Project
Inflammatory response time course, HUVEC (Wada, 2009)
Navigation
Downloads
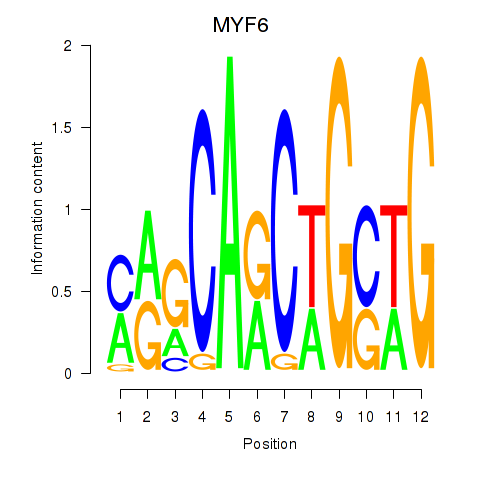
Results for MYF6
Z-value: 2.03
Transcription factors associated with MYF6
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
MYF6
|
ENSG00000111046.4 | MYF6 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| MYF6 | hg38_v1_chr12_+_80707625_80707642 | -0.08 | 7.2e-01 | Click! |
Activity profile of MYF6 motif
Sorted Z-values of MYF6 motif
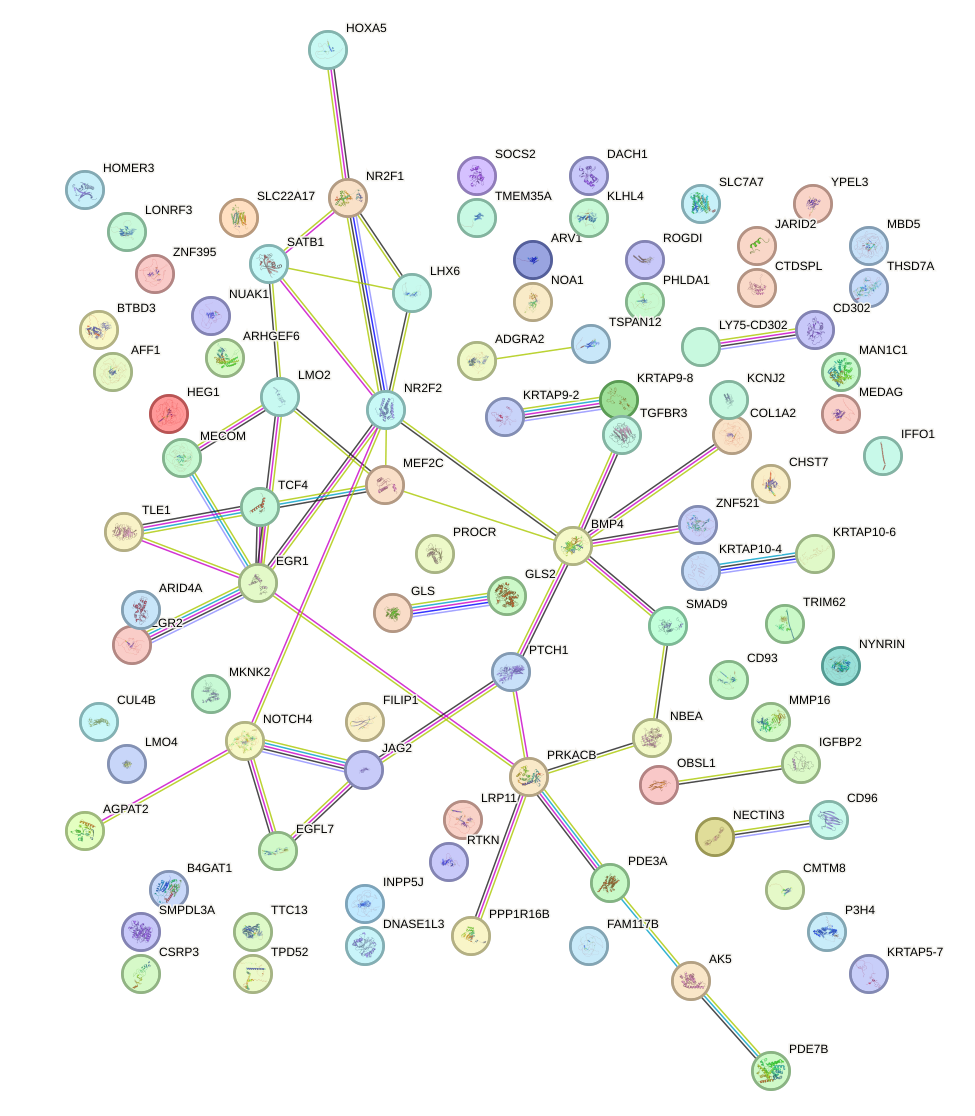
Network of associatons between targets according to the STRING database.
First level regulatory network of MYF6
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.9 | 23.7 | GO:0060399 | positive regulation of growth hormone receptor signaling pathway(GO:0060399) |
| 3.7 | 11.0 | GO:2000007 | apoptotic process involved in endocardial cushion morphogenesis(GO:0003277) intermediate mesoderm morphogenesis(GO:0048390) intermediate mesoderm formation(GO:0048391) intermediate mesodermal cell differentiation(GO:0048392) regulation of cardiac muscle fiber development(GO:0055018) positive regulation of cardiac muscle fiber development(GO:0055020) bud dilation involved in lung branching(GO:0060503) BMP signaling pathway involved in ureter morphogenesis(GO:0061149) renal system segmentation(GO:0061150) BMP signaling pathway involved in renal system segmentation(GO:0061151) pulmonary artery endothelial tube morphogenesis(GO:0061155) regulation of transcription from RNA polymerase II promoter involved in mesonephros development(GO:0061216) BMP signaling pathway involved in nephric duct formation(GO:0071893) negative regulation of branch elongation involved in ureteric bud branching(GO:0072096) negative regulation of branch elongation involved in ureteric bud branching by BMP signaling pathway(GO:0072097) anterior/posterior pattern specification involved in ureteric bud development(GO:0072099) specification of ureteric bud anterior/posterior symmetry(GO:0072100) specification of ureteric bud anterior/posterior symmetry by BMP signaling pathway(GO:0072101) ureter epithelial cell differentiation(GO:0072192) negative regulation of mesenchymal cell proliferation involved in ureter development(GO:0072200) positive regulation of cell proliferation involved in outflow tract morphogenesis(GO:1901964) cardiac jelly development(GO:1905072) regulation of metanephric S-shaped body morphogenesis(GO:2000004) negative regulation of metanephric S-shaped body morphogenesis(GO:2000005) regulation of metanephric comma-shaped body morphogenesis(GO:2000006) negative regulation of metanephric comma-shaped body morphogenesis(GO:2000007) |
| 3.1 | 9.4 | GO:0021593 | rhombomere morphogenesis(GO:0021593) rhombomere 3 morphogenesis(GO:0021658) |
| 2.7 | 24.4 | GO:0060244 | negative regulation of cell proliferation involved in contact inhibition(GO:0060244) |
| 1.9 | 7.7 | GO:1904381 | Golgi apparatus mannose trimming(GO:1904381) |
| 1.6 | 4.9 | GO:0050720 | interleukin-1 beta biosynthetic process(GO:0050720) |
| 1.6 | 9.6 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 1.6 | 4.7 | GO:0060435 | bronchiole development(GO:0060435) |
| 1.5 | 6.0 | GO:0007181 | transforming growth factor beta receptor complex assembly(GO:0007181) |
| 1.4 | 5.8 | GO:0009956 | radial pattern formation(GO:0009956) |
| 1.4 | 9.6 | GO:0090210 | regulation of establishment of blood-brain barrier(GO:0090210) |
| 1.3 | 3.9 | GO:0090341 | negative regulation of secretion of lysosomal enzymes(GO:0090341) |
| 1.2 | 3.7 | GO:1902460 | regulation of mesenchymal stem cell proliferation(GO:1902460) positive regulation of mesenchymal stem cell proliferation(GO:1902462) |
| 1.2 | 4.9 | GO:0090076 | relaxation of skeletal muscle(GO:0090076) |
| 1.0 | 3.0 | GO:0002728 | negative regulation of natural killer cell cytokine production(GO:0002728) |
| 0.9 | 3.5 | GO:0006172 | ADP biosynthetic process(GO:0006172) purine deoxyribonucleoside diphosphate biosynthetic process(GO:0009183) |
| 0.9 | 8.6 | GO:0071321 | cellular response to cGMP(GO:0071321) |
| 0.9 | 1.7 | GO:0033159 | negative regulation of protein import into nucleus, translocation(GO:0033159) |
| 0.8 | 4.0 | GO:0032185 | septin cytoskeleton organization(GO:0032185) |
| 0.8 | 2.3 | GO:1902309 | negative regulation of peptidyl-serine dephosphorylation(GO:1902309) |
| 0.7 | 1.5 | GO:0016476 | regulation of embryonic cell shape(GO:0016476) |
| 0.7 | 2.7 | GO:0055011 | atrial cardiac muscle cell differentiation(GO:0055011) atrial cardiac muscle cell development(GO:0055014) |
| 0.7 | 2.0 | GO:0090271 | positive regulation of fibroblast growth factor production(GO:0090271) |
| 0.7 | 3.9 | GO:0021773 | striatal medium spiny neuron differentiation(GO:0021773) |
| 0.6 | 1.9 | GO:0090031 | positive regulation of steroid hormone biosynthetic process(GO:0090031) |
| 0.6 | 3.1 | GO:0015891 | iron chelate transport(GO:0015688) siderophore transport(GO:0015891) |
| 0.6 | 2.4 | GO:0010157 | response to chlorate(GO:0010157) |
| 0.5 | 11.0 | GO:0007220 | Notch receptor processing(GO:0007220) |
| 0.5 | 1.6 | GO:0006667 | sphinganine metabolic process(GO:0006667) |
| 0.5 | 1.6 | GO:1990641 | regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002589) negative regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002590) regulation of T cell antigen processing and presentation(GO:0002625) positive regulation of iron ion transport(GO:0034758) positive regulation of iron ion transmembrane transport(GO:0034761) regulation of iron ion import(GO:1900390) regulation of ferrous iron import into cell(GO:1903989) positive regulation of ferrous iron import into cell(GO:1903991) regulation of ferrous iron binding(GO:1904432) positive regulation of ferrous iron binding(GO:1904434) regulation of transferrin receptor binding(GO:1904435) positive regulation of transferrin receptor binding(GO:1904437) regulation of ferrous iron import across plasma membrane(GO:1904438) positive regulation of ferrous iron import across plasma membrane(GO:1904440) response to iron ion starvation(GO:1990641) |
| 0.5 | 4.2 | GO:0003172 | primary heart field specification(GO:0003138) sinoatrial valve development(GO:0003172) sinoatrial valve morphogenesis(GO:0003185) |
| 0.5 | 1.6 | GO:0002582 | positive regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002582) positive regulation of antigen processing and presentation of peptide antigen(GO:0002585) positive regulation of antigen processing and presentation of peptide antigen via MHC class II(GO:0002588) |
| 0.5 | 3.7 | GO:0032380 | regulation of intracellular lipid transport(GO:0032377) regulation of intracellular sterol transport(GO:0032380) regulation of intracellular cholesterol transport(GO:0032383) |
| 0.5 | 2.6 | GO:0036378 | calcitriol biosynthetic process from calciol(GO:0036378) |
| 0.5 | 4.0 | GO:0097026 | dendritic cell dendrite assembly(GO:0097026) |
| 0.5 | 2.5 | GO:0097368 | establishment of Sertoli cell barrier(GO:0097368) |
| 0.5 | 3.5 | GO:0000821 | regulation of arginine metabolic process(GO:0000821) |
| 0.5 | 0.5 | GO:0031443 | fast-twitch skeletal muscle fiber contraction(GO:0031443) |
| 0.5 | 2.9 | GO:2000563 | positive regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000563) |
| 0.5 | 2.9 | GO:2000676 | positive regulation of type B pancreatic cell apoptotic process(GO:2000676) |
| 0.5 | 2.9 | GO:0030311 | poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.5 | 1.4 | GO:0021793 | chemorepulsion of branchiomotor axon(GO:0021793) |
| 0.5 | 1.4 | GO:0061580 | colon epithelial cell migration(GO:0061580) |
| 0.4 | 1.3 | GO:0014876 | response to injury involved in regulation of muscle adaptation(GO:0014876) |
| 0.4 | 2.5 | GO:0071461 | cellular response to redox state(GO:0071461) |
| 0.4 | 5.0 | GO:0001766 | membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) |
| 0.4 | 1.1 | GO:0035494 | SNARE complex disassembly(GO:0035494) |
| 0.4 | 8.1 | GO:0070914 | UV-damage excision repair(GO:0070914) |
| 0.4 | 1.1 | GO:0018243 | protein O-linked glycosylation via threonine(GO:0018243) |
| 0.4 | 1.4 | GO:1903998 | regulation of eating behavior(GO:1903998) |
| 0.4 | 5.4 | GO:0015074 | DNA integration(GO:0015074) |
| 0.4 | 1.1 | GO:0036510 | trimming of terminal mannose on C branch(GO:0036510) |
| 0.3 | 1.3 | GO:0003150 | muscular septum morphogenesis(GO:0003150) |
| 0.3 | 1.0 | GO:0050915 | sensory perception of sour taste(GO:0050915) |
| 0.3 | 3.3 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.3 | 4.3 | GO:1902414 | protein localization to cell junction(GO:1902414) |
| 0.3 | 2.0 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
| 0.3 | 1.0 | GO:0042939 | glutathione transport(GO:0034635) tripeptide transport(GO:0042939) |
| 0.3 | 4.5 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.3 | 0.3 | GO:0002339 | B cell selection(GO:0002339) |
| 0.3 | 2.1 | GO:0097338 | response to clozapine(GO:0097338) |
| 0.3 | 1.2 | GO:1903984 | rhythmic synaptic transmission(GO:0060024) negative regulation of ribosome biogenesis(GO:0090071) positive regulation of TRAIL-activated apoptotic signaling pathway(GO:1903984) |
| 0.3 | 5.3 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.3 | 3.5 | GO:0061366 | behavioral response to chemical pain(GO:0061366) behavioral response to formalin induced pain(GO:0061368) |
| 0.3 | 0.9 | GO:0010808 | positive regulation of synaptic vesicle priming(GO:0010808) |
| 0.3 | 1.7 | GO:0048014 | Tie signaling pathway(GO:0048014) |
| 0.3 | 0.8 | GO:0032826 | natural killer cell differentiation involved in immune response(GO:0002325) negative regulation of natural killer cell differentiation(GO:0032824) regulation of natural killer cell differentiation involved in immune response(GO:0032826) negative regulation of natural killer cell differentiation involved in immune response(GO:0032827) positive regulation of cytolysis in other organism(GO:0051714) |
| 0.3 | 6.0 | GO:0010603 | regulation of cytoplasmic mRNA processing body assembly(GO:0010603) |
| 0.3 | 0.8 | GO:0072720 | response to dithiothreitol(GO:0072720) |
| 0.3 | 1.0 | GO:0061357 | positive regulation of Wnt protein secretion(GO:0061357) |
| 0.3 | 2.1 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.2 | 4.2 | GO:0042659 | spinal cord association neuron differentiation(GO:0021527) regulation of cell fate specification(GO:0042659) |
| 0.2 | 1.2 | GO:0071727 | toll-like receptor TLR1:TLR2 signaling pathway(GO:0038123) response to triacyl bacterial lipopeptide(GO:0071725) cellular response to triacyl bacterial lipopeptide(GO:0071727) |
| 0.2 | 2.1 | GO:0035507 | regulation of myosin-light-chain-phosphatase activity(GO:0035507) |
| 0.2 | 2.3 | GO:0043589 | skin morphogenesis(GO:0043589) |
| 0.2 | 1.4 | GO:0034165 | positive regulation of toll-like receptor 9 signaling pathway(GO:0034165) |
| 0.2 | 2.5 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.2 | 0.5 | GO:0070172 | positive regulation of tooth mineralization(GO:0070172) |
| 0.2 | 4.0 | GO:0009313 | oligosaccharide catabolic process(GO:0009313) |
| 0.2 | 0.7 | GO:0001714 | endodermal cell fate specification(GO:0001714) |
| 0.2 | 1.5 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.2 | 2.0 | GO:0016139 | glycoside catabolic process(GO:0016139) |
| 0.2 | 1.5 | GO:0002725 | negative regulation of T cell cytokine production(GO:0002725) |
| 0.2 | 0.6 | GO:0035691 | macrophage migration inhibitory factor signaling pathway(GO:0035691) |
| 0.2 | 0.8 | GO:0006669 | sphinganine-1-phosphate biosynthetic process(GO:0006669) |
| 0.2 | 0.8 | GO:0021966 | corticospinal neuron axon guidance(GO:0021966) |
| 0.2 | 1.2 | GO:0048050 | post-embryonic eye morphogenesis(GO:0048050) |
| 0.2 | 1.0 | GO:1902847 | regulation of neuronal signal transduction(GO:1902847) positive regulation of neurofibrillary tangle assembly(GO:1902998) |
| 0.2 | 0.6 | GO:0052047 | interaction with other organism via secreted substance involved in symbiotic interaction(GO:0052047) |
| 0.2 | 11.6 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.2 | 1.8 | GO:0046600 | negative regulation of centriole replication(GO:0046600) |
| 0.2 | 1.0 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.2 | 1.8 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.2 | 1.2 | GO:0070829 | response to vitamin B2(GO:0033274) heterochromatin maintenance(GO:0070829) |
| 0.2 | 4.7 | GO:0007130 | synaptonemal complex assembly(GO:0007130) |
| 0.2 | 2.3 | GO:0021800 | cerebral cortex tangential migration(GO:0021800) |
| 0.2 | 3.2 | GO:0010623 | programmed cell death involved in cell development(GO:0010623) |
| 0.2 | 2.5 | GO:0009414 | response to water deprivation(GO:0009414) |
| 0.2 | 4.0 | GO:0016024 | CDP-diacylglycerol biosynthetic process(GO:0016024) |
| 0.2 | 0.6 | GO:0019858 | cytosine metabolic process(GO:0019858) |
| 0.2 | 0.4 | GO:0060932 | His-Purkinje system cell differentiation(GO:0060932) |
| 0.2 | 1.7 | GO:0038026 | reelin-mediated signaling pathway(GO:0038026) |
| 0.2 | 0.2 | GO:0016539 | intein-mediated protein splicing(GO:0016539) protein splicing(GO:0030908) |
| 0.2 | 2.0 | GO:0042985 | negative regulation of amyloid precursor protein biosynthetic process(GO:0042985) |
| 0.2 | 2.0 | GO:0033227 | dsRNA transport(GO:0033227) |
| 0.2 | 0.5 | GO:0090675 | intermicrovillar adhesion(GO:0090675) |
| 0.2 | 1.6 | GO:0097491 | sympathetic neuron projection extension(GO:0097490) sympathetic neuron projection guidance(GO:0097491) |
| 0.2 | 1.8 | GO:0008343 | adult feeding behavior(GO:0008343) |
| 0.2 | 1.2 | GO:0006154 | adenosine catabolic process(GO:0006154) inosine biosynthetic process(GO:0046103) |
| 0.2 | 0.5 | GO:0002232 | leukocyte chemotaxis involved in inflammatory response(GO:0002232) cell surface pattern recognition receptor signaling pathway(GO:0002752) |
| 0.2 | 1.9 | GO:0051791 | medium-chain fatty acid metabolic process(GO:0051791) |
| 0.2 | 0.7 | GO:0030200 | heparan sulfate proteoglycan catabolic process(GO:0030200) |
| 0.2 | 2.7 | GO:0051574 | positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.2 | 0.5 | GO:1903937 | response to acrylamide(GO:1903937) |
| 0.2 | 0.5 | GO:0061086 | negative regulation of histone H3-K27 methylation(GO:0061086) |
| 0.2 | 1.2 | GO:0035754 | B cell chemotaxis(GO:0035754) |
| 0.2 | 1.2 | GO:0000255 | allantoin metabolic process(GO:0000255) |
| 0.2 | 0.5 | GO:0090274 | positive regulation of pancreatic juice secretion(GO:0090187) regulation of somatostatin secretion(GO:0090273) positive regulation of somatostatin secretion(GO:0090274) |
| 0.2 | 0.6 | GO:0005986 | sucrose biosynthetic process(GO:0005986) |
| 0.2 | 0.5 | GO:2000646 | positive regulation of receptor catabolic process(GO:2000646) |
| 0.2 | 2.0 | GO:0007042 | lysosomal lumen acidification(GO:0007042) |
| 0.2 | 0.8 | GO:0035802 | adrenal cortex development(GO:0035801) adrenal cortex formation(GO:0035802) embryonic heart tube left/right pattern formation(GO:0060971) negative regulation of transcription from RNA polymerase II promoter in response to hypoxia(GO:0061428) |
| 0.2 | 2.5 | GO:2000774 | positive regulation of cellular senescence(GO:2000774) |
| 0.2 | 0.8 | GO:0021590 | cerebellum maturation(GO:0021590) cerebellar cortex maturation(GO:0021699) |
| 0.2 | 1.8 | GO:0032836 | glomerular basement membrane development(GO:0032836) |
| 0.1 | 3.7 | GO:0042789 | mRNA transcription from RNA polymerase II promoter(GO:0042789) |
| 0.1 | 4.7 | GO:2000811 | negative regulation of anoikis(GO:2000811) |
| 0.1 | 1.7 | GO:0016081 | synaptic vesicle docking(GO:0016081) |
| 0.1 | 1.4 | GO:0042426 | choline catabolic process(GO:0042426) |
| 0.1 | 0.5 | GO:0002182 | cytoplasmic translational elongation(GO:0002182) regulation of cytoplasmic translational elongation(GO:1900247) negative regulation of cytoplasmic translational elongation(GO:1900248) |
| 0.1 | 1.6 | GO:0090336 | positive regulation of brown fat cell differentiation(GO:0090336) |
| 0.1 | 1.9 | GO:1902659 | regulation of glucose mediated signaling pathway(GO:1902659) |
| 0.1 | 2.9 | GO:0006044 | N-acetylglucosamine metabolic process(GO:0006044) |
| 0.1 | 0.3 | GO:0097676 | histone H3-K36 dimethylation(GO:0097676) |
| 0.1 | 1.6 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.1 | 0.4 | GO:0071314 | cellular response to cocaine(GO:0071314) |
| 0.1 | 1.0 | GO:0060754 | positive regulation of mast cell chemotaxis(GO:0060754) |
| 0.1 | 4.8 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.1 | 0.8 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.1 | 0.8 | GO:0010757 | negative regulation of plasminogen activation(GO:0010757) |
| 0.1 | 1.3 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.1 | 3.4 | GO:0010226 | response to lithium ion(GO:0010226) |
| 0.1 | 2.8 | GO:0031115 | negative regulation of microtubule polymerization(GO:0031115) |
| 0.1 | 0.7 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.1 | 0.3 | GO:1902996 | regulation of neurofibrillary tangle assembly(GO:1902996) |
| 0.1 | 0.5 | GO:1902473 | regulation of protein localization to synapse(GO:1902473) positive regulation of protein localization to synapse(GO:1902474) |
| 0.1 | 0.9 | GO:0010825 | positive regulation of centrosome duplication(GO:0010825) |
| 0.1 | 0.9 | GO:0071386 | cellular response to corticosterone stimulus(GO:0071386) |
| 0.1 | 2.6 | GO:0097094 | craniofacial suture morphogenesis(GO:0097094) |
| 0.1 | 1.4 | GO:0050713 | negative regulation of interleukin-1 beta secretion(GO:0050713) |
| 0.1 | 1.1 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.1 | 0.5 | GO:1902268 | negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.1 | 0.4 | GO:0021615 | glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.1 | 0.7 | GO:0002318 | myeloid progenitor cell differentiation(GO:0002318) |
| 0.1 | 2.1 | GO:0040015 | negative regulation of multicellular organism growth(GO:0040015) |
| 0.1 | 2.5 | GO:0043457 | regulation of cellular respiration(GO:0043457) |
| 0.1 | 1.2 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.1 | 0.4 | GO:1990167 | protein K6-linked deubiquitination(GO:0044313) protein K27-linked deubiquitination(GO:1990167) |
| 0.1 | 2.8 | GO:0007216 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.1 | 1.8 | GO:0000712 | resolution of meiotic recombination intermediates(GO:0000712) |
| 0.1 | 0.5 | GO:0000379 | tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 0.1 | 1.8 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.1 | 0.6 | GO:1900264 | regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.1 | 0.3 | GO:0072061 | inner medullary collecting duct development(GO:0072061) |
| 0.1 | 1.0 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.1 | 0.8 | GO:0007144 | female meiosis I(GO:0007144) |
| 0.1 | 2.0 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.1 | 0.3 | GO:0030576 | Cajal body organization(GO:0030576) |
| 0.1 | 0.4 | GO:0033088 | negative regulation of immature T cell proliferation in thymus(GO:0033088) |
| 0.1 | 0.4 | GO:0097411 | hypoxia-inducible factor-1alpha signaling pathway(GO:0097411) |
| 0.1 | 0.8 | GO:0051005 | negative regulation of lipoprotein lipase activity(GO:0051005) |
| 0.1 | 0.5 | GO:0000432 | regulation of transcription from RNA polymerase II promoter by glucose(GO:0000430) positive regulation of transcription from RNA polymerase II promoter by glucose(GO:0000432) late viral transcription(GO:0019086) positive regulation of transcription by glucose(GO:0046016) |
| 0.1 | 0.5 | GO:0015680 | intracellular copper ion transport(GO:0015680) |
| 0.1 | 0.2 | GO:1902683 | regulation of receptor localization to synapse(GO:1902683) |
| 0.1 | 0.3 | GO:0036518 | chemorepulsion of dopaminergic neuron axon(GO:0036518) chemorepulsion of axon(GO:0061643) |
| 0.1 | 2.1 | GO:0071243 | cellular response to arsenic-containing substance(GO:0071243) |
| 0.1 | 0.4 | GO:0070295 | transformation of host cell by virus(GO:0019087) renal water absorption(GO:0070295) |
| 0.1 | 0.3 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.1 | 1.0 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.1 | 0.9 | GO:1904424 | regulation of GTP binding(GO:1904424) |
| 0.1 | 1.4 | GO:0010839 | negative regulation of keratinocyte proliferation(GO:0010839) |
| 0.1 | 0.4 | GO:2000617 | regulation of stem cell division(GO:2000035) positive regulation of histone H3-K9 acetylation(GO:2000617) |
| 0.1 | 1.1 | GO:0033962 | cytoplasmic mRNA processing body assembly(GO:0033962) |
| 0.1 | 0.2 | GO:0042369 | vitamin D catabolic process(GO:0042369) |
| 0.1 | 1.0 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.1 | 0.8 | GO:0031936 | negative regulation of chromatin silencing(GO:0031936) |
| 0.1 | 0.4 | GO:0006296 | nucleotide-excision repair, DNA incision, 5'-to lesion(GO:0006296) |
| 0.1 | 0.6 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 0.1 | 0.7 | GO:0001887 | selenium compound metabolic process(GO:0001887) |
| 0.1 | 0.5 | GO:0051599 | response to hydrostatic pressure(GO:0051599) |
| 0.1 | 1.0 | GO:0098719 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.1 | 0.5 | GO:0033629 | negative regulation of cell adhesion mediated by integrin(GO:0033629) |
| 0.1 | 0.5 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.1 | 0.9 | GO:0097113 | AMPA glutamate receptor clustering(GO:0097113) glutamate receptor clustering(GO:0097688) |
| 0.1 | 0.2 | GO:2000255 | negative regulation of male germ cell proliferation(GO:2000255) |
| 0.1 | 0.6 | GO:0015939 | pantothenate metabolic process(GO:0015939) |
| 0.1 | 0.3 | GO:0021539 | subthalamus development(GO:0021539) |
| 0.1 | 1.1 | GO:0045475 | locomotor rhythm(GO:0045475) |
| 0.1 | 1.5 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.1 | 0.6 | GO:2000252 | negative regulation of feeding behavior(GO:2000252) |
| 0.1 | 1.3 | GO:0006704 | glucocorticoid biosynthetic process(GO:0006704) |
| 0.1 | 2.5 | GO:0071539 | protein localization to centrosome(GO:0071539) |
| 0.1 | 2.4 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.1 | 1.7 | GO:0051290 | protein heterotetramerization(GO:0051290) |
| 0.1 | 0.1 | GO:0035698 | CD8-positive, alpha-beta cytotoxic T cell extravasation(GO:0035698) regulation of CD8-positive, alpha-beta cytotoxic T cell extravasation(GO:2000452) |
| 0.1 | 0.4 | GO:0035860 | glial cell-derived neurotrophic factor receptor signaling pathway(GO:0035860) |
| 0.1 | 0.3 | GO:1900738 | positive regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900738) |
| 0.1 | 1.6 | GO:0043537 | negative regulation of blood vessel endothelial cell migration(GO:0043537) |
| 0.1 | 2.3 | GO:0046596 | regulation of viral entry into host cell(GO:0046596) |
| 0.1 | 0.4 | GO:0097319 | fructose transport(GO:0015755) fructose import(GO:0032445) carbohydrate import into cell(GO:0097319) carbohydrate import across plasma membrane(GO:0098704) fructose import across plasma membrane(GO:1990539) |
| 0.1 | 0.5 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.1 | 0.3 | GO:0001992 | regulation of systemic arterial blood pressure by vasopressin(GO:0001992) |
| 0.1 | 0.3 | GO:0015862 | uridine transport(GO:0015862) |
| 0.1 | 0.3 | GO:0048496 | adherens junction maintenance(GO:0034334) maintenance of organ identity(GO:0048496) |
| 0.1 | 1.7 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 0.1 | 0.2 | GO:0002184 | cytoplasmic translational termination(GO:0002184) |
| 0.1 | 1.2 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.1 | 1.0 | GO:0035246 | peptidyl-arginine N-methylation(GO:0035246) |
| 0.1 | 0.7 | GO:0072711 | response to hydroxyurea(GO:0072710) cellular response to hydroxyurea(GO:0072711) |
| 0.1 | 0.3 | GO:0030950 | establishment or maintenance of actin cytoskeleton polarity(GO:0030950) |
| 0.1 | 0.4 | GO:0071316 | cellular response to nicotine(GO:0071316) |
| 0.1 | 2.3 | GO:0006308 | DNA catabolic process(GO:0006308) |
| 0.1 | 0.5 | GO:0038145 | macrophage colony-stimulating factor signaling pathway(GO:0038145) |
| 0.1 | 0.2 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 0.1 | 0.6 | GO:0030321 | transepithelial chloride transport(GO:0030321) |
| 0.1 | 0.4 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) |
| 0.1 | 0.8 | GO:0051546 | keratinocyte migration(GO:0051546) |
| 0.1 | 9.9 | GO:0007286 | spermatid development(GO:0007286) |
| 0.1 | 0.6 | GO:0016127 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.1 | 0.6 | GO:0071397 | cellular response to cholesterol(GO:0071397) |
| 0.0 | 0.8 | GO:0003215 | cardiac right ventricle morphogenesis(GO:0003215) |
| 0.0 | 0.3 | GO:2000781 | positive regulation of double-strand break repair(GO:2000781) |
| 0.0 | 1.1 | GO:0008209 | androgen metabolic process(GO:0008209) |
| 0.0 | 0.3 | GO:0006642 | triglyceride mobilization(GO:0006642) |
| 0.0 | 1.9 | GO:0045599 | negative regulation of fat cell differentiation(GO:0045599) |
| 0.0 | 0.3 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.2 | GO:0015770 | disaccharide transport(GO:0015766) sucrose transport(GO:0015770) oligosaccharide transport(GO:0015772) |
| 0.0 | 0.5 | GO:0010668 | ectodermal cell differentiation(GO:0010668) |
| 0.0 | 2.9 | GO:0051693 | actin filament capping(GO:0051693) |
| 0.0 | 0.4 | GO:2000271 | positive regulation of fibroblast apoptotic process(GO:2000271) |
| 0.0 | 0.1 | GO:0019303 | D-ribose catabolic process(GO:0019303) |
| 0.0 | 0.8 | GO:0072698 | protein localization to microtubule cytoskeleton(GO:0072698) |
| 0.0 | 0.6 | GO:0001514 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.0 | 1.2 | GO:0036150 | phosphatidylserine acyl-chain remodeling(GO:0036150) |
| 0.0 | 1.2 | GO:0043555 | regulation of translation in response to stress(GO:0043555) |
| 0.0 | 0.5 | GO:0043981 | histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.0 | 1.5 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.0 | 9.8 | GO:0065004 | protein-DNA complex assembly(GO:0065004) |
| 0.0 | 0.2 | GO:0038060 | nitric oxide-cGMP-mediated signaling pathway(GO:0038060) |
| 0.0 | 0.4 | GO:0002903 | negative regulation of B cell apoptotic process(GO:0002903) |
| 0.0 | 0.1 | GO:0070843 | misfolded protein transport(GO:0070843) polyubiquitinated protein transport(GO:0070844) polyubiquitinated misfolded protein transport(GO:0070845) Hsp90 deacetylation(GO:0070846) positive regulation of hydrogen peroxide-mediated programmed cell death(GO:1901300) |
| 0.0 | 0.2 | GO:0043117 | positive regulation of vascular permeability(GO:0043117) |
| 0.0 | 0.3 | GO:0051415 | interphase microtubule nucleation by interphase microtubule organizing center(GO:0051415) microtubule nucleation by microtubule organizing center(GO:0051418) |
| 0.0 | 0.9 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.0 | 0.6 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.0 | 0.3 | GO:0008215 | spermine metabolic process(GO:0008215) |
| 0.0 | 0.4 | GO:0098535 | de novo centriole assembly(GO:0098535) |
| 0.0 | 0.4 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.0 | 2.9 | GO:0043966 | histone H3 acetylation(GO:0043966) |
| 0.0 | 4.3 | GO:2000134 | negative regulation of G1/S transition of mitotic cell cycle(GO:2000134) |
| 0.0 | 0.2 | GO:0055064 | chloride ion homeostasis(GO:0055064) |
| 0.0 | 0.4 | GO:2000344 | positive regulation of acrosome reaction(GO:2000344) |
| 0.0 | 0.2 | GO:1900121 | negative regulation of receptor binding(GO:1900121) |
| 0.0 | 0.9 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 0.0 | 0.2 | GO:0070164 | negative regulation of adiponectin secretion(GO:0070164) |
| 0.0 | 0.1 | GO:0006121 | mitochondrial electron transport, succinate to ubiquinone(GO:0006121) |
| 0.0 | 0.7 | GO:1900151 | regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:1900151) positive regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:1900153) |
| 0.0 | 0.3 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.0 | 0.5 | GO:0045956 | positive regulation of calcium ion-dependent exocytosis(GO:0045956) |
| 0.0 | 0.1 | GO:0032661 | regulation of interleukin-18 production(GO:0032661) negative regulation of calcium-transporting ATPase activity(GO:1901895) |
| 0.0 | 0.1 | GO:0044861 | protein transport into plasma membrane raft(GO:0044861) |
| 0.0 | 1.9 | GO:0007040 | lysosome organization(GO:0007040) lytic vacuole organization(GO:0080171) |
| 0.0 | 0.0 | GO:0044209 | AMP salvage(GO:0044209) |
| 0.0 | 0.4 | GO:0035563 | positive regulation of chromatin binding(GO:0035563) |
| 0.0 | 0.2 | GO:0032455 | renal response to blood flow involved in circulatory renin-angiotensin regulation of systemic arterial blood pressure(GO:0001999) renin secretion into blood stream(GO:0002001) nerve growth factor processing(GO:0032455) |
| 0.0 | 0.8 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.0 | 0.3 | GO:0044387 | negative regulation of protein kinase activity by regulation of protein phosphorylation(GO:0044387) |
| 0.0 | 0.6 | GO:0048935 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.0 | 0.5 | GO:0033139 | regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033139) |
| 0.0 | 0.2 | GO:0061143 | alveolar primary septum development(GO:0061143) paramesonephric duct development(GO:0061205) |
| 0.0 | 0.7 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.0 | 0.2 | GO:0033327 | Leydig cell differentiation(GO:0033327) |
| 0.0 | 0.7 | GO:0048536 | spleen development(GO:0048536) |
| 0.0 | 0.7 | GO:0030208 | dermatan sulfate biosynthetic process(GO:0030208) |
| 0.0 | 0.1 | GO:0045650 | negative regulation of macrophage differentiation(GO:0045650) |
| 0.0 | 0.4 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.0 | 0.1 | GO:0045110 | intermediate filament bundle assembly(GO:0045110) |
| 0.0 | 0.1 | GO:2001013 | epithelial cell proliferation involved in renal tubule morphogenesis(GO:2001013) |
| 0.0 | 0.5 | GO:0031116 | positive regulation of microtubule polymerization(GO:0031116) |
| 0.0 | 0.1 | GO:0035610 | protein side chain deglutamylation(GO:0035610) |
| 0.0 | 0.4 | GO:0036295 | cellular response to increased oxygen levels(GO:0036295) |
| 0.0 | 0.8 | GO:0044458 | motile cilium assembly(GO:0044458) |
| 0.0 | 0.2 | GO:0010756 | positive regulation of plasminogen activation(GO:0010756) |
| 0.0 | 0.3 | GO:0045736 | negative regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045736) |
| 0.0 | 0.2 | GO:1901409 | positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.0 | 2.0 | GO:0006911 | phagocytosis, engulfment(GO:0006911) |
| 0.0 | 0.4 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.0 | 0.6 | GO:0010832 | negative regulation of myotube differentiation(GO:0010832) |
| 0.0 | 0.1 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.0 | 0.2 | GO:0070166 | enamel mineralization(GO:0070166) |
| 0.0 | 0.2 | GO:1904714 | regulation of chaperone-mediated autophagy(GO:1904714) |
| 0.0 | 1.5 | GO:0030183 | B cell differentiation(GO:0030183) |
| 0.0 | 0.5 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.0 | 0.9 | GO:0046716 | muscle cell cellular homeostasis(GO:0046716) |
| 0.0 | 0.6 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.7 | GO:0002026 | regulation of the force of heart contraction(GO:0002026) |
| 0.0 | 0.1 | GO:2000002 | negative regulation of cell cycle checkpoint(GO:1901977) negative regulation of DNA damage checkpoint(GO:2000002) |
| 0.0 | 0.7 | GO:0032007 | negative regulation of TOR signaling(GO:0032007) |
| 0.0 | 1.1 | GO:0010955 | negative regulation of protein processing(GO:0010955) negative regulation of protein maturation(GO:1903318) |
| 0.0 | 1.4 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.0 | 0.3 | GO:0051256 | mitotic spindle midzone assembly(GO:0051256) |
| 0.0 | 0.2 | GO:0071847 | TNFSF11-mediated signaling pathway(GO:0071847) |
| 0.0 | 0.1 | GO:0031937 | positive regulation of chromatin silencing(GO:0031937) |
| 0.0 | 0.0 | GO:0036102 | leukotriene catabolic process(GO:0036100) leukotriene B4 catabolic process(GO:0036101) leukotriene B4 metabolic process(GO:0036102) icosanoid catabolic process(GO:1901523) fatty acid derivative catabolic process(GO:1901569) |
| 0.0 | 0.2 | GO:0000715 | nucleotide-excision repair, DNA damage recognition(GO:0000715) |
| 0.0 | 3.8 | GO:0031032 | actomyosin structure organization(GO:0031032) |
| 0.0 | 0.2 | GO:0014883 | transition between fast and slow fiber(GO:0014883) |
| 0.0 | 0.1 | GO:0019557 | histidine catabolic process to glutamate and formamide(GO:0019556) histidine catabolic process to glutamate and formate(GO:0019557) formamide metabolic process(GO:0043606) |
| 0.0 | 0.2 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 0.0 | 1.4 | GO:0042116 | macrophage activation(GO:0042116) |
| 0.0 | 0.0 | GO:0002322 | B cell proliferation involved in immune response(GO:0002322) |
| 0.0 | 0.3 | GO:0097062 | dendritic spine maintenance(GO:0097062) |
| 0.0 | 0.2 | GO:0070459 | prolactin secretion(GO:0070459) |
| 0.0 | 1.7 | GO:0001676 | long-chain fatty acid metabolic process(GO:0001676) |
| 0.0 | 0.7 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.0 | 0.2 | GO:0030210 | heparin metabolic process(GO:0030202) heparin biosynthetic process(GO:0030210) |
| 0.0 | 0.6 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.0 | 0.3 | GO:0000920 | cell separation after cytokinesis(GO:0000920) |
| 0.0 | 0.2 | GO:0019388 | galactose catabolic process(GO:0019388) |
| 0.0 | 1.5 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.0 | 4.0 | GO:0000086 | G2/M transition of mitotic cell cycle(GO:0000086) |
| 0.0 | 1.1 | GO:0001895 | retina homeostasis(GO:0001895) |
| 0.0 | 0.2 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.0 | 1.2 | GO:0042795 | snRNA transcription(GO:0009301) snRNA transcription from RNA polymerase II promoter(GO:0042795) |
| 0.0 | 1.3 | GO:0043647 | inositol phosphate metabolic process(GO:0043647) |
| 0.0 | 0.2 | GO:0071157 | negative regulation of cell cycle arrest(GO:0071157) |
| 0.0 | 0.7 | GO:0045600 | positive regulation of fat cell differentiation(GO:0045600) |
| 0.0 | 0.1 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
| 0.0 | 0.2 | GO:0050962 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.0 | 0.3 | GO:0060314 | regulation of ryanodine-sensitive calcium-release channel activity(GO:0060314) |
| 0.0 | 0.3 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.0 | 0.0 | GO:0042264 | peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 0.0 | 0.1 | GO:0097398 | response to interleukin-17(GO:0097396) cellular response to interleukin-17(GO:0097398) |
| 0.0 | 0.6 | GO:0006303 | double-strand break repair via nonhomologous end joining(GO:0006303) |
| 0.0 | 0.5 | GO:0006409 | tRNA export from nucleus(GO:0006409) tRNA-containing ribonucleoprotein complex export from nucleus(GO:0071431) |
| 0.0 | 0.2 | GO:0006853 | carnitine shuttle(GO:0006853) |
| 0.0 | 0.4 | GO:0072661 | protein targeting to plasma membrane(GO:0072661) |
| 0.0 | 1.0 | GO:0006687 | glycosphingolipid metabolic process(GO:0006687) |
| 0.0 | 0.7 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.2 | GO:0033006 | regulation of mast cell activation involved in immune response(GO:0033006) regulation of mast cell degranulation(GO:0043304) |
| 0.0 | 0.6 | GO:0045921 | positive regulation of exocytosis(GO:0045921) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 6.0 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.9 | 24.2 | GO:0010369 | chromocenter(GO:0010369) |
| 0.8 | 3.3 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.7 | 8.1 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.5 | 0.5 | GO:1990667 | PCSK9-AnxA2 complex(GO:1990667) |
| 0.4 | 1.8 | GO:1990851 | Wnt-Frizzled-LRP5/6 complex(GO:1990851) |
| 0.4 | 1.2 | GO:0070195 | growth hormone receptor complex(GO:0070195) |
| 0.4 | 15.6 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 0.4 | 2.3 | GO:1990393 | 3M complex(GO:1990393) |
| 0.4 | 1.8 | GO:0034680 | integrin alpha10-beta1 complex(GO:0034680) |
| 0.3 | 1.6 | GO:0072558 | NLRP1 inflammasome complex(GO:0072558) |
| 0.3 | 1.8 | GO:0016035 | zeta DNA polymerase complex(GO:0016035) |
| 0.3 | 1.7 | GO:0031417 | NatC complex(GO:0031417) |
| 0.3 | 0.3 | GO:0034667 | integrin alpha3-beta1 complex(GO:0034667) |
| 0.3 | 1.6 | GO:1990357 | terminal web(GO:1990357) |
| 0.3 | 2.7 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.3 | 2.3 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.3 | 0.8 | GO:0070931 | Golgi-associated vesicle lumen(GO:0070931) |
| 0.3 | 0.8 | GO:0071062 | alphav-beta3 integrin-vitronectin complex(GO:0071062) |
| 0.2 | 2.9 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.2 | 2.5 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.2 | 2.0 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.2 | 3.0 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.2 | 12.0 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.2 | 2.0 | GO:0016011 | dystroglycan complex(GO:0016011) |
| 0.2 | 1.4 | GO:0072559 | NLRP3 inflammasome complex(GO:0072559) |
| 0.2 | 0.5 | GO:0097224 | sperm connecting piece(GO:0097224) |
| 0.2 | 0.7 | GO:0071821 | FANCM-MHF complex(GO:0071821) |
| 0.2 | 3.0 | GO:0008091 | spectrin(GO:0008091) |
| 0.2 | 3.6 | GO:0043220 | Schmidt-Lanterman incisure(GO:0043220) |
| 0.2 | 2.1 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.1 | 0.8 | GO:0016589 | NURF complex(GO:0016589) |
| 0.1 | 0.8 | GO:0097013 | phagocytic vesicle lumen(GO:0097013) |
| 0.1 | 1.1 | GO:0000110 | nucleotide-excision repair factor 1 complex(GO:0000110) |
| 0.1 | 0.5 | GO:0038039 | G-protein coupled receptor heterodimeric complex(GO:0038039) |
| 0.1 | 1.2 | GO:0031211 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.1 | 1.2 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.1 | 1.0 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.1 | 0.5 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 0.1 | 1.1 | GO:0043190 | ATP-binding cassette (ABC) transporter complex(GO:0043190) |
| 0.1 | 0.7 | GO:0044305 | calyx of Held(GO:0044305) |
| 0.1 | 0.5 | GO:0005971 | ribonucleoside-diphosphate reductase complex(GO:0005971) |
| 0.1 | 5.3 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.1 | 1.0 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.1 | 0.5 | GO:0044214 | spanning component of plasma membrane(GO:0044214) spanning component of membrane(GO:0089717) |
| 0.1 | 0.4 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.1 | 3.8 | GO:0035869 | ciliary transition zone(GO:0035869) |
| 0.1 | 1.6 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.1 | 0.5 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.1 | 0.5 | GO:0016342 | catenin complex(GO:0016342) |
| 0.1 | 5.4 | GO:0005844 | polysome(GO:0005844) |
| 0.1 | 1.0 | GO:0042583 | chromaffin granule(GO:0042583) |
| 0.1 | 4.7 | GO:0000795 | synaptonemal complex(GO:0000795) |
| 0.1 | 39.7 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.1 | 1.6 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.1 | 1.5 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 0.1 | 5.1 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.1 | 11.4 | GO:0030175 | filopodium(GO:0030175) |
| 0.1 | 4.9 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.1 | 2.0 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.1 | 1.2 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.1 | 1.5 | GO:0044232 | organelle membrane contact site(GO:0044232) |
| 0.1 | 0.5 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.1 | 0.6 | GO:0031390 | Ctf18 RFC-like complex(GO:0031390) |
| 0.1 | 1.7 | GO:0005605 | basal lamina(GO:0005605) |
| 0.1 | 0.9 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.1 | 1.7 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.1 | 1.2 | GO:0031045 | dense core granule(GO:0031045) |
| 0.1 | 0.2 | GO:0018444 | translation release factor complex(GO:0018444) |
| 0.1 | 1.2 | GO:0001527 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.1 | 0.4 | GO:0098536 | deuterosome(GO:0098536) |
| 0.1 | 1.1 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.1 | 0.6 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.1 | 0.4 | GO:0032279 | asymmetric synapse(GO:0032279) |
| 0.1 | 1.4 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 1.1 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.0 | 0.2 | GO:0005958 | DNA-dependent protein kinase-DNA ligase 4 complex(GO:0005958) |
| 0.0 | 1.5 | GO:0005720 | nuclear heterochromatin(GO:0005720) |
| 0.0 | 0.6 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.0 | 4.3 | GO:0045178 | basal part of cell(GO:0045178) |
| 0.0 | 0.8 | GO:0090665 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 0.4 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.0 | 5.7 | GO:0035578 | azurophil granule lumen(GO:0035578) |
| 0.0 | 0.7 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.0 | 0.9 | GO:0005652 | nuclear lamina(GO:0005652) |
| 0.0 | 0.6 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.0 | 4.3 | GO:0101003 | ficolin-1-rich granule membrane(GO:0101003) |
| 0.0 | 0.7 | GO:0033202 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 0.0 | 4.9 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 0.5 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.0 | 2.1 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.0 | 3.1 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.0 | 0.9 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.0 | 5.1 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 0.3 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.0 | 2.0 | GO:0031907 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 0.6 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 4.3 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 0.2 | GO:0044194 | cytolytic granule(GO:0044194) |
| 0.0 | 0.7 | GO:0016471 | vacuolar proton-transporting V-type ATPase complex(GO:0016471) |
| 0.0 | 0.9 | GO:0044322 | endoplasmic reticulum quality control compartment(GO:0044322) |
| 0.0 | 0.1 | GO:0005854 | nascent polypeptide-associated complex(GO:0005854) |
| 0.0 | 2.8 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.0 | 0.6 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.0 | 8.5 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 0.1 | GO:0045257 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.0 | 0.7 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.8 | GO:0099738 | cell cortex region(GO:0099738) |
| 0.0 | 0.8 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.0 | 0.3 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.0 | 0.3 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.0 | 2.1 | GO:0031903 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 1.4 | GO:0031430 | M band(GO:0031430) |
| 0.0 | 1.6 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 2.3 | GO:0044291 | cell-cell contact zone(GO:0044291) |
| 0.0 | 1.0 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 0.2 | GO:0044352 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.0 | 0.9 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 0.5 | GO:0071339 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.0 | 0.4 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.7 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.0 | 0.2 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 1.4 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.0 | 9.2 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 1.0 | GO:0016591 | DNA-directed RNA polymerase II, holoenzyme(GO:0016591) |
| 0.0 | 0.2 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.0 | 0.5 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.0 | 0.9 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 0.4 | GO:0030137 | COPI-coated vesicle(GO:0030137) |
| 0.0 | 0.1 | GO:0032009 | early phagosome(GO:0032009) |
| 0.0 | 0.5 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 0.3 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 1.0 | GO:0016529 | sarcoplasmic reticulum(GO:0016529) |
| 0.0 | 0.6 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 0.5 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 0.2 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.0 | 0.7 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.0 | 0.2 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 0.7 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.3 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.9 | 8.6 | GO:0004119 | cGMP-inhibited cyclic-nucleotide phosphodiesterase activity(GO:0004119) |
| 1.6 | 9.6 | GO:0004359 | glutaminase activity(GO:0004359) |
| 1.5 | 24.1 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 1.2 | 3.6 | GO:0051717 | inositol-1,3,4,5-tetrakisphosphate 3-phosphatase activity(GO:0051717) |
| 1.1 | 14.4 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 1.0 | 8.8 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.9 | 3.5 | GO:0016262 | protein N-acetylglucosaminyltransferase activity(GO:0016262) |
| 0.8 | 3.9 | GO:0032810 | sterol response element binding(GO:0032810) |
| 0.6 | 2.6 | GO:0030343 | vitamin D3 25-hydroxylase activity(GO:0030343) vitamin D 25-hydroxylase activity(GO:0070643) |
| 0.6 | 1.8 | GO:1904928 | coreceptor activity involved in canonical Wnt signaling pathway(GO:1904928) |
| 0.6 | 12.0 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.5 | 3.2 | GO:0044729 | hemi-methylated DNA-binding(GO:0044729) |
| 0.5 | 4.0 | GO:0015923 | mannosidase activity(GO:0015923) |
| 0.5 | 6.0 | GO:0005114 | type II transforming growth factor beta receptor binding(GO:0005114) |
| 0.5 | 2.4 | GO:0005119 | smoothened binding(GO:0005119) hedgehog family protein binding(GO:0097108) |
| 0.5 | 4.4 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.5 | 0.5 | GO:0034189 | very-low-density lipoprotein particle binding(GO:0034189) |
| 0.5 | 2.8 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.5 | 1.9 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.4 | 2.9 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 0.4 | 1.1 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.4 | 4.5 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.4 | 1.4 | GO:0032089 | NACHT domain binding(GO:0032089) |
| 0.4 | 3.2 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.3 | 3.4 | GO:0001162 | RNA polymerase II intronic transcription regulatory region sequence-specific DNA binding(GO:0001162) |
| 0.3 | 1.4 | GO:0003943 | N-acetylgalactosamine-4-sulfatase activity(GO:0003943) |
| 0.3 | 2.0 | GO:0016019 | peptidoglycan receptor activity(GO:0016019) |
| 0.3 | 1.7 | GO:0038025 | reelin receptor activity(GO:0038025) |
| 0.3 | 2.0 | GO:0051033 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.3 | 1.6 | GO:0016454 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.3 | 2.8 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.3 | 11.6 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.3 | 0.9 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.3 | 7.7 | GO:0005112 | Notch binding(GO:0005112) |
| 0.3 | 0.9 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.3 | 6.8 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.3 | 1.9 | GO:0001165 | RNA polymerase I upstream control element sequence-specific DNA binding(GO:0001165) |
| 0.3 | 4.3 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.3 | 3.2 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.3 | 2.1 | GO:0008269 | JAK pathway signal transduction adaptor activity(GO:0008269) |
| 0.3 | 11.6 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.2 | 2.5 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.2 | 4.7 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.2 | 1.5 | GO:0010465 | nerve growth factor receptor activity(GO:0010465) |
| 0.2 | 1.9 | GO:0016406 | carnitine O-acyltransferase activity(GO:0016406) |
| 0.2 | 4.1 | GO:0005072 | transforming growth factor beta receptor, cytoplasmic mediator activity(GO:0005072) |
| 0.2 | 0.7 | GO:0004566 | beta-glucuronidase activity(GO:0004566) heparanase activity(GO:0030305) |
| 0.2 | 1.8 | GO:0098639 | collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.2 | 0.7 | GO:0008511 | sodium:potassium:chloride symporter activity(GO:0008511) |
| 0.2 | 3.5 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.2 | 2.1 | GO:0034485 | phosphatidylinositol-3,4,5-trisphosphate 5-phosphatase activity(GO:0034485) |
| 0.2 | 0.6 | GO:0001226 | RNA polymerase II transcription corepressor binding(GO:0001226) |
| 0.2 | 1.5 | GO:0048408 | epidermal growth factor binding(GO:0048408) |
| 0.2 | 0.6 | GO:0004980 | melanocyte-stimulating hormone receptor activity(GO:0004980) |
| 0.2 | 1.6 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.2 | 1.0 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) |
| 0.2 | 1.8 | GO:0043426 | MRF binding(GO:0043426) |
| 0.2 | 6.1 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.2 | 1.2 | GO:0004489 | methylenetetrahydrofolate reductase (NAD(P)H) activity(GO:0004489) |
| 0.2 | 0.8 | GO:0008798 | beta-aspartyl-peptidase activity(GO:0008798) |
| 0.2 | 1.1 | GO:0010521 | telomerase inhibitor activity(GO:0010521) |
| 0.2 | 0.9 | GO:0004666 | prostaglandin-endoperoxide synthase activity(GO:0004666) |
| 0.2 | 1.4 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.2 | 2.1 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.2 | 1.2 | GO:0031685 | adenosine receptor binding(GO:0031685) |
| 0.2 | 13.1 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.2 | 0.5 | GO:0005011 | macrophage colony-stimulating factor receptor activity(GO:0005011) |
| 0.2 | 5.8 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.2 | 0.5 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.2 | 0.8 | GO:0086062 | voltage-gated sodium channel activity involved in Purkinje myocyte action potential(GO:0086062) |
| 0.2 | 1.6 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.2 | 0.6 | GO:0042132 | fructose 1,6-bisphosphate 1-phosphatase activity(GO:0042132) |
| 0.2 | 0.5 | GO:0030549 | acetylcholine receptor activator activity(GO:0030549) |
| 0.2 | 4.0 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.2 | 1.1 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 0.2 | 0.6 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.2 | 3.1 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.2 | 0.6 | GO:0004677 | DNA-dependent protein kinase activity(GO:0004677) |
| 0.1 | 1.3 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.1 | 3.9 | GO:0098811 | transcriptional activator activity, RNA polymerase II transcription factor binding(GO:0001190) transcriptional repressor activity, RNA polymerase II activating transcription factor binding(GO:0098811) |
| 0.1 | 2.4 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.1 | 0.8 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.1 | 1.6 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.1 | 0.4 | GO:0003845 | 11-beta-hydroxysteroid dehydrogenase [NAD(P)] activity(GO:0003845) |
| 0.1 | 1.6 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.1 | 0.6 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.1 | 1.8 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.1 | 1.6 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.1 | 1.3 | GO:0035877 | death effector domain binding(GO:0035877) caspase binding(GO:0089720) |
| 0.1 | 0.1 | GO:0035650 | AP-1 adaptor complex binding(GO:0035650) |
| 0.1 | 0.5 | GO:0047493 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.1 | 3.9 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.1 | 1.4 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.1 | 0.7 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.1 | 4.0 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.1 | 0.9 | GO:0031811 | G-protein coupled nucleotide receptor binding(GO:0031811) P2Y1 nucleotide receptor binding(GO:0031812) |
| 0.1 | 1.2 | GO:0034618 | arginine binding(GO:0034618) |
| 0.1 | 0.9 | GO:0035368 | selenocysteine insertion sequence binding(GO:0035368) |
| 0.1 | 0.4 | GO:0050253 | retinyl-palmitate esterase activity(GO:0050253) |
| 0.1 | 0.5 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.1 | 0.5 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.1 | 0.5 | GO:0016728 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.1 | 1.4 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.1 | 1.6 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.1 | 0.6 | GO:0045029 | UDP-activated nucleotide receptor activity(GO:0045029) |
| 0.1 | 0.2 | GO:1990763 | arrestin family protein binding(GO:1990763) |
| 0.1 | 6.1 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.1 | 0.5 | GO:0016532 | superoxide dismutase copper chaperone activity(GO:0016532) |
| 0.1 | 3.0 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.1 | 1.7 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 0.1 | 2.4 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.1 | 1.3 | GO:0003857 | 3-hydroxyacyl-CoA dehydrogenase activity(GO:0003857) |
| 0.1 | 0.6 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.1 | 1.0 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.1 | 2.6 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.1 | 0.9 | GO:0032395 | MHC class II receptor activity(GO:0032395) |
| 0.1 | 0.2 | GO:0070576 | vitamin D 24-hydroxylase activity(GO:0070576) |
| 0.1 | 3.3 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.1 | 2.8 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.1 | 2.6 | GO:0004407 | histone deacetylase activity(GO:0004407) |
| 0.1 | 8.6 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.1 | 2.9 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.1 | 0.7 | GO:0001537 | N-acetylgalactosamine 4-O-sulfotransferase activity(GO:0001537) |
| 0.1 | 0.6 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.1 | 0.9 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.1 | 0.6 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.1 | 2.6 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.1 | 0.5 | GO:0031994 | insulin-like growth factor I binding(GO:0031994) |
| 0.1 | 8.5 | GO:0035326 | enhancer binding(GO:0035326) |
| 0.1 | 1.0 | GO:0016273 | arginine N-methyltransferase activity(GO:0016273) protein-arginine N-methyltransferase activity(GO:0016274) |
| 0.1 | 1.6 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.1 | 0.4 | GO:0016167 | glial cell-derived neurotrophic factor receptor activity(GO:0016167) |
| 0.1 | 0.4 | GO:0005353 | fructose transmembrane transporter activity(GO:0005353) |
| 0.1 | 2.7 | GO:0032452 | histone demethylase activity(GO:0032452) |
| 0.1 | 1.0 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.1 | 0.5 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.1 | 2.2 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.1 | 1.1 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.1 | 0.7 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.1 | 0.4 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.1 | 0.8 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.1 | 1.6 | GO:0070001 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.1 | 0.5 | GO:0000213 | tRNA-intron endonuclease activity(GO:0000213) |
| 0.1 | 2.4 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.1 | 0.3 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 0.0 | 4.1 | GO:0004536 | deoxyribonuclease activity(GO:0004536) |
| 0.0 | 2.1 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.2 | GO:0005113 | patched binding(GO:0005113) |
| 0.0 | 1.0 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.5 | GO:0043425 | bHLH transcription factor binding(GO:0043425) |
| 0.0 | 0.2 | GO:0015154 | sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.0 | 0.2 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.0 | 0.2 | GO:0050119 | N-acetylglucosamine deacetylase activity(GO:0050119) |
| 0.0 | 0.4 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.1 | GO:0044715 | 8-oxo-dGDP phosphatase activity(GO:0044715) |
| 0.0 | 0.5 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.0 | 0.5 | GO:0043996 | histone acetyltransferase activity (H4-K5 specific)(GO:0043995) histone acetyltransferase activity (H4-K8 specific)(GO:0043996) histone acetyltransferase activity (H4-K16 specific)(GO:0046972) |
| 0.0 | 0.2 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.0 | 0.5 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.0 | 0.5 | GO:0008430 | selenium binding(GO:0008430) |
| 0.0 | 2.1 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 0.3 | GO:0086083 | cell adhesive protein binding involved in bundle of His cell-Purkinje myocyte communication(GO:0086083) |
| 0.0 | 0.3 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.0 | 0.8 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 0.3 | GO:0097603 | temperature-gated ion channel activity(GO:0097603) |
| 0.0 | 0.3 | GO:0099583 | neurotransmitter receptor activity involved in regulation of postsynaptic cytosolic calcium ion concentration(GO:0099583) |
| 0.0 | 0.9 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.0 | 0.6 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.0 | 0.3 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.0 | 0.2 | GO:0000217 | DNA secondary structure binding(GO:0000217) |
| 0.0 | 0.4 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.0 | 0.3 | GO:0016681 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors(GO:0016679) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.0 | 1.2 | GO:0042288 | MHC class I protein binding(GO:0042288) |
| 0.0 | 0.2 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.0 | 0.6 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.0 | 0.8 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.0 | 0.5 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.0 | 1.1 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.0 | 0.7 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.0 | 0.8 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.0 | 1.0 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 0.1 | GO:0045322 | unmethylated CpG binding(GO:0045322) |
| 0.0 | 0.2 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.0 | 0.7 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 1.8 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
| 0.0 | 0.1 | GO:0034353 | RNA pyrophosphohydrolase activity(GO:0034353) |
| 0.0 | 0.9 | GO:0019865 | immunoglobulin binding(GO:0019865) |
| 0.0 | 0.4 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 0.8 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 1.2 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 3.2 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.0 | 0.1 | GO:0000104 | succinate dehydrogenase activity(GO:0000104) |
| 0.0 | 1.2 | GO:0051539 | 4 iron, 4 sulfur cluster binding(GO:0051539) |
| 0.0 | 0.2 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.0 | 0.3 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.0 | 0.6 | GO:0000339 | RNA cap binding(GO:0000339) |
| 0.0 | 0.4 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 0.5 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.0 | 0.3 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.0 | 1.4 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 1.3 | GO:0008146 | sulfotransferase activity(GO:0008146) |
| 0.0 | 0.8 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 1.6 | GO:0016655 | oxidoreductase activity, acting on NAD(P)H, quinone or similar compound as acceptor(GO:0016655) |
| 0.0 | 0.4 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.0 | 0.4 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 2.6 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 0.3 | GO:0031404 | chloride ion binding(GO:0031404) |
| 0.0 | 0.6 | GO:0004180 | carboxypeptidase activity(GO:0004180) |
| 0.0 | 0.4 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.0 | 9.6 | GO:0003682 | chromatin binding(GO:0003682) |
| 0.0 | 1.2 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 0.5 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 1.0 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
| 0.0 | 0.5 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 1.1 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.0 | 1.8 | GO:0004553 | hydrolase activity, hydrolyzing O-glycosyl compounds(GO:0004553) |
| 0.0 | 0.1 | GO:0004704 | NF-kappaB-inducing kinase activity(GO:0004704) |
| 0.0 | 0.1 | GO:0030346 | protein phosphatase 2B binding(GO:0030346) |
| 0.0 | 0.6 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.5 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.0 | 0.2 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.0 | 0.2 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.0 | 0.1 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.0 | 0.2 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.0 | 6.9 | GO:0046982 | protein heterodimerization activity(GO:0046982) |
| 0.0 | 0.0 | GO:0035402 | histone kinase activity (H3-T11 specific)(GO:0035402) |
| 0.0 | 0.9 | GO:0050681 | androgen receptor binding(GO:0050681) |
| 0.0 | 0.3 | GO:0015114 | phosphate ion transmembrane transporter activity(GO:0015114) |
| 0.0 | 3.9 | GO:0001077 | transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) |
| 0.0 | 1.0 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 0.1 | GO:0043008 | U4 snRNA binding(GO:0030621) ATP-dependent protein binding(GO:0043008) |
| 0.0 | 0.1 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.0 | 0.1 | GO:0016812 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amides(GO:0016812) |
| 0.0 | 0.0 | GO:0052871 | tocopherol omega-hydroxylase activity(GO:0052870) alpha-tocopherol omega-hydroxylase activity(GO:0052871) 20-hydroxy-leukotriene B4 omega oxidase activity(GO:0097258) 20-aldehyde-leukotriene B4 20-monooxygenase activity(GO:0097259) |
| 0.0 | 0.2 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 2.2 | GO:0003713 | transcription coactivator activity(GO:0003713) |
| 0.0 | 0.4 | GO:0048487 | beta-tubulin binding(GO:0048487) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 20.6 | PID BMP PATHWAY | BMP receptor signaling |
| 0.2 | 14.2 | ST DIFFERENTIATION PATHWAY IN PC12 CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
| 0.2 | 2.7 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.1 | 11.5 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.1 | 2.5 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.1 | 12.2 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.1 | 1.2 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.1 | 2.2 | PID LYMPH ANGIOGENESIS PATHWAY | VEGFR3 signaling in lymphatic endothelium |
| 0.1 | 12.4 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.1 | 2.4 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.1 | 8.0 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.1 | 5.7 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.1 | 3.7 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.1 | 3.6 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.1 | 0.8 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.1 | 2.5 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.1 | 5.0 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.1 | 4.3 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.1 | 2.9 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.1 | 2.6 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.1 | 2.1 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.1 | 4.5 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.1 | 2.5 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.1 | 1.2 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.1 | 0.8 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.1 | 1.4 | PID IL2 1PATHWAY | IL2-mediated signaling events |
| 0.1 | 1.8 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 4.1 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 0.7 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.0 | 1.4 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 1.8 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 0.3 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.0 | 1.3 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 1.3 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 1.6 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 2.7 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.0 | 1.4 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 0.2 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 0.6 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.0 | 0.6 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.0 | 0.2 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 0.5 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 10.1 | REACTOME SIGNALING BY NOTCH4 | Genes involved in Signaling by NOTCH4 |
| 0.3 | 9.6 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.2 | 1.9 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.2 | 3.2 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.2 | 11.9 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.1 | 1.8 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.1 | 5.8 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.1 | 0.4 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.1 | 2.1 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.1 | 1.5 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.1 | 4.4 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.1 | 3.9 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.1 | 4.0 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.1 | 2.8 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.1 | 13.6 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.1 | 4.0 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.1 | 1.6 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.1 | 4.1 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.1 | 2.2 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.1 | 18.9 | REACTOME G ALPHA S SIGNALLING EVENTS | Genes involved in G alpha (s) signalling events |
| 0.1 | 2.8 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.1 | 1.6 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.1 | 5.0 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.1 | 0.4 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.1 | 1.9 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.1 | 1.6 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.1 | 1.2 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.1 | 1.2 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS | Genes involved in Synthesis of bile acids and bile salts |
| 0.1 | 1.1 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.1 | 1.6 | REACTOME GLUCAGON SIGNALING IN METABOLIC REGULATION | Genes involved in Glucagon signaling in metabolic regulation |
| 0.1 | 1.2 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.1 | 1.9 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.1 | 2.5 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.1 | 1.2 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.1 | 0.1 | REACTOME SHC MEDIATED SIGNALLING | Genes involved in SHC-mediated signalling |
| 0.1 | 1.3 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.1 | 3.1 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.1 | 3.4 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.1 | 2.1 | REACTOME GROWTH HORMONE RECEPTOR SIGNALING | Genes involved in Growth hormone receptor signaling |
| 0.1 | 1.3 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.1 | 4.1 | REACTOME SIGNALING BY NOTCH1 | Genes involved in Signaling by NOTCH1 |
| 0.1 | 1.0 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 1.1 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 0.2 | REACTOME NUCLEAR SIGNALING BY ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.0 | 1.4 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 1.7 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.0 | 0.7 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.0 | 0.7 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 3.9 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.6 | REACTOME GRB2 SOS PROVIDES LINKAGE TO MAPK SIGNALING FOR INTERGRINS | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |
| 0.0 | 0.9 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 1.7 | REACTOME GLOBAL GENOMIC NER GG NER | Genes involved in Global Genomic NER (GG-NER) |
| 0.0 | 0.9 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.0 | 0.6 | REACTOME SIGNALLING TO P38 VIA RIT AND RIN | Genes involved in Signalling to p38 via RIT and RIN |
| 0.0 | 2.7 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 0.6 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 2.2 | REACTOME PHASE1 FUNCTIONALIZATION OF COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |
| 0.0 | 0.5 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 1.0 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 1.0 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.0 | 0.8 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 0.2 | REACTOME PEPTIDE HORMONE BIOSYNTHESIS | Genes involved in Peptide hormone biosynthesis |
| 0.0 | 1.7 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.7 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.0 | 0.7 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 1.8 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.7 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.8 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.0 | 0.4 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 1.2 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 0.4 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.0 | 0.3 | REACTOME REGULATION OF INSULIN SECRETION BY GLUCAGON LIKE PEPTIDE1 | Genes involved in Regulation of Insulin Secretion by Glucagon-like Peptide-1 |
| 0.0 | 2.5 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.6 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.0 | 1.0 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.3 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.0 | 0.4 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.0 | 0.6 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 0.4 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.0 | 0.3 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 0.2 | REACTOME FORMATION OF RNA POL II ELONGATION COMPLEX | Genes involved in Formation of RNA Pol II elongation complex |
| 0.0 | 0.1 | REACTOME RIP MEDIATED NFKB ACTIVATION VIA DAI | Genes involved in RIP-mediated NFkB activation via DAI |
| 0.0 | 0.2 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 0.5 | REACTOME REGULATION OF GLUCOKINASE BY GLUCOKINASE REGULATORY PROTEIN | Genes involved in Regulation of Glucokinase by Glucokinase Regulatory Protein |
| 0.0 | 1.1 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 1.7 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 0.1 | REACTOME SEMA3A PAK DEPENDENT AXON REPULSION | Genes involved in Sema3A PAK dependent Axon repulsion |