Project
Inflammatory response time course, HUVEC (Wada, 2009)
Navigation
Downloads
Results for MSX1
Z-value: 0.46
Transcription factors associated with MSX1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
MSX1
|
ENSG00000163132.8 | MSX1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| MSX1 | hg38_v1_chr4_+_4859658_4859672 | -0.36 | 7.9e-02 | Click! |
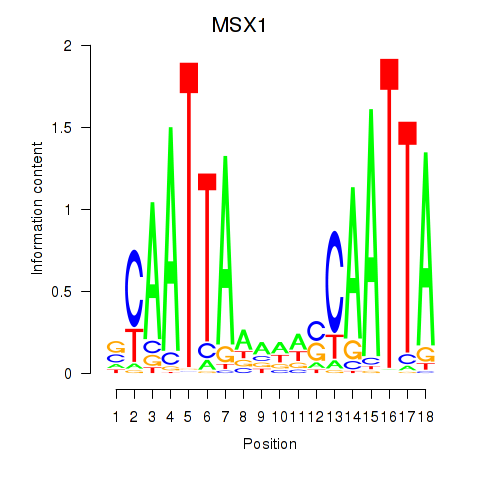
Activity profile of MSX1 motif
Sorted Z-values of MSX1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of MSX1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_-_123620496 | 1.27 |
ENST00000578202.1
|
MYLK
|
myosin light chain kinase |
| chr3_-_123620571 | 1.24 |
ENST00000583087.5
|
MYLK
|
myosin light chain kinase |
| chr10_-_99620401 | 1.00 |
ENST00000370495.6
|
SLC25A28
|
solute carrier family 25 member 28 |
| chr12_-_52517929 | 0.97 |
ENST00000548409.5
|
KRT5
|
keratin 5 |
| chr1_-_149936324 | 0.88 |
ENST00000369140.7
|
MTMR11
|
myotubularin related protein 11 |
| chr1_-_150765785 | 0.87 |
ENST00000680311.1
ENST00000681728.1 ENST00000680288.1 |
CTSS
|
cathepsin S |
| chr2_+_167135901 | 0.85 |
ENST00000628543.2
|
XIRP2
|
xin actin binding repeat containing 2 |
| chr21_+_42199686 | 0.85 |
ENST00000398457.6
|
ABCG1
|
ATP binding cassette subfamily G member 1 |
| chr22_+_41301514 | 0.73 |
ENST00000352645.5
|
ZC3H7B
|
zinc finger CCCH-type containing 7B |
| chr1_+_206635521 | 0.64 |
ENST00000367109.8
|
DYRK3
|
dual specificity tyrosine phosphorylation regulated kinase 3 |
| chr3_-_15440560 | 0.49 |
ENST00000595627.5
ENST00000597949.1 ENST00000494875.3 ENST00000595975.1 ENST00000598878.1 |
EAF1-AS1
METTL6
|
EAF1 antisense RNA 1 methyltransferase like 6 |
| chr8_+_30387064 | 0.46 |
ENST00000523115.5
ENST00000519647.5 |
RBPMS
|
RNA binding protein, mRNA processing factor |
| chr17_-_39778213 | 0.45 |
ENST00000583368.1
|
IKZF3
|
IKAROS family zinc finger 3 |
| chr14_+_77116562 | 0.43 |
ENST00000557408.5
|
TMEM63C
|
transmembrane protein 63C |
| chr11_+_45804038 | 0.40 |
ENST00000442528.2
ENST00000526817.1 |
SLC35C1
|
solute carrier family 35 member C1 |
| chr11_-_104968533 | 0.40 |
ENST00000444739.7
|
CASP4
|
caspase 4 |
| chr22_+_50738198 | 0.39 |
ENST00000216139.10
ENST00000529621.1 |
ACR
|
acrosin |
| chr13_-_37598750 | 0.38 |
ENST00000379743.8
ENST00000379742.4 ENST00000379749.8 ENST00000379747.9 ENST00000541179.5 ENST00000541481.5 |
POSTN
|
periostin |
| chr11_-_55936400 | 0.37 |
ENST00000301532.3
|
OR5I1
|
olfactory receptor family 5 subfamily I member 1 |
| chr14_-_74923234 | 0.35 |
ENST00000556776.1
ENST00000557413.6 ENST00000555647.5 |
RPS6KL1
|
ribosomal protein S6 kinase like 1 |
| chr1_-_158426237 | 0.34 |
ENST00000641042.1
|
OR10K2
|
olfactory receptor family 10 subfamily K member 2 |
| chr11_-_102625332 | 0.32 |
ENST00000260228.3
|
MMP20
|
matrix metallopeptidase 20 |
| chr3_+_171843337 | 0.30 |
ENST00000334567.9
ENST00000619900.4 ENST00000450693.1 |
TMEM212
|
transmembrane protein 212 |
| chr12_-_12684490 | 0.28 |
ENST00000540510.1
|
GPR19
|
G protein-coupled receptor 19 |
| chr18_+_63970029 | 0.28 |
ENST00000353706.6
ENST00000542677.5 ENST00000397985.7 ENST00000636430.1 ENST00000397988.7 ENST00000448851.5 |
SERPINB8
|
serpin family B member 8 |
| chr2_+_147845020 | 0.28 |
ENST00000241416.12
|
ACVR2A
|
activin A receptor type 2A |
| chr11_+_55635113 | 0.28 |
ENST00000641760.1
|
OR4P4
|
olfactory receptor family 4 subfamily P member 4 |
| chr16_+_56935371 | 0.27 |
ENST00000568358.1
|
HERPUD1
|
homocysteine inducible ER protein with ubiquitin like domain 1 |
| chr7_-_32299287 | 0.26 |
ENST00000396193.5
|
PDE1C
|
phosphodiesterase 1C |
| chr18_-_5396265 | 0.25 |
ENST00000579951.2
|
EPB41L3
|
erythrocyte membrane protein band 4.1 like 3 |
| chr17_-_31901658 | 0.24 |
ENST00000261708.9
|
UTP6
|
UTP6 small subunit processome component |
| chr18_-_50195138 | 0.24 |
ENST00000285039.12
|
MYO5B
|
myosin VB |
| chr17_+_57085092 | 0.23 |
ENST00000575322.1
ENST00000337714.8 |
AKAP1
|
A-kinase anchoring protein 1 |
| chr19_-_53254688 | 0.22 |
ENST00000594517.5
ENST00000601413.5 ENST00000594681.5 |
ZNF677
|
zinc finger protein 677 |
| chr2_-_208163588 | 0.22 |
ENST00000304502.5
|
CRYGA
|
crystallin gamma A |
| chr20_+_6007245 | 0.22 |
ENST00000378868.4
|
CRLS1
|
cardiolipin synthase 1 |
| chr8_+_116950951 | 0.21 |
ENST00000427715.2
|
SLC30A8
|
solute carrier family 30 member 8 |
| chr1_-_167518583 | 0.21 |
ENST00000392122.3
|
CD247
|
CD247 molecule |
| chr19_-_53254841 | 0.20 |
ENST00000601828.5
ENST00000599012.5 ENST00000598513.6 ENST00000598806.5 |
ZNF677
|
zinc finger protein 677 |
| chr5_-_147782518 | 0.19 |
ENST00000507386.5
|
JAKMIP2
|
janus kinase and microtubule interacting protein 2 |
| chr4_+_118888918 | 0.19 |
ENST00000434046.6
|
SYNPO2
|
synaptopodin 2 |
| chr5_+_83471736 | 0.19 |
ENST00000265077.8
|
VCAN
|
versican |
| chr9_+_113150991 | 0.18 |
ENST00000259392.8
|
SLC31A2
|
solute carrier family 31 member 2 |
| chr1_+_110873135 | 0.18 |
ENST00000271324.6
|
CD53
|
CD53 molecule |
| chr1_-_167518521 | 0.18 |
ENST00000362089.10
|
CD247
|
CD247 molecule |
| chr11_-_93197932 | 0.18 |
ENST00000326402.9
|
SLC36A4
|
solute carrier family 36 member 4 |
| chr12_+_104986292 | 0.18 |
ENST00000552951.7
ENST00000637147.1 ENST00000622317.5 ENST00000280749.5 |
C12orf45
|
chromosome 12 open reading frame 45 |
| chr12_+_9827472 | 0.18 |
ENST00000617793.4
ENST00000617889.5 ENST00000354855.7 ENST00000279545.7 |
KLRF1
|
killer cell lectin like receptor F1 |
| chr5_+_83471764 | 0.18 |
ENST00000512590.6
ENST00000513960.5 ENST00000513984.5 |
VCAN
|
versican |
| chr6_+_96563117 | 0.18 |
ENST00000450218.6
|
FHL5
|
four and a half LIM domains 5 |
| chrX_-_11265975 | 0.17 |
ENST00000303025.10
ENST00000657361.1 |
ARHGAP6
|
Rho GTPase activating protein 6 |
| chr12_+_106357729 | 0.17 |
ENST00000228347.9
|
POLR3B
|
RNA polymerase III subunit B |
| chr1_+_224183197 | 0.17 |
ENST00000323699.9
|
DEGS1
|
delta 4-desaturase, sphingolipid 1 |
| chr2_+_95165784 | 0.16 |
ENST00000622059.4
ENST00000614034.5 ENST00000611147.1 |
ZNF2
|
zinc finger protein 2 |
| chr9_+_127264740 | 0.16 |
ENST00000373387.9
|
GARNL3
|
GTPase activating Rap/RanGAP domain like 3 |
| chr1_-_72100930 | 0.16 |
ENST00000306821.3
|
NEGR1
|
neuronal growth regulator 1 |
| chr7_+_148590760 | 0.16 |
ENST00000307003.3
|
C7orf33
|
chromosome 7 open reading frame 33 |
| chr16_+_69311339 | 0.16 |
ENST00000254950.13
|
VPS4A
|
vacuolar protein sorting 4 homolog A |
| chr15_+_51377247 | 0.15 |
ENST00000396399.6
|
GLDN
|
gliomedin |
| chr5_+_83471668 | 0.15 |
ENST00000342785.8
ENST00000343200.9 |
VCAN
|
versican |
| chr16_+_21678514 | 0.15 |
ENST00000286149.8
ENST00000388958.8 |
OTOA
|
otoancorin |
| chr17_-_50707855 | 0.14 |
ENST00000285243.7
|
ANKRD40
|
ankyrin repeat domain 40 |
| chr17_-_8210203 | 0.14 |
ENST00000578549.5
ENST00000582368.5 |
AURKB
|
aurora kinase B |
| chr15_-_76012390 | 0.13 |
ENST00000394907.8
|
NRG4
|
neuregulin 4 |
| chr1_+_158845798 | 0.13 |
ENST00000438394.1
|
MNDA
|
myeloid cell nuclear differentiation antigen |
| chr21_+_32298849 | 0.13 |
ENST00000303645.10
|
MRAP
|
melanocortin 2 receptor accessory protein |
| chr7_-_20217342 | 0.13 |
ENST00000400331.10
ENST00000332878.8 |
MACC1
|
MET transcriptional regulator MACC1 |
| chr17_-_40994159 | 0.12 |
ENST00000391586.3
|
KRTAP3-3
|
keratin associated protein 3-3 |
| chr4_+_112647059 | 0.12 |
ENST00000511529.1
|
LARP7
|
La ribonucleoprotein 7, transcriptional regulator |
| chr21_+_32298945 | 0.12 |
ENST00000339944.4
|
MRAP
|
melanocortin 2 receptor accessory protein |
| chr9_-_128656649 | 0.12 |
ENST00000372715.7
|
DYNC2I2
|
dynein 2 intermediate chain 2 |
| chr18_-_3874247 | 0.12 |
ENST00000581699.5
|
DLGAP1
|
DLG associated protein 1 |
| chr1_+_244835616 | 0.12 |
ENST00000366528.3
ENST00000411948.7 |
COX20
|
cytochrome c oxidase assembly factor COX20 |
| chr5_+_122129597 | 0.12 |
ENST00000514925.1
|
ENSG00000250803.6
|
novel zinc finger protein |
| chr5_+_122129533 | 0.11 |
ENST00000296600.5
ENST00000504912.1 ENST00000505843.1 |
ZNF474
|
zinc finger protein 474 |
| chr10_-_32378720 | 0.11 |
ENST00000375110.6
|
EPC1
|
enhancer of polycomb homolog 1 |
| chr15_-_77420135 | 0.11 |
ENST00000560626.6
|
PEAK1
|
pseudopodium enriched atypical kinase 1 |
| chr6_+_29170907 | 0.11 |
ENST00000641417.1
|
OR2J2
|
olfactory receptor family 2 subfamily J member 2 |
| chr1_+_74235377 | 0.10 |
ENST00000326637.8
|
TNNI3K
|
TNNI3 interacting kinase |
| chr2_+_157257687 | 0.10 |
ENST00000259056.5
|
GALNT5
|
polypeptide N-acetylgalactosaminyltransferase 5 |
| chr16_-_29923237 | 0.10 |
ENST00000568995.1
ENST00000566413.1 |
KCTD13
|
potassium channel tetramerization domain containing 13 |
| chr15_-_42208153 | 0.10 |
ENST00000348544.4
ENST00000318006.10 |
VPS39
|
VPS39 subunit of HOPS complex |
| chr18_-_3874270 | 0.10 |
ENST00000400149.7
ENST00000400155.5 ENST00000400150.7 |
DLGAP1
|
DLG associated protein 1 |
| chr11_-_129024157 | 0.09 |
ENST00000392657.7
|
ARHGAP32
|
Rho GTPase activating protein 32 |
| chr10_+_27532521 | 0.09 |
ENST00000683924.1
|
RAB18
|
RAB18, member RAS oncogene family |
| chr8_-_25424260 | 0.09 |
ENST00000421054.7
|
GNRH1
|
gonadotropin releasing hormone 1 |
| chr3_-_21751189 | 0.09 |
ENST00000281523.8
|
ZNF385D
|
zinc finger protein 385D |
| chr15_-_77420087 | 0.09 |
ENST00000564328.5
ENST00000682557.1 ENST00000558305.5 |
PEAK1
|
pseudopodium enriched atypical kinase 1 |
| chr7_+_139133744 | 0.09 |
ENST00000430935.5
ENST00000495038.5 ENST00000474035.6 ENST00000478836.6 ENST00000464848.5 ENST00000343187.8 |
TTC26
|
tetratricopeptide repeat domain 26 |
| chr9_-_92482350 | 0.09 |
ENST00000375543.2
|
ASPN
|
asporin |
| chr2_+_32277883 | 0.08 |
ENST00000238831.9
|
YIPF4
|
Yip1 domain family member 4 |
| chr12_-_42144823 | 0.08 |
ENST00000398675.8
|
GXYLT1
|
glucoside xylosyltransferase 1 |
| chr5_-_139404050 | 0.08 |
ENST00000514983.5
ENST00000507779.2 ENST00000451821.6 ENST00000509959.5 ENST00000302091.9 ENST00000450845.7 |
SPATA24
|
spermatogenesis associated 24 |
| chr10_+_96304425 | 0.08 |
ENST00000371174.5
|
DNTT
|
DNA nucleotidylexotransferase |
| chr12_+_7711418 | 0.08 |
ENST00000345088.3
|
DPPA3
|
developmental pluripotency associated 3 |
| chr19_-_7747559 | 0.08 |
ENST00000394173.8
|
CD209
|
CD209 molecule |
| chr14_-_51096029 | 0.08 |
ENST00000298355.7
|
TRIM9
|
tripartite motif containing 9 |
| chr20_+_54208072 | 0.07 |
ENST00000371419.7
|
PFDN4
|
prefoldin subunit 4 |
| chr5_+_173889337 | 0.07 |
ENST00000520867.5
ENST00000334035.9 |
CPEB4
|
cytoplasmic polyadenylation element binding protein 4 |
| chr10_+_112283399 | 0.07 |
ENST00000643850.1
ENST00000646139.2 ENST00000645243.1 |
TECTB
|
tectorin beta |
| chr1_-_160523204 | 0.07 |
ENST00000368055.1
ENST00000368057.8 ENST00000368059.7 |
SLAMF6
|
SLAM family member 6 |
| chr21_+_29130630 | 0.07 |
ENST00000399926.5
ENST00000399928.6 |
MAP3K7CL
|
MAP3K7 C-terminal like |
| chr11_-_124320197 | 0.06 |
ENST00000624618.2
|
OR8D2
|
olfactory receptor family 8 subfamily D member 2 |
| chr6_+_29173303 | 0.06 |
ENST00000377167.3
|
OR2J2
|
olfactory receptor family 2 subfamily J member 2 |
| chr1_+_202122910 | 0.06 |
ENST00000682545.1
ENST00000367282.6 ENST00000682887.1 |
GPR37L1
|
G protein-coupled receptor 37 like 1 |
| chr1_-_205994439 | 0.06 |
ENST00000617991.4
|
RAB7B
|
RAB7B, member RAS oncogene family |
| chr3_+_108589667 | 0.06 |
ENST00000361582.8
ENST00000486815.5 |
DZIP3
|
DAZ interacting zinc finger protein 3 |
| chr6_+_29100609 | 0.06 |
ENST00000377171.3
|
OR2J1
|
olfactory receptor family 2 subfamily J member 1 |
| chr10_-_29634964 | 0.06 |
ENST00000375398.6
ENST00000355867.8 |
SVIL
|
supervillin |
| chr14_-_106791226 | 0.06 |
ENST00000433072.2
|
IGHV3-72
|
immunoglobulin heavy variable 3-72 |
| chr3_+_114294020 | 0.05 |
ENST00000383671.8
|
TIGIT
|
T cell immunoreceptor with Ig and ITIM domains |
| chrX_-_123623155 | 0.05 |
ENST00000618150.4
|
THOC2
|
THO complex 2 |
| chr4_+_158521714 | 0.05 |
ENST00000613319.4
ENST00000423548.5 ENST00000448688.6 |
RXFP1
|
relaxin family peptide receptor 1 |
| chr6_+_26240385 | 0.05 |
ENST00000244537.6
|
H4C6
|
H4 clustered histone 6 |
| chr6_-_32816910 | 0.05 |
ENST00000447394.1
ENST00000438763.7 |
HLA-DOB
|
major histocompatibility complex, class II, DO beta |
| chr4_+_158521872 | 0.05 |
ENST00000307765.10
|
RXFP1
|
relaxin family peptide receptor 1 |
| chr15_+_36579782 | 0.04 |
ENST00000437989.6
|
CDIN1
|
CDAN1 interacting nuclease 1 |
| chr4_+_118888829 | 0.04 |
ENST00000448416.6
ENST00000307142.9 ENST00000429713.7 |
SYNPO2
|
synaptopodin 2 |
| chr18_-_72865680 | 0.04 |
ENST00000397929.5
|
NETO1
|
neuropilin and tolloid like 1 |
| chr20_+_58389197 | 0.04 |
ENST00000475243.6
ENST00000395802.7 |
VAPB
|
VAMP associated protein B and C |
| chr8_-_71361860 | 0.04 |
ENST00000303824.11
ENST00000645451.1 |
EYA1
|
EYA transcriptional coactivator and phosphatase 1 |
| chrX_-_123622809 | 0.04 |
ENST00000441692.5
|
THOC2
|
THO complex 2 |
| chr19_-_53159004 | 0.04 |
ENST00000599096.1
ENST00000597183.5 ENST00000601804.5 ENST00000334197.12 ENST00000601469.2 ENST00000452676.6 |
ZNF347
|
zinc finger protein 347 |
| chrX_-_14873027 | 0.03 |
ENST00000452869.1
ENST00000398334.5 ENST00000650831.1 ENST00000324138.7 |
FANCB
|
FA complementation group B |
| chr2_-_168890368 | 0.03 |
ENST00000282074.7
|
SPC25
|
SPC25 component of NDC80 kinetochore complex |
| chr16_-_71484543 | 0.03 |
ENST00000565100.6
|
ZNF19
|
zinc finger protein 19 |
| chr14_+_55027200 | 0.03 |
ENST00000395472.2
ENST00000555846.2 |
SOCS4
|
suppressor of cytokine signaling 4 |
| chr6_+_28281555 | 0.03 |
ENST00000259883.3
ENST00000682144.1 |
PGBD1
|
piggyBac transposable element derived 1 |
| chr14_+_55027576 | 0.03 |
ENST00000339298.2
|
SOCS4
|
suppressor of cytokine signaling 4 |
| chr2_-_77522347 | 0.03 |
ENST00000409093.1
ENST00000409884.6 ENST00000409088.3 |
LRRTM4
|
leucine rich repeat transmembrane neuronal 4 |
| chr1_-_247536440 | 0.02 |
ENST00000366487.4
ENST00000641802.1 |
OR2C3
|
olfactory receptor family 2 subfamily C member 3 |
| chr11_+_126327863 | 0.02 |
ENST00000648516.1
|
DCPS
|
decapping enzyme, scavenger |
| chr14_+_93185304 | 0.02 |
ENST00000415050.3
|
TMEM251
|
transmembrane protein 251 |
| chr10_+_88586762 | 0.02 |
ENST00000371939.7
|
LIPJ
|
lipase family member J |
| chr6_-_109483208 | 0.02 |
ENST00000230122.4
|
ZBTB24
|
zinc finger and BTB domain containing 24 |
| chr14_-_73558945 | 0.02 |
ENST00000563329.1
ENST00000553558.6 ENST00000334988.2 |
HEATR4
|
HEAT repeat containing 4 |
| chr17_+_76732978 | 0.01 |
ENST00000587459.1
|
ENSG00000267168.1
|
novel protein |
| chr13_-_85799400 | 0.01 |
ENST00000647374.2
|
SLITRK6
|
SLIT and NTRK like family member 6 |
| chr9_-_83978429 | 0.01 |
ENST00000351839.7
|
HNRNPK
|
heterogeneous nuclear ribonucleoprotein K |
| chr7_+_99828010 | 0.01 |
ENST00000631161.2
ENST00000354829.7 ENST00000342499.8 ENST00000417625.5 ENST00000415413.5 ENST00000444905.5 ENST00000222382.5 ENST00000312017.9 |
CYP3A43
|
cytochrome P450 family 3 subfamily A member 43 |
| chr6_+_118548289 | 0.01 |
ENST00000357525.6
|
PLN
|
phospholamban |
| chr6_+_6588082 | 0.01 |
ENST00000379953.6
|
LY86
|
lymphocyte antigen 86 |
| chr4_-_99219230 | 0.01 |
ENST00000394897.5
ENST00000508558.1 ENST00000394899.6 |
ADH6
|
alcohol dehydrogenase 6 (class V) |
| chr4_-_163613505 | 0.01 |
ENST00000339875.9
|
MARCHF1
|
membrane associated ring-CH-type finger 1 |
| chr19_-_9107475 | 0.00 |
ENST00000641081.1
|
OR7G2
|
olfactory receptor family 7 subfamily G member 2 |
| chr14_+_51860632 | 0.00 |
ENST00000555472.5
ENST00000556766.5 |
GNG2
|
G protein subunit gamma 2 |
| chrX_+_11293411 | 0.00 |
ENST00000348912.4
ENST00000380714.7 ENST00000380712.7 |
AMELX
|
amelogenin X-linked |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 2.5 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.3 | 1.0 | GO:0048250 | mitochondrial iron ion transport(GO:0048250) |
| 0.3 | 0.9 | GO:0034769 | basement membrane disassembly(GO:0034769) |
| 0.3 | 0.8 | GO:0009720 | detection of hormone stimulus(GO:0009720) |
| 0.1 | 0.6 | GO:0035617 | stress granule disassembly(GO:0035617) |
| 0.1 | 0.4 | GO:0015783 | GDP-fucose transport(GO:0015783) purine nucleotide-sugar transport(GO:0036079) |
| 0.1 | 0.4 | GO:1990523 | bone regeneration(GO:1990523) |
| 0.1 | 0.4 | GO:0007341 | penetration of zona pellucida(GO:0007341) |
| 0.1 | 0.2 | GO:1903774 | positive regulation of viral budding via host ESCRT complex(GO:1903774) |
| 0.0 | 0.4 | GO:1903265 | positive regulation of tumor necrosis factor-mediated signaling pathway(GO:1903265) |
| 0.0 | 0.3 | GO:0060011 | Sertoli cell proliferation(GO:0060011) |
| 0.0 | 1.0 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.2 | GO:0032439 | endosome localization(GO:0032439) |
| 0.0 | 0.4 | GO:0070170 | regulation of tooth mineralization(GO:0070170) |
| 0.0 | 0.2 | GO:0002175 | protein localization to paranode region of axon(GO:0002175) |
| 0.0 | 0.1 | GO:0044878 | mitotic cytokinesis checkpoint(GO:0044878) |
| 0.0 | 0.1 | GO:0044725 | chromatin reprogramming in the zygote(GO:0044725) |
| 0.0 | 0.2 | GO:0015808 | L-alanine transport(GO:0015808) |
| 0.0 | 0.2 | GO:0097068 | response to thyroxine(GO:0097068) response to L-phenylalanine derivative(GO:1904386) |
| 0.0 | 0.5 | GO:0060391 | positive regulation of SMAD protein import into nucleus(GO:0060391) |
| 0.0 | 0.1 | GO:1990637 | response to prolactin(GO:1990637) |
| 0.0 | 0.1 | GO:0035544 | negative regulation of SNARE complex assembly(GO:0035544) |
| 0.0 | 0.5 | GO:0030208 | dermatan sulfate biosynthetic process(GO:0030208) |
| 0.0 | 0.0 | GO:0002581 | negative regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002581) |
| 0.0 | 0.2 | GO:0061088 | regulation of sequestering of zinc ion(GO:0061088) |
| 0.0 | 0.3 | GO:1903071 | positive regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903071) |
| 0.0 | 0.3 | GO:0090136 | epithelial cell-cell adhesion(GO:0090136) |
| 0.0 | 0.1 | GO:0034164 | negative regulation of toll-like receptor 9 signaling pathway(GO:0034164) |
| 0.0 | 0.1 | GO:1902774 | late endosome to lysosome transport(GO:1902774) |
| 0.0 | 0.2 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.0 | 0.4 | GO:0045577 | regulation of B cell differentiation(GO:0045577) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0043159 | acrosomal matrix(GO:0043159) |
| 0.1 | 0.9 | GO:0036021 | endolysosome lumen(GO:0036021) |
| 0.1 | 0.4 | GO:0097169 | IPAF inflammasome complex(GO:0072557) AIM2 inflammasome complex(GO:0097169) |
| 0.0 | 0.3 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.0 | 0.3 | GO:1990037 | Lewy body core(GO:1990037) |
| 0.0 | 0.4 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.0 | 0.2 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.0 | 3.6 | GO:0097517 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 0.2 | GO:0045179 | apical cortex(GO:0045179) |
| 0.0 | 0.5 | GO:0005685 | U1 snRNP(GO:0005685) |
| 0.0 | 0.1 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 0.0 | 0.1 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 1.1 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 0.2 | GO:0033270 | paranode region of axon(GO:0033270) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.5 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.2 | 0.8 | GO:0034041 | sterol-transporting ATPase activity(GO:0034041) |
| 0.1 | 0.4 | GO:0036080 | GDP-fucose transmembrane transporter activity(GO:0005457) purine nucleotide-sugar transmembrane transporter activity(GO:0036080) |
| 0.1 | 0.4 | GO:0004040 | amidase activity(GO:0004040) |
| 0.1 | 0.5 | GO:0035614 | snRNA stem-loop binding(GO:0035614) |
| 0.1 | 0.3 | GO:0031780 | corticotropin hormone receptor binding(GO:0031780) type 5 melanocortin receptor binding(GO:0031783) |
| 0.1 | 1.0 | GO:0005381 | iron ion transmembrane transporter activity(GO:0005381) |
| 0.1 | 0.2 | GO:0015173 | aromatic amino acid transmembrane transporter activity(GO:0015173) |
| 0.1 | 0.2 | GO:0042284 | sphingolipid delta-4 desaturase activity(GO:0042284) |
| 0.0 | 0.3 | GO:0098821 | BMP receptor activity(GO:0098821) |
| 0.0 | 0.4 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.0 | 0.9 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.0 | 0.2 | GO:0047144 | 2-acylglycerol-3-phosphate O-acyltransferase activity(GO:0047144) |
| 0.0 | 0.1 | GO:0035174 | histone serine kinase activity(GO:0035174) |
| 0.0 | 0.4 | GO:0005549 | odorant binding(GO:0005549) |
| 0.0 | 0.5 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.2 | GO:0032393 | MHC class I receptor activity(GO:0032393) |
| 0.0 | 0.1 | GO:0031013 | troponin I binding(GO:0031013) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 2.6 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 0.5 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 1.0 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.0 | 2.5 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.8 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.0 | 0.4 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.0 | 0.5 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 0.0 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |