Project
Inflammatory response time course, HUVEC (Wada, 2009)
Navigation
Downloads
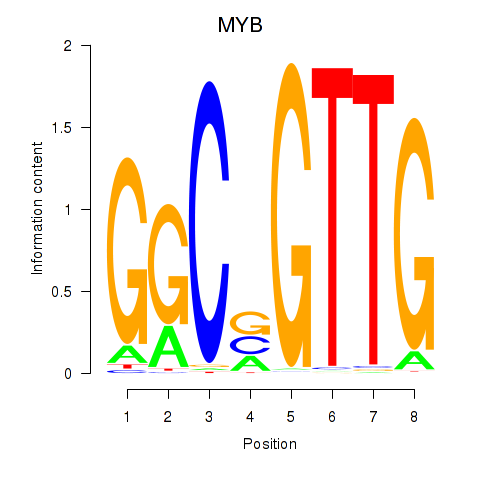
Results for MYB
Z-value: 1.39
Transcription factors associated with MYB
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
MYB
|
ENSG00000118513.19 | MYB |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| MYB | hg38_v1_chr6_+_135181323_135181328, hg38_v1_chr6_+_135181268_135181322 | -0.26 | 2.2e-01 | Click! |
Activity profile of MYB motif
Sorted Z-values of MYB motif
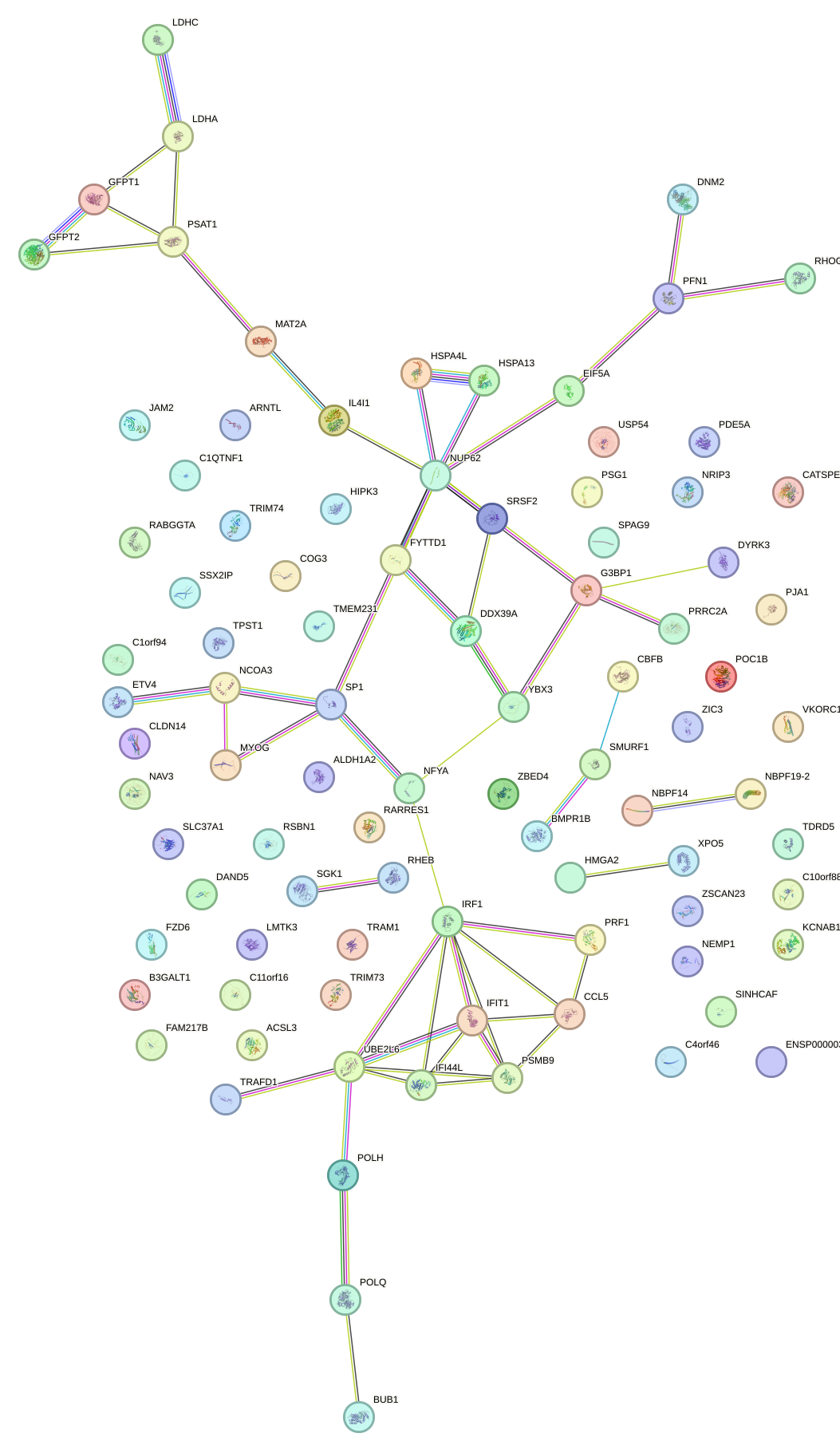
Network of associatons between targets according to the STRING database.
First level regulatory network of MYB
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_+_78620722 | 3.71 |
ENST00000679848.1
|
IFI44L
|
interferon induced protein 44 like |
| chr17_+_7307579 | 3.56 |
ENST00000572815.5
|
EIF5A
|
eukaryotic translation initiation factor 5A |
| chr5_+_151771884 | 2.94 |
ENST00000627077.2
ENST00000678976.1 ENST00000677408.1 ENST00000678070.1 ENST00000678964.1 ENST00000678925.1 ENST00000394123.7 ENST00000522761.6 ENST00000676827.1 |
G3BP1
|
G3BP stress granule assembly factor 1 |
| chr17_-_35880350 | 2.72 |
ENST00000605140.6
ENST00000651122.1 ENST00000603197.6 |
CCL5
|
C-C motif chemokine ligand 5 |
| chr5_+_151771943 | 2.30 |
ENST00000520177.6
ENST00000678854.1 ENST00000520006.2 ENST00000677105.1 ENST00000678101.1 ENST00000678910.1 ENST00000356245.8 ENST00000678646.1 ENST00000677433.1 ENST00000677369.1 ENST00000677284.1 ENST00000678564.1 |
G3BP1
|
G3BP stress granule assembly factor 1 |
| chr12_+_77830886 | 2.17 |
ENST00000397909.7
ENST00000549464.5 |
NAV3
|
neuron navigator 3 |
| chr3_-_158732442 | 2.14 |
ENST00000479756.1
ENST00000237696.10 |
RARRES1
|
retinoic acid receptor responder 1 |
| chr17_+_7307602 | 2.13 |
ENST00000573542.5
ENST00000336458.13 |
EIF5A
|
eukaryotic translation initiation factor 5A |
| chr5_-_150290093 | 2.12 |
ENST00000672479.1
|
CAMK2A
|
calcium/calmodulin dependent protein kinase II alpha |
| chr6_-_134318097 | 1.95 |
ENST00000367858.10
ENST00000533224.1 |
SGK1
|
serum/glucocorticoid regulated kinase 1 |
| chr16_-_75556214 | 1.90 |
ENST00000568377.5
ENST00000565067.5 ENST00000258173.11 |
TMEM231
|
transmembrane protein 231 |
| chr6_+_32844789 | 1.78 |
ENST00000414474.5
|
PSMB9
|
proteasome 20S subunit beta 9 |
| chr12_-_57078739 | 1.74 |
ENST00000379391.7
|
NEMP1
|
nuclear envelope integral membrane protein 1 |
| chr12_-_57078784 | 1.71 |
ENST00000300128.9
|
NEMP1
|
nuclear envelope integral membrane protein 1 |
| chr1_+_206635521 | 1.70 |
ENST00000367109.8
|
DYRK3
|
dual specificity tyrosine phosphorylation regulated kinase 3 |
| chr6_-_134174853 | 1.68 |
ENST00000475719.6
ENST00000367857.9 ENST00000237305.11 |
SGK1
|
serum/glucocorticoid regulated kinase 1 |
| chr17_+_7307961 | 1.68 |
ENST00000419711.6
ENST00000571955.5 ENST00000573714.5 |
EIF5A
|
eukaryotic translation initiation factor 5A |
| chr14_-_91244669 | 1.63 |
ENST00000650645.1
|
GPR68
|
G protein-coupled receptor 68 |
| chr17_-_51120734 | 1.61 |
ENST00000505279.5
|
SPAG9
|
sperm associated antigen 9 |
| chr11_+_18412292 | 1.58 |
ENST00000541669.6
ENST00000280704.8 |
LDHC
|
lactate dehydrogenase C |
| chr19_-_49929525 | 1.56 |
ENST00000596437.5
ENST00000341114.7 ENST00000595948.5 |
NUP62
IL4I1
|
nucleoporin 62 interleukin 4 induced 1 |
| chr2_-_69387130 | 1.53 |
ENST00000674438.1
|
GFPT1
|
glutamine--fructose-6-phosphate transaminase 1 |
| chr5_-_150289764 | 1.50 |
ENST00000671881.1
ENST00000672752.1 ENST00000510347.2 ENST00000672829.1 ENST00000348628.11 |
CAMK2A
|
calcium/calmodulin dependent protein kinase II alpha |
| chr5_-_180353317 | 1.50 |
ENST00000253778.13
|
GFPT2
|
glutamine-fructose-6-phosphate transaminase 2 |
| chr17_+_79024142 | 1.49 |
ENST00000579760.6
ENST00000339142.6 |
C1QTNF1
|
C1q and TNF related 1 |
| chr1_-_148679734 | 1.49 |
ENST00000606877.2
ENST00000593495.3 |
NBPF14
NOTCH2NLB
|
NBPF member 14 notch 2 N-terminal like B |
| chr11_-_57568276 | 1.48 |
ENST00000340573.8
|
UBE2L6
|
ubiquitin conjugating enzyme E2 L6 |
| chr5_-_150289941 | 1.47 |
ENST00000682786.1
|
CAMK2A
|
calcium/calmodulin dependent protein kinase II alpha |
| chr1_+_206635573 | 1.43 |
ENST00000367108.7
|
DYRK3
|
dual specificity tyrosine phosphorylation regulated kinase 3 |
| chr1_+_206635772 | 1.41 |
ENST00000441486.5
ENST00000367106.1 |
DYRK3
|
dual specificity tyrosine phosphorylation regulated kinase 3 |
| chr14_-_91244508 | 1.38 |
ENST00000535815.5
ENST00000529102.1 |
GPR68
|
G protein-coupled receptor 68 |
| chr10_+_89392546 | 1.37 |
ENST00000546318.2
ENST00000371804.4 |
IFIT1
|
interferon induced protein with tetratricopeptide repeats 1 |
| chr7_-_127392310 | 1.35 |
ENST00000393312.5
|
ZNF800
|
zinc finger protein 800 |
| chr16_+_67029359 | 1.32 |
ENST00000565389.1
|
CBFB
|
core-binding factor subunit beta |
| chr4_+_95051671 | 1.26 |
ENST00000440890.7
|
BMPR1B
|
bone morphogenetic protein receptor type 1B |
| chr1_+_34176900 | 1.21 |
ENST00000488417.2
|
C1orf94
|
chromosome 1 open reading frame 94 |
| chr20_+_59933761 | 1.21 |
ENST00000358293.7
|
FAM217B
|
family with sequence similarity 217 member B |
| chr12_+_65824475 | 1.21 |
ENST00000403681.7
|
HMGA2
|
high mobility group AT-hook 2 |
| chr5_-_132490750 | 1.21 |
ENST00000437654.6
ENST00000245414.9 ENST00000680139.1 ENST00000680352.1 ENST00000679440.1 ENST00000680903.1 |
IRF1
|
interferon regulatory factor 1 |
| chr6_+_43576119 | 1.18 |
ENST00000372236.9
|
POLH
|
DNA polymerase eta |
| chr1_+_206635703 | 1.16 |
ENST00000649163.1
|
DYRK3
|
dual specificity tyrosine phosphorylation regulated kinase 3 |
| chr19_+_48321454 | 1.14 |
ENST00000599704.5
|
EMP3
|
epithelial membrane protein 3 |
| chr17_-_51120855 | 1.13 |
ENST00000618113.4
ENST00000357122.8 ENST00000262013.12 |
SPAG9
|
sperm associated antigen 9 |
| chr11_+_18396266 | 1.12 |
ENST00000540430.5
ENST00000379412.9 |
LDHA
|
lactate dehydrogenase A |
| chr16_+_67029093 | 1.12 |
ENST00000561924.6
|
CBFB
|
core-binding factor subunit beta |
| chr7_+_75395063 | 1.10 |
ENST00000450434.5
|
TRIM73
|
tripartite motif containing 73 |
| chr20_-_31723902 | 1.10 |
ENST00000676942.1
ENST00000450273.2 ENST00000678563.1 ENST00000456404.6 ENST00000420488.6 ENST00000439267.2 |
BCL2L1
|
BCL2 like 1 |
| chr17_+_7308339 | 1.10 |
ENST00000416016.2
|
EIF5A
|
eukaryotic translation initiation factor 5A |
| chr2_-_69387188 | 1.09 |
ENST00000674507.1
ENST00000357308.9 |
GFPT1
|
glutamine--fructose-6-phosphate transaminase 1 |
| chr16_+_50153248 | 1.06 |
ENST00000561678.7
|
TENT4B
|
terminal nucleotidyltransferase 4B |
| chr16_+_50152900 | 1.05 |
ENST00000436909.8
|
TENT4B
|
terminal nucleotidyltransferase 4B |
| chr3_+_88059231 | 1.02 |
ENST00000636215.2
|
ZNF654
|
zinc finger protein 654 |
| chr21_+_25639251 | 1.02 |
ENST00000480456.6
|
JAM2
|
junctional adhesion molecule 2 |
| chr10_-_122954184 | 1.01 |
ENST00000481909.2
|
C10orf88
|
chromosome 10 open reading frame 88 |
| chr7_-_72969466 | 1.01 |
ENST00000285805.3
|
TRIM74
|
tripartite motif containing 74 |
| chr21_-_14383125 | 1.01 |
ENST00000285667.4
|
HSPA13
|
heat shock protein family A (Hsp70) member 13 |
| chr12_+_112125531 | 1.00 |
ENST00000549358.5
ENST00000257604.9 ENST00000548092.5 ENST00000412615.7 ENST00000552896.1 |
TRAFD1
|
TRAF-type zinc finger domain containing 1 |
| chrX_+_140091415 | 1.00 |
ENST00000453380.2
|
ENSG00000230707.2
|
novel protein (LOC389895) |
| chr6_+_43576205 | 1.00 |
ENST00000372226.1
ENST00000443535.1 |
POLH
|
DNA polymerase eta |
| chr2_-_69387241 | 0.99 |
ENST00000361060.5
|
GFPT1
|
glutamine--fructose-6-phosphate transaminase 1 |
| chr2_-_110678033 | 0.99 |
ENST00000447014.5
ENST00000420328.5 ENST00000302759.11 ENST00000535254.6 ENST00000409311.5 |
BUB1
|
BUB1 mitotic checkpoint serine/threonine kinase |
| chr17_+_7307531 | 0.98 |
ENST00000576930.5
|
EIF5A
|
eukaryotic translation initiation factor 5A |
| chr8_+_103298836 | 0.97 |
ENST00000523739.5
ENST00000358755.5 |
FZD6
|
frizzled class receptor 6 |
| chr4_+_127781815 | 0.97 |
ENST00000508776.5
|
HSPA4L
|
heat shock protein family A (Hsp70) member 4 like |
| chr5_-_150289625 | 0.95 |
ENST00000683332.1
ENST00000398376.8 ENST00000672785.1 ENST00000672396.1 |
CAMK2A
|
calcium/calmodulin dependent protein kinase II alpha |
| chr6_+_63572472 | 0.91 |
ENST00000370651.7
ENST00000626021.3 |
ENSG00000285976.2
PTP4A1
|
novel protein protein tyrosine phosphatase 4A1 |
| chr6_+_31620701 | 0.91 |
ENST00000376033.3
ENST00000376007.8 |
PRRC2A
|
proline rich coiled-coil 2A |
| chr12_-_10723307 | 0.90 |
ENST00000279550.11
ENST00000228251.9 |
YBX3
|
Y-box binding protein 3 |
| chr8_+_103298433 | 0.88 |
ENST00000522566.5
|
FZD6
|
frizzled class receptor 6 |
| chr7_+_75395629 | 0.88 |
ENST00000323819.7
ENST00000430211.5 |
TRIM73
|
tripartite motif containing 73 |
| chr6_-_43575966 | 0.87 |
ENST00000265351.12
|
XPO5
|
exportin 5 |
| chr21_+_42499600 | 0.87 |
ENST00000398341.7
|
SLC37A1
|
solute carrier family 37 member 1 |
| chr12_+_53380639 | 0.85 |
ENST00000426431.2
|
SP1
|
Sp1 transcription factor |
| chr4_-_119627631 | 0.85 |
ENST00000264805.9
|
PDE5A
|
phosphodiesterase 5A |
| chr11_-_3840829 | 0.84 |
ENST00000396978.1
|
RHOG
|
ras homolog family member G |
| chr15_-_58065703 | 0.83 |
ENST00000249750.9
|
ALDH1A2
|
aldehyde dehydrogenase 1 family member A2 |
| chr7_+_66205325 | 0.83 |
ENST00000304842.6
ENST00000649664.1 |
TPST1
|
tyrosylprotein sulfotransferase 1 |
| chr7_+_65873324 | 0.83 |
ENST00000434382.2
|
VKORC1L1
|
vitamin K epoxide reductase complex subunit 1 like 1 |
| chr2_+_222861059 | 0.82 |
ENST00000681697.1
ENST00000680921.1 |
ACSL3
|
acyl-CoA synthetase long chain family member 3 |
| chr22_+_30080460 | 0.81 |
ENST00000336726.11
|
HORMAD2
|
HORMA domain containing 2 |
| chr4_+_127782270 | 0.81 |
ENST00000508549.5
ENST00000296464.9 |
HSPA4L
|
heat shock protein family A (Hsp70) member 4 like |
| chr8_+_76683779 | 0.81 |
ENST00000523885.2
|
ZFHX4
|
zinc finger homeobox 4 |
| chr3_+_197749855 | 0.81 |
ENST00000241502.9
|
FYTTD1
|
forty-two-three domain containing 1 |
| chrX_-_69165430 | 0.80 |
ENST00000374584.3
ENST00000590146.1 ENST00000374571.5 |
PJA1
|
praja ring finger ubiquitin ligase 1 |
| chr11_+_33257265 | 0.79 |
ENST00000303296.9
ENST00000379016.7 |
HIPK3
|
homeodomain interacting protein kinase 3 |
| chr19_-_14419331 | 0.77 |
ENST00000242776.9
|
DDX39A
|
DExD-box helicase 39A |
| chrX_-_69165509 | 0.77 |
ENST00000361478.1
|
PJA1
|
praja ring finger ubiquitin ligase 1 |
| chr21_-_36480060 | 0.77 |
ENST00000399137.5
|
CLDN14
|
claudin 14 |
| chr19_+_38335775 | 0.76 |
ENST00000410018.5
ENST00000409235.8 ENST00000409410.6 |
CATSPERG
|
cation channel sperm associated auxiliary subunit gamma |
| chr2_+_222860942 | 0.76 |
ENST00000392066.7
ENST00000680251.1 ENST00000679541.1 ENST00000679545.1 ENST00000680395.1 |
ACSL3
|
acyl-CoA synthetase long chain family member 3 |
| chr11_-_8932944 | 0.76 |
ENST00000326053.10
ENST00000525780.5 |
C11orf16
|
chromosome 11 open reading frame 16 |
| chr6_+_1389553 | 0.75 |
ENST00000645481.2
|
FOXF2
|
forkhead box F2 |
| chr17_-_43546323 | 0.75 |
ENST00000545954.5
ENST00000319349.10 |
ETV4
|
ETS variant transcription factor 4 |
| chr1_-_84690255 | 0.75 |
ENST00000603677.1
|
SSX2IP
|
SSX family member 2 interacting protein |
| chr4_-_158671843 | 0.75 |
ENST00000379205.5
ENST00000508457.1 |
C4orf46
|
chromosome 4 open reading frame 46 |
| chr1_-_100895132 | 0.73 |
ENST00000535414.5
|
EXTL2
|
exostosin like glycosyltransferase 2 |
| chr17_-_4948519 | 0.73 |
ENST00000225655.6
|
PFN1
|
profilin 1 |
| chr7_-_99144053 | 0.73 |
ENST00000361125.1
ENST00000361368.7 |
SMURF1
|
SMAD specific E3 ubiquitin protein ligase 1 |
| chr19_+_10718114 | 0.72 |
ENST00000408974.8
|
DNM2
|
dynamin 2 |
| chr2_+_85539158 | 0.72 |
ENST00000306434.8
|
MAT2A
|
methionine adenosyltransferase 2A |
| chr3_+_156291834 | 0.72 |
ENST00000389634.5
|
KCNAB1
|
potassium voltage-gated channel subfamily A member regulatory beta subunit 1 |
| chr6_-_28443463 | 0.72 |
ENST00000289788.4
|
ZSCAN23
|
zinc finger and SCAN domain containing 23 |
| chr12_-_31326171 | 0.72 |
ENST00000542983.1
|
SINHCAF
|
SIN3-HDAC complex associated factor |
| chr14_-_24271503 | 0.72 |
ENST00000216840.11
ENST00000399409.7 |
RABGGTA
|
Rab geranylgeranyltransferase subunit alpha |
| chr17_-_43545891 | 0.71 |
ENST00000591713.5
ENST00000538265.5 |
ETV4
|
ETS variant transcription factor 4 |
| chr1_-_113812448 | 0.70 |
ENST00000612242.4
ENST00000261441.9 |
RSBN1
|
round spermatid basic protein 1 |
| chr11_-_9003994 | 0.69 |
ENST00000309166.8
ENST00000525100.1 ENST00000531090.5 |
NRIP3
|
nuclear receptor interacting protein 3 |
| chr10_-_73625951 | 0.69 |
ENST00000433394.1
ENST00000422491.7 |
USP54
|
ubiquitin specific peptidase 54 |
| chr1_-_100894818 | 0.69 |
ENST00000370114.8
|
EXTL2
|
exostosin like glycosyltransferase 2 |
| chr2_+_222861005 | 0.69 |
ENST00000680147.1
ENST00000681009.1 ENST00000679514.1 ENST00000357430.8 |
ACSL3
|
acyl-CoA synthetase long chain family member 3 |
| chr1_-_100894775 | 0.69 |
ENST00000416479.1
ENST00000370113.7 |
EXTL2
|
exostosin like glycosyltransferase 2 |
| chr7_-_151520080 | 0.69 |
ENST00000496004.5
|
RHEB
|
Ras homolog, mTORC1 binding |
| chr1_+_179591876 | 0.69 |
ENST00000294848.12
|
TDRD5
|
tudor domain containing 5 |
| chr11_+_13277639 | 0.68 |
ENST00000527998.5
ENST00000529388.6 ENST00000401424.6 ENST00000403510.8 ENST00000403290.6 ENST00000533520.5 ENST00000389707.8 ENST00000673817.1 |
ARNTL
|
aryl hydrocarbon receptor nuclear translocator like |
| chr8_+_32647080 | 0.68 |
ENST00000520502.7
ENST00000523041.2 ENST00000650819.1 |
NRG1
|
neuregulin 1 |
| chrX_+_137566119 | 0.68 |
ENST00000287538.10
|
ZIC3
|
Zic family member 3 |
| chr12_-_89525444 | 0.68 |
ENST00000549035.1
ENST00000393179.8 |
POC1B
|
POC1 centriolar protein B |
| chr19_-_48513161 | 0.68 |
ENST00000673139.1
|
LMTK3
|
lemur tyrosine kinase 3 |
| chr6_+_41072939 | 0.67 |
ENST00000341376.11
ENST00000353205.5 |
NFYA
|
nuclear transcription factor Y subunit alpha |
| chr2_+_167868948 | 0.67 |
ENST00000392690.3
|
B3GALT1
|
beta-1,3-galactosyltransferase 1 |
| chr9_+_78297143 | 0.67 |
ENST00000347159.6
|
PSAT1
|
phosphoserine aminotransferase 1 |
| chr12_+_53380141 | 0.66 |
ENST00000551969.5
ENST00000327443.9 |
SP1
|
Sp1 transcription factor |
| chr13_+_45464995 | 0.66 |
ENST00000617493.1
|
COG3
|
component of oligomeric golgi complex 3 |
| chr1_-_203086001 | 0.66 |
ENST00000241651.5
|
MYOG
|
myogenin |
| chr17_-_76737321 | 0.66 |
ENST00000359995.10
ENST00000508921.7 ENST00000583836.1 ENST00000358156.6 ENST00000392485.2 |
SRSF2
|
serine and arginine rich splicing factor 2 |
| chr22_+_49853801 | 0.65 |
ENST00000216268.6
|
ZBED4
|
zinc finger BED-type containing 4 |
| chr8_-_70608407 | 0.64 |
ENST00000262213.7
|
TRAM1
|
translocation associated membrane protein 1 |
| chr19_+_10654327 | 0.64 |
ENST00000407004.7
ENST00000589998.5 ENST00000589600.5 |
ILF3
|
interleukin enhancer binding factor 3 |
| chr1_+_179591819 | 0.64 |
ENST00000444136.6
|
TDRD5
|
tudor domain containing 5 |
| chr18_-_59359245 | 0.64 |
ENST00000251047.6
|
LMAN1
|
lectin, mannose binding 1 |
| chr12_-_132761814 | 0.64 |
ENST00000357997.10
|
ANKLE2
|
ankyrin repeat and LEM domain containing 2 |
| chr6_-_32844643 | 0.64 |
ENST00000374881.3
|
PSMB8
|
proteasome 20S subunit beta 8 |
| chr2_-_39120383 | 0.63 |
ENST00000395038.6
|
SOS1
|
SOS Ras/Rac guanine nucleotide exchange factor 1 |
| chr2_+_105851748 | 0.63 |
ENST00000425756.1
ENST00000393349.2 |
NCK2
|
NCK adaptor protein 2 |
| chr1_-_68497030 | 0.63 |
ENST00000456315.7
ENST00000525124.1 ENST00000370966.9 |
DEPDC1
|
DEP domain containing 1 |
| chr7_-_106112205 | 0.63 |
ENST00000470347.1
ENST00000455385.7 |
SYPL1
|
synaptophysin like 1 |
| chr8_+_85107172 | 0.63 |
ENST00000360375.8
|
LRRCC1
|
leucine rich repeat and coiled-coil centrosomal protein 1 |
| chr11_-_65900413 | 0.63 |
ENST00000448083.6
ENST00000531493.5 ENST00000532401.1 |
FOSL1
|
FOS like 1, AP-1 transcription factor subunit |
| chr16_+_67666770 | 0.62 |
ENST00000403458.9
ENST00000602365.1 |
C16orf86
|
chromosome 16 open reading frame 86 |
| chrX_+_50204753 | 0.62 |
ENST00000376042.6
|
CCNB3
|
cyclin B3 |
| chr12_-_57633136 | 0.62 |
ENST00000341156.9
ENST00000550764.5 ENST00000551220.1 |
B4GALNT1
|
beta-1,4-N-acetyl-galactosaminyltransferase 1 |
| chr10_+_21534440 | 0.62 |
ENST00000650772.1
ENST00000652497.1 |
MLLT10
|
MLLT10 histone lysine methyltransferase DOT1L cofactor |
| chr6_-_35921047 | 0.62 |
ENST00000361690.7
ENST00000512445.5 |
SRPK1
|
SRSF protein kinase 1 |
| chr17_+_49132763 | 0.62 |
ENST00000393354.7
|
B4GALNT2
|
beta-1,4-N-acetyl-galactosaminyltransferase 2 |
| chr2_+_185738798 | 0.62 |
ENST00000424728.6
|
FSIP2
|
fibrous sheath interacting protein 2 |
| chrX_-_120115909 | 0.61 |
ENST00000217999.3
|
RHOXF1
|
Rhox homeobox family member 1 |
| chr12_+_65824403 | 0.61 |
ENST00000393578.7
ENST00000425208.6 ENST00000536545.5 ENST00000354636.7 |
HMGA2
|
high mobility group AT-hook 2 |
| chr2_+_17878637 | 0.61 |
ENST00000304101.9
|
KCNS3
|
potassium voltage-gated channel modifier subfamily S member 3 |
| chr16_+_67029133 | 0.60 |
ENST00000290858.11
ENST00000412916.7 ENST00000564034.6 |
CBFB
|
core-binding factor subunit beta |
| chr17_+_35587478 | 0.60 |
ENST00000618940.4
|
AP2B1
|
adaptor related protein complex 2 subunit beta 1 |
| chr4_-_184649436 | 0.59 |
ENST00000308394.9
ENST00000517513.5 ENST00000447121.2 ENST00000393588.8 ENST00000523916.5 |
CASP3
|
caspase 3 |
| chr19_+_10655023 | 0.59 |
ENST00000590009.5
|
ILF3
|
interleukin enhancer binding factor 3 |
| chr10_-_13707536 | 0.59 |
ENST00000632570.1
ENST00000477221.2 |
FRMD4A
|
FERM domain containing 4A |
| chr6_+_33391805 | 0.59 |
ENST00000428849.7
ENST00000450504.1 |
KIFC1
|
kinesin family member C1 |
| chr6_+_69232406 | 0.59 |
ENST00000238918.12
|
ADGRB3
|
adhesion G protein-coupled receptor B3 |
| chr16_+_56638659 | 0.58 |
ENST00000290705.12
|
MT1A
|
metallothionein 1A |
| chr1_+_156338619 | 0.58 |
ENST00000481479.5
ENST00000368252.5 ENST00000466306.5 ENST00000368251.1 |
TSACC
|
TSSK6 activating cochaperone |
| chr11_+_34052210 | 0.58 |
ENST00000532820.5
|
CAPRIN1
|
cell cycle associated protein 1 |
| chr9_+_15422704 | 0.57 |
ENST00000380821.7
ENST00000610884.4 ENST00000421710.5 |
SNAPC3
|
small nuclear RNA activating complex polypeptide 3 |
| chr6_-_158819355 | 0.57 |
ENST00000367075.4
|
EZR
|
ezrin |
| chr2_+_232633551 | 0.57 |
ENST00000264059.8
|
EFHD1
|
EF-hand domain family member D1 |
| chr6_-_35921128 | 0.57 |
ENST00000510290.5
ENST00000423325.6 |
SRPK1
|
SRSF protein kinase 1 |
| chr6_-_35921079 | 0.56 |
ENST00000507909.1
ENST00000373825.7 |
SRPK1
|
SRSF protein kinase 1 |
| chr12_-_119877300 | 0.56 |
ENST00000392521.7
|
CIT
|
citron rho-interacting serine/threonine kinase |
| chr20_+_48921775 | 0.56 |
ENST00000681021.1
ENST00000679436.1 |
ARFGEF2
|
ADP ribosylation factor guanine nucleotide exchange factor 2 |
| chr17_-_40937445 | 0.56 |
ENST00000436344.7
ENST00000485751.1 |
KRT23
|
keratin 23 |
| chr14_+_77458032 | 0.56 |
ENST00000535854.6
ENST00000555517.1 ENST00000216479.8 |
AHSA1
|
activator of HSP90 ATPase activity 1 |
| chr5_-_115626161 | 0.55 |
ENST00000282382.8
|
TMED7-TICAM2
|
TMED7-TICAM2 readthrough |
| chr5_-_1886938 | 0.55 |
ENST00000613726.4
|
IRX4
|
iroquois homeobox 4 |
| chr16_+_30650728 | 0.55 |
ENST00000568754.5
|
PRR14
|
proline rich 14 |
| chr13_-_72782096 | 0.55 |
ENST00000545453.5
|
DIS3
|
DIS3 homolog, exosome endoribonuclease and 3'-5' exoribonuclease |
| chr20_-_31723491 | 0.55 |
ENST00000676582.1
ENST00000422920.2 |
BCL2L1
|
BCL2 like 1 |
| chr5_+_172641241 | 0.54 |
ENST00000369800.6
ENST00000520919.5 ENST00000522853.5 |
NEURL1B
|
neuralized E3 ubiquitin protein ligase 1B |
| chr19_-_58558871 | 0.54 |
ENST00000595957.5
|
UBE2M
|
ubiquitin conjugating enzyme E2 M |
| chr18_-_5543960 | 0.54 |
ENST00000400111.8
|
EPB41L3
|
erythrocyte membrane protein band 4.1 like 3 |
| chr9_+_78297117 | 0.54 |
ENST00000376588.4
|
PSAT1
|
phosphoserine aminotransferase 1 |
| chr16_+_56451513 | 0.54 |
ENST00000562150.5
ENST00000561646.5 ENST00000566157.6 ENST00000568397.1 |
OGFOD1
|
2-oxoglutarate and iron dependent oxygenase domain containing 1 |
| chrX_+_132023580 | 0.53 |
ENST00000496850.1
|
STK26
|
serine/threonine kinase 26 |
| chr14_+_77457861 | 0.53 |
ENST00000555133.5
|
AHSA1
|
activator of HSP90 ATPase activity 1 |
| chr19_+_46347063 | 0.53 |
ENST00000012443.9
ENST00000391919.1 |
PPP5C
|
protein phosphatase 5 catalytic subunit |
| chr8_+_17922974 | 0.53 |
ENST00000517730.5
ENST00000518537.5 ENST00000523055.5 ENST00000519253.5 |
PCM1
|
pericentriolar material 1 |
| chr17_+_2593925 | 0.53 |
ENST00000674717.1
ENST00000676353.1 ENST00000675202.1 ENST00000674608.1 ENST00000676098.1 |
PAFAH1B1
|
platelet activating factor acetylhydrolase 1b regulatory subunit 1 |
| chr20_-_49915509 | 0.53 |
ENST00000289431.10
|
SPATA2
|
spermatogenesis associated 2 |
| chr19_-_49929454 | 0.53 |
ENST00000597723.5
ENST00000599788.1 ENST00000352066.8 ENST00000596217.1 ENST00000593652.5 ENST00000599567.5 ENST00000600935.1 ENST00000596011.1 ENST00000596022.5 ENST00000597295.5 |
NUP62
IL4I1
|
nucleoporin 62 interleukin 4 induced 1 |
| chr17_-_82273424 | 0.53 |
ENST00000398519.9
ENST00000580446.1 |
CSNK1D
|
casein kinase 1 delta |
| chr2_-_55050442 | 0.53 |
ENST00000337526.11
|
RTN4
|
reticulon 4 |
| chr10_-_133358006 | 0.52 |
ENST00000278025.9
ENST00000368552.7 |
FUOM
|
fucose mutarotase |
| chr1_-_111563956 | 0.52 |
ENST00000369717.8
|
TMIGD3
|
transmembrane and immunoglobulin domain containing 3 |
| chr10_-_43409160 | 0.52 |
ENST00000337970.7
ENST00000682386.1 |
HNRNPF
|
heterogeneous nuclear ribonucleoprotein F |
| chr5_-_115625972 | 0.52 |
ENST00000333314.3
ENST00000456936.4 |
TMED7-TICAM2
TMED7
|
TMED7-TICAM2 readthrough transmembrane p24 trafficking protein 7 |
| chr18_+_12254354 | 0.52 |
ENST00000320477.10
|
CIDEA
|
cell death inducing DFFA like effector a |
| chr3_-_160399512 | 0.52 |
ENST00000498409.5
ENST00000475677.5 ENST00000478536.1 |
IFT80
|
intraflagellar transport 80 |
| chr7_-_151519891 | 0.51 |
ENST00000262187.10
|
RHEB
|
Ras homolog, mTORC1 binding |
| chr7_-_141973773 | 0.51 |
ENST00000547270.1
|
TAS2R38
|
taste 2 receptor member 38 |
| chr17_+_2594148 | 0.51 |
ENST00000675331.1
|
PAFAH1B1
|
platelet activating factor acetylhydrolase 1b regulatory subunit 1 |
| chr12_+_110281231 | 0.51 |
ENST00000539276.7
|
ATP2A2
|
ATPase sarcoplasmic/endoplasmic reticulum Ca2+ transporting 2 |
| chr19_-_58558561 | 0.51 |
ENST00000253023.8
|
UBE2M
|
ubiquitin conjugating enzyme E2 M |
| chr3_-_187139477 | 0.50 |
ENST00000455270.5
ENST00000296277.9 |
RPL39L
|
ribosomal protein L39 like |
| chr7_-_106112525 | 0.50 |
ENST00000011473.6
|
SYPL1
|
synaptophysin like 1 |
| chr19_-_49929396 | 0.50 |
ENST00000596680.5
ENST00000594673.5 ENST00000597029.5 |
NUP62
|
nucleoporin 62 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 9.5 | GO:0006452 | translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 0.9 | 5.7 | GO:0035617 | stress granule disassembly(GO:0035617) |
| 0.7 | 2.7 | GO:0033634 | positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
| 0.7 | 0.7 | GO:0014873 | response to muscle activity involved in regulation of muscle adaptation(GO:0014873) |
| 0.7 | 2.6 | GO:1903490 | regulation of cytokinetic process(GO:0032954) regulation of mitotic cytokinetic process(GO:1903436) positive regulation of mitotic cytokinetic process(GO:1903438) positive regulation of mitotic cytokinesis(GO:1903490) |
| 0.6 | 2.3 | GO:2001247 | positive regulation of phosphatidylcholine biosynthetic process(GO:2001247) |
| 0.5 | 2.5 | GO:1904693 | midbrain morphogenesis(GO:1904693) |
| 0.5 | 2.7 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.5 | 1.8 | GO:0035978 | mesodermal-endodermal cell signaling(GO:0003131) programmed DNA elimination(GO:0031049) chromosome breakage(GO:0031052) histone H2A-S139 phosphorylation(GO:0035978) positive regulation of cellular response to X-ray(GO:2000685) |
| 0.4 | 2.2 | GO:0071494 | cellular response to UV-C(GO:0071494) |
| 0.4 | 1.2 | GO:1903526 | negative regulation of membrane tubulation(GO:1903526) |
| 0.4 | 3.6 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 0.4 | 1.2 | GO:0008614 | pyridoxine metabolic process(GO:0008614) pyridoxine biosynthetic process(GO:0008615) vitamin B6 biosynthetic process(GO:0042819) |
| 0.4 | 1.6 | GO:0019249 | lactate biosynthetic process(GO:0019249) |
| 0.4 | 2.7 | GO:0090074 | negative regulation of protein homodimerization activity(GO:0090074) |
| 0.4 | 1.5 | GO:0036166 | phenotypic switching(GO:0036166) regulation of phenotypic switching(GO:1900239) |
| 0.4 | 1.1 | GO:0030719 | oocyte construction(GO:0007308) oocyte axis specification(GO:0007309) oocyte anterior/posterior axis specification(GO:0007314) pole plasm assembly(GO:0007315) maternal determination of anterior/posterior axis, embryo(GO:0008358) P granule organization(GO:0030719) |
| 0.3 | 1.0 | GO:0090176 | microtubule cytoskeleton organization involved in establishment of planar polarity(GO:0090176) |
| 0.3 | 3.0 | GO:2001206 | positive regulation of osteoclast development(GO:2001206) |
| 0.3 | 1.2 | GO:0035740 | CD8-positive, alpha-beta T cell proliferation(GO:0035740) regulation of CD8-positive, alpha-beta T cell proliferation(GO:2000564) |
| 0.3 | 5.1 | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.3 | 0.9 | GO:0035281 | pre-miRNA export from nucleus(GO:0035281) |
| 0.3 | 6.0 | GO:0010666 | positive regulation of striated muscle cell apoptotic process(GO:0010663) positive regulation of cardiac muscle cell apoptotic process(GO:0010666) |
| 0.3 | 0.8 | GO:0006478 | peptidyl-tyrosine sulfation(GO:0006478) |
| 0.3 | 0.8 | GO:1903233 | regulation of calcium ion-dependent exocytosis of neurotransmitter(GO:1903233) |
| 0.3 | 0.8 | GO:0051097 | negative regulation of helicase activity(GO:0051097) |
| 0.2 | 0.7 | GO:0090403 | oxidative stress-induced premature senescence(GO:0090403) |
| 0.2 | 2.2 | GO:0051177 | meiotic sister chromatid cohesion(GO:0051177) |
| 0.2 | 1.5 | GO:2000860 | positive regulation of mineralocorticoid secretion(GO:2000857) positive regulation of aldosterone secretion(GO:2000860) |
| 0.2 | 0.8 | GO:1900127 | positive regulation of hyaluronan biosynthetic process(GO:1900127) |
| 0.2 | 1.6 | GO:0060154 | cellular process regulating host cell cycle in response to virus(GO:0060154) |
| 0.2 | 0.7 | GO:0072302 | negative regulation of metanephric glomerulus development(GO:0072299) negative regulation of metanephric glomerular mesangial cell proliferation(GO:0072302) |
| 0.2 | 0.6 | GO:0048560 | establishment of anatomical structure orientation(GO:0048560) |
| 0.2 | 0.5 | GO:0030974 | thiamine pyrophosphate transport(GO:0030974) |
| 0.2 | 0.7 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.2 | 1.3 | GO:0001550 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) negative regulation of chondrocyte proliferation(GO:1902731) |
| 0.2 | 1.3 | GO:0060282 | positive regulation of oocyte development(GO:0060282) |
| 0.2 | 1.0 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.2 | 0.8 | GO:0071034 | CUT catabolic process(GO:0071034) CUT metabolic process(GO:0071043) |
| 0.2 | 0.5 | GO:0071962 | mitotic sister chromatid cohesion, centromeric(GO:0071962) |
| 0.2 | 1.0 | GO:0030259 | lipid glycosylation(GO:0030259) |
| 0.2 | 1.9 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.2 | 0.6 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.2 | 0.5 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.2 | 0.9 | GO:0071169 | establishment of protein localization to chromatin(GO:0071169) |
| 0.2 | 0.5 | GO:0001546 | preantral ovarian follicle growth(GO:0001546) multi-layer follicle stage(GO:0048162) |
| 0.1 | 1.5 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.1 | 0.6 | GO:0099558 | maintenance of synapse structure(GO:0099558) |
| 0.1 | 0.4 | GO:1901492 | positive regulation of lymphangiogenesis(GO:1901492) |
| 0.1 | 1.1 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.1 | 1.8 | GO:0035092 | sperm chromatin condensation(GO:0035092) |
| 0.1 | 1.2 | GO:2000074 | regulation of type B pancreatic cell development(GO:2000074) |
| 0.1 | 0.4 | GO:0061182 | negative regulation of chondrocyte development(GO:0061182) |
| 0.1 | 0.5 | GO:0072369 | regulation of lipid transport by positive regulation of transcription from RNA polymerase II promoter(GO:0072369) |
| 0.1 | 0.4 | GO:0072233 | thick ascending limb development(GO:0072023) metanephric thick ascending limb development(GO:0072233) |
| 0.1 | 0.4 | GO:0060623 | regulation of chromosome condensation(GO:0060623) |
| 0.1 | 1.0 | GO:0035469 | determination of pancreatic left/right asymmetry(GO:0035469) |
| 0.1 | 0.9 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.1 | 1.5 | GO:0008627 | intrinsic apoptotic signaling pathway in response to osmotic stress(GO:0008627) |
| 0.1 | 0.6 | GO:1902896 | terminal web assembly(GO:1902896) |
| 0.1 | 0.7 | GO:0047484 | regulation of response to osmotic stress(GO:0047484) |
| 0.1 | 0.6 | GO:1902412 | regulation of mitotic cytokinesis(GO:1902412) |
| 0.1 | 0.6 | GO:0010814 | substance P catabolic process(GO:0010814) calcitonin catabolic process(GO:0010816) endothelin maturation(GO:0034959) |
| 0.1 | 1.0 | GO:0002175 | protein localization to paranode region of axon(GO:0002175) |
| 0.1 | 0.9 | GO:0015760 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.1 | 0.6 | GO:0036493 | positive regulation of translation in response to endoplasmic reticulum stress(GO:0036493) |
| 0.1 | 0.3 | GO:0030997 | regulation of centriole-centriole cohesion(GO:0030997) |
| 0.1 | 0.4 | GO:0009236 | cobalamin biosynthetic process(GO:0009236) |
| 0.1 | 0.7 | GO:0022027 | interkinetic nuclear migration(GO:0022027) |
| 0.1 | 0.3 | GO:0046271 | phenylpropanoid catabolic process(GO:0046271) |
| 0.1 | 0.4 | GO:0007499 | ectoderm and mesoderm interaction(GO:0007499) |
| 0.1 | 0.9 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 0.1 | 0.3 | GO:2000195 | negative regulation of female gonad development(GO:2000195) |
| 0.1 | 0.3 | GO:1904204 | regulation of skeletal muscle hypertrophy(GO:1904204) |
| 0.1 | 0.8 | GO:0060715 | syncytiotrophoblast cell differentiation involved in labyrinthine layer development(GO:0060715) |
| 0.1 | 0.8 | GO:1902748 | positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.1 | 0.8 | GO:0042905 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.1 | 0.5 | GO:0099590 | neurotransmitter receptor internalization(GO:0099590) |
| 0.1 | 0.3 | GO:0032470 | regulation of fast-twitch skeletal muscle fiber contraction(GO:0031446) positive regulation of fast-twitch skeletal muscle fiber contraction(GO:0031448) positive regulation of endoplasmic reticulum calcium ion concentration(GO:0032470) |
| 0.1 | 0.6 | GO:2000035 | regulation of stem cell division(GO:2000035) |
| 0.1 | 0.4 | GO:0098937 | dendritic transport(GO:0098935) anterograde dendritic transport(GO:0098937) |
| 0.1 | 0.6 | GO:0070562 | regulation of vitamin D receptor signaling pathway(GO:0070562) |
| 0.1 | 1.9 | GO:0043508 | negative regulation of JUN kinase activity(GO:0043508) |
| 0.1 | 1.1 | GO:0038129 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) ERBB3 signaling pathway(GO:0038129) |
| 0.1 | 0.3 | GO:1900247 | cytoplasmic translational elongation(GO:0002182) regulation of cytoplasmic translational elongation(GO:1900247) negative regulation of cytoplasmic translational elongation(GO:1900248) |
| 0.1 | 0.3 | GO:0060152 | peroxisome localization(GO:0060151) microtubule-based peroxisome localization(GO:0060152) |
| 0.1 | 0.9 | GO:0050882 | voluntary musculoskeletal movement(GO:0050882) |
| 0.1 | 0.3 | GO:0046833 | positive regulation of RNA export from nucleus(GO:0046833) |
| 0.1 | 0.3 | GO:1904562 | phosphatidylinositol 5-phosphate metabolic process(GO:1904562) |
| 0.1 | 0.5 | GO:0010825 | positive regulation of centrosome duplication(GO:0010825) |
| 0.1 | 0.8 | GO:1902525 | regulation of protein monoubiquitination(GO:1902525) |
| 0.1 | 3.0 | GO:2000810 | regulation of bicellular tight junction assembly(GO:2000810) |
| 0.1 | 0.4 | GO:0006449 | regulation of translational termination(GO:0006449) |
| 0.1 | 0.7 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.1 | 0.4 | GO:0045204 | MAPK export from nucleus(GO:0045204) |
| 0.1 | 0.2 | GO:0006106 | fumarate metabolic process(GO:0006106) aspartate biosynthetic process(GO:0006532) aspartate catabolic process(GO:0006533) |
| 0.1 | 0.7 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.1 | 0.2 | GO:0042668 | auditory receptor cell fate determination(GO:0042668) |
| 0.1 | 0.5 | GO:2000324 | positive regulation of glucocorticoid receptor signaling pathway(GO:2000324) |
| 0.1 | 0.3 | GO:0042538 | hyperosmotic salinity response(GO:0042538) |
| 0.1 | 0.4 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.1 | 1.1 | GO:0006089 | lactate metabolic process(GO:0006089) |
| 0.1 | 0.4 | GO:1904565 | response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904565) cellular response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904566) |
| 0.1 | 0.4 | GO:0010903 | negative regulation of very-low-density lipoprotein particle remodeling(GO:0010903) |
| 0.1 | 0.6 | GO:0006682 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.1 | 0.4 | GO:0071393 | cellular response to progesterone stimulus(GO:0071393) |
| 0.1 | 0.3 | GO:1990108 | protein linear deubiquitination(GO:1990108) |
| 0.1 | 0.2 | GO:0097680 | double-strand break repair via classical nonhomologous end joining(GO:0097680) |
| 0.1 | 0.8 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.1 | 0.5 | GO:0060011 | Sertoli cell proliferation(GO:0060011) |
| 0.1 | 0.4 | GO:1903435 | positive regulation of constitutive secretory pathway(GO:1903435) |
| 0.1 | 0.4 | GO:0051106 | positive regulation of DNA ligation(GO:0051106) |
| 0.1 | 0.3 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.1 | 0.4 | GO:0070086 | ubiquitin-dependent endocytosis(GO:0070086) |
| 0.1 | 0.3 | GO:0035986 | senescence-associated heterochromatin focus assembly(GO:0035986) |
| 0.1 | 0.3 | GO:0007341 | penetration of zona pellucida(GO:0007341) |
| 0.1 | 1.0 | GO:1900029 | positive regulation of ruffle assembly(GO:1900029) |
| 0.1 | 0.8 | GO:2000623 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.1 | 0.4 | GO:2000819 | regulation of nucleotide-excision repair(GO:2000819) |
| 0.1 | 0.2 | GO:0070836 | caveola assembly(GO:0070836) |
| 0.1 | 0.8 | GO:0042373 | vitamin K metabolic process(GO:0042373) |
| 0.1 | 0.2 | GO:0006147 | guanine catabolic process(GO:0006147) |
| 0.1 | 0.1 | GO:1900113 | negative regulation of histone H3-K9 trimethylation(GO:1900113) |
| 0.1 | 0.2 | GO:1902630 | regulation of membrane hyperpolarization(GO:1902630) |
| 0.1 | 0.5 | GO:0010890 | positive regulation of sequestering of triglyceride(GO:0010890) |
| 0.1 | 0.3 | GO:0043456 | regulation of pentose-phosphate shunt(GO:0043456) |
| 0.1 | 0.2 | GO:0006172 | ADP biosynthetic process(GO:0006172) |
| 0.1 | 0.1 | GO:1990918 | double-strand break repair involved in meiotic recombination(GO:1990918) |
| 0.1 | 0.3 | GO:0080154 | regulation of fertilization(GO:0080154) |
| 0.1 | 1.1 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.1 | 1.0 | GO:0060180 | female mating behavior(GO:0060180) |
| 0.1 | 0.6 | GO:0035095 | behavioral response to nicotine(GO:0035095) |
| 0.1 | 0.2 | GO:0002023 | reduction of food intake in response to dietary excess(GO:0002023) |
| 0.0 | 0.6 | GO:1904153 | negative regulation of protein exit from endoplasmic reticulum(GO:0070862) negative regulation of retrograde protein transport, ER to cytosol(GO:1904153) |
| 0.0 | 0.3 | GO:0044314 | protein K27-linked ubiquitination(GO:0044314) |
| 0.0 | 5.6 | GO:0032508 | DNA duplex unwinding(GO:0032508) |
| 0.0 | 0.5 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.0 | 0.8 | GO:0001675 | acrosome assembly(GO:0001675) |
| 0.0 | 0.2 | GO:0036100 | leukotriene catabolic process(GO:0036100) leukotriene B4 catabolic process(GO:0036101) leukotriene B4 metabolic process(GO:0036102) icosanoid catabolic process(GO:1901523) fatty acid derivative catabolic process(GO:1901569) |
| 0.0 | 0.7 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.0 | 0.3 | GO:0060770 | negative regulation of epithelial cell proliferation involved in prostate gland development(GO:0060770) |
| 0.0 | 0.8 | GO:1901978 | positive regulation of cell cycle checkpoint(GO:1901978) |
| 0.0 | 0.1 | GO:0007343 | egg activation(GO:0007343) |
| 0.0 | 0.5 | GO:0060052 | neurofilament cytoskeleton organization(GO:0060052) |
| 0.0 | 0.2 | GO:1905224 | clathrin-coated pit assembly(GO:1905224) |
| 0.0 | 0.2 | GO:0046642 | negative regulation of alpha-beta T cell proliferation(GO:0046642) |
| 0.0 | 0.0 | GO:0046010 | positive regulation of circadian sleep/wake cycle, non-REM sleep(GO:0046010) |
| 0.0 | 0.2 | GO:0060480 | lung goblet cell differentiation(GO:0060480) |
| 0.0 | 0.3 | GO:0072180 | mesonephric duct morphogenesis(GO:0072180) |
| 0.0 | 1.2 | GO:1900027 | regulation of ruffle assembly(GO:1900027) |
| 0.0 | 0.5 | GO:0071786 | endoplasmic reticulum tubular network organization(GO:0071786) |
| 0.0 | 0.5 | GO:1902018 | negative regulation of cilium assembly(GO:1902018) |
| 0.0 | 0.2 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.0 | 0.9 | GO:0010867 | positive regulation of triglyceride biosynthetic process(GO:0010867) |
| 0.0 | 0.1 | GO:0003245 | cardiac muscle tissue growth involved in heart morphogenesis(GO:0003245) |
| 0.0 | 0.1 | GO:0048022 | negative regulation of melanin biosynthetic process(GO:0048022) negative regulation of secondary metabolite biosynthetic process(GO:1900377) |
| 0.0 | 0.2 | GO:0001831 | trophectodermal cellular morphogenesis(GO:0001831) |
| 0.0 | 0.2 | GO:1900535 | medium-chain fatty-acyl-CoA catabolic process(GO:0036114) long-chain fatty-acyl-CoA catabolic process(GO:0036116) palmitic acid metabolic process(GO:1900533) palmitic acid biosynthetic process(GO:1900535) |
| 0.0 | 0.7 | GO:1902260 | negative regulation of delayed rectifier potassium channel activity(GO:1902260) negative regulation of voltage-gated potassium channel activity(GO:1903817) |
| 0.0 | 0.3 | GO:0060701 | negative regulation of ribonuclease activity(GO:0060701) negative regulation of endoribonuclease activity(GO:0060702) |
| 0.0 | 0.4 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 0.0 | 0.7 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.0 | 0.1 | GO:2000118 | regulation of sodium-dependent phosphate transport(GO:2000118) |
| 0.0 | 0.2 | GO:0003290 | atrial septum secundum morphogenesis(GO:0003290) |
| 0.0 | 0.2 | GO:0043335 | protein unfolding(GO:0043335) |
| 0.0 | 0.3 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.0 | 0.3 | GO:0007000 | nucleolus organization(GO:0007000) |
| 0.0 | 0.2 | GO:1900126 | negative regulation of hyaluronan biosynthetic process(GO:1900126) |
| 0.0 | 0.1 | GO:0036292 | DNA rewinding(GO:0036292) |
| 0.0 | 0.1 | GO:0009386 | translational attenuation(GO:0009386) |
| 0.0 | 0.3 | GO:0051988 | regulation of attachment of spindle microtubules to kinetochore(GO:0051988) |
| 0.0 | 0.7 | GO:0042249 | establishment of planar polarity of embryonic epithelium(GO:0042249) |
| 0.0 | 0.1 | GO:0097476 | spinal cord motor neuron migration(GO:0097476) |
| 0.0 | 0.1 | GO:0035744 | T-helper 1 cell cytokine production(GO:0035744) |
| 0.0 | 0.1 | GO:1903238 | positive regulation of leukocyte tethering or rolling(GO:1903238) |
| 0.0 | 0.5 | GO:0010909 | regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) positive regulation of proteoglycan biosynthetic process(GO:1902730) |
| 0.0 | 0.4 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) tRNA transcription(GO:0009304) |
| 0.0 | 0.3 | GO:0010835 | regulation of protein ADP-ribosylation(GO:0010835) |
| 0.0 | 0.4 | GO:0003352 | regulation of cilium movement(GO:0003352) |
| 0.0 | 0.2 | GO:0060370 | susceptibility to T cell mediated cytotoxicity(GO:0060370) |
| 0.0 | 0.3 | GO:0017185 | peptidyl-lysine hydroxylation(GO:0017185) |
| 0.0 | 0.1 | GO:0045759 | negative regulation of action potential(GO:0045759) |
| 0.0 | 0.1 | GO:1903719 | regulation of I-kappaB phosphorylation(GO:1903719) positive regulation of I-kappaB phosphorylation(GO:1903721) |
| 0.0 | 0.9 | GO:0030033 | microvillus assembly(GO:0030033) |
| 0.0 | 0.1 | GO:0006288 | base-excision repair, DNA ligation(GO:0006288) |
| 0.0 | 0.2 | GO:0060613 | fat pad development(GO:0060613) |
| 0.0 | 0.4 | GO:0060075 | regulation of resting membrane potential(GO:0060075) |
| 0.0 | 0.5 | GO:2000052 | positive regulation of non-canonical Wnt signaling pathway(GO:2000052) |
| 0.0 | 0.6 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.0 | 0.3 | GO:0046598 | positive regulation of viral entry into host cell(GO:0046598) |
| 0.0 | 0.7 | GO:0070935 | 3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 0.0 | 0.2 | GO:0035610 | protein side chain deglutamylation(GO:0035610) |
| 0.0 | 1.2 | GO:0035428 | hexose transmembrane transport(GO:0035428) glucose transmembrane transport(GO:1904659) |
| 0.0 | 0.4 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.0 | 0.2 | GO:0032000 | positive regulation of fatty acid beta-oxidation(GO:0032000) |
| 0.0 | 0.5 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.0 | 0.1 | GO:0030950 | establishment or maintenance of actin cytoskeleton polarity(GO:0030950) |
| 0.0 | 0.4 | GO:0080182 | histone H3-K4 trimethylation(GO:0080182) |
| 0.0 | 0.2 | GO:0060161 | peptidyl-arginine methylation, to symmetrical-dimethyl arginine(GO:0019918) positive regulation of dopamine receptor signaling pathway(GO:0060161) |
| 0.0 | 0.4 | GO:0070544 | histone H3-K36 demethylation(GO:0070544) |
| 0.0 | 0.3 | GO:1903944 | regulation of hepatocyte apoptotic process(GO:1903943) negative regulation of hepatocyte apoptotic process(GO:1903944) |
| 0.0 | 0.5 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.0 | 0.1 | GO:1903251 | multi-ciliated epithelial cell differentiation(GO:1903251) |
| 0.0 | 2.4 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.0 | 2.8 | GO:0051436 | negative regulation of ubiquitin-protein ligase activity involved in mitotic cell cycle(GO:0051436) |
| 0.0 | 1.1 | GO:0007080 | mitotic metaphase plate congression(GO:0007080) |
| 0.0 | 0.5 | GO:0035635 | entry of bacterium into host cell(GO:0035635) |
| 0.0 | 0.1 | GO:1904526 | regulation of microtubule binding(GO:1904526) |
| 0.0 | 0.6 | GO:0071294 | cellular response to zinc ion(GO:0071294) |
| 0.0 | 0.1 | GO:0034154 | toll-like receptor 7 signaling pathway(GO:0034154) |
| 0.0 | 0.1 | GO:0010513 | positive regulation of phosphatidylinositol biosynthetic process(GO:0010513) |
| 0.0 | 0.2 | GO:0035414 | negative regulation of catenin import into nucleus(GO:0035414) |
| 0.0 | 2.0 | GO:0032465 | regulation of cytokinesis(GO:0032465) |
| 0.0 | 0.8 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.0 | 0.3 | GO:0090168 | Golgi reassembly(GO:0090168) |
| 0.0 | 0.2 | GO:1990481 | mRNA pseudouridine synthesis(GO:1990481) |
| 0.0 | 1.0 | GO:0001881 | receptor recycling(GO:0001881) |
| 0.0 | 1.0 | GO:0032570 | response to progesterone(GO:0032570) |
| 0.0 | 0.2 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.0 | 0.4 | GO:0098877 | neurotransmitter receptor transport to plasma membrane(GO:0098877) neurotransmitter receptor transport to postsynaptic membrane(GO:0098969) protein localization to postsynaptic membrane(GO:1903539) establishment of protein localization to postsynaptic membrane(GO:1903540) |
| 0.0 | 0.3 | GO:0061158 | 3'-UTR-mediated mRNA destabilization(GO:0061158) |
| 0.0 | 0.3 | GO:1990126 | retrograde transport, endosome to plasma membrane(GO:1990126) |
| 0.0 | 0.4 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.0 | 0.2 | GO:0002155 | thyroid hormone mediated signaling pathway(GO:0002154) regulation of thyroid hormone mediated signaling pathway(GO:0002155) |
| 0.0 | 0.5 | GO:0071493 | cellular response to UV-B(GO:0071493) |
| 0.0 | 0.7 | GO:0022011 | myelination in peripheral nervous system(GO:0022011) peripheral nervous system axon ensheathment(GO:0032292) |
| 0.0 | 0.4 | GO:0060285 | cilium-dependent cell motility(GO:0060285) |
| 0.0 | 0.4 | GO:0007130 | synaptonemal complex assembly(GO:0007130) |
| 0.0 | 0.4 | GO:0035020 | regulation of Rac protein signal transduction(GO:0035020) |
| 0.0 | 0.1 | GO:0014886 | transition between slow and fast fiber(GO:0014886) |
| 0.0 | 0.2 | GO:0048630 | skeletal muscle tissue growth(GO:0048630) |
| 0.0 | 0.2 | GO:1904776 | regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.0 | 0.3 | GO:0002291 | T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell(GO:0002291) |
| 0.0 | 0.3 | GO:0033182 | regulation of histone ubiquitination(GO:0033182) |
| 0.0 | 0.1 | GO:0072429 | response to intra-S DNA damage checkpoint signaling(GO:0072429) |
| 0.0 | 0.3 | GO:1904869 | regulation of protein localization to Cajal body(GO:1904869) positive regulation of protein localization to Cajal body(GO:1904871) |
| 0.0 | 0.1 | GO:0002357 | defense response to tumor cell(GO:0002357) |
| 0.0 | 0.3 | GO:0001514 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.0 | 0.2 | GO:0006335 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 | 0.2 | GO:0000098 | sulfur amino acid catabolic process(GO:0000098) |
| 0.0 | 0.1 | GO:2000255 | negative regulation of male germ cell proliferation(GO:2000255) |
| 0.0 | 0.0 | GO:2000426 | negative regulation of apoptotic cell clearance(GO:2000426) |
| 0.0 | 0.4 | GO:2001224 | positive regulation of neuron migration(GO:2001224) |
| 0.0 | 0.2 | GO:0090161 | Golgi ribbon formation(GO:0090161) |
| 0.0 | 0.5 | GO:0090129 | positive regulation of synapse maturation(GO:0090129) |
| 0.0 | 0.1 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.0 | 0.4 | GO:0035641 | locomotory exploration behavior(GO:0035641) |
| 0.0 | 0.1 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.0 | 0.1 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.0 | 0.3 | GO:0045109 | intermediate filament organization(GO:0045109) |
| 0.0 | 0.1 | GO:0070863 | regulation of protein exit from endoplasmic reticulum(GO:0070861) positive regulation of protein exit from endoplasmic reticulum(GO:0070863) |
| 0.0 | 0.1 | GO:1904263 | positive regulation of TORC1 signaling(GO:1904263) |
| 0.0 | 0.3 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.0 | 0.5 | GO:0045737 | positive regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045737) |
| 0.0 | 0.5 | GO:0030970 | retrograde protein transport, ER to cytosol(GO:0030970) |
| 0.0 | 0.5 | GO:0021542 | dentate gyrus development(GO:0021542) |
| 0.0 | 0.1 | GO:0032971 | regulation of muscle filament sliding(GO:0032971) |
| 0.0 | 0.1 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.0 | 0.6 | GO:0030252 | growth hormone secretion(GO:0030252) |
| 0.0 | 0.4 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.0 | 0.4 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 0.0 | 1.3 | GO:0045071 | negative regulation of viral genome replication(GO:0045071) |
| 0.0 | 0.2 | GO:0042754 | negative regulation of circadian rhythm(GO:0042754) |
| 0.0 | 0.9 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.0 | 0.4 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 0.2 | GO:0042921 | glucocorticoid receptor signaling pathway(GO:0042921) |
| 0.0 | 0.1 | GO:1903546 | receptor localization to nonmotile primary cilium(GO:0097500) protein localization to photoreceptor outer segment(GO:1903546) |
| 0.0 | 0.1 | GO:0036010 | protein localization to endosome(GO:0036010) |
| 0.0 | 0.2 | GO:0034433 | steroid esterification(GO:0034433) sterol esterification(GO:0034434) cholesterol esterification(GO:0034435) |
| 0.0 | 0.4 | GO:0000301 | retrograde transport, vesicle recycling within Golgi(GO:0000301) |
| 0.0 | 0.7 | GO:0007190 | activation of adenylate cyclase activity(GO:0007190) |
| 0.0 | 0.2 | GO:0098779 | mitophagy in response to mitochondrial depolarization(GO:0098779) |
| 0.0 | 0.9 | GO:0007528 | neuromuscular junction development(GO:0007528) |
| 0.0 | 0.5 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.0 | 0.1 | GO:0033211 | adiponectin-activated signaling pathway(GO:0033211) |
| 0.0 | 0.1 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.0 | 0.1 | GO:0060391 | positive regulation of SMAD protein import into nucleus(GO:0060391) |
| 0.0 | 0.2 | GO:2001241 | positive regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001241) |
| 0.0 | 0.3 | GO:0060236 | regulation of mitotic spindle organization(GO:0060236) |
| 0.0 | 0.1 | GO:0008635 | activation of cysteine-type endopeptidase activity involved in apoptotic process by cytochrome c(GO:0008635) |
| 0.0 | 0.5 | GO:0045824 | negative regulation of innate immune response(GO:0045824) |
| 0.0 | 0.8 | GO:0045540 | regulation of cholesterol biosynthetic process(GO:0045540) |
| 0.0 | 0.1 | GO:0031086 | nuclear-transcribed mRNA catabolic process, deadenylation-independent decay(GO:0031086) deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.0 | 0.1 | GO:0042670 | retinal cone cell differentiation(GO:0042670) retinal cone cell development(GO:0046549) |
| 0.0 | 0.3 | GO:0032967 | positive regulation of collagen biosynthetic process(GO:0032967) |
| 0.0 | 0.2 | GO:0042135 | neurotransmitter catabolic process(GO:0042135) |
| 0.0 | 0.1 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.0 | 0.0 | GO:0071874 | cellular response to norepinephrine stimulus(GO:0071874) |
| 0.0 | 0.3 | GO:1900115 | extracellular regulation of signal transduction(GO:1900115) extracellular negative regulation of signal transduction(GO:1900116) |
| 0.0 | 0.2 | GO:0042753 | positive regulation of circadian rhythm(GO:0042753) |
| 0.0 | 0.3 | GO:0071480 | cellular response to gamma radiation(GO:0071480) |
| 0.0 | 0.1 | GO:0070345 | negative regulation of fat cell proliferation(GO:0070345) |
| 0.0 | 0.4 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.0 | 0.2 | GO:2000696 | regulation of epithelial cell differentiation involved in kidney development(GO:2000696) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 12.1 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.3 | 2.4 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.3 | 0.9 | GO:0042565 | RNA nuclear export complex(GO:0042565) |
| 0.3 | 0.8 | GO:0044609 | DBIRD complex(GO:0044609) |
| 0.2 | 0.7 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.2 | 2.2 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.2 | 1.0 | GO:0000942 | condensed nuclear chromosome outer kinetochore(GO:0000942) |
| 0.2 | 0.9 | GO:0071595 | Nem1-Spo7 phosphatase complex(GO:0071595) |
| 0.2 | 0.9 | GO:0036398 | TCR signalosome(GO:0036398) |
| 0.2 | 0.7 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.2 | 0.8 | GO:0035061 | interchromatin granule(GO:0035061) |
| 0.2 | 2.4 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.2 | 0.5 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.2 | 1.3 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.2 | 13.7 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.2 | 0.6 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.1 | 1.0 | GO:0032280 | symmetric synapse(GO:0032280) |
| 0.1 | 0.4 | GO:0001674 | female germ cell nucleus(GO:0001674) |
| 0.1 | 1.6 | GO:0000235 | astral microtubule(GO:0000235) aster(GO:0005818) |
| 0.1 | 1.6 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.1 | 1.0 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.1 | 0.3 | GO:1990590 | ATF1-ATF4 transcription factor complex(GO:1990590) |
| 0.1 | 0.6 | GO:0031302 | intrinsic component of endosome membrane(GO:0031302) |
| 0.1 | 0.3 | GO:0043159 | acrosomal matrix(GO:0043159) |
| 0.1 | 0.4 | GO:0071817 | MMXD complex(GO:0071817) |
| 0.1 | 0.3 | GO:0035370 | UBC13-UEV1A complex(GO:0035370) |
| 0.1 | 0.4 | GO:0000126 | transcription factor TFIIIB complex(GO:0000126) |
| 0.1 | 0.4 | GO:0071001 | U4/U6 snRNP(GO:0071001) |
| 0.1 | 0.4 | GO:0034365 | discoidal high-density lipoprotein particle(GO:0034365) |
| 0.1 | 1.3 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.1 | 0.7 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 0.1 | 0.3 | GO:0005745 | m-AAA complex(GO:0005745) |
| 0.1 | 1.9 | GO:0036038 | MKS complex(GO:0036038) |
| 0.1 | 0.2 | GO:0000799 | nuclear condensin complex(GO:0000799) |
| 0.1 | 0.5 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.1 | 0.7 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.1 | 0.3 | GO:0070435 | Shc-EGFR complex(GO:0070435) |
| 0.1 | 0.6 | GO:0000938 | GARP complex(GO:0000938) |
| 0.1 | 0.8 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.1 | 0.6 | GO:0030430 | host cell cytoplasm(GO:0030430) host cell cytoplasm part(GO:0033655) |
| 0.1 | 0.8 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.1 | 0.3 | GO:0071008 | U2-type post-mRNA release spliceosomal complex(GO:0071008) |
| 0.1 | 0.5 | GO:0031415 | NatA complex(GO:0031415) |
| 0.1 | 0.5 | GO:0043203 | axon hillock(GO:0043203) |
| 0.1 | 1.0 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.1 | 0.3 | GO:0032311 | angiogenin-PRI complex(GO:0032311) |
| 0.1 | 0.1 | GO:0070695 | FHF complex(GO:0070695) |
| 0.1 | 0.7 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.1 | 0.2 | GO:0031933 | telomeric heterochromatin(GO:0031933) |
| 0.1 | 1.4 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 0.1 | 0.8 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.1 | 1.9 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.1 | 1.9 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.1 | 0.4 | GO:0098536 | deuterosome(GO:0098536) |
| 0.1 | 0.7 | GO:0008278 | cohesin complex(GO:0008278) |
| 0.1 | 0.5 | GO:0071204 | histone pre-mRNA 3'end processing complex(GO:0071204) |
| 0.0 | 0.3 | GO:0000801 | central element(GO:0000801) |
| 0.0 | 7.3 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.0 | 2.1 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 1.1 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 0.2 | GO:0030906 | retromer, cargo-selective complex(GO:0030906) |
| 0.0 | 0.4 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.0 | 0.4 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.0 | 0.1 | GO:0033011 | perinuclear theca(GO:0033011) |
| 0.0 | 0.2 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.0 | 1.5 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.6 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.0 | 0.2 | GO:0032807 | DNA ligase IV complex(GO:0032807) |
| 0.0 | 0.3 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.0 | 1.6 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 0.4 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.0 | 0.9 | GO:0031264 | death-inducing signaling complex(GO:0031264) |
| 0.0 | 0.6 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.0 | 0.3 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.0 | 0.3 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.0 | 0.1 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.0 | 0.5 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 0.4 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.0 | 0.3 | GO:0042567 | insulin-like growth factor ternary complex(GO:0042567) |
| 0.0 | 0.4 | GO:0044613 | nuclear pore central transport channel(GO:0044613) |
| 0.0 | 0.6 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.0 | 0.8 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 0.2 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.0 | 1.0 | GO:0097228 | sperm principal piece(GO:0097228) |
| 0.0 | 0.3 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.0 | 0.3 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.0 | 1.1 | GO:0000795 | synaptonemal complex(GO:0000795) |
| 0.0 | 0.3 | GO:0002199 | zona pellucida receptor complex(GO:0002199) |
| 0.0 | 0.3 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.0 | 0.1 | GO:0030132 | clathrin coat of coated pit(GO:0030132) |
| 0.0 | 0.5 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 2.5 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 1.8 | GO:0043657 | host(GO:0018995) host cell(GO:0043657) |
| 0.0 | 0.5 | GO:0016461 | unconventional myosin complex(GO:0016461) |
| 0.0 | 0.4 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.2 | GO:0045179 | apical cortex(GO:0045179) |
| 0.0 | 0.2 | GO:0070436 | Grb2-EGFR complex(GO:0070436) |
| 0.0 | 0.5 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.0 | 1.6 | GO:0097014 | axoneme(GO:0005930) ciliary plasm(GO:0097014) |
| 0.0 | 1.7 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.0 | 0.6 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 0.2 | GO:0098554 | cytoplasmic side of endoplasmic reticulum membrane(GO:0098554) |
| 0.0 | 0.1 | GO:1990131 | Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.0 | 0.2 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.0 | 0.3 | GO:0034709 | methylosome(GO:0034709) |
| 0.0 | 0.3 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.0 | 0.1 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.0 | 0.1 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 0.0 | 2.1 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 3.1 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.0 | 0.2 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.0 | 0.1 | GO:1990075 | periciliary membrane compartment(GO:1990075) |
| 0.0 | 0.6 | GO:0044298 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.0 | 0.3 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 0.0 | GO:0055087 | Ski complex(GO:0055087) |
| 0.0 | 0.1 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.0 | 0.5 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 0.8 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 3.1 | GO:0005681 | spliceosomal complex(GO:0005681) |
| 0.0 | 0.1 | GO:0070761 | pre-snoRNP complex(GO:0070761) |
| 0.0 | 0.5 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 0.1 | GO:0048476 | Holliday junction resolvase complex(GO:0048476) |
| 0.0 | 0.2 | GO:0030897 | HOPS complex(GO:0030897) |
| 0.0 | 1.1 | GO:0005798 | Golgi-associated vesicle(GO:0005798) |
| 0.0 | 1.2 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.0 | 0.4 | GO:0035267 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.0 | 1.4 | GO:0031514 | motile cilium(GO:0031514) |
| 0.0 | 0.2 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 0.1 | GO:0019907 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) |
| 0.0 | 1.5 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 0.2 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.0 | 0.2 | GO:0005849 | mRNA cleavage factor complex(GO:0005849) |
| 0.0 | 0.1 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 0.0 | 0.1 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.0 | 0.1 | GO:0070776 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 5.1 | GO:0004360 | glutamine-fructose-6-phosphate transaminase (isomerizing) activity(GO:0004360) |
| 0.7 | 2.1 | GO:0035248 | alpha-1,4-N-acetylgalactosaminyltransferase activity(GO:0035248) |
| 0.7 | 2.1 | GO:0001716 | L-amino-acid oxidase activity(GO:0001716) |
| 0.5 | 2.7 | GO:0030298 | receptor signaling protein tyrosine kinase activator activity(GO:0030298) CCR1 chemokine receptor binding(GO:0031726) |
| 0.5 | 9.8 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.5 | 1.8 | GO:0035501 | MH1 domain binding(GO:0035501) |
| 0.4 | 2.7 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.3 | 6.0 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.3 | 0.9 | GO:0090631 | pre-miRNA transporter activity(GO:0090631) |
| 0.3 | 0.8 | GO:0008476 | protein-tyrosine sulfotransferase activity(GO:0008476) |
| 0.2 | 1.2 | GO:0031749 | D2 dopamine receptor binding(GO:0031749) |
| 0.2 | 1.3 | GO:0042806 | fucose binding(GO:0042806) |
| 0.2 | 1.5 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 0.2 | 2.7 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.2 | 0.5 | GO:0090422 | thiamine pyrophosphate transporter activity(GO:0090422) |
| 0.2 | 0.9 | GO:0015315 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 0.2 | 0.5 | GO:0071207 | histone pre-mRNA stem-loop binding(GO:0071207) |
| 0.2 | 0.5 | GO:0052858 | peptidyl-lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0052858) |
| 0.1 | 0.7 | GO:0004663 | Rab geranylgeranyltransferase activity(GO:0004663) |
| 0.1 | 3.6 | GO:0017081 | chloride channel regulator activity(GO:0017081) |
| 0.1 | 1.6 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.1 | 0.7 | GO:0044729 | hemi-methylated DNA-binding(GO:0044729) |
| 0.1 | 1.1 | GO:0019788 | NEDD8 transferase activity(GO:0019788) |
| 0.1 | 0.9 | GO:0050733 | RS domain binding(GO:0050733) |
| 0.1 | 0.8 | GO:0016900 | oxidoreductase activity, acting on the CH-OH group of donors, disulfide as acceptor(GO:0016900) vitamin-K-epoxide reductase (warfarin-sensitive) activity(GO:0047057) |
| 0.1 | 0.5 | GO:0031544 | peptidyl-proline 3-dioxygenase activity(GO:0031544) |
| 0.1 | 0.9 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.1 | 0.5 | GO:0003875 | ADP-ribosylarginine hydrolase activity(GO:0003875) |
| 0.1 | 5.9 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.1 | 1.3 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.1 | 0.4 | GO:0001026 | TFIIIB-type transcription factor activity(GO:0001026) |
| 0.1 | 2.3 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.1 | 0.5 | GO:0033906 | hyaluronoglucuronidase activity(GO:0033906) |
| 0.1 | 0.4 | GO:0051765 | inositol tetrakisphosphate kinase activity(GO:0051765) |
| 0.1 | 0.4 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.1 | 0.3 | GO:0060422 | peptidyl-dipeptidase inhibitor activity(GO:0060422) |
| 0.1 | 0.5 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.1 | 0.2 | GO:0034736 | sterol O-acyltransferase activity(GO:0004772) cholesterol O-acyltransferase activity(GO:0034736) |
| 0.1 | 0.4 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.1 | 0.3 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 0.1 | 0.5 | GO:0098821 | BMP receptor activity(GO:0098821) |
| 0.1 | 0.7 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.1 | 2.3 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.1 | 0.2 | GO:0080130 | L-aspartate:2-oxoglutarate aminotransferase activity(GO:0004069) L-phenylalanine:2-oxoglutarate aminotransferase activity(GO:0080130) |
| 0.1 | 0.2 | GO:0097259 | tocopherol omega-hydroxylase activity(GO:0052870) alpha-tocopherol omega-hydroxylase activity(GO:0052871) 20-hydroxy-leukotriene B4 omega oxidase activity(GO:0097258) 20-aldehyde-leukotriene B4 20-monooxygenase activity(GO:0097259) |
| 0.1 | 0.4 | GO:0070653 | high-density lipoprotein particle receptor binding(GO:0070653) |
| 0.1 | 1.9 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.1 | 0.8 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.1 | 1.0 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.1 | 2.4 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.1 | 0.3 | GO:0061676 | importin-alpha family protein binding(GO:0061676) |
| 0.1 | 0.5 | GO:0035473 | lipase binding(GO:0035473) |
| 0.1 | 5.4 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.1 | 0.2 | GO:0060001 | minus-end directed microfilament motor activity(GO:0060001) |
| 0.1 | 1.1 | GO:0030297 | ErbB-2 class receptor binding(GO:0005176) transmembrane receptor protein tyrosine kinase activator activity(GO:0030297) |
| 0.1 | 0.9 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.1 | 0.7 | GO:0048185 | activin binding(GO:0048185) |
| 0.1 | 0.3 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.1 | 0.2 | GO:0008892 | guanine deaminase activity(GO:0008892) |
| 0.1 | 0.7 | GO:0005168 | neurotrophin TRKA receptor binding(GO:0005168) |
| 0.1 | 0.3 | GO:0004619 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 0.1 | 0.6 | GO:0008428 | ribonuclease inhibitor activity(GO:0008428) |
| 0.1 | 1.5 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.1 | 0.5 | GO:0051425 | PTB domain binding(GO:0051425) |
| 0.1 | 0.5 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.1 | 0.4 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.1 | 1.4 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.1 | 0.3 | GO:0004301 | epoxide hydrolase activity(GO:0004301) |
| 0.0 | 1.0 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.0 | 0.2 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.0 | 0.4 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) |
| 0.0 | 1.7 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.0 | 0.9 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.0 | 1.7 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.0 | 0.1 | GO:0004040 | amidase activity(GO:0004040) |
| 0.0 | 0.4 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.0 | 0.2 | GO:0000026 | alpha-1,2-mannosyltransferase activity(GO:0000026) |
| 0.0 | 0.7 | GO:0019864 | IgG binding(GO:0019864) |
| 0.0 | 0.9 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.0 | 0.5 | GO:0055131 | C3HC4-type RING finger domain binding(GO:0055131) |
| 0.0 | 0.4 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) |
| 0.0 | 0.0 | GO:0016420 | [acyl-carrier-protein] S-malonyltransferase activity(GO:0004314) S-malonyltransferase activity(GO:0016419) malonyltransferase activity(GO:0016420) |
| 0.0 | 0.3 | GO:0030284 | estrogen receptor activity(GO:0030284) |
| 0.0 | 0.1 | GO:0050146 | nucleoside phosphotransferase activity(GO:0050146) |
| 0.0 | 0.2 | GO:0031208 | POZ domain binding(GO:0031208) |
| 0.0 | 0.9 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 2.0 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.0 | 0.3 | GO:0003910 | DNA ligase (ATP) activity(GO:0003910) |
| 0.0 | 1.3 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.0 | 0.3 | GO:0000285 | 1-phosphatidylinositol-3-phosphate 5-kinase activity(GO:0000285) |
| 0.0 | 1.7 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 0.6 | GO:0008499 | UDP-galactose:beta-N-acetylglucosamine beta-1,3-galactosyltransferase activity(GO:0008499) |
| 0.0 | 0.1 | GO:0004949 | cannabinoid receptor activity(GO:0004949) |
| 0.0 | 0.1 | GO:0005174 | CD40 receptor binding(GO:0005174) |
| 0.0 | 0.4 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 0.3 | GO:0034511 | U3 snoRNA binding(GO:0034511) |
| 0.0 | 0.8 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.0 | 0.8 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.0 | 0.1 | GO:0050254 | rhodopsin kinase activity(GO:0050254) |
| 0.0 | 0.8 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 0.4 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.0 | 0.2 | GO:0035243 | protein-arginine omega-N symmetric methyltransferase activity(GO:0035243) |
| 0.0 | 0.5 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.0 | 0.4 | GO:0016723 | oxidoreductase activity, oxidizing metal ions, NAD or NADP as acceptor(GO:0016723) |
| 0.0 | 0.7 | GO:0070402 | NADPH binding(GO:0070402) |
| 0.0 | 1.0 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 0.2 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.0 | 0.3 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.0 | 0.8 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.0 | 0.2 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.0 | 0.3 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.0 | 0.9 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.0 | 1.1 | GO:0030159 | receptor signaling complex scaffold activity(GO:0030159) |
| 0.0 | 0.6 | GO:0098748 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.0 | 2.5 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.0 | 1.0 | GO:0000175 | 3'-5'-exoribonuclease activity(GO:0000175) |
| 0.0 | 0.3 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.0 | 0.6 | GO:0031005 | filamin binding(GO:0031005) |
| 0.0 | 0.5 | GO:0005537 | mannose binding(GO:0005537) |
| 0.0 | 0.5 | GO:0008408 | 3'-5' exonuclease activity(GO:0008408) |
| 0.0 | 0.8 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 0.2 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.0 | 0.4 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.5 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.0 | 0.2 | GO:0030911 | TPR domain binding(GO:0030911) |
| 0.0 | 0.6 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.4 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.0 | 0.2 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 0.0 | 0.6 | GO:0008656 | cysteine-type endopeptidase activator activity involved in apoptotic process(GO:0008656) |
| 0.0 | 0.8 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.0 | 0.2 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.2 | GO:0016015 | morphogen activity(GO:0016015) |
| 0.0 | 0.1 | GO:0071936 | coreceptor activity involved in Wnt signaling pathway(GO:0071936) coreceptor activity involved in Wnt signaling pathway, planar cell polarity pathway(GO:1904929) |
| 0.0 | 0.2 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.0 | 0.6 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 0.7 | GO:0030553 | cGMP binding(GO:0030553) |
| 0.0 | 0.3 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.0 | 1.3 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.5 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.0 | 0.1 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.0 | 0.2 | GO:0042799 | histone methyltransferase activity (H4-K20 specific)(GO:0042799) |
| 0.0 | 0.9 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.2 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.0 | 0.1 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.0 | 0.1 | GO:1990446 | U1 snRNP binding(GO:1990446) |
| 0.0 | 0.4 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 0.7 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.0 | GO:0001601 | peptide YY receptor activity(GO:0001601) |
| 0.0 | 0.4 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.0 | 0.3 | GO:0030275 | LRR domain binding(GO:0030275) |
| 0.0 | 0.6 | GO:0008376 | acetylgalactosaminyltransferase activity(GO:0008376) |
| 0.0 | 0.2 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.2 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 0.4 | GO:0001871 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.0 | 0.7 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 1.5 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 1.9 | GO:0017048 | Rho GTPase binding(GO:0017048) |
| 0.0 | 1.7 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.7 | GO:0000980 | RNA polymerase II distal enhancer sequence-specific DNA binding(GO:0000980) |
| 0.0 | 0.1 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.1 | GO:0046625 | sphingolipid binding(GO:0046625) |
| 0.0 | 0.1 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.0 | 1.8 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.3 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.0 | 0.4 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.0 | 0.3 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.0 | GO:0031696 | alpha-2C adrenergic receptor binding(GO:0031696) |
| 0.0 | 0.3 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.0 | 0.3 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.0 | 0.4 | GO:0008175 | tRNA methyltransferase activity(GO:0008175) |
| 0.0 | 0.2 | GO:0070300 | phosphatidic acid binding(GO:0070300) |
| 0.0 | 0.1 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 6.9 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.1 | 5.9 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.1 | 1.6 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.1 | 0.7 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.1 | 2.1 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.1 | 4.1 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.1 | 0.8 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.0 | 3.6 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.0 | 2.8 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 0.2 | PID TCPTP PATHWAY | Signaling events mediated by TCPTP |
| 0.0 | 3.9 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 2.7 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.0 | 1.5 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 0.8 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.0 | 1.5 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 0.7 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 1.1 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 1.1 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.2 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.0 | 0.1 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 0.3 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 1.9 | ST FAS SIGNALING PATHWAY | Fas Signaling Pathway |
| 0.0 | 0.7 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 0.3 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.0 | 0.5 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.0 | 0.7 | PID IFNG PATHWAY | IFN-gamma pathway |
| 0.0 | 0.6 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.0 | 0.3 | SIG PIP3 SIGNALING IN CARDIAC MYOCTES | Genes related to PIP3 signaling in cardiac myocytes |
| 0.0 | 2.0 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 0.7 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.0 | 1.2 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.0 | 0.4 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
| 0.0 | 1.0 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.0 | 0.5 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 0.3 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.0 | 0.3 | ST P38 MAPK PATHWAY | p38 MAPK Pathway |
| 0.0 | 0.6 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 0.2 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.0 | 1.0 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 0.9 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 1.0 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 0.6 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.0 | 1.5 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 0.7 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 0.4 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 0.1 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.0 | 1.7 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 0.8 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.3 | PID ATM PATHWAY | ATM pathway |
| 0.0 | 2.9 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.5 | PID RAC1 PATHWAY | RAC1 signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.2 | REACTOME APC CDC20 MEDIATED DEGRADATION OF NEK2A | Genes involved in APC-Cdc20 mediated degradation of Nek2A |
| 0.2 | 9.5 | REACTOME PTM GAMMA CARBOXYLATION HYPUSINE FORMATION AND ARYLSULFATASE ACTIVATION | Genes involved in PTM: gamma carboxylation, hypusine formation and arylsulfatase activation |
| 0.2 | 6.6 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF CAMKII | Genes involved in CREB phosphorylation through the activation of CaMKII |
| 0.1 | 0.2 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.1 | 1.5 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.1 | 1.1 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.1 | 0.2 | REACTOME SIGNALING BY EGFR IN CANCER | Genes involved in Signaling by EGFR in Cancer |
| 0.1 | 0.7 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.1 | 1.4 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.1 | 1.4 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.1 | 2.6 | REACTOME REGULATION OF GLUCOKINASE BY GLUCOKINASE REGULATORY PROTEIN | Genes involved in Regulation of Glucokinase by Glucokinase Regulatory Protein |
| 0.1 | 1.2 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.1 | 0.8 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.1 | 2.5 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.1 | 0.6 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.1 | 1.6 | REACTOME INFLAMMASOMES | Genes involved in Inflammasomes |
| 0.0 | 1.4 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 0.7 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 1.5 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 3.1 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.0 | 1.1 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 2.7 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 0.5 | REACTOME ENERGY DEPENDENT REGULATION OF MTOR BY LKB1 AMPK | Genes involved in Energy dependent regulation of mTOR by LKB1-AMPK |
| 0.0 | 4.5 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 3.1 | REACTOME DESTABILIZATION OF MRNA BY AUF1 HNRNP D0 | Genes involved in Destabilization of mRNA by AUF1 (hnRNP D0) |
| 0.0 | 0.6 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.0 | 1.1 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.0 | 1.1 | REACTOME PYRUVATE METABOLISM | Genes involved in Pyruvate metabolism |
| 0.0 | 1.3 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 1.7 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 1.2 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 1.0 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 1.4 | REACTOME CLEAVAGE OF GROWING TRANSCRIPT IN THE TERMINATION REGION | Genes involved in Cleavage of Growing Transcript in the Termination Region |
| 0.0 | 1.3 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.0 | 0.1 | REACTOME SEMA4D IN SEMAPHORIN SIGNALING | Genes involved in Sema4D in semaphorin signaling |
| 0.0 | 0.5 | REACTOME SCFSKP2 MEDIATED DEGRADATION OF P27 P21 | Genes involved in SCF(Skp2)-mediated degradation of p27/p21 |
| 0.0 | 2.0 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 0.5 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.0 | 0.7 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 0.8 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.0 | 0.6 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 0.8 | REACTOME REGULATION OF MRNA STABILITY BY PROTEINS THAT BIND AU RICH ELEMENTS | Genes involved in Regulation of mRNA Stability by Proteins that Bind AU-rich Elements |
| 0.0 | 0.4 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 0.8 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 0.1 | REACTOME SCF BETA TRCP MEDIATED DEGRADATION OF EMI1 | Genes involved in SCF-beta-TrCP mediated degradation of Emi1 |
| 0.0 | 1.4 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.5 | REACTOME APC C CDH1 MEDIATED DEGRADATION OF CDC20 AND OTHER APC C CDH1 TARGETED PROTEINS IN LATE MITOSIS EARLY G1 | Genes involved in APC/C:Cdh1 mediated degradation of Cdc20 and other APC/C:Cdh1 targeted proteins in late mitosis/early G1 |
| 0.0 | 0.4 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 0.4 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.0 | 2.3 | REACTOME P75 NTR RECEPTOR MEDIATED SIGNALLING | Genes involved in p75 NTR receptor-mediated signalling |
| 0.0 | 0.5 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.0 | 0.8 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 1.9 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 1.2 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.4 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.3 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 3.9 | REACTOME G ALPHA Q SIGNALLING EVENTS | Genes involved in G alpha (q) signalling events |
| 0.0 | 0.2 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 0.5 | REACTOME MICRORNA MIRNA BIOGENESIS | Genes involved in MicroRNA (miRNA) Biogenesis |
| 0.0 | 0.4 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 0.1 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 0.6 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |
| 0.0 | 0.4 | REACTOME ACTIVATION OF KAINATE RECEPTORS UPON GLUTAMATE BINDING | Genes involved in Activation of Kainate Receptors upon glutamate binding |
| 0.0 | 0.7 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.0 | 0.2 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.0 | 0.5 | REACTOME CIRCADIAN CLOCK | Genes involved in Circadian Clock |
| 0.0 | 0.4 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.2 | REACTOME NUCLEAR EVENTS KINASE AND TRANSCRIPTION FACTOR ACTIVATION | Genes involved in Nuclear Events (kinase and transcription factor activation) |
| 0.0 | 0.2 | REACTOME N GLYCAN TRIMMING IN THE ER AND CALNEXIN CALRETICULIN CYCLE | Genes involved in N-glycan trimming in the ER and Calnexin/Calreticulin cycle |
| 0.0 | 0.3 | REACTOME GRB2 SOS PROVIDES LINKAGE TO MAPK SIGNALING FOR INTERGRINS | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |
| 0.0 | 0.6 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.4 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.0 | 0.2 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 0.3 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 2.3 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 0.4 | REACTOME ACTIVATION OF ATR IN RESPONSE TO REPLICATION STRESS | Genes involved in Activation of ATR in response to replication stress |
| 0.0 | 0.3 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |