chr19_+_35000426 | 0.36 |
ENST00000411896.6
ENST00000424536.2 |
GRAMD1A
|
GRAM domain containing 1A |
| chr1_+_156728442 | 0.36 |
ENST00000368218.8
ENST00000368216.9 |
RRNAD1
|
ribosomal RNA adenine dimethylase domain containing 1 |
| chr11_-_6655788 | 0.36 |
ENST00000299441.5
|
DCHS1
|
dachsous cadherin-related 1 |
| chr9_+_128276222 | 0.36 |
ENST00000608796.6
ENST00000419867.7 ENST00000418976.2 |
SWI5
|
SWI5 homologous recombination repair protein |
| chr7_-_29990113 | 0.36 |
ENST00000426154.5
ENST00000421434.5 ENST00000434476.6 |
SCRN1
|
secernin 1 |
| chr20_-_31870510 | 0.36 |
ENST00000339738.10
|
DUSP15
|
dual specificity phosphatase 15 |
| chr5_-_81751085 | 0.35 |
ENST00000515395.5
|
SSBP2
|
single stranded DNA binding protein 2 |
| chr12_+_6766353 | 0.35 |
ENST00000309083.8
ENST00000389462.8 |
PTMS
|
parathymosin |
| chr1_+_32741779 | 0.35 |
ENST00000401073.7
|
KIAA1522
|
KIAA1522 |
| chr19_-_31349408 | 0.35 |
ENST00000240587.5
|
TSHZ3
|
teashirt zinc finger homeobox 3 |
| chr7_-_29989774 | 0.35 |
ENST00000242059.10
|
SCRN1
|
secernin 1 |
| chr1_-_165355746 | 0.35 |
ENST00000367893.4
|
LMX1A
|
LIM homeobox transcription factor 1 alpha |
| chr9_-_34589716 | 0.35 |
ENST00000378980.8
|
CNTFR
|
ciliary neurotrophic factor receptor |
| chr5_+_141637398 | 0.35 |
ENST00000518856.1
|
RELL2
|
RELT like 2 |
| chr2_-_219229309 | 0.35 |
ENST00000443140.5
ENST00000432520.5 ENST00000409618.5 |
ATG9A
|
autophagy related 9A |
| chr1_+_210232776 | 0.35 |
ENST00000367012.4
|
SERTAD4
|
SERTA domain containing 4 |
| chr9_+_36036899 | 0.34 |
ENST00000377966.4
|
RECK
|
reversion inducing cysteine rich protein with kazal motifs |
| chr2_-_219229571 | 0.34 |
ENST00000436856.5
ENST00000428226.5 ENST00000409422.5 ENST00000361242.9 ENST00000431715.5 ENST00000457841.5 ENST00000439812.5 ENST00000396761.6 |
ATG9A
|
autophagy related 9A |
| chr9_-_127755243 | 0.34 |
ENST00000629203.2
ENST00000420366.5 |
SH2D3C
|
SH2 domain containing 3C |
| chr2_+_181891904 | 0.34 |
ENST00000320370.11
|
ITPRID2
|
ITPR interacting domain containing 2 |
| chr11_+_62613236 | 0.34 |
ENST00000278833.4
|
ROM1
|
retinal outer segment membrane protein 1 |
| chr19_-_6279921 | 0.34 |
ENST00000252674.9
|
MLLT1
|
MLLT1 super elongation complex subunit |
| chr9_-_34589701 | 0.34 |
ENST00000351266.8
|
CNTFR
|
ciliary neurotrophic factor receptor |
| chr9_+_35605277 | 0.33 |
ENST00000620767.4
|
TESK1
|
testis associated actin remodelling kinase 1 |
| chr12_-_124863902 | 0.33 |
ENST00000339570.9
ENST00000680556.1 |
SCARB1
|
scavenger receptor class B member 1 |
| chr9_+_128275343 | 0.33 |
ENST00000495313.5
ENST00000372898.6 |
SWI5
|
SWI5 homologous recombination repair protein |
| chr19_-_14090695 | 0.33 |
ENST00000533683.7
|
SAMD1
|
sterile alpha motif domain containing 1 |
| chr2_+_181891697 | 0.33 |
ENST00000431877.7
|
ITPRID2
|
ITPR interacting domain containing 2 |
| chr9_+_125747345 | 0.33 |
ENST00000342287.9
ENST00000373489.10 ENST00000373487.8 |
PBX3
|
PBX homeobox 3 |
| chr9_+_35605234 | 0.33 |
ENST00000336395.6
|
TESK1
|
testis associated actin remodelling kinase 1 |
| chr12_-_79934920 | 0.33 |
ENST00000550107.5
|
PPP1R12A
|
protein phosphatase 1 regulatory subunit 12A |
| chr16_+_788614 | 0.32 |
ENST00000262315.14
ENST00000455171.6 ENST00000317063.10 |
CHTF18
|
chromosome transmission fidelity factor 18 |
| chr17_-_3964291 | 0.31 |
ENST00000359983.7
ENST00000352011.7 |
ATP2A3
|
ATPase sarcoplasmic/endoplasmic reticulum Ca2+ transporting 3 |
| chr1_+_155323911 | 0.31 |
ENST00000368347.8
|
RUSC1
|
RUN and SH3 domain containing 1 |
| chr4_-_2262082 | 0.31 |
ENST00000337190.7
|
MXD4
|
MAX dimerization protein 4 |
| chr3_-_184361594 | 0.31 |
ENST00000344937.11
ENST00000434054.6 ENST00000457512.1 ENST00000265593.9 |
CLCN2
|
chloride voltage-gated channel 2 |
| chr11_-_19240936 | 0.31 |
ENST00000250024.9
|
E2F8
|
E2F transcription factor 8 |
| chr11_-_62612725 | 0.31 |
ENST00000419857.1
ENST00000394773.7 |
EML3
|
EMAP like 3 |
| chr4_+_112145445 | 0.30 |
ENST00000309733.6
|
FAM241A
|
family with sequence similarity 241 member A |
| chr19_-_55180242 | 0.30 |
ENST00000592470.1
ENST00000354308.8 |
SYT5
|
synaptotagmin 5 |
| chr19_+_33796846 | 0.30 |
ENST00000590771.5
ENST00000589786.5 ENST00000284006.10 ENST00000683859.1 ENST00000588881.5 |
KCTD15
|
potassium channel tetramerization domain containing 15 |
| chr12_-_7018465 | 0.30 |
ENST00000261407.9
|
LPCAT3
|
lysophosphatidylcholine acyltransferase 3 |
| chr5_-_180291967 | 0.30 |
ENST00000425491.6
ENST00000523583.1 ENST00000393360.7 ENST00000455781.5 ENST00000452135.7 ENST00000347470.8 ENST00000343111.10 |
MAPK9
|
mitogen-activated protein kinase 9 |
| chr15_-_82647960 | 0.30 |
ENST00000615198.4
|
CPEB1
|
cytoplasmic polyadenylation element binding protein 1 |
| chr17_-_63842663 | 0.30 |
ENST00000613943.4
ENST00000448276.7 |
SMARCD2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 2 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.6 | GO:1904761 | negative regulation of myofibroblast differentiation(GO:1904761) |
| 0.5 | 0.5 | GO:1901187 | regulation of ephrin receptor signaling pathway(GO:1901187) |
| 0.4 | 1.3 | GO:0061027 | umbilical cord morphogenesis(GO:0036304) umbilical cord development(GO:0061027) |
| 0.4 | 1.5 | GO:0090427 | embryonic nail plate morphogenesis(GO:0035880) activation of meiosis(GO:0090427) |
| 0.4 | 1.4 | GO:1904428 | negative regulation of tubulin deacetylation(GO:1904428) |
| 0.3 | 0.8 | GO:0060266 | negative regulation of respiratory burst involved in inflammatory response(GO:0060266) |
| 0.3 | 2.4 | GO:1904627 | response to phorbol 13-acetate 12-myristate(GO:1904627) cellular response to phorbol 13-acetate 12-myristate(GO:1904628) |
| 0.3 | 0.3 | GO:1902958 | neuron intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:0036482) positive regulation of mitochondrial electron transport, NADH to ubiquinone(GO:1902958) regulation of hydrogen peroxide-induced neuron intrinsic apoptotic signaling pathway(GO:1903383) negative regulation of hydrogen peroxide-induced neuron intrinsic apoptotic signaling pathway(GO:1903384) |
| 0.2 | 0.7 | GO:0045360 | regulation of interleukin-1 biosynthetic process(GO:0045360) positive regulation of interleukin-1 biosynthetic process(GO:0045362) interleukin-1 beta biosynthetic process(GO:0050720) response to interleukin-8(GO:0098758) cellular response to interleukin-8(GO:0098759) regulation of progesterone biosynthetic process(GO:2000182) |
| 0.2 | 1.0 | GO:2000170 | positive regulation of peptidyl-cysteine S-nitrosylation(GO:2000170) |
| 0.2 | 2.0 | GO:0089700 | protein kinase D signaling(GO:0089700) |
| 0.2 | 0.9 | GO:1905205 | positive regulation of connective tissue replacement(GO:1905205) |
| 0.2 | 0.6 | GO:0032877 | positive regulation of DNA endoreduplication(GO:0032877) |
| 0.2 | 0.6 | GO:0043973 | histone H3-K4 acetylation(GO:0043973) |
| 0.2 | 1.7 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 0.2 | 0.6 | GO:0090271 | positive regulation of fibroblast growth factor production(GO:0090271) |
| 0.2 | 3.1 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.2 | 0.7 | GO:0002541 | activation of plasma proteins involved in acute inflammatory response(GO:0002541) |
| 0.2 | 4.2 | GO:1901897 | regulation of relaxation of cardiac muscle(GO:1901897) |
| 0.2 | 0.7 | GO:0044805 | late nucleophagy(GO:0044805) |
| 0.2 | 1.2 | GO:0060478 | acrosomal vesicle exocytosis(GO:0060478) |
| 0.2 | 0.5 | GO:0043397 | corticotropin-releasing hormone secretion(GO:0043396) regulation of corticotropin-releasing hormone secretion(GO:0043397) |
| 0.2 | 0.5 | GO:0019542 | acetate biosynthetic process(GO:0019413) acetyl-CoA biosynthetic process from acetate(GO:0019427) propionate biosynthetic process(GO:0019542) |
| 0.2 | 0.8 | GO:0016480 | negative regulation of transcription from RNA polymerase III promoter(GO:0016480) |
| 0.1 | 0.6 | GO:0009956 | radial pattern formation(GO:0009956) |
| 0.1 | 0.4 | GO:0014740 | negative regulation of muscle hyperplasia(GO:0014740) |
| 0.1 | 1.0 | GO:0036486 | trunk segmentation(GO:0035290) trunk neural crest cell migration(GO:0036484) ventral trunk neural crest cell migration(GO:0036486) |
| 0.1 | 0.5 | GO:1903294 | regulation of glutamate secretion, neurotransmission(GO:1903294) positive regulation of glutamate secretion, neurotransmission(GO:1903296) |
| 0.1 | 0.8 | GO:0061687 | detoxification of inorganic compound(GO:0061687) stress response to metal ion(GO:0097501) |
| 0.1 | 0.4 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.1 | 0.2 | GO:0035905 | ascending aorta development(GO:0035905) ascending aorta morphogenesis(GO:0035910) |
| 0.1 | 0.6 | GO:0006344 | optic cup formation involved in camera-type eye development(GO:0003408) maintenance of chromatin silencing(GO:0006344) |
| 0.1 | 0.2 | GO:0071603 | endothelial cell-cell adhesion(GO:0071603) |
| 0.1 | 0.6 | GO:0035992 | tendon cell differentiation(GO:0035990) tendon formation(GO:0035992) |
| 0.1 | 0.3 | GO:0015920 | lipopolysaccharide transport(GO:0015920) |
| 0.1 | 0.3 | GO:0003192 | mitral valve formation(GO:0003192) |
| 0.1 | 0.5 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 0.1 | 0.3 | GO:0060689 | cell differentiation involved in salivary gland development(GO:0060689) |
| 0.1 | 0.4 | GO:0002501 | peptide antigen assembly with MHC protein complex(GO:0002501) |
| 0.1 | 2.5 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.1 | 0.4 | GO:0015772 | disaccharide transport(GO:0015766) sucrose transport(GO:0015770) oligosaccharide transport(GO:0015772) |
| 0.1 | 0.3 | GO:1904395 | positive regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904395) |
| 0.1 | 0.7 | GO:0000389 | mRNA 3'-splice site recognition(GO:0000389) |
| 0.1 | 0.4 | GO:0035625 | negative regulation of epinephrine secretion(GO:0032811) epidermal growth factor-activated receptor transactivation by G-protein coupled receptor signaling pathway(GO:0035625) |
| 0.1 | 0.5 | GO:0061304 | retinal blood vessel morphogenesis(GO:0061304) |
| 0.1 | 0.8 | GO:0035507 | regulation of myosin-light-chain-phosphatase activity(GO:0035507) |
| 0.1 | 0.2 | GO:0038162 | erythropoietin-mediated signaling pathway(GO:0038162) |
| 0.1 | 1.0 | GO:0061368 | behavioral response to chemical pain(GO:0061366) behavioral response to formalin induced pain(GO:0061368) |
| 0.1 | 0.5 | GO:0060754 | positive regulation of mast cell chemotaxis(GO:0060754) |
| 0.1 | 0.4 | GO:0019075 | virus maturation(GO:0019075) |
| 0.1 | 1.2 | GO:0043983 | histone H4-K12 acetylation(GO:0043983) |
| 0.1 | 0.3 | GO:1904017 | cellular response to Thyroglobulin triiodothyronine(GO:1904017) |
| 0.1 | 0.2 | GO:0061300 | cerebellum vasculature development(GO:0061300) |
| 0.1 | 0.5 | GO:0071557 | histone H3-K27 demethylation(GO:0071557) |
| 0.1 | 0.2 | GO:0048633 | positive regulation of skeletal muscle tissue growth(GO:0048633) |
| 0.1 | 0.5 | GO:0060399 | positive regulation of growth hormone receptor signaling pathway(GO:0060399) |
| 0.1 | 0.2 | GO:1900169 | regulation of glucocorticoid mediated signaling pathway(GO:1900169) |
| 0.1 | 5.2 | GO:0007566 | embryo implantation(GO:0007566) |
| 0.1 | 2.7 | GO:0060065 | uterus development(GO:0060065) |
| 0.1 | 0.7 | GO:0003360 | brainstem development(GO:0003360) |
| 0.1 | 2.1 | GO:0043153 | entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.1 | 0.1 | GO:2000820 | negative regulation of transcription from RNA polymerase II promoter involved in smooth muscle cell differentiation(GO:2000820) |
| 0.1 | 0.3 | GO:0060010 | Sertoli cell fate commitment(GO:0060010) |
| 0.1 | 0.7 | GO:0060120 | auditory receptor cell fate commitment(GO:0009912) gamma-delta T cell differentiation(GO:0042492) inner ear receptor cell fate commitment(GO:0060120) |
| 0.1 | 0.4 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.1 | 1.5 | GO:1902894 | negative regulation of pri-miRNA transcription from RNA polymerase II promoter(GO:1902894) |
| 0.1 | 0.3 | GO:0003150 | muscular septum morphogenesis(GO:0003150) |
| 0.1 | 0.5 | GO:0033132 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.1 | 0.2 | GO:0038033 | positive regulation of endothelial cell chemotaxis by VEGF-activated vascular endothelial growth factor receptor signaling pathway(GO:0038033) |
| 0.1 | 0.2 | GO:0061537 | glycine secretion(GO:0061536) glycine secretion, neurotransmission(GO:0061537) |
| 0.1 | 0.2 | GO:1902683 | regulation of receptor localization to synapse(GO:1902683) |
| 0.1 | 0.2 | GO:0016340 | calcium-dependent cell-matrix adhesion(GO:0016340) regulation of embryonic cell shape(GO:0016476) |
| 0.1 | 0.5 | GO:0051151 | negative regulation of smooth muscle cell differentiation(GO:0051151) |
| 0.1 | 0.5 | GO:0018231 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.1 | 0.3 | GO:0071934 | thiamine transmembrane transport(GO:0071934) |
| 0.1 | 0.2 | GO:0014022 | neural plate elongation(GO:0014022) convergent extension involved in neural plate elongation(GO:0022007) |
| 0.1 | 0.2 | GO:2001287 | negative regulation of caveolin-mediated endocytosis(GO:2001287) |
| 0.1 | 0.4 | GO:1901896 | positive regulation of calcium-transporting ATPase activity(GO:1901896) |
| 0.1 | 0.3 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.1 | 0.1 | GO:0001560 | regulation of cell growth by extracellular stimulus(GO:0001560) |
| 0.1 | 0.3 | GO:0045048 | protein insertion into ER membrane(GO:0045048) |
| 0.1 | 0.3 | GO:0097155 | fasciculation of sensory neuron axon(GO:0097155) |
| 0.1 | 1.3 | GO:0018027 | peptidyl-lysine dimethylation(GO:0018027) |
| 0.1 | 0.2 | GO:0003420 | regulation of growth plate cartilage chondrocyte proliferation(GO:0003420) histone H3-R2 methylation(GO:0034970) |
| 0.0 | 0.1 | GO:1902824 | positive regulation of late endosome to lysosome transport(GO:1902824) |
| 0.0 | 0.1 | GO:0070846 | misfolded protein transport(GO:0070843) polyubiquitinated protein transport(GO:0070844) polyubiquitinated misfolded protein transport(GO:0070845) Hsp90 deacetylation(GO:0070846) |
| 0.0 | 0.1 | GO:2000777 | positive regulation of oocyte maturation(GO:1900195) positive regulation of proteasomal ubiquitin-dependent protein catabolic process involved in cellular response to hypoxia(GO:2000777) |
| 0.0 | 0.2 | GO:0051138 | positive regulation of NK T cell differentiation(GO:0051138) |
| 0.0 | 0.2 | GO:1902559 | 3'-phosphoadenosine 5'-phosphosulfate transport(GO:0046963) 3'-phospho-5'-adenylyl sulfate transmembrane transport(GO:1902559) |
| 0.0 | 0.7 | GO:0045003 | DNA recombinase assembly(GO:0000730) double-strand break repair via synthesis-dependent strand annealing(GO:0045003) |
| 0.0 | 0.3 | GO:1900262 | regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.0 | 0.4 | GO:1990504 | dense core granule exocytosis(GO:1990504) |
| 0.0 | 0.1 | GO:0060979 | vasculogenesis involved in coronary vascular morphogenesis(GO:0060979) |
| 0.0 | 0.2 | GO:0031456 | glycine betaine biosynthetic process from choline(GO:0019285) glycine betaine metabolic process(GO:0031455) glycine betaine biosynthetic process(GO:0031456) |
| 0.0 | 0.6 | GO:0070257 | positive regulation of mucus secretion(GO:0070257) |
| 0.0 | 0.3 | GO:0090170 | regulation of Golgi inheritance(GO:0090170) |
| 0.0 | 0.5 | GO:0042048 | olfactory behavior(GO:0042048) |
| 0.0 | 0.6 | GO:0001542 | ovulation from ovarian follicle(GO:0001542) |
| 0.0 | 0.1 | GO:0051228 | mitotic spindle disassembly(GO:0051228) spindle disassembly(GO:0051230) |
| 0.0 | 0.4 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 0.0 | 0.2 | GO:1990737 | regulation of endoplasmic reticulum stress-induced eIF2 alpha phosphorylation(GO:0060734) response to manganese-induced endoplasmic reticulum stress(GO:1990737) |
| 0.0 | 0.5 | GO:0039536 | negative regulation of RIG-I signaling pathway(GO:0039536) |
| 0.0 | 0.2 | GO:0042426 | choline catabolic process(GO:0042426) |
| 0.0 | 0.4 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.0 | 0.2 | GO:1902268 | negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.0 | 0.2 | GO:0033274 | response to vitamin B2(GO:0033274) heterochromatin maintenance(GO:0070829) |
| 0.0 | 0.1 | GO:0042040 | molybdenum incorporation into molybdenum-molybdopterin complex(GO:0018315) metal incorporation into metallo-molybdopterin complex(GO:0042040) glycine receptor clustering(GO:0072579) |
| 0.0 | 0.2 | GO:1902075 | cellular response to salt(GO:1902075) |
| 0.0 | 0.5 | GO:1902414 | protein localization to cell junction(GO:1902414) |
| 0.0 | 0.3 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.0 | 0.4 | GO:0043981 | histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.0 | 0.1 | GO:0000415 | negative regulation of histone H3-K36 methylation(GO:0000415) |
| 0.0 | 0.2 | GO:0006931 | substrate-dependent cell migration, cell attachment to substrate(GO:0006931) |
| 0.0 | 0.2 | GO:0070345 | negative regulation of fat cell proliferation(GO:0070345) |
| 0.0 | 0.1 | GO:1903984 | rhythmic synaptic transmission(GO:0060024) negative regulation of ribosome biogenesis(GO:0090071) regulation of TRAIL-activated apoptotic signaling pathway(GO:1903121) positive regulation of TRAIL-activated apoptotic signaling pathway(GO:1903984) |
| 0.0 | 0.6 | GO:0021514 | ventral spinal cord interneuron differentiation(GO:0021514) |
| 0.0 | 0.5 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.0 | 0.2 | GO:0033029 | regulation of neutrophil apoptotic process(GO:0033029) |
| 0.0 | 0.5 | GO:0010991 | negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.0 | 0.6 | GO:0050884 | neuromuscular process controlling posture(GO:0050884) |
| 0.0 | 0.4 | GO:0061469 | regulation of type B pancreatic cell proliferation(GO:0061469) |
| 0.0 | 0.1 | GO:1904379 | protein localization to cytosolic proteasome complex(GO:1904327) protein localization to cytosolic proteasome complex involved in ERAD pathway(GO:1904379) |
| 0.0 | 0.2 | GO:0034499 | late endosome to Golgi transport(GO:0034499) |
| 0.0 | 0.2 | GO:0038108 | negative regulation of appetite by leptin-mediated signaling pathway(GO:0038108) |
| 0.0 | 0.3 | GO:2000786 | positive regulation of autophagosome assembly(GO:2000786) |
| 0.0 | 0.1 | GO:1904381 | Golgi apparatus mannose trimming(GO:1904381) |
| 0.0 | 0.2 | GO:0070164 | negative regulation of adiponectin secretion(GO:0070164) |
| 0.0 | 0.2 | GO:0046543 | development of secondary female sexual characteristics(GO:0046543) |
| 0.0 | 0.1 | GO:0006121 | mitochondrial electron transport, succinate to ubiquinone(GO:0006121) respiratory chain complex II assembly(GO:0034552) mitochondrial respiratory chain complex II assembly(GO:0034553) mitochondrial respiratory chain complex II biogenesis(GO:0097032) |
| 0.0 | 0.2 | GO:0050966 | detection of mechanical stimulus involved in sensory perception of pain(GO:0050966) |
| 0.0 | 0.1 | GO:1901078 | mastication(GO:0071626) learned vocalization behavior(GO:0098583) negative regulation of relaxation of muscle(GO:1901078) |
| 0.0 | 0.1 | GO:2000691 | negative regulation of cardioblast differentiation(GO:0051892) regulation of cardiac muscle cell myoblast differentiation(GO:2000690) negative regulation of cardiac muscle cell myoblast differentiation(GO:2000691) |
| 0.0 | 0.3 | GO:0021942 | radial glia guided migration of Purkinje cell(GO:0021942) |
| 0.0 | 0.2 | GO:0090336 | positive regulation of brown fat cell differentiation(GO:0090336) |
| 0.0 | 0.3 | GO:0048625 | myoblast fate commitment(GO:0048625) |
| 0.0 | 0.4 | GO:0035518 | histone H2A monoubiquitination(GO:0035518) |
| 0.0 | 0.1 | GO:0010746 | regulation of plasma membrane long-chain fatty acid transport(GO:0010746) negative regulation of plasma membrane long-chain fatty acid transport(GO:0010748) |
| 0.0 | 0.2 | GO:1902459 | positive regulation of stem cell population maintenance(GO:1902459) |
| 0.0 | 0.1 | GO:0003278 | apoptotic process involved in heart morphogenesis(GO:0003278) |
| 0.0 | 0.1 | GO:0010609 | mRNA localization resulting in posttranscriptional regulation of gene expression(GO:0010609) |
| 0.0 | 0.2 | GO:0043562 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 | 0.3 | GO:0033235 | positive regulation of protein sumoylation(GO:0033235) |
| 0.0 | 0.2 | GO:2000580 | regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.0 | 0.1 | GO:0043418 | homocysteine catabolic process(GO:0043418) |
| 0.0 | 0.1 | GO:0045629 | negative regulation of T-helper 2 cell differentiation(GO:0045629) |
| 0.0 | 0.1 | GO:0080154 | regulation of fertilization(GO:0080154) |
| 0.0 | 1.1 | GO:1990090 | cellular response to nerve growth factor stimulus(GO:1990090) |
| 0.0 | 0.1 | GO:0060448 | dichotomous subdivision of terminal units involved in lung branching(GO:0060448) |
| 0.0 | 0.4 | GO:0060219 | camera-type eye photoreceptor cell differentiation(GO:0060219) |
| 0.0 | 0.6 | GO:0051084 | 'de novo' posttranslational protein folding(GO:0051084) |
| 0.0 | 0.2 | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.0 | 0.1 | GO:0060068 | vagina development(GO:0060068) |
| 0.0 | 0.0 | GO:0021526 | medial motor column neuron differentiation(GO:0021526) |
| 0.0 | 0.1 | GO:0030421 | defecation(GO:0030421) |
| 0.0 | 0.3 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.0 | 0.7 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.0 | 0.1 | GO:0051586 | glial cell-derived neurotrophic factor secretion(GO:0044467) positive regulation of neurotransmitter uptake(GO:0051582) positive regulation of dopamine uptake involved in synaptic transmission(GO:0051586) positive regulation of catecholamine uptake involved in synaptic transmission(GO:0051944) regulation of glial cell-derived neurotrophic factor secretion(GO:1900166) positive regulation of glial cell-derived neurotrophic factor secretion(GO:1900168) |
| 0.0 | 0.2 | GO:0035871 | protein K11-linked deubiquitination(GO:0035871) |
| 0.0 | 0.1 | GO:0018076 | N-terminal peptidyl-lysine acetylation(GO:0018076) |
| 0.0 | 0.1 | GO:0030950 | establishment or maintenance of actin cytoskeleton polarity(GO:0030950) |
| 0.0 | 0.0 | GO:0034729 | histone H3-K79 methylation(GO:0034729) regulation of genetic imprinting(GO:2000653) |
| 0.0 | 0.3 | GO:0045898 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) |
| 0.0 | 0.3 | GO:0001675 | acrosome assembly(GO:0001675) |
| 0.0 | 0.2 | GO:0060539 | diaphragm development(GO:0060539) |
| 0.0 | 0.2 | GO:0009396 | folic acid-containing compound biosynthetic process(GO:0009396) |
| 0.0 | 0.1 | GO:0035610 | protein side chain deglutamylation(GO:0035610) |
| 0.0 | 0.4 | GO:0031167 | rRNA methylation(GO:0031167) |
| 0.0 | 0.2 | GO:2000427 | positive regulation of apoptotic cell clearance(GO:2000427) |
| 0.0 | 0.3 | GO:0030497 | fatty acid elongation(GO:0030497) |
| 0.0 | 0.4 | GO:1901522 | positive regulation of transcription from RNA polymerase II promoter involved in cellular response to chemical stimulus(GO:1901522) |
| 0.0 | 0.6 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.0 | 0.3 | GO:0036152 | phosphatidylethanolamine acyl-chain remodeling(GO:0036152) |
| 0.0 | 0.0 | GO:0046671 | negative regulation of cellular pH reduction(GO:0032848) CD8-positive, alpha-beta T cell lineage commitment(GO:0043375) negative regulation of retinal cell programmed cell death(GO:0046671) |
| 0.0 | 0.2 | GO:0060628 | regulation of ER to Golgi vesicle-mediated transport(GO:0060628) |
| 0.0 | 0.0 | GO:1900194 | negative regulation of oocyte maturation(GO:1900194) |
| 0.0 | 0.0 | GO:0036215 | response to stem cell factor(GO:0036215) cellular response to stem cell factor stimulus(GO:0036216) Kit signaling pathway(GO:0038109) |
| 0.0 | 0.1 | GO:0015862 | uridine transport(GO:0015862) |
| 0.0 | 0.0 | GO:1900126 | negative regulation of hyaluronan biosynthetic process(GO:1900126) |
| 0.0 | 0.1 | GO:2000574 | regulation of microtubule motor activity(GO:2000574) |
| 0.0 | 0.2 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.0 | 0.3 | GO:0043555 | regulation of translation in response to stress(GO:0043555) |
| 0.0 | 0.3 | GO:0007064 | mitotic sister chromatid cohesion(GO:0007064) |
| 0.0 | 0.1 | GO:1902723 | negative regulation of skeletal muscle cell proliferation(GO:0014859) negative regulation of skeletal muscle satellite cell proliferation(GO:1902723) |
| 0.0 | 0.1 | GO:0031119 | tRNA pseudouridine synthesis(GO:0031119) |
| 0.0 | 0.5 | GO:0090102 | cochlea development(GO:0090102) |
| 0.0 | 0.1 | GO:1901908 | diadenosine polyphosphate metabolic process(GO:0015959) diadenosine polyphosphate catabolic process(GO:0015961) diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 2.0 | GO:0032449 | CBM complex(GO:0032449) |
| 0.2 | 0.7 | GO:0032798 | Swi5-Sfr1 complex(GO:0032798) |
| 0.2 | 0.7 | GO:0097059 | CNTFR-CLCF1 complex(GO:0097059) |
| 0.1 | 1.3 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.1 | 1.2 | GO:0016589 | NURF complex(GO:0016589) |
| 0.1 | 0.6 | GO:1990769 | proximal neuron projection(GO:1990769) |
| 0.1 | 1.7 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.1 | 0.5 | GO:1990851 | Wnt-Frizzled-LRP5/6 complex(GO:1990851) |
| 0.1 | 0.3 | GO:0071148 | TEAD-1-YAP complex(GO:0071148) |
| 0.1 | 0.4 | GO:0031510 | SUMO activating enzyme complex(GO:0031510) |
| 0.1 | 0.5 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.1 | 3.1 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.1 | 0.8 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.1 | 0.7 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.1 | 0.3 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.1 | 0.8 | GO:0070775 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.1 | 0.5 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.1 | 0.6 | GO:0031390 | Ctf18 RFC-like complex(GO:0031390) |
| 0.1 | 4.2 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.1 | 1.0 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.1 | 1.1 | GO:0033276 | transcription factor TFTC complex(GO:0033276) |
| 0.0 | 0.1 | GO:0014802 | terminal cisterna(GO:0014802) |
| 0.0 | 0.1 | GO:0000805 | X chromosome(GO:0000805) autosome(GO:0030849) |
| 0.0 | 0.2 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.0 | 1.5 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 0.0 | 0.7 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.0 | 0.2 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 0.0 | 0.4 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.0 | 0.6 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.0 | 0.4 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 0.5 | GO:0070938 | contractile ring(GO:0070938) |
| 0.0 | 0.5 | GO:0016461 | unconventional myosin complex(GO:0016461) |
| 0.0 | 0.2 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 0.0 | 0.3 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 0.0 | 0.1 | GO:0031501 | mannosyltransferase complex(GO:0031501) |
| 0.0 | 0.1 | GO:0060987 | lipid tube(GO:0060987) |
| 0.0 | 0.3 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 0.1 | GO:0036398 | TCR signalosome(GO:0036398) |
| 0.0 | 0.7 | GO:0031233 | intrinsic component of external side of plasma membrane(GO:0031233) |
| 0.0 | 0.3 | GO:0097449 | astrocyte projection(GO:0097449) |
| 0.0 | 1.6 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 0.5 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.0 | 0.3 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.0 | 0.1 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.4 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.1 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.0 | 0.0 | GO:0031084 | BLOC-2 complex(GO:0031084) |
| 0.0 | 0.1 | GO:0044327 | dendritic spine head(GO:0044327) |
| 0.0 | 0.1 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 0.2 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 0.3 | GO:0010369 | chromocenter(GO:0010369) |
| 0.0 | 0.1 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.0 | 0.4 | GO:0044665 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 3.1 | GO:0004513 | neolactotetraosylceramide alpha-2,3-sialyltransferase activity(GO:0004513) lactosylceramide alpha-2,3-sialyltransferase activity(GO:0047291) |
| 0.4 | 5.1 | GO:0004723 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) |
| 0.3 | 1.3 | GO:0035939 | microsatellite binding(GO:0035939) |
| 0.2 | 1.2 | GO:0008240 | tripeptidyl-peptidase activity(GO:0008240) |
| 0.2 | 0.7 | GO:0045131 | pre-mRNA branch point binding(GO:0045131) |
| 0.2 | 0.5 | GO:1904928 | coreceptor activity involved in canonical Wnt signaling pathway(GO:1904928) |
| 0.1 | 0.6 | GO:0050510 | N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase activity(GO:0050510) |
| 0.1 | 2.8 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.1 | 1.2 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.1 | 0.6 | GO:0035276 | ethanol binding(GO:0035276) |
| 0.1 | 0.7 | GO:0044729 | hemi-methylated DNA-binding(GO:0044729) |
| 0.1 | 0.4 | GO:0004938 | alpha2-adrenergic receptor activity(GO:0004938) |
| 0.1 | 0.5 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.1 | 0.3 | GO:0070506 | high-density lipoprotein particle receptor activity(GO:0070506) |
| 0.1 | 0.5 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.1 | 0.3 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.1 | 0.3 | GO:0015403 | thiamine uptake transmembrane transporter activity(GO:0015403) |
| 0.1 | 0.4 | GO:0015154 | sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.1 | 0.3 | GO:0017095 | heparan sulfate 6-O-sulfotransferase activity(GO:0017095) |
| 0.1 | 0.4 | GO:0001010 | transcription factor activity, sequence-specific DNA binding transcription factor recruiting(GO:0001010) |
| 0.1 | 0.6 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) |
| 0.1 | 0.5 | GO:0097001 | glycosphingolipid binding(GO:0043208) ceramide binding(GO:0097001) |
| 0.1 | 0.7 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.1 | 0.3 | GO:0017099 | very-long-chain-acyl-CoA dehydrogenase activity(GO:0017099) |
| 0.1 | 0.3 | GO:0033142 | progesterone receptor binding(GO:0033142) |
| 0.1 | 0.5 | GO:0010465 | neurotrophin p75 receptor binding(GO:0005166) nerve growth factor receptor activity(GO:0010465) |
| 0.1 | 0.2 | GO:0003826 | alpha-ketoacid dehydrogenase activity(GO:0003826) 3-methyl-2-oxobutanoate dehydrogenase (2-methylpropanoyl-transferring) activity(GO:0003863) |
| 0.1 | 0.5 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.1 | 0.4 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.1 | 0.4 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.1 | 0.7 | GO:0004897 | ciliary neurotrophic factor receptor activity(GO:0004897) |
| 0.1 | 0.3 | GO:0008427 | calcium-dependent protein kinase inhibitor activity(GO:0008427) |
| 0.1 | 1.7 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.1 | 0.3 | GO:0042903 | tubulin deacetylase activity(GO:0042903) |
| 0.1 | 0.2 | GO:0030272 | 5-formyltetrahydrofolate cyclo-ligase activity(GO:0030272) |
| 0.1 | 0.8 | GO:0048273 | mitogen-activated protein kinase p38 binding(GO:0048273) |
| 0.1 | 0.3 | GO:0050436 | microfibril binding(GO:0050436) |
| 0.0 | 0.1 | GO:0004584 | dolichyl-phosphate-mannose-glycolipid alpha-mannosyltransferase activity(GO:0004584) |
| 0.0 | 0.2 | GO:0046964 | 3'-phosphoadenosine 5'-phosphosulfate transmembrane transporter activity(GO:0046964) |
| 0.0 | 0.2 | GO:0008426 | protein kinase C inhibitor activity(GO:0008426) |
| 0.0 | 0.2 | GO:0015375 | glycine:sodium symporter activity(GO:0015375) |
| 0.0 | 0.8 | GO:0005402 | sugar:proton symporter activity(GO:0005351) cation:sugar symporter activity(GO:0005402) |
| 0.0 | 0.4 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.0 | 0.6 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 0.3 | GO:0047184 | 1-acylglycerophosphocholine O-acyltransferase activity(GO:0047184) |
| 0.0 | 0.7 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 0.4 | GO:0044388 | small protein activating enzyme binding(GO:0044388) |
| 0.0 | 0.9 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 0.1 | GO:0051717 | inositol-1,3,4,5-tetrakisphosphate 3-phosphatase activity(GO:0051717) phosphatidylinositol-3,4-bisphosphate 3-phosphatase activity(GO:0051800) |
| 0.0 | 0.2 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.0 | 0.8 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 0.0 | 0.2 | GO:0004489 | methylenetetrahydrofolate reductase (NAD(P)H) activity(GO:0004489) |
| 0.0 | 0.4 | GO:0000403 | Y-form DNA binding(GO:0000403) |
| 0.0 | 0.1 | GO:0061598 | nitrate reductase activity(GO:0008940) molybdopterin adenylyltransferase activity(GO:0061598) molybdopterin molybdotransferase activity(GO:0061599) |
| 0.0 | 0.3 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.0 | 0.3 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.0 | 0.5 | GO:0003857 | 3-hydroxyacyl-CoA dehydrogenase activity(GO:0003857) |
| 0.0 | 0.4 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.7 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.0 | 0.1 | GO:0010385 | double-stranded methylated DNA binding(GO:0010385) |
| 0.0 | 0.5 | GO:0034483 | heparan sulfate sulfotransferase activity(GO:0034483) |
| 0.0 | 0.1 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 0.4 | GO:0046972 | histone acetyltransferase activity (H4-K5 specific)(GO:0043995) histone acetyltransferase activity (H4-K8 specific)(GO:0043996) histone acetyltransferase activity (H4-K16 specific)(GO:0046972) |
| 0.0 | 0.5 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.0 | 0.7 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 0.7 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.0 | 0.3 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.0 | 0.2 | GO:0086083 | cell adhesive protein binding involved in bundle of His cell-Purkinje myocyte communication(GO:0086083) |
| 0.0 | 0.1 | GO:0051766 | inositol trisphosphate kinase activity(GO:0051766) |
| 0.0 | 0.4 | GO:0010857 | calcium-dependent protein kinase activity(GO:0010857) |
| 0.0 | 0.2 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.0 | 0.2 | GO:0030021 | extracellular matrix structural constituent conferring compression resistance(GO:0030021) structural constituent of tooth enamel(GO:0030345) |
| 0.0 | 0.1 | GO:0071535 | RING-like zinc finger domain binding(GO:0071535) |
| 0.0 | 1.7 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 0.2 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.0 | 0.2 | GO:0070679 | inositol 1,4,5 trisphosphate binding(GO:0070679) |
| 0.0 | 0.3 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.0 | 0.9 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.0 | 0.5 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.1 | GO:0042610 | CD8 receptor binding(GO:0042610) |
| 0.0 | 0.2 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.0 | 0.6 | GO:0050750 | low-density lipoprotein particle receptor binding(GO:0050750) |
| 0.0 | 0.2 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.0 | 0.4 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.0 | 0.6 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.0 | 0.2 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 0.0 | 1.9 | GO:0008094 | DNA-dependent ATPase activity(GO:0008094) |
| 0.0 | 0.3 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.0 | 2.9 | GO:0035326 | enhancer binding(GO:0035326) |
| 0.0 | 0.2 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.0 | 0.3 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.0 | 0.5 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.9 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.0 | 0.1 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.0 | 0.1 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.0 | 0.5 | GO:0001530 | lipopolysaccharide binding(GO:0001530) |
| 0.0 | 0.5 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.3 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.1 | GO:0071253 | connexin binding(GO:0071253) |
| 0.0 | 0.5 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.2 | GO:0001134 | transcription factor activity, transcription factor recruiting(GO:0001134) |
| 0.0 | 0.3 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.0 | 0.1 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.0 | 0.3 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.1 | GO:0035252 | UDP-xylosyltransferase activity(GO:0035252) xylosyltransferase activity(GO:0042285) |
| 0.0 | 0.3 | GO:0019789 | SUMO transferase activity(GO:0019789) |
| 0.0 | 0.7 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.0 | 0.1 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.0 | 0.1 | GO:0086062 | voltage-gated sodium channel activity involved in Purkinje myocyte action potential(GO:0086062) |
| 0.0 | 0.9 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.0 | 0.8 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 0.2 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 0.1 | GO:0034432 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) inositol diphosphate tetrakisphosphate diphosphatase activity(GO:0052840) inositol bisdiphosphate tetrakisphosphate diphosphatase activity(GO:0052841) inositol diphosphate pentakisphosphate diphosphatase activity(GO:0052842) inositol-1-diphosphate-2,3,4,5,6-pentakisphosphate diphosphatase activity(GO:0052843) inositol-3-diphosphate-1,2,4,5,6-pentakisphosphate diphosphatase activity(GO:0052844) inositol-5-diphosphate-1,2,3,4,6-pentakisphosphate diphosphatase activity(GO:0052845) inositol-1,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 1-diphosphatase activity(GO:0052846) inositol-1,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 5-diphosphatase activity(GO:0052847) inositol-3,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 5-diphosphatase activity(GO:0052848) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 4.4 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.1 | 0.2 | PID EPO PATHWAY | EPO signaling pathway |
| 0.1 | 2.5 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 4.4 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.0 | 0.1 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.0 | 1.9 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 1.0 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 1.7 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 0.3 | ST STAT3 PATHWAY | STAT3 Pathway |
| 0.0 | 0.4 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 0.5 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 0.9 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.0 | 0.5 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.0 | 0.7 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 1.0 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.0 | 0.3 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.0 | 1.3 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
| 0.0 | 0.1 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 0.1 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 0.7 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 0.8 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 0.1 | ST G ALPHA I PATHWAY | G alpha i Pathway |
| 0.0 | 0.6 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.3 | PID P53 REGULATION PATHWAY | p53 pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.1 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 1.1 | REACTOME DOPAMINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 0.0 | 0.4 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.0 | 0.6 | REACTOME SIGNALING BY NOTCH3 | Genes involved in Signaling by NOTCH3 |
| 0.0 | 0.5 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.0 | 0.8 | REACTOME SIGNALLING TO P38 VIA RIT AND RIN | Genes involved in Signalling to p38 via RIT and RIN |
| 0.0 | 0.5 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 1.2 | REACTOME RNA POL I TRANSCRIPTION INITIATION | Genes involved in RNA Polymerase I Transcription Initiation |
| 0.0 | 0.6 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 0.4 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 0.8 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.2 | REACTOME REGULATION OF ORNITHINE DECARBOXYLASE ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |
| 0.0 | 1.0 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 0.4 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.5 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.0 | 0.1 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 0.4 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.0 | 0.7 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.5 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.0 | 1.3 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 1.0 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.3 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.0 | 0.1 | REACTOME PRE NOTCH EXPRESSION AND PROCESSING | Genes involved in Pre-NOTCH Expression and Processing |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 0.6 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.0 | 0.2 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.0 | 0.3 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 0.3 | REACTOME GROWTH HORMONE RECEPTOR SIGNALING | Genes involved in Growth hormone receptor signaling |
| 0.0 | 0.1 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.0 | 0.3 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 0.5 | REACTOME FORMATION OF FIBRIN CLOT CLOTTING CASCADE | Genes involved in Formation of Fibrin Clot (Clotting Cascade) |
| 0.0 | 0.3 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |