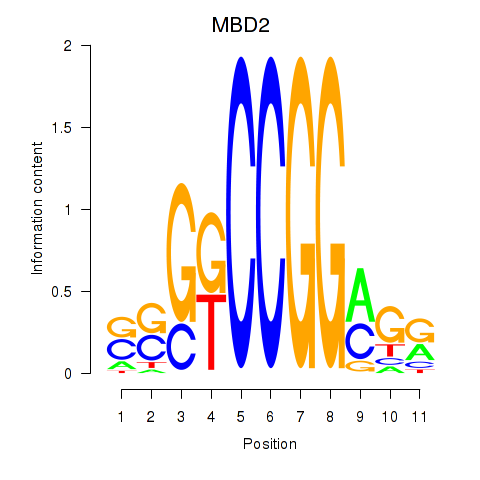
|
chr17_+_67825664
Show fit
|
3.40 |
ENST00000321892.8
|
BPTF
|
bromodomain PHD finger transcription factor
|
|
chr17_+_67825494
Show fit
|
3.32 |
ENST00000306378.11
ENST00000544778.6
|
BPTF
|
bromodomain PHD finger transcription factor
|
|
chr18_-_25352116
Show fit
|
2.26 |
ENST00000584787.5
ENST00000538137.6
ENST00000361524.8
|
ZNF521
|
zinc finger protein 521
|
|
chr1_+_81800906
Show fit
|
2.20 |
ENST00000674393.1
ENST00000674208.1
|
ADGRL2
|
adhesion G protein-coupled receptor L2
|
|
chr8_+_37796850
Show fit
|
2.14 |
ENST00000412232.3
|
ADGRA2
|
adhesion G protein-coupled receptor A2
|
|
chr8_+_37796906
Show fit
|
2.08 |
ENST00000315215.11
|
ADGRA2
|
adhesion G protein-coupled receptor A2
|
|
chr8_-_28386073
Show fit
|
2.01 |
ENST00000523095.5
ENST00000522795.1
|
ZNF395
|
zinc finger protein 395
|
|
chr3_-_18424533
Show fit
|
1.89 |
ENST00000417717.6
|
SATB1
|
SATB homeobox 1
|
|
chr8_-_28386417
Show fit
|
1.83 |
ENST00000521185.5
ENST00000520290.5
ENST00000344423.10
|
ZNF395
|
zinc finger protein 395
|
|
chr1_-_154502402
Show fit
|
1.74 |
ENST00000304760.3
|
SHE
|
Src homology 2 domain containing E
|
|
chr17_-_64919456
Show fit
|
1.68 |
ENST00000339474.9
ENST00000584306.6
ENST00000581368.5
|
LRRC37A3
|
leucine rich repeat containing 37 member A3
|
|
chr1_+_81800368
Show fit
|
1.64 |
ENST00000674489.1
ENST00000674442.1
ENST00000674419.1
ENST00000674407.1
ENST00000674168.1
ENST00000674307.1
ENST00000674209.1
ENST00000370715.5
ENST00000370713.5
ENST00000319517.10
ENST00000627151.2
ENST00000370717.6
|
ADGRL2
|
adhesion G protein-coupled receptor L2
|
|
chr19_-_14090695
Show fit
|
1.57 |
ENST00000533683.7
|
SAMD1
|
sterile alpha motif domain containing 1
|
|
chr14_+_52553273
Show fit
|
1.55 |
ENST00000542169.6
ENST00000555622.1
|
GPR137C
|
G protein-coupled receptor 137C
|
|
chr14_+_52552830
Show fit
|
1.52 |
ENST00000321662.11
|
GPR137C
|
G protein-coupled receptor 137C
|
|
chr12_+_19129779
Show fit
|
1.49 |
ENST00000539256.5
ENST00000299275.10
ENST00000538714.5
|
PLEKHA5
|
pleckstrin homology domain containing A5
|
|
chr9_+_136712531
Show fit
|
1.44 |
ENST00000371692.9
|
DIPK1B
|
divergent protein kinase domain 1B
|
|
chr4_-_25862979
Show fit
|
1.42 |
ENST00000399878.8
|
SEL1L3
|
SEL1L family member 3
|
|
chr12_+_27244222
Show fit
|
1.40 |
ENST00000545470.5
ENST00000389032.8
ENST00000540996.5
|
STK38L
|
serine/threonine kinase 38 like
|
|
chr15_-_68820861
Show fit
|
1.38 |
ENST00000560303.1
ENST00000465139.6
|
ANP32A
|
acidic nuclear phosphoprotein 32 family member A
|
|
chr1_+_6785518
Show fit
|
1.27 |
ENST00000467404.6
|
CAMTA1
|
calmodulin binding transcription activator 1
|
|
chr6_+_37353972
Show fit
|
1.17 |
ENST00000373479.9
|
RNF8
|
ring finger protein 8
|
|
chr3_+_37861926
Show fit
|
1.16 |
ENST00000443503.6
|
CTDSPL
|
CTD small phosphatase like
|
|
chr11_+_44726811
Show fit
|
1.16 |
ENST00000533202.5
ENST00000520358.7
ENST00000533080.5
ENST00000520999.6
|
TSPAN18
|
tetraspanin 18
|
|
chr6_+_15248855
Show fit
|
1.16 |
ENST00000397311.4
|
JARID2
|
jumonji and AT-rich interaction domain containing 2
|
|
chr15_-_64775574
Show fit
|
1.14 |
ENST00000300069.5
|
RBPMS2
|
RNA binding protein, mRNA processing factor 2
|
|
chr3_-_125055987
Show fit
|
1.14 |
ENST00000311127.9
|
HEG1
|
heart development protein with EGF like domains 1
|
|
chr11_-_2139382
Show fit
|
1.14 |
ENST00000416167.7
|
IGF2
|
insulin like growth factor 2
|
|
chr6_-_16761447
Show fit
|
1.12 |
ENST00000244769.8
ENST00000436367.6
|
ATXN1
|
ataxin 1
|
|
chr3_-_18425295
Show fit
|
1.09 |
ENST00000338745.11
ENST00000450898.1
|
SATB1
|
SATB homeobox 1
|
|
chr7_-_28958321
Show fit
|
1.07 |
ENST00000539664.3
|
TRIL
|
TLR4 interactor with leucine rich repeats
|
|
chr18_-_55588535
Show fit
|
1.06 |
ENST00000566286.5
ENST00000566279.5
ENST00000626595.2
ENST00000564999.5
ENST00000616053.4
ENST00000356073.8
|
TCF4
|
transcription factor 4
|
|
chr2_+_47403116
Show fit
|
1.06 |
ENST00000645506.1
ENST00000406134.5
ENST00000233146.7
|
MSH2
|
mutS homolog 2
|
|
chr1_+_108560031
Show fit
|
1.04 |
ENST00000405454.1
ENST00000370035.8
|
FAM102B
|
family with sequence similarity 102 member B
|
|
chr11_-_75525925
Show fit
|
1.03 |
ENST00000336898.8
|
GDPD5
|
glycerophosphodiester phosphodiesterase domain containing 5
|
|
chr11_+_74748831
Show fit
|
1.01 |
ENST00000299563.5
|
RNF169
|
ring finger protein 169
|
|
chr11_-_89491131
Show fit
|
1.00 |
ENST00000343727.9
ENST00000531342.5
ENST00000375979.7
|
NOX4
|
NADPH oxidase 4
|
|
chr1_+_84078043
Show fit
|
0.97 |
ENST00000370689.6
ENST00000370688.7
|
PRKACB
|
protein kinase cAMP-activated catalytic subunit beta
|
|
chr2_+_42048012
Show fit
|
0.96 |
ENST00000294964.6
|
PKDCC
|
protein kinase domain containing, cytoplasmic
|
|
chr5_-_180649613
Show fit
|
0.94 |
ENST00000393347.7
ENST00000619105.4
|
FLT4
|
fms related receptor tyrosine kinase 4
|
|
chr19_-_11262499
Show fit
|
0.94 |
ENST00000294618.12
|
DOCK6
|
dedicator of cytokinesis 6
|
|
chr14_-_89417148
Show fit
|
0.93 |
ENST00000557258.6
|
FOXN3
|
forkhead box N3
|
|
chr11_-_89491320
Show fit
|
0.92 |
ENST00000534731.5
ENST00000527626.5
ENST00000525196.5
ENST00000263317.9
|
NOX4
|
NADPH oxidase 4
|
|
chr10_+_128047559
Show fit
|
0.90 |
ENST00000306042.9
|
PTPRE
|
protein tyrosine phosphatase receptor type E
|
|
chrX_-_63755187
Show fit
|
0.90 |
ENST00000635729.1
ENST00000623566.3
|
ARHGEF9
|
Cdc42 guanine nucleotide exchange factor 9
|
|
chr13_+_20703677
Show fit
|
0.89 |
ENST00000682841.1
|
IL17D
|
interleukin 17D
|
|
chrX_-_63754664
Show fit
|
0.88 |
ENST00000677315.1
ENST00000636392.1
ENST00000637040.1
ENST00000637178.1
ENST00000637557.1
ENST00000636048.1
ENST00000638021.1
ENST00000672513.1
|
ENSG00000288661.1
ARHGEF9
|
novel protein
Cdc42 guanine nucleotide exchange factor 9
|
|
chr10_-_124744280
Show fit
|
0.88 |
ENST00000337318.8
|
FAM53B
|
family with sequence similarity 53 member B
|
|
chr20_-_48827992
Show fit
|
0.88 |
ENST00000371941.4
|
PREX1
|
phosphatidylinositol-3,4,5-trisphosphate dependent Rac exchange factor 1
|
|
chr9_+_87498491
Show fit
|
0.87 |
ENST00000622514.4
|
DAPK1
|
death associated protein kinase 1
|
|
chr22_+_24952705
Show fit
|
0.87 |
ENST00000358431.8
|
KIAA1671
|
KIAA1671
|
|
chrX_-_63755032
Show fit
|
0.86 |
ENST00000624538.2
ENST00000636276.1
ENST00000624843.3
ENST00000671907.1
ENST00000624210.3
ENST00000374870.8
ENST00000635967.1
ENST00000253401.10
ENST00000672194.1
ENST00000637723.2
ENST00000637417.1
ENST00000637520.1
ENST00000374872.4
ENST00000636926.1
|
ARHGEF9
|
Cdc42 guanine nucleotide exchange factor 9
|
|
chr19_-_14090963
Show fit
|
0.83 |
ENST00000269724.5
|
SAMD1
|
sterile alpha motif domain containing 1
|
|
chr19_-_18940289
Show fit
|
0.83 |
ENST00000542541.6
ENST00000433218.6
|
HOMER3
|
homer scaffold protein 3
|
|
chr6_+_89433164
Show fit
|
0.81 |
ENST00000369408.9
ENST00000447838.6
|
ANKRD6
|
ankyrin repeat domain 6
|
|
chr18_-_55588184
Show fit
|
0.80 |
ENST00000354452.8
ENST00000565908.6
ENST00000635822.2
|
TCF4
|
transcription factor 4
|
|
chr5_-_180649579
Show fit
|
0.80 |
ENST00000261937.11
ENST00000502649.5
|
FLT4
|
fms related receptor tyrosine kinase 4
|
|
chr19_+_39413528
Show fit
|
0.79 |
ENST00000438123.5
ENST00000409797.6
ENST00000451354.6
|
PLEKHG2
|
pleckstrin homology and RhoGEF domain containing G2
|
|
chr19_-_42255119
Show fit
|
0.79 |
ENST00000222329.9
ENST00000594664.1
|
ERF
ENSG00000268643.2
|
ETS2 repressor factor
novel protein
|
|
chr1_+_154502207
Show fit
|
0.79 |
ENST00000368482.8
|
TDRD10
|
tudor domain containing 10
|
|
chr19_-_17245889
Show fit
|
0.78 |
ENST00000291442.4
|
NR2F6
|
nuclear receptor subfamily 2 group F member 6
|
|
chr18_+_79395856
Show fit
|
0.78 |
ENST00000253506.9
ENST00000591814.5
ENST00000427363.7
|
NFATC1
|
nuclear factor of activated T cells 1
|
|
chr16_-_89720861
Show fit
|
0.78 |
ENST00000389386.8
|
VPS9D1
|
VPS9 domain containing 1
|
|
chr17_+_19648723
Show fit
|
0.77 |
ENST00000672357.1
ENST00000584332.6
ENST00000176643.11
ENST00000339618.8
ENST00000579855.5
ENST00000671878.1
ENST00000395575.7
|
ALDH3A2
|
aldehyde dehydrogenase 3 family member A2
|
|
chr16_-_49856105
Show fit
|
0.76 |
ENST00000563137.7
|
ZNF423
|
zinc finger protein 423
|
|
chr6_+_89433127
Show fit
|
0.74 |
ENST00000339746.9
|
ANKRD6
|
ankyrin repeat domain 6
|
|
chr11_-_27700472
Show fit
|
0.74 |
ENST00000418212.5
ENST00000533246.5
|
BDNF
|
brain derived neurotrophic factor
|
|
chr19_-_11197516
Show fit
|
0.73 |
ENST00000592903.5
ENST00000586659.6
ENST00000589359.5
ENST00000588724.5
|
KANK2
|
KN motif and ankyrin repeat domains 2
|
|
chr8_-_51899002
Show fit
|
0.73 |
ENST00000522514.6
|
PCMTD1
|
protein-L-isoaspartate (D-aspartate) O-methyltransferase domain containing 1
|
|
chr2_+_180980347
Show fit
|
0.73 |
ENST00000602959.5
ENST00000602479.5
ENST00000392415.6
ENST00000602291.5
|
UBE2E3
|
ubiquitin conjugating enzyme E2 E3
|
|
chr11_-_27700447
Show fit
|
0.73 |
ENST00000356660.9
|
BDNF
|
brain derived neurotrophic factor
|
|
chr1_+_33256479
Show fit
|
0.70 |
ENST00000539719.6
ENST00000483388.5
|
ZNF362
|
zinc finger protein 362
|
|
chr2_+_10911924
Show fit
|
0.70 |
ENST00000295082.3
|
KCNF1
|
potassium voltage-gated channel modifier subfamily F member 1
|
|
chr2_+_45651650
Show fit
|
0.69 |
ENST00000306156.8
|
PRKCE
|
protein kinase C epsilon
|
|
chr12_-_57430778
Show fit
|
0.69 |
ENST00000448732.1
ENST00000634871.1
|
R3HDM2
|
R3H domain containing 2
|
|
chr12_-_57430956
Show fit
|
0.68 |
ENST00000347140.7
ENST00000402412.5
|
R3HDM2
|
R3H domain containing 2
|
|
chr18_+_79395942
Show fit
|
0.68 |
ENST00000397790.6
|
NFATC1
|
nuclear factor of activated T cells 1
|
|
chr13_-_28495079
Show fit
|
0.68 |
ENST00000615840.4
ENST00000282397.9
ENST00000541932.5
ENST00000539099.1
ENST00000639477.1
|
FLT1
|
fms related receptor tyrosine kinase 1
|
|
chr5_+_123512214
Show fit
|
0.67 |
ENST00000511130.6
ENST00000512718.7
|
CSNK1G3
|
casein kinase 1 gamma 3
|
|
chr6_+_37354046
Show fit
|
0.67 |
ENST00000487950.1
ENST00000469731.5
|
RNF8
|
ring finger protein 8
|
|
chr3_-_185825029
Show fit
|
0.66 |
ENST00000382199.7
|
IGF2BP2
|
insulin like growth factor 2 mRNA binding protein 2
|
|
chr19_+_35000426
Show fit
|
0.66 |
ENST00000411896.6
ENST00000424536.2
|
GRAMD1A
|
GRAM domain containing 1A
|
|
chr9_-_127854636
Show fit
|
0.65 |
ENST00000344849.4
ENST00000373203.9
|
ENG
|
endoglin
|
|
chr3_-_128487916
Show fit
|
0.65 |
ENST00000430265.6
|
GATA2
|
GATA binding protein 2
|
|
chr3_-_185824966
Show fit
|
0.64 |
ENST00000457616.6
ENST00000346192.7
|
IGF2BP2
|
insulin like growth factor 2 mRNA binding protein 2
|
|
chr18_-_36828771
Show fit
|
0.63 |
ENST00000589049.5
ENST00000587129.5
ENST00000590842.5
|
TPGS2
|
tubulin polyglutamylase complex subunit 2
|
|
chr11_-_63768762
Show fit
|
0.63 |
ENST00000433688.2
|
C11orf95
|
chromosome 11 open reading frame 95
|
|
chrX_-_54496212
Show fit
|
0.63 |
ENST00000375135.4
|
FGD1
|
FYVE, RhoGEF and PH domain containing 1
|
|
chr4_-_77819356
Show fit
|
0.62 |
ENST00000649644.1
ENST00000504123.6
ENST00000515441.2
|
CNOT6L
|
CCR4-NOT transcription complex subunit 6 like
|
|
chr22_+_19714450
Show fit
|
0.62 |
ENST00000455784.7
ENST00000406395.5
|
SEPTIN5
|
septin 5
|
|
chr17_+_19648915
Show fit
|
0.62 |
ENST00000672567.1
ENST00000672709.1
|
ALDH3A2
|
aldehyde dehydrogenase 3 family member A2
|
|
chr19_+_35000275
Show fit
|
0.62 |
ENST00000317991.10
ENST00000680623.1
|
GRAMD1A
|
GRAM domain containing 1A
|
|
chr17_-_28951285
Show fit
|
0.62 |
ENST00000577226.5
|
PHF12
|
PHD finger protein 12
|
|
chr16_+_2969270
Show fit
|
0.61 |
ENST00000293978.12
|
PAQR4
|
progestin and adipoQ receptor family member 4
|
|
chr16_+_2969307
Show fit
|
0.61 |
ENST00000576565.1
ENST00000318782.9
|
PAQR4
|
progestin and adipoQ receptor family member 4
|
|
chr18_-_36829154
Show fit
|
0.61 |
ENST00000614939.4
ENST00000610723.4
|
TPGS2
|
tubulin polyglutamylase complex subunit 2
|
|
chr4_+_26860778
Show fit
|
0.61 |
ENST00000467011.6
|
STIM2
|
stromal interaction molecule 2
|
|
chr16_+_2969548
Show fit
|
0.60 |
ENST00000572687.1
|
PAQR4
|
progestin and adipoQ receptor family member 4
|
|
chr2_+_216633411
Show fit
|
0.60 |
ENST00000233809.9
|
IGFBP2
|
insulin like growth factor binding protein 2
|
|
chr11_-_47176851
Show fit
|
0.60 |
ENST00000629231.2
ENST00000526342.5
ENST00000528444.5
ENST00000530596.5
ENST00000525398.5
ENST00000524782.6
ENST00000527927.5
ENST00000525314.5
|
ARFGAP2
|
ADP ribosylation factor GTPase activating protein 2
|
|
chr3_-_184017863
Show fit
|
0.60 |
ENST00000427120.6
ENST00000334444.11
ENST00000392579.6
ENST00000382494.6
ENST00000265586.10
ENST00000446941.2
|
ABCC5
|
ATP binding cassette subfamily C member 5
|
|
chr12_+_57089094
Show fit
|
0.59 |
ENST00000342556.6
ENST00000300131.8
|
NAB2
|
NGFI-A binding protein 2
|
|
chr12_+_52948840
Show fit
|
0.58 |
ENST00000388837.6
ENST00000550600.5
|
KRT18
|
keratin 18
|
|
chr6_+_2765361
Show fit
|
0.58 |
ENST00000380773.9
ENST00000380771.8
ENST00000618555.4
|
WRNIP1
|
WRN helicase interacting protein 1
|
|
chr7_+_69599588
Show fit
|
0.58 |
ENST00000403018.3
|
AUTS2
|
activator of transcription and developmental regulator AUTS2
|
|
chr11_+_123430259
Show fit
|
0.57 |
ENST00000533341.3
ENST00000635736.2
|
GRAMD1B
|
GRAM domain containing 1B
|
|
chr1_+_215567279
Show fit
|
0.57 |
ENST00000259154.9
|
KCTD3
|
potassium channel tetramerization domain containing 3
|
|
chr20_+_325536
Show fit
|
0.56 |
ENST00000342665.5
|
SOX12
|
SRY-box transcription factor 12
|
|
chr18_-_812230
Show fit
|
0.56 |
ENST00000314574.5
|
YES1
|
YES proto-oncogene 1, Src family tyrosine kinase
|
|
chr3_+_33114007
Show fit
|
0.56 |
ENST00000320954.11
|
CRTAP
|
cartilage associated protein
|
|
chr2_-_53786916
Show fit
|
0.55 |
ENST00000406687.5
ENST00000263634.8
ENST00000394717.3
|
ASB3
|
ankyrin repeat and SOCS box containing 3
|
|
chr17_-_48613468
Show fit
|
0.54 |
ENST00000498634.2
|
HOXB8
|
homeobox B8
|
|
chr17_-_63842663
Show fit
|
0.54 |
ENST00000613943.4
ENST00000448276.7
|
SMARCD2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 2
|
|
chr9_+_128068172
Show fit
|
0.54 |
ENST00000373068.6
ENST00000373069.10
|
SLC25A25
|
solute carrier family 25 member 25
|
|
chr13_+_34942263
Show fit
|
0.53 |
ENST00000379939.7
ENST00000400445.7
|
NBEA
|
neurobeachin
|
|
chr11_-_119340544
Show fit
|
0.53 |
ENST00000530681.2
|
C1QTNF5
|
C1q and TNF related 5
|
|
chr1_+_109466527
Show fit
|
0.53 |
ENST00000369872.4
|
SYPL2
|
synaptophysin like 2
|
|
chr2_+_180980566
Show fit
|
0.53 |
ENST00000410062.9
|
UBE2E3
|
ubiquitin conjugating enzyme E2 E3
|
|
chr1_+_52142044
Show fit
|
0.52 |
ENST00000287727.8
ENST00000371591.2
|
ZFYVE9
|
zinc finger FYVE-type containing 9
|
|
chr10_-_102120246
Show fit
|
0.51 |
ENST00000425280.2
|
LDB1
|
LIM domain binding 1
|
|
chr18_-_812516
Show fit
|
0.51 |
ENST00000584307.5
|
YES1
|
YES proto-oncogene 1, Src family tyrosine kinase
|
|
chr17_+_19648792
Show fit
|
0.50 |
ENST00000630662.2
|
ALDH3A2
|
aldehyde dehydrogenase 3 family member A2
|
|
chr11_-_117098415
Show fit
|
0.50 |
ENST00000445177.6
ENST00000375300.6
ENST00000446921.6
|
SIK3
|
SIK family kinase 3
|
|
chr17_+_56593685
Show fit
|
0.49 |
ENST00000332822.6
|
NOG
|
noggin
|
|
chr4_+_26860809
Show fit
|
0.49 |
ENST00000465503.6
ENST00000467087.7
|
STIM2
|
stromal interaction molecule 2
|
|
chr17_+_40121955
Show fit
|
0.49 |
ENST00000398532.9
|
MSL1
|
MSL complex subunit 1
|
|
chr7_+_87152531
Show fit
|
0.49 |
ENST00000432366.6
ENST00000423590.6
ENST00000394703.9
|
DMTF1
|
cyclin D binding myb like transcription factor 1
|
|
chr17_+_76868396
Show fit
|
0.48 |
ENST00000569840.7
|
MGAT5B
|
alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase B
|
|
chr2_+_112275588
Show fit
|
0.48 |
ENST00000409871.6
ENST00000343936.4
|
ZC3H6
|
zinc finger CCCH-type containing 6
|
|
chr5_+_168529299
Show fit
|
0.47 |
ENST00000338333.5
|
FBLL1
|
fibrillarin like 1
|
|
chr1_-_6485433
Show fit
|
0.47 |
ENST00000535355.6
|
PLEKHG5
|
pleckstrin homology and RhoGEF domain containing G5
|
|
chr8_-_73746830
Show fit
|
0.47 |
ENST00000524300.6
ENST00000523558.5
ENST00000521210.5
ENST00000355780.9
ENST00000524104.5
ENST00000521736.5
ENST00000521447.5
ENST00000517542.5
ENST00000521451.5
ENST00000521419.5
ENST00000518502.5
|
STAU2
|
staufen double-stranded RNA binding protein 2
|
|
chr10_+_23439060
Show fit
|
0.47 |
ENST00000376495.5
|
OTUD1
|
OTU deubiquitinase 1
|
|
chr16_+_58025745
Show fit
|
0.47 |
ENST00000219271.4
|
MMP15
|
matrix metallopeptidase 15
|
|
chr11_+_43358908
Show fit
|
0.47 |
ENST00000039989.9
ENST00000299240.10
|
TTC17
|
tetratricopeptide repeat domain 17
|
|
chr12_-_57006476
Show fit
|
0.47 |
ENST00000300101.3
|
ZBTB39
|
zinc finger and BTB domain containing 39
|
|
chr13_-_80341100
Show fit
|
0.46 |
ENST00000377104.4
|
SPRY2
|
sprouty RTK signaling antagonist 2
|
|
chr17_-_28951443
Show fit
|
0.46 |
ENST00000268756.7
ENST00000332830.9
ENST00000584685.1
|
PHF12
|
PHD finger protein 12
|
|
chr7_+_87152409
Show fit
|
0.46 |
ENST00000413276.6
ENST00000446796.6
ENST00000420131.5
ENST00000414630.6
ENST00000453049.5
ENST00000428819.5
ENST00000448598.5
ENST00000449088.7
ENST00000430405.7
ENST00000331242.12
|
DMTF1
|
cyclin D binding myb like transcription factor 1
|
|
chr4_-_6472548
Show fit
|
0.46 |
ENST00000382599.9
|
PPP2R2C
|
protein phosphatase 2 regulatory subunit Bgamma
|
|
chr17_+_29593118
Show fit
|
0.45 |
ENST00000394859.8
|
ANKRD13B
|
ankyrin repeat domain 13B
|
|
chr1_-_46719074
Show fit
|
0.45 |
ENST00000371933.8
ENST00000672422.2
ENST00000674263.1
ENST00000674415.1
ENST00000674435.1
|
EFCAB14
|
EF-hand calcium binding domain 14
|
|
chr17_+_75525682
Show fit
|
0.45 |
ENST00000392550.8
ENST00000167462.9
ENST00000375227.8
ENST00000578363.5
ENST00000579392.5
|
LLGL2
|
LLGL scribble cell polarity complex component 2
|
|
chr4_-_16226460
Show fit
|
0.44 |
ENST00000405303.7
|
TAPT1
|
transmembrane anterior posterior transformation 1
|
|
chr18_+_7567266
Show fit
|
0.44 |
ENST00000580170.6
|
PTPRM
|
protein tyrosine phosphatase receptor type M
|
|
chr16_+_53434430
Show fit
|
0.44 |
ENST00000262133.11
|
RBL2
|
RB transcriptional corepressor like 2
|
|
chr10_+_79068955
Show fit
|
0.43 |
ENST00000334512.10
|
ZMIZ1
|
zinc finger MIZ-type containing 1
|
|
chr16_+_70114306
Show fit
|
0.43 |
ENST00000288050.9
ENST00000398122.7
ENST00000568530.5
|
PDPR
|
pyruvate dehydrogenase phosphatase regulatory subunit
|
|
chr7_-_94655993
Show fit
|
0.43 |
ENST00000647110.1
ENST00000647048.1
ENST00000643020.1
ENST00000644682.1
ENST00000646119.1
ENST00000646265.1
ENST00000645445.1
ENST00000647334.1
ENST00000645262.1
ENST00000428696.7
ENST00000647096.1
ENST00000642394.1
ENST00000645725.1
ENST00000647351.1
ENST00000646943.1
ENST00000648936.2
ENST00000642707.1
ENST00000645109.1
ENST00000646489.1
ENST00000642933.1
ENST00000643128.1
ENST00000445866.7
ENST00000645101.1
ENST00000644116.1
ENST00000642441.1
ENST00000646879.1
ENST00000647018.1
|
SGCE
|
sarcoglycan epsilon
|
|
chr16_+_29806519
Show fit
|
0.42 |
ENST00000322945.11
ENST00000562337.5
ENST00000566906.6
ENST00000563402.1
ENST00000219782.10
|
MAZ
|
MYC associated zinc finger protein
|
|
chr11_+_45146631
Show fit
|
0.42 |
ENST00000534751.3
ENST00000683152.1
|
PRDM11
|
PR/SET domain 11
|
|
chr14_-_39432414
Show fit
|
0.42 |
ENST00000554932.1
ENST00000298097.7
|
FBXO33
|
F-box protein 33
|
|
chr1_-_153946652
Show fit
|
0.41 |
ENST00000361217.9
|
DENND4B
|
DENN domain containing 4B
|
|
chrX_+_153072454
Show fit
|
0.41 |
ENST00000421798.5
|
PNMA6A
|
PNMA family member 6A
|
|
chr7_-_150978284
Show fit
|
0.41 |
ENST00000262186.10
|
KCNH2
|
potassium voltage-gated channel subfamily H member 2
|
|
chr17_+_66964638
Show fit
|
0.40 |
ENST00000262138.4
|
CACNG4
|
calcium voltage-gated channel auxiliary subunit gamma 4
|
|
chr6_+_156779363
Show fit
|
0.40 |
ENST00000494260.2
ENST00000636748.1
ENST00000636607.1
ENST00000414678.7
ENST00000638000.1
|
ARID1B
|
AT-rich interaction domain 1B
|
|
chr7_-_94656197
Show fit
|
0.40 |
ENST00000643903.1
ENST00000644122.1
ENST00000447873.6
ENST00000644816.1
ENST00000644375.1
|
SGCE
|
sarcoglycan epsilon
|
|
chr3_-_134250744
Show fit
|
0.40 |
ENST00000620660.4
|
RYK
|
receptor like tyrosine kinase
|
|
chr11_+_118359572
Show fit
|
0.39 |
ENST00000252108.8
ENST00000431736.6
|
UBE4A
|
ubiquitination factor E4A
|
|
chr2_+_177392734
Show fit
|
0.39 |
ENST00000680770.1
ENST00000637633.2
ENST00000679459.1
ENST00000409888.1
ENST00000264167.11
ENST00000642466.2
|
AGPS
|
alkylglycerone phosphate synthase
|
|
chr10_-_17617326
Show fit
|
0.39 |
ENST00000326961.6
ENST00000361271.8
|
HACD1
|
3-hydroxyacyl-CoA dehydratase 1
|
|
chr7_-_17940468
Show fit
|
0.39 |
ENST00000611725.4
ENST00000409604.1
ENST00000428135.7
|
SNX13
|
sorting nexin 13
|
|
chr21_+_41168142
Show fit
|
0.39 |
ENST00000330333.11
|
BACE2
|
beta-secretase 2
|
|
chr7_-_94656160
Show fit
|
0.39 |
ENST00000644609.1
ENST00000643272.1
ENST00000646137.1
ENST00000646098.1
ENST00000643193.1
ENST00000437425.7
ENST00000644551.1
ENST00000415788.3
|
SGCE
|
sarcoglycan epsilon
|
|
chr10_+_115093404
Show fit
|
0.38 |
ENST00000527407.5
|
ATRNL1
|
attractin like 1
|
|
chr1_-_35557378
Show fit
|
0.38 |
ENST00000325722.8
ENST00000469892.5
|
KIAA0319L
|
KIAA0319 like
|
|
chr2_-_219060914
Show fit
|
0.38 |
ENST00000295731.7
|
IHH
|
Indian hedgehog signaling molecule
|
|
chr3_+_14947680
Show fit
|
0.37 |
ENST00000435454.5
ENST00000323373.10
|
NR2C2
|
nuclear receptor subfamily 2 group C member 2
|
|
chr15_+_40323683
Show fit
|
0.37 |
ENST00000638170.2
|
INAFM2
|
InaF motif containing 2
|
|
chr9_+_22446808
Show fit
|
0.37 |
ENST00000325870.3
|
DMRTA1
|
DMRT like family A1
|
|
chr3_+_170357647
Show fit
|
0.36 |
ENST00000476188.5
ENST00000259119.9
ENST00000426052.6
|
SKIL
|
SKI like proto-oncogene
|
|
chr12_-_31590967
Show fit
|
0.36 |
ENST00000354285.8
|
DENND5B
|
DENN domain containing 5B
|
|
chr10_-_102120318
Show fit
|
0.36 |
ENST00000673968.1
|
LDB1
|
LIM domain binding 1
|
|
chr5_+_176810552
Show fit
|
0.36 |
ENST00000329542.9
|
UNC5A
|
unc-5 netrin receptor A
|
|
chr18_+_62523002
Show fit
|
0.35 |
ENST00000269499.10
|
ZCCHC2
|
zinc finger CCHC-type containing 2
|
|
chr1_-_6485941
Show fit
|
0.35 |
ENST00000676287.1
ENST00000400913.6
|
PLEKHG5
|
pleckstrin homology and RhoGEF domain containing G5
|
|
chr12_+_122078740
Show fit
|
0.35 |
ENST00000319080.12
|
MLXIP
|
MLX interacting protein
|
|
chr14_-_103057509
Show fit
|
0.35 |
ENST00000361246.7
|
CDC42BPB
|
CDC42 binding protein kinase beta
|
|
chr22_-_20016807
Show fit
|
0.35 |
ENST00000263207.8
|
ARVCF
|
ARVCF delta catenin family member
|
|
chr7_+_94656325
Show fit
|
0.35 |
ENST00000482108.1
ENST00000488574.5
ENST00000612748.1
ENST00000613043.1
|
PEG10
|
paternally expressed 10
|
|
chr21_+_41167774
Show fit
|
0.35 |
ENST00000328735.10
ENST00000347667.5
|
BACE2
|
beta-secretase 2
|
|
chr3_-_38029604
Show fit
|
0.35 |
ENST00000334661.5
|
PLCD1
|
phospholipase C delta 1
|
|
chr12_-_49828394
Show fit
|
0.34 |
ENST00000335999.7
|
NCKAP5L
|
NCK associated protein 5 like
|
|
chr3_+_14947568
Show fit
|
0.34 |
ENST00000413118.5
ENST00000425241.5
|
NR2C2
|
nuclear receptor subfamily 2 group C member 2
|
|
chr4_+_1793776
Show fit
|
0.34 |
ENST00000352904.6
|
FGFR3
|
fibroblast growth factor receptor 3
|
|
chr10_+_102644462
Show fit
|
0.33 |
ENST00000643721.2
ENST00000302424.12
|
TRIM8
|
tripartite motif containing 8
|
|
chr5_+_176810498
Show fit
|
0.33 |
ENST00000509580.2
|
UNC5A
|
unc-5 netrin receptor A
|
|
chr22_-_31346143
Show fit
|
0.33 |
ENST00000405309.7
ENST00000351933.8
|
PATZ1
|
POZ/BTB and AT hook containing zinc finger 1
|
|
chr2_-_99489955
Show fit
|
0.33 |
ENST00000393445.7
ENST00000258428.8
|
REV1
|
REV1 DNA directed polymerase
|
|
chr1_-_17011891
Show fit
|
0.33 |
ENST00000341676.9
ENST00000326735.13
ENST00000452699.5
|
ATP13A2
|
ATPase cation transporting 13A2
|
|
chr4_+_173168800
Show fit
|
0.33 |
ENST00000512285.5
ENST00000265000.9
|
GALNT7
|
polypeptide N-acetylgalactosaminyltransferase 7
|
|
chr5_-_141682192
Show fit
|
0.33 |
ENST00000508305.5
|
ARAP3
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 3
|
|
chr14_-_93115270
Show fit
|
0.32 |
ENST00000555553.5
ENST00000555495.5
ENST00000554999.5
|
ITPK1
|
inositol-tetrakisphosphate 1-kinase
|
|
chr10_+_102420823
Show fit
|
0.32 |
ENST00000369956.8
ENST00000425536.1
|
FBXL15
|
F-box and leucine rich repeat protein 15
|
|
chr14_+_73950489
Show fit
|
0.32 |
ENST00000554320.1
|
COQ6
|
coenzyme Q6, monooxygenase
|
|
chr17_+_29593468
Show fit
|
0.32 |
ENST00000614878.4
|
ANKRD13B
|
ankyrin repeat domain 13B
|
|
chr11_-_6655788
Show fit
|
0.32 |
ENST00000299441.5
|
DCHS1
|
dachsous cadherin-related 1
|
|
chr1_-_53328053
Show fit
|
0.32 |
ENST00000371454.6
ENST00000667377.1
ENST00000306052.12
ENST00000668448.1
|
LRP8
|
LDL receptor related protein 8
|
|
chr12_+_2877193
Show fit
|
0.32 |
ENST00000538636.5
ENST00000461997.5
ENST00000618250.4
ENST00000366285.5
ENST00000538700.2
ENST00000489288.7
|
RHNO1
|
RAD9-HUS1-RAD1 interacting nuclear orphan 1
|