Project
Inflammatory response time course, HUVEC (Wada, 2009)
Navigation
Downloads
Results for MEF2D_MEF2A
Z-value: 0.63
Transcription factors associated with MEF2D_MEF2A
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
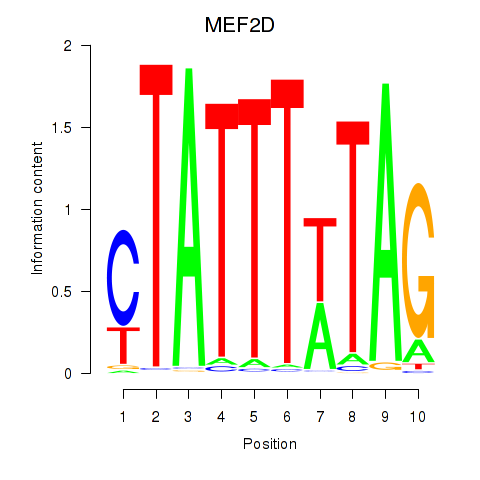
MEF2D
|
ENSG00000116604.18 | MEF2D |
|
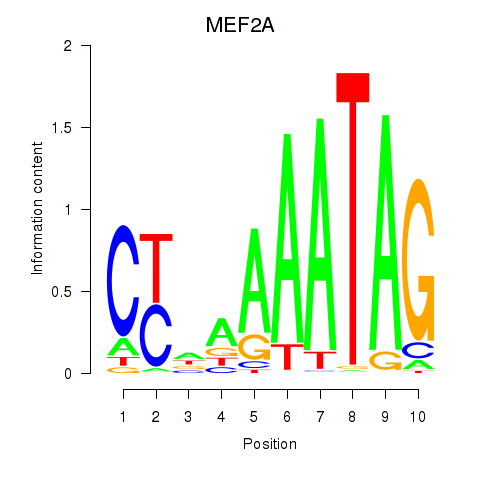
MEF2A
|
ENSG00000068305.17 | MEF2A |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| MEF2D | hg38_v1_chr1_-_156500763_156500851 | 0.39 | 5.7e-02 | Click! |
| MEF2A | hg38_v1_chr15_+_99566066_99566087 | -0.23 | 2.8e-01 | Click! |
Activity profile of MEF2D_MEF2A motif
Sorted Z-values of MEF2D_MEF2A motif
Network of associatons between targets according to the STRING database.
First level regulatory network of MEF2D_MEF2A
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_+_52051402 | 3.28 |
ENST00000243050.5
ENST00000550763.1 ENST00000394825.6 ENST00000394824.2 ENST00000548232.1 ENST00000562373.1 |
NR4A1
|
nuclear receptor subfamily 4 group A member 1 |
| chr2_-_156342348 | 1.70 |
ENST00000409572.5
|
NR4A2
|
nuclear receptor subfamily 4 group A member 2 |
| chr19_+_45469841 | 1.27 |
ENST00000592811.5
ENST00000586615.5 |
FOSB
|
FosB proto-oncogene, AP-1 transcription factor subunit |
| chr21_-_26843012 | 0.83 |
ENST00000517777.6
|
ADAMTS1
|
ADAM metallopeptidase with thrombospondin type 1 motif 1 |
| chr21_-_26843063 | 0.83 |
ENST00000678221.1
|
ADAMTS1
|
ADAM metallopeptidase with thrombospondin type 1 motif 1 |
| chr1_-_169734064 | 0.81 |
ENST00000333360.12
|
SELE
|
selectin E |
| chr6_+_135851681 | 0.79 |
ENST00000308191.11
|
PDE7B
|
phosphodiesterase 7B |
| chr4_-_148442342 | 0.73 |
ENST00000358102.8
|
NR3C2
|
nuclear receptor subfamily 3 group C member 2 |
| chr18_+_23135452 | 0.71 |
ENST00000580153.5
ENST00000256925.12 |
CABLES1
|
Cdk5 and Abl enzyme substrate 1 |
| chr12_+_12785652 | 0.70 |
ENST00000356591.5
|
APOLD1
|
apolipoprotein L domain containing 1 |
| chr8_+_40153475 | 0.70 |
ENST00000315792.5
|
TCIM
|
transcriptional and immune response regulator |
| chr3_+_194136138 | 0.69 |
ENST00000232424.4
|
HES1
|
hes family bHLH transcription factor 1 |
| chr2_-_207166818 | 0.66 |
ENST00000423015.5
|
KLF7
|
Kruppel like factor 7 |
| chr5_+_54455661 | 0.60 |
ENST00000302005.3
|
HSPB3
|
heat shock protein family B (small) member 3 |
| chr9_+_124291935 | 0.60 |
ENST00000546191.5
|
NEK6
|
NIMA related kinase 6 |
| chr9_-_94593810 | 0.60 |
ENST00000375337.4
|
FBP2
|
fructose-bisphosphatase 2 |
| chr2_+_238138661 | 0.58 |
ENST00000409223.2
|
KLHL30
|
kelch like family member 30 |
| chr5_+_173889337 | 0.58 |
ENST00000520867.5
ENST00000334035.9 |
CPEB4
|
cytoplasmic polyadenylation element binding protein 4 |
| chr1_-_58784035 | 0.54 |
ENST00000371222.4
|
JUN
|
Jun proto-oncogene, AP-1 transcription factor subunit |
| chr5_-_111756245 | 0.53 |
ENST00000447165.6
|
NREP
|
neuronal regeneration related protein |
| chr2_-_105438503 | 0.52 |
ENST00000393352.7
ENST00000607522.1 |
FHL2
|
four and a half LIM domains 2 |
| chr17_+_7855055 | 0.51 |
ENST00000574668.1
ENST00000301599.7 |
TMEM88
|
transmembrane protein 88 |
| chr3_+_8501846 | 0.51 |
ENST00000454244.4
|
LMCD1
|
LIM and cysteine rich domains 1 |
| chr7_-_41703062 | 0.50 |
ENST00000242208.5
|
INHBA
|
inhibin subunit beta A |
| chr11_-_19202004 | 0.50 |
ENST00000648719.1
|
CSRP3
|
cysteine and glycine rich protein 3 |
| chr11_+_9575496 | 0.50 |
ENST00000681684.1
|
WEE1
|
WEE1 G2 checkpoint kinase |
| chr5_-_138139382 | 0.50 |
ENST00000265191.4
|
NME5
|
NME/NM23 family member 5 |
| chr7_+_154305256 | 0.48 |
ENST00000619756.4
|
DPP6
|
dipeptidyl peptidase like 6 |
| chr19_+_35138993 | 0.46 |
ENST00000612146.4
ENST00000589209.5 |
FXYD1
|
FXYD domain containing ion transport regulator 1 |
| chr19_+_35139440 | 0.45 |
ENST00000455515.6
|
FXYD1
|
FXYD domain containing ion transport regulator 1 |
| chr2_-_207165923 | 0.45 |
ENST00000309446.11
|
KLF7
|
Kruppel like factor 7 |
| chr4_-_185775890 | 0.44 |
ENST00000437304.6
|
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr2_+_148021404 | 0.42 |
ENST00000638043.2
|
MBD5
|
methyl-CpG binding domain protein 5 |
| chr10_-_3785197 | 0.42 |
ENST00000497571.6
|
KLF6
|
Kruppel like factor 6 |
| chr1_+_84164962 | 0.41 |
ENST00000614872.4
ENST00000394839.6 |
PRKACB
|
protein kinase cAMP-activated catalytic subunit beta |
| chr13_+_108629605 | 0.41 |
ENST00000457511.7
|
MYO16
|
myosin XVI |
| chr9_+_113594118 | 0.40 |
ENST00000620489.1
|
RGS3
|
regulator of G protein signaling 3 |
| chr11_+_7576975 | 0.40 |
ENST00000684215.1
ENST00000650027.1 |
PPFIBP2
|
PPFIA binding protein 2 |
| chr2_+_33436304 | 0.40 |
ENST00000402538.7
|
RASGRP3
|
RAS guanyl releasing protein 3 |
| chr10_-_3785179 | 0.39 |
ENST00000469435.1
|
KLF6
|
Kruppel like factor 6 |
| chr7_-_151187772 | 0.39 |
ENST00000377867.7
|
ASB10
|
ankyrin repeat and SOCS box containing 10 |
| chr10_-_3785225 | 0.39 |
ENST00000542957.1
|
KLF6
|
Kruppel like factor 6 |
| chr8_-_69833338 | 0.38 |
ENST00000524945.5
|
SLCO5A1
|
solute carrier organic anion transporter family member 5A1 |
| chr5_-_88883701 | 0.38 |
ENST00000636998.1
|
MEF2C
|
myocyte enhancer factor 2C |
| chr6_-_75493629 | 0.38 |
ENST00000393004.6
|
FILIP1
|
filamin A interacting protein 1 |
| chr10_-_73655984 | 0.37 |
ENST00000394810.3
|
SYNPO2L
|
synaptopodin 2 like |
| chr20_+_43945677 | 0.37 |
ENST00000358131.5
|
TOX2
|
TOX high mobility group box family member 2 |
| chr4_-_185775271 | 0.37 |
ENST00000430503.5
ENST00000319454.10 ENST00000450341.5 |
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr3_-_157160751 | 0.35 |
ENST00000461804.5
|
CCNL1
|
cyclin L1 |
| chr8_+_2045037 | 0.34 |
ENST00000262113.9
|
MYOM2
|
myomesin 2 |
| chr7_-_151187212 | 0.34 |
ENST00000420175.3
ENST00000275838.5 |
ASB10
|
ankyrin repeat and SOCS box containing 10 |
| chr17_+_70169516 | 0.33 |
ENST00000243457.4
|
KCNJ2
|
potassium inwardly rectifying channel subfamily J member 2 |
| chr11_-_31804067 | 0.33 |
ENST00000639548.1
ENST00000640125.1 ENST00000481563.6 ENST00000639079.1 ENST00000638762.1 ENST00000638346.1 |
PAX6
|
paired box 6 |
| chr9_-_131276499 | 0.33 |
ENST00000372271.4
|
FAM78A
|
family with sequence similarity 78 member A |
| chr1_+_203128279 | 0.32 |
ENST00000367235.1
ENST00000618295.1 |
ADORA1
|
adenosine A1 receptor |
| chr3_+_8501807 | 0.32 |
ENST00000426878.2
ENST00000397386.7 ENST00000415597.5 ENST00000157600.8 |
LMCD1
|
LIM and cysteine rich domains 1 |
| chr4_-_142305935 | 0.32 |
ENST00000511838.5
|
INPP4B
|
inositol polyphosphate-4-phosphatase type II B |
| chr8_+_2045058 | 0.30 |
ENST00000523438.1
|
MYOM2
|
myomesin 2 |
| chr4_-_142305826 | 0.30 |
ENST00000514525.1
|
INPP4B
|
inositol polyphosphate-4-phosphatase type II B |
| chrX_+_105948429 | 0.29 |
ENST00000540278.1
|
NRK
|
Nik related kinase |
| chr14_-_64942720 | 0.29 |
ENST00000557049.1
ENST00000389614.6 |
GPX2
|
glutathione peroxidase 2 |
| chr14_-_64942783 | 0.28 |
ENST00000612794.1
|
GPX2
|
glutathione peroxidase 2 |
| chr15_-_70702273 | 0.28 |
ENST00000558758.5
ENST00000379983.6 ENST00000560441.5 |
UACA
|
uveal autoantigen with coiled-coil domains and ankyrin repeats |
| chr11_-_47378391 | 0.28 |
ENST00000227163.8
|
SPI1
|
Spi-1 proto-oncogene |
| chr2_+_167187364 | 0.27 |
ENST00000672671.1
|
XIRP2
|
xin actin binding repeat containing 2 |
| chr22_+_25742141 | 0.27 |
ENST00000536101.5
ENST00000335473.12 ENST00000407587.6 |
MYO18B
|
myosin XVIIIB |
| chr11_-_47378527 | 0.27 |
ENST00000378538.8
|
SPI1
|
Spi-1 proto-oncogene |
| chr15_-_52652031 | 0.27 |
ENST00000546305.6
|
FAM214A
|
family with sequence similarity 214 member A |
| chr19_+_35138778 | 0.27 |
ENST00000351325.9
ENST00000586871.5 ENST00000592174.1 |
FXYD1
ENSG00000221857.7
|
FXYD domain containing ion transport regulator 1 novel transcript |
| chr1_+_34792990 | 0.27 |
ENST00000450137.1
ENST00000342280.5 |
GJA4
|
gap junction protein alpha 4 |
| chr2_+_167187283 | 0.27 |
ENST00000409605.1
ENST00000409273.6 |
XIRP2
|
xin actin binding repeat containing 2 |
| chr3_+_138348592 | 0.27 |
ENST00000621127.4
ENST00000494949.5 |
MRAS
|
muscle RAS oncogene homolog |
| chr7_-_37448845 | 0.27 |
ENST00000310758.9
|
ELMO1
|
engulfment and cell motility 1 |
| chr3_-_30894661 | 0.26 |
ENST00000282538.10
ENST00000454381.3 |
GADL1
|
glutamate decarboxylase like 1 |
| chr4_-_75902444 | 0.26 |
ENST00000286719.12
|
PPEF2
|
protein phosphatase with EF-hand domain 2 |
| chr11_-_31803663 | 0.26 |
ENST00000638802.1
ENST00000638878.1 |
PAX6
|
paired box 6 |
| chr16_+_31214088 | 0.26 |
ENST00000613872.1
|
TRIM72
|
tripartite motif containing 72 |
| chr7_-_22220226 | 0.26 |
ENST00000420196.5
|
RAPGEF5
|
Rap guanine nucleotide exchange factor 5 |
| chr10_-_73651013 | 0.26 |
ENST00000372873.8
|
SYNPO2L
|
synaptopodin 2 like |
| chr3_+_138348570 | 0.25 |
ENST00000423968.7
|
MRAS
|
muscle RAS oncogene homolog |
| chr1_-_145910031 | 0.25 |
ENST00000369304.8
|
ITGA10
|
integrin subunit alpha 10 |
| chr1_-_145910066 | 0.25 |
ENST00000539363.2
|
ITGA10
|
integrin subunit alpha 10 |
| chr11_-_31803620 | 0.25 |
ENST00000639006.1
|
PAX6
|
paired box 6 |
| chr16_-_31428325 | 0.24 |
ENST00000287490.5
|
COX6A2
|
cytochrome c oxidase subunit 6A2 |
| chr1_+_113979391 | 0.24 |
ENST00000393300.6
ENST00000369551.5 |
OLFML3
|
olfactomedin like 3 |
| chr6_-_6006878 | 0.24 |
ENST00000244766.7
|
NRN1
|
neuritin 1 |
| chr1_+_113979460 | 0.24 |
ENST00000320334.5
|
OLFML3
|
olfactomedin like 3 |
| chr6_-_2744126 | 0.24 |
ENST00000647417.1
|
MYLK4
|
myosin light chain kinase family member 4 |
| chr5_-_67196791 | 0.23 |
ENST00000256447.5
|
CD180
|
CD180 molecule |
| chr2_+_178284907 | 0.23 |
ENST00000409631.5
|
OSBPL6
|
oxysterol binding protein like 6 |
| chr16_+_31214111 | 0.23 |
ENST00000322122.8
|
TRIM72
|
tripartite motif containing 72 |
| chr11_-_47378494 | 0.22 |
ENST00000533030.1
|
SPI1
|
Spi-1 proto-oncogene |
| chr1_-_201421718 | 0.22 |
ENST00000367312.5
ENST00000555340.6 ENST00000361379.9 ENST00000622580.4 |
TNNI1
|
troponin I1, slow skeletal type |
| chr11_-_18012883 | 0.22 |
ENST00000530613.5
ENST00000532389.5 ENST00000529728.5 ENST00000532265.5 |
SERGEF
|
secretion regulating guanine nucleotide exchange factor |
| chr12_+_40224956 | 0.21 |
ENST00000298910.12
ENST00000343742.6 |
LRRK2
|
leucine rich repeat kinase 2 |
| chr9_+_5450503 | 0.21 |
ENST00000381573.8
ENST00000381577.4 |
CD274
|
CD274 molecule |
| chr12_+_78864768 | 0.21 |
ENST00000261205.9
ENST00000457153.6 |
SYT1
|
synaptotagmin 1 |
| chr12_-_21941225 | 0.20 |
ENST00000538350.5
|
ABCC9
|
ATP binding cassette subfamily C member 9 |
| chr1_+_101238090 | 0.20 |
ENST00000475289.2
|
S1PR1
|
sphingosine-1-phosphate receptor 1 |
| chr12_-_9115907 | 0.20 |
ENST00000318602.12
|
A2M
|
alpha-2-macroglobulin |
| chr1_+_228208024 | 0.20 |
ENST00000570156.7
ENST00000680850.1 |
OBSCN
|
obscurin, cytoskeletal calmodulin and titin-interacting RhoGEF |
| chr11_-_19201976 | 0.19 |
ENST00000647990.1
ENST00000649235.1 ENST00000265968.9 ENST00000649842.1 |
CSRP3
|
cysteine and glycine rich protein 3 |
| chr5_-_16508812 | 0.19 |
ENST00000683414.1
|
RETREG1
|
reticulophagy regulator 1 |
| chr5_+_191477 | 0.19 |
ENST00000328278.4
|
LRRC14B
|
leucine rich repeat containing 14B |
| chr7_+_30911845 | 0.19 |
ENST00000652696.1
ENST00000311813.11 |
AQP1
|
aquaporin 1 (Colton blood group) |
| chr7_-_134459089 | 0.19 |
ENST00000285930.9
|
AKR1B1
|
aldo-keto reductase family 1 member B |
| chr9_-_33402449 | 0.19 |
ENST00000377425.8
|
AQP7
|
aquaporin 7 |
| chr22_+_31093358 | 0.18 |
ENST00000404574.5
|
SMTN
|
smoothelin |
| chr5_-_16508951 | 0.18 |
ENST00000682628.1
|
RETREG1
|
reticulophagy regulator 1 |
| chr9_-_83267230 | 0.18 |
ENST00000328788.5
|
FRMD3
|
FERM domain containing 3 |
| chr4_-_142846275 | 0.18 |
ENST00000513000.5
ENST00000509777.5 ENST00000503927.5 |
INPP4B
|
inositol polyphosphate-4-phosphatase type II B |
| chrX_+_71301742 | 0.18 |
ENST00000373829.8
ENST00000538820.1 |
ITGB1BP2
|
integrin subunit beta 1 binding protein 2 |
| chr5_-_16508858 | 0.18 |
ENST00000684456.1
|
RETREG1
|
reticulophagy regulator 1 |
| chr1_-_156490599 | 0.18 |
ENST00000360595.7
|
MEF2D
|
myocyte enhancer factor 2D |
| chr17_+_7558296 | 0.18 |
ENST00000438470.5
ENST00000436057.5 |
TNFSF13
|
TNF superfamily member 13 |
| chr1_-_25430147 | 0.17 |
ENST00000349320.7
|
RHCE
|
Rh blood group CcEe antigens |
| chr10_-_6062290 | 0.17 |
ENST00000256876.10
ENST00000379954.5 |
IL2RA
|
interleukin 2 receptor subunit alpha |
| chr3_+_35642159 | 0.17 |
ENST00000187397.8
|
ARPP21
|
cAMP regulated phosphoprotein 21 |
| chr8_-_42501224 | 0.17 |
ENST00000520262.6
ENST00000517366.1 |
SLC20A2
|
solute carrier family 20 member 2 |
| chr14_+_23372809 | 0.17 |
ENST00000397242.2
ENST00000329715.2 |
IL25
|
interleukin 25 |
| chr12_+_6226136 | 0.17 |
ENST00000676764.1
ENST00000646407.1 |
CD9
|
CD9 molecule |
| chr20_+_46029165 | 0.16 |
ENST00000616201.4
ENST00000616202.4 ENST00000616933.4 ENST00000626937.2 |
SLC12A5
|
solute carrier family 12 member 5 |
| chr8_-_21812320 | 0.16 |
ENST00000517328.5
|
GFRA2
|
GDNF family receptor alpha 2 |
| chr1_-_223665685 | 0.16 |
ENST00000366872.10
|
CAPN8
|
calpain 8 |
| chr6_+_27810041 | 0.16 |
ENST00000369163.3
|
H3C10
|
H3 clustered histone 10 |
| chr8_-_13514821 | 0.16 |
ENST00000276297.9
|
DLC1
|
DLC1 Rho GTPase activating protein |
| chr18_+_3252208 | 0.16 |
ENST00000578562.6
|
MYL12A
|
myosin light chain 12A |
| chr13_+_75804169 | 0.16 |
ENST00000526371.1
ENST00000526528.1 |
LMO7
|
LIM domain 7 |
| chr11_-_31803960 | 0.15 |
ENST00000640872.1
ENST00000639109.1 ENST00000638629.1 ENST00000639386.2 |
PAX6
|
paired box 6 |
| chr14_+_103123452 | 0.15 |
ENST00000558056.1
ENST00000560869.6 |
TNFAIP2
|
TNF alpha induced protein 2 |
| chr13_-_96994350 | 0.15 |
ENST00000298440.5
ENST00000543457.6 ENST00000541038.2 |
OXGR1
|
oxoglutarate receptor 1 |
| chr5_-_142325001 | 0.15 |
ENST00000344120.4
ENST00000434127.3 |
SPRY4
|
sprouty RTK signaling antagonist 4 |
| chr4_-_113761068 | 0.15 |
ENST00000342666.9
ENST00000514328.5 ENST00000515496.5 ENST00000508738.5 ENST00000379773.6 |
CAMK2D
|
calcium/calmodulin dependent protein kinase II delta |
| chr4_-_39638893 | 0.15 |
ENST00000511809.5
ENST00000505729.1 |
SMIM14
|
small integral membrane protein 14 |
| chr20_+_46029206 | 0.15 |
ENST00000243964.7
|
SLC12A5
|
solute carrier family 12 member 5 |
| chr5_-_42811884 | 0.15 |
ENST00000514985.6
ENST00000511224.5 ENST00000507920.5 ENST00000510965.1 |
SELENOP
|
selenoprotein P |
| chr10_-_6062349 | 0.15 |
ENST00000379959.8
|
IL2RA
|
interleukin 2 receptor subunit alpha |
| chr20_+_34977625 | 0.14 |
ENST00000618182.6
|
MYH7B
|
myosin heavy chain 7B |
| chr1_+_26021768 | 0.14 |
ENST00000374280.4
|
EXTL1
|
exostosin like glycosyltransferase 1 |
| chrX_-_21758097 | 0.14 |
ENST00000379494.4
|
SMPX
|
small muscle protein X-linked |
| chr4_+_41360759 | 0.14 |
ENST00000508501.5
ENST00000512946.5 ENST00000313860.11 ENST00000512632.5 ENST00000512820.5 |
LIMCH1
|
LIM and calponin homology domains 1 |
| chr12_+_80099535 | 0.14 |
ENST00000646859.1
ENST00000547103.7 |
OTOGL
|
otogelin like |
| chr19_-_49423441 | 0.14 |
ENST00000270631.2
|
PTH2
|
parathyroid hormone 2 |
| chr17_-_41055211 | 0.14 |
ENST00000542910.1
ENST00000398477.1 |
KRTAP2-2
|
keratin associated protein 2-2 |
| chr9_-_33402551 | 0.14 |
ENST00000297988.6
ENST00000624075.3 ENST00000625032.1 ENST00000625109.3 |
AQP7
|
aquaporin 7 |
| chr5_-_157345645 | 0.14 |
ENST00000312349.5
|
FNDC9
|
fibronectin type III domain containing 9 |
| chr1_+_202010575 | 0.13 |
ENST00000367283.7
ENST00000367284.10 |
ELF3
|
E74 like ETS transcription factor 3 |
| chr6_+_151807319 | 0.13 |
ENST00000443427.5
|
ESR1
|
estrogen receptor 1 |
| chr18_+_44680093 | 0.13 |
ENST00000426838.8
ENST00000677068.1 |
SETBP1
|
SET binding protein 1 |
| chr14_+_99684283 | 0.13 |
ENST00000261835.8
|
CYP46A1
|
cytochrome P450 family 46 subfamily A member 1 |
| chr3_-_196515315 | 0.13 |
ENST00000397537.3
|
SMCO1
|
single-pass membrane protein with coiled-coil domains 1 |
| chr17_-_69244846 | 0.13 |
ENST00000269081.8
|
ABCA10
|
ATP binding cassette subfamily A member 10 |
| chr15_-_82806054 | 0.13 |
ENST00000541889.1
ENST00000334574.12 ENST00000561368.1 |
FSD2
|
fibronectin type III and SPRY domain containing 2 |
| chr20_+_45407207 | 0.13 |
ENST00000372712.6
|
DBNDD2
|
dysbindin domain containing 2 |
| chr7_-_11832190 | 0.13 |
ENST00000423059.9
ENST00000617773.1 |
THSD7A
|
thrombospondin type 1 domain containing 7A |
| chr12_+_4269771 | 0.13 |
ENST00000676411.1
|
CCND2
|
cyclin D2 |
| chr5_-_36151853 | 0.12 |
ENST00000296603.5
|
LMBRD2
|
LMBR1 domain containing 2 |
| chr14_-_102238439 | 0.12 |
ENST00000522534.5
|
MOK
|
MOK protein kinase |
| chr13_-_44474296 | 0.12 |
ENST00000611198.4
|
TSC22D1
|
TSC22 domain family member 1 |
| chr12_-_262828 | 0.12 |
ENST00000343164.9
ENST00000436453.1 ENST00000445055.6 ENST00000546319.5 |
SLC6A13
|
solute carrier family 6 member 13 |
| chr16_-_71577082 | 0.12 |
ENST00000355962.5
|
TAT
|
tyrosine aminotransferase |
| chr15_+_84817346 | 0.12 |
ENST00000258888.6
|
ALPK3
|
alpha kinase 3 |
| chr12_-_54973683 | 0.12 |
ENST00000532804.5
ENST00000531122.5 ENST00000533446.5 |
TESPA1
|
thymocyte expressed, positive selection associated 1 |
| chr14_+_71586261 | 0.11 |
ENST00000358550.6
|
SIPA1L1
|
signal induced proliferation associated 1 like 1 |
| chr4_+_118888918 | 0.11 |
ENST00000434046.6
|
SYNPO2
|
synaptopodin 2 |
| chr17_+_7884783 | 0.11 |
ENST00000380358.9
|
CHD3
|
chromodomain helicase DNA binding protein 3 |
| chr2_-_218831791 | 0.11 |
ENST00000439262.6
ENST00000430489.1 |
PRKAG3
|
protein kinase AMP-activated non-catalytic subunit gamma 3 |
| chr15_+_80072559 | 0.11 |
ENST00000560228.5
ENST00000559835.5 ENST00000559775.5 ENST00000558688.5 ENST00000560392.5 ENST00000560976.5 ENST00000558272.5 ENST00000558390.5 |
ZFAND6
|
zinc finger AN1-type containing 6 |
| chr2_+_112645930 | 0.11 |
ENST00000272542.8
|
SLC20A1
|
solute carrier family 20 member 1 |
| chr20_+_1894145 | 0.11 |
ENST00000400068.7
|
SIRPA
|
signal regulatory protein alpha |
| chr1_+_212608628 | 0.11 |
ENST00000613954.4
ENST00000341491.9 ENST00000366985.5 |
ATF3
|
activating transcription factor 3 |
| chr6_+_44227025 | 0.10 |
ENST00000371708.1
|
SLC29A1
|
solute carrier family 29 member 1 (Augustine blood group) |
| chrX_-_30309387 | 0.10 |
ENST00000378970.5
|
NR0B1
|
nuclear receptor subfamily 0 group B member 1 |
| chr1_+_26159071 | 0.10 |
ENST00000374268.5
|
FAM110D
|
family with sequence similarity 110 member D |
| chr3_+_16884942 | 0.10 |
ENST00000615277.5
|
PLCL2
|
phospholipase C like 2 |
| chr16_-_15643105 | 0.10 |
ENST00000548025.5
ENST00000551742.5 ENST00000396368.8 |
MARF1
|
meiosis regulator and mRNA stability factor 1 |
| chr8_-_48921419 | 0.10 |
ENST00000020945.4
|
SNAI2
|
snail family transcriptional repressor 2 |
| chr8_+_106270161 | 0.10 |
ENST00000517566.7
ENST00000531443.6 |
OXR1
|
oxidation resistance 1 |
| chr8_+_73991345 | 0.10 |
ENST00000284818.7
ENST00000518893.1 |
LY96
|
lymphocyte antigen 96 |
| chr2_+_178320228 | 0.10 |
ENST00000315022.2
|
OSBPL6
|
oxysterol binding protein like 6 |
| chr17_+_7281711 | 0.10 |
ENST00000317370.13
ENST00000571308.5 |
SLC2A4
|
solute carrier family 2 member 4 |
| chr16_+_8674605 | 0.10 |
ENST00000268251.13
|
ABAT
|
4-aminobutyrate aminotransferase |
| chr11_+_129375841 | 0.10 |
ENST00000281437.6
|
BARX2
|
BARX homeobox 2 |
| chr7_-_124929938 | 0.10 |
ENST00000668382.1
ENST00000655761.1 ENST00000393329.5 |
POT1
|
protection of telomeres 1 |
| chr1_+_89633086 | 0.10 |
ENST00000370454.9
|
LRRC8C
|
leucine rich repeat containing 8 VRAC subunit C |
| chr20_+_1894462 | 0.10 |
ENST00000622179.4
|
SIRPA
|
signal regulatory protein alpha |
| chr1_+_95117923 | 0.10 |
ENST00000455656.1
ENST00000604534.5 |
TLCD4
TLCD4-RWDD3
|
TLC domain containing 4 TLCD4-RWDD3 readthrough |
| chr16_-_15643024 | 0.10 |
ENST00000540441.6
|
MARF1
|
meiosis regulator and mRNA stability factor 1 |
| chr6_+_7107941 | 0.10 |
ENST00000379938.7
ENST00000467782.5 ENST00000334984.10 ENST00000349384.10 |
RREB1
|
ras responsive element binding protein 1 |
| chr21_-_36980789 | 0.10 |
ENST00000675307.1
ENST00000612277.4 |
HLCS
|
holocarboxylase synthetase |
| chr3_+_188947199 | 0.09 |
ENST00000433971.5
|
TPRG1
|
tumor protein p63 regulated 1 |
| chr1_-_39672080 | 0.09 |
ENST00000235628.2
|
NT5C1A
|
5'-nucleotidase, cytosolic IA |
| chr15_+_78266181 | 0.09 |
ENST00000446172.2
|
DNAJA4
|
DnaJ heat shock protein family (Hsp40) member A4 |
| chr10_+_69802424 | 0.09 |
ENST00000673802.2
ENST00000517713.5 ENST00000520133.5 ENST00000522165.5 ENST00000673641.2 ENST00000673628.2 |
COL13A1
|
collagen type XIII alpha 1 chain |
| chr18_-_55403682 | 0.09 |
ENST00000564228.5
ENST00000630828.2 |
TCF4
|
transcription factor 4 |
| chrX_-_110440218 | 0.09 |
ENST00000372057.1
ENST00000372054.3 |
AMMECR1
GNG5P2
|
AMMECR nuclear protein 1 G protein subunit gamma 5 pseudogene 2 |
| chr7_-_124929801 | 0.09 |
ENST00000653241.1
ENST00000664366.1 ENST00000446993.6 ENST00000654766.1 ENST00000357628.8 ENST00000609702.5 |
POT1
|
protection of telomeres 1 |
| chr22_+_19723525 | 0.09 |
ENST00000366425.4
|
GP1BB
|
glycoprotein Ib platelet subunit beta |
| chr7_-_27662836 | 0.09 |
ENST00000265395.7
|
HIBADH
|
3-hydroxyisobutyrate dehydrogenase |
| chr18_+_13611764 | 0.09 |
ENST00000585931.5
|
LDLRAD4
|
low density lipoprotein receptor class A domain containing 4 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.7 | GO:0021986 | epithalamus development(GO:0021538) habenula development(GO:0021986) |
| 0.5 | 3.3 | GO:0043435 | response to corticotropin-releasing hormone(GO:0043435) cellular response to corticotropin-releasing hormone stimulus(GO:0071376) |
| 0.2 | 0.7 | GO:0045608 | trochlear nerve development(GO:0021558) auditory receptor cell fate determination(GO:0042668) negative regulation of auditory receptor cell differentiation(GO:0045608) regulation of timing of neuron differentiation(GO:0060164) negative regulation of pro-B cell differentiation(GO:2000974) |
| 0.2 | 1.2 | GO:0010731 | protein glutathionylation(GO:0010731) regulation of protein glutathionylation(GO:0010732) negative regulation of protein glutathionylation(GO:0010734) |
| 0.2 | 0.5 | GO:0051365 | leading edge cell differentiation(GO:0035026) cellular response to potassium ion starvation(GO:0051365) |
| 0.2 | 0.5 | GO:0060279 | positive regulation of ovulation(GO:0060279) |
| 0.2 | 0.8 | GO:0044027 | hypermethylation of CpG island(GO:0044027) |
| 0.1 | 0.6 | GO:0005986 | sucrose biosynthetic process(GO:0005986) |
| 0.1 | 0.5 | GO:0055014 | atrial cardiac muscle cell differentiation(GO:0055011) atrial cardiac muscle cell development(GO:0055014) |
| 0.1 | 0.3 | GO:0070256 | negative regulation of circadian sleep/wake cycle, non-REM sleep(GO:0042323) negative regulation of mucus secretion(GO:0070256) |
| 0.1 | 0.3 | GO:0060721 | spongiotrophoblast cell proliferation(GO:0060720) regulation of spongiotrophoblast cell proliferation(GO:0060721) cell proliferation involved in embryonic placenta development(GO:0060722) regulation of cell proliferation involved in embryonic placenta development(GO:0060723) |
| 0.1 | 0.6 | GO:0060399 | positive regulation of growth hormone receptor signaling pathway(GO:0060399) |
| 0.1 | 1.6 | GO:0001542 | ovulation from ovarian follicle(GO:0001542) |
| 0.1 | 0.3 | GO:0090076 | relaxation of skeletal muscle(GO:0090076) |
| 0.1 | 0.2 | GO:1902823 | negative regulation of late endosome to lysosome transport(GO:1902823) regulation of protein processing involved in protein targeting to mitochondrion(GO:1903216) negative regulation of protein processing involved in protein targeting to mitochondrion(GO:1903217) |
| 0.1 | 0.6 | GO:0061709 | reticulophagy(GO:0061709) |
| 0.1 | 0.2 | GO:0098746 | fast, calcium ion-dependent exocytosis of neurotransmitter(GO:0098746) |
| 0.1 | 0.5 | GO:0097338 | response to clozapine(GO:0097338) |
| 0.1 | 0.2 | GO:0003245 | cardiac muscle tissue growth involved in heart morphogenesis(GO:0003245) |
| 0.1 | 0.2 | GO:0001868 | regulation of complement activation, lectin pathway(GO:0001868) negative regulation of complement activation, lectin pathway(GO:0001869) |
| 0.1 | 0.3 | GO:0046013 | regulation of T cell homeostatic proliferation(GO:0046013) |
| 0.1 | 0.2 | GO:0060382 | regulation of DNA strand elongation(GO:0060382) |
| 0.1 | 0.2 | GO:0072019 | nitric oxide transport(GO:0030185) cerebrospinal fluid secretion(GO:0033326) proximal convoluted tubule development(GO:0072019) metanephric proximal convoluted tubule development(GO:0072229) |
| 0.1 | 0.2 | GO:0019401 | hexitol metabolic process(GO:0006059) alditol biosynthetic process(GO:0019401) |
| 0.1 | 1.0 | GO:0003322 | pancreatic A cell development(GO:0003322) |
| 0.1 | 0.1 | GO:1901420 | negative regulation of response to alcohol(GO:1901420) |
| 0.0 | 0.3 | GO:0002322 | B cell proliferation involved in immune response(GO:0002322) |
| 0.0 | 0.4 | GO:0003172 | primary heart field specification(GO:0003138) sinoatrial valve development(GO:0003172) sinoatrial valve morphogenesis(GO:0003185) |
| 0.0 | 1.3 | GO:0051412 | response to corticosterone(GO:0051412) |
| 0.0 | 0.3 | GO:0015793 | glycerol transport(GO:0015793) |
| 0.0 | 0.5 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.0 | 0.1 | GO:0061394 | regulation of transcription from RNA polymerase II promoter in response to arsenic-containing substance(GO:0061394) |
| 0.0 | 0.2 | GO:2001181 | positive regulation of interleukin-10 secretion(GO:2001181) |
| 0.0 | 0.6 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.0 | 0.2 | GO:0035995 | detection of muscle stretch(GO:0035995) |
| 0.0 | 0.0 | GO:0090214 | spongiotrophoblast layer developmental growth(GO:0090214) |
| 0.0 | 0.1 | GO:1903691 | positive regulation of wound healing, spreading of epidermal cells(GO:1903691) |
| 0.0 | 0.3 | GO:1901409 | positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.0 | 0.8 | GO:0002523 | leukocyte migration involved in inflammatory response(GO:0002523) |
| 0.0 | 0.1 | GO:0010387 | COP9 signalosome assembly(GO:0010387) |
| 0.0 | 0.1 | GO:0044313 | protein K6-linked deubiquitination(GO:0044313) |
| 0.0 | 0.1 | GO:1900042 | positive regulation of interleukin-2 secretion(GO:1900042) |
| 0.0 | 0.8 | GO:0070886 | positive regulation of calcineurin-NFAT signaling cascade(GO:0070886) |
| 0.0 | 0.2 | GO:0030644 | cellular chloride ion homeostasis(GO:0030644) |
| 0.0 | 0.1 | GO:0014053 | negative regulation of gamma-aminobutyric acid secretion(GO:0014053) aspartate secretion(GO:0061528) regulation of aspartate secretion(GO:1904448) positive regulation of aspartate secretion(GO:1904450) |
| 0.0 | 0.3 | GO:0036309 | protein localization to M-band(GO:0036309) |
| 0.0 | 0.5 | GO:0006228 | UTP biosynthetic process(GO:0006228) |
| 0.0 | 0.7 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.0 | 0.1 | GO:0009305 | protein biotinylation(GO:0009305) histone biotinylation(GO:0071110) |
| 0.0 | 0.3 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.0 | 0.6 | GO:0003301 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.0 | 0.1 | GO:0038155 | interleukin-23-mediated signaling pathway(GO:0038155) |
| 0.0 | 0.2 | GO:0030222 | eosinophil differentiation(GO:0030222) |
| 0.0 | 0.1 | GO:0015862 | uridine transport(GO:0015862) |
| 0.0 | 0.1 | GO:0032497 | detection of lipopolysaccharide(GO:0032497) |
| 0.0 | 0.1 | GO:1903595 | positive regulation of histamine secretion by mast cell(GO:1903595) |
| 0.0 | 0.1 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 0.0 | 0.2 | GO:0009414 | response to water deprivation(GO:0009414) |
| 0.0 | 0.1 | GO:0044858 | plasma membrane raft distribution(GO:0044855) plasma membrane raft localization(GO:0044856) plasma membrane raft polarization(GO:0044858) regulation of plasma membrane raft polarization(GO:1903906) |
| 0.0 | 0.1 | GO:0036058 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.0 | 0.1 | GO:0060745 | mammary gland branching involved in pregnancy(GO:0060745) epithelial cell proliferation involved in mammary gland duct elongation(GO:0060750) |
| 0.0 | 0.1 | GO:0006574 | valine catabolic process(GO:0006574) |
| 0.0 | 0.1 | GO:0001887 | selenium compound metabolic process(GO:0001887) |
| 0.0 | 0.0 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.0 | 0.0 | GO:0014813 | skeletal muscle satellite cell commitment(GO:0014813) |
| 0.0 | 0.1 | GO:0051643 | endoplasmic reticulum localization(GO:0051643) |
| 0.0 | 0.2 | GO:1901897 | regulation of relaxation of cardiac muscle(GO:1901897) |
| 0.0 | 0.0 | GO:0010808 | positive regulation of synaptic vesicle priming(GO:0010808) |
| 0.0 | 0.2 | GO:0035860 | glial cell-derived neurotrophic factor receptor signaling pathway(GO:0035860) |
| 0.0 | 0.6 | GO:2000772 | regulation of cellular senescence(GO:2000772) |
| 0.0 | 0.1 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.0 | 0.8 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.0 | 0.1 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.0 | 0.1 | GO:0060023 | soft palate development(GO:0060023) |
| 0.0 | 0.2 | GO:0048298 | positive regulation of isotype switching to IgA isotypes(GO:0048298) |
| 0.0 | 0.1 | GO:2001295 | malonyl-CoA biosynthetic process(GO:2001295) |
| 0.0 | 0.1 | GO:0060056 | mammary gland involution(GO:0060056) |
| 0.0 | 0.1 | GO:0016127 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.0 | 0.2 | GO:1900119 | positive regulation of execution phase of apoptosis(GO:1900119) |
| 0.0 | 0.1 | GO:0099640 | axo-dendritic protein transport(GO:0099640) |
| 0.0 | 0.3 | GO:0010801 | negative regulation of peptidyl-threonine phosphorylation(GO:0010801) |
| 0.0 | 0.5 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0043511 | inhibin complex(GO:0043511) inhibin A complex(GO:0043512) |
| 0.1 | 0.5 | GO:0034680 | integrin alpha10-beta1 complex(GO:0034680) |
| 0.1 | 0.2 | GO:0044753 | amphisome(GO:0044753) |
| 0.1 | 0.5 | GO:0035976 | AP1 complex(GO:0035976) |
| 0.1 | 0.2 | GO:0005600 | collagen type XIII trimer(GO:0005600) transmembrane collagen trimer(GO:0030936) |
| 0.1 | 1.2 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.1 | 0.1 | GO:1990622 | CHOP-ATF3 complex(GO:1990622) |
| 0.0 | 0.2 | GO:0020003 | symbiont-containing vacuole(GO:0020003) |
| 0.0 | 0.2 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 0.0 | 0.2 | GO:0060201 | clathrin-sculpted acetylcholine transport vesicle(GO:0060200) clathrin-sculpted acetylcholine transport vesicle membrane(GO:0060201) |
| 0.0 | 0.2 | GO:0097454 | Schwann cell microvillus(GO:0097454) |
| 0.0 | 0.7 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.1 | GO:0005592 | collagen type XI trimer(GO:0005592) |
| 0.0 | 0.1 | GO:0032144 | 4-aminobutyrate transaminase complex(GO:0032144) |
| 0.0 | 0.1 | GO:0072536 | interleukin-23 receptor complex(GO:0072536) |
| 0.0 | 0.5 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.1 | GO:0097129 | cyclin D2-CDK4 complex(GO:0097129) |
| 0.0 | 0.6 | GO:0010369 | chromocenter(GO:0010369) |
| 0.0 | 0.6 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.2 | GO:0070187 | telosome(GO:0070187) |
| 0.0 | 0.0 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.0 | 0.2 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.0 | 0.2 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.0 | 0.1 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.0 | 0.1 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.0 | 0.3 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 0.1 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.0 | 0.2 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0042132 | fructose 1,6-bisphosphate 1-phosphatase activity(GO:0042132) |
| 0.1 | 0.8 | GO:0052828 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) inositol-1,3,4-trisphosphate 4-phosphatase activity(GO:0017161) phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) inositol-3,4-bisphosphate 4-phosphatase activity(GO:0052828) |
| 0.1 | 0.3 | GO:0004782 | sulfinoalanine decarboxylase activity(GO:0004782) |
| 0.1 | 0.3 | GO:0004911 | interleukin-2 receptor activity(GO:0004911) interleukin-2 binding(GO:0019976) |
| 0.1 | 0.2 | GO:1904713 | beta-catenin destruction complex binding(GO:1904713) |
| 0.1 | 0.8 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.1 | 0.7 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.1 | 0.2 | GO:0019959 | interleukin-8 binding(GO:0019959) |
| 0.1 | 0.3 | GO:0032795 | heterotrimeric G-protein binding(GO:0032795) |
| 0.1 | 0.5 | GO:0015168 | glycerol transmembrane transporter activity(GO:0015168) |
| 0.1 | 0.5 | GO:0098639 | collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.1 | 0.5 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.1 | 0.3 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) |
| 0.1 | 1.3 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.7 | GO:0031433 | telethonin binding(GO:0031433) |
| 0.0 | 0.1 | GO:0080130 | L-phenylalanine:2-oxoglutarate aminotransferase activity(GO:0080130) |
| 0.0 | 0.1 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.0 | 0.3 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.2 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.0 | 0.2 | GO:0010521 | telomerase inhibitor activity(GO:0010521) |
| 0.0 | 0.6 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.0 | 0.5 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 2.8 | GO:0004879 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) transcription factor activity, direct ligand regulated sequence-specific DNA binding(GO:0098531) |
| 0.0 | 0.2 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.0 | 0.8 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.4 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 0.1 | GO:0003867 | 4-aminobutyrate transaminase activity(GO:0003867) succinate-semialdehyde dehydrogenase binding(GO:0032145) (S)-3-amino-2-methylpropionate transaminase activity(GO:0047298) |
| 0.0 | 0.1 | GO:0018271 | biotin-[acetyl-CoA-carboxylase] ligase activity(GO:0004077) biotin-[methylcrotonoyl-CoA-carboxylase] ligase activity(GO:0004078) biotin-[methylmalonyl-CoA-carboxytransferase] ligase activity(GO:0004079) biotin-[propionyl-CoA-carboxylase (ATP-hydrolyzing)] ligase activity(GO:0004080) biotin-protein ligase activity(GO:0018271) |
| 0.0 | 0.2 | GO:0016167 | glial cell-derived neurotrophic factor receptor activity(GO:0016167) |
| 0.0 | 0.7 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.0 | 0.3 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.0 | 0.1 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 0.0 | 0.1 | GO:1904288 | BAT3 complex binding(GO:1904288) |
| 0.0 | 0.1 | GO:0042019 | interleukin-23 binding(GO:0042019) interleukin-23 receptor activity(GO:0042020) |
| 0.0 | 0.5 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 0.1 | GO:0004667 | prostaglandin-D synthase activity(GO:0004667) |
| 0.0 | 0.1 | GO:0004949 | cannabinoid receptor activity(GO:0004949) |
| 0.0 | 0.1 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.0 | 0.2 | GO:0004032 | alditol:NADP+ 1-oxidoreductase activity(GO:0004032) |
| 0.0 | 0.4 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.0 | 1.2 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 0.1 | GO:0002046 | opsin binding(GO:0002046) |
| 0.0 | 0.8 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.3 | GO:0086008 | voltage-gated potassium channel activity involved in cardiac muscle cell action potential repolarization(GO:0086008) |
| 0.0 | 0.7 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.0 | 0.6 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 0.6 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.0 | 0.2 | GO:0046625 | sphingolipid binding(GO:0046625) |
| 0.0 | 0.1 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.0 | 0.5 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.0 | 0.1 | GO:0008430 | selenium binding(GO:0008430) |
| 0.0 | 0.1 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.1 | GO:0008426 | protein kinase C inhibitor activity(GO:0008426) |
| 0.0 | 0.0 | GO:0017099 | very-long-chain-acyl-CoA dehydrogenase activity(GO:0017099) |
| 0.0 | 0.1 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) |
| 0.0 | 0.3 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 0.4 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 0.1 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.0 | 0.1 | GO:0003989 | acetyl-CoA carboxylase activity(GO:0003989) |
| 0.0 | 0.5 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.0 | 0.1 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 0.0 | 0.5 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 1.7 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.1 | GO:0033695 | oxidoreductase activity, acting on CH or CH2 groups, quinone or similar compound as acceptor(GO:0033695) caffeine oxidase activity(GO:0034875) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.3 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.5 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 0.5 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 1.2 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.0 | 1.3 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 1.3 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.0 | 0.5 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 0.5 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 1.5 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 0.4 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 0.5 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
| 0.0 | 0.4 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 0.7 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.2 | REACTOME PIP3 ACTIVATES AKT SIGNALING | Genes involved in PIP3 activates AKT signaling |
| 0.0 | 0.5 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.0 | 0.8 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.0 | 0.5 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 0.5 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.0 | 1.0 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.0 | 0.7 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.0 | 0.5 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 1.9 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.2 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.0 | 0.5 | REACTOME NUCLEOTIDE LIKE PURINERGIC RECEPTORS | Genes involved in Nucleotide-like (purinergic) receptors |
| 0.0 | 0.5 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.0 | 0.3 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.0 | 0.3 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.0 | 0.1 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.0 | 0.2 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |