Project
Inflammatory response time course, HUVEC (Wada, 2009)
Navigation
Downloads
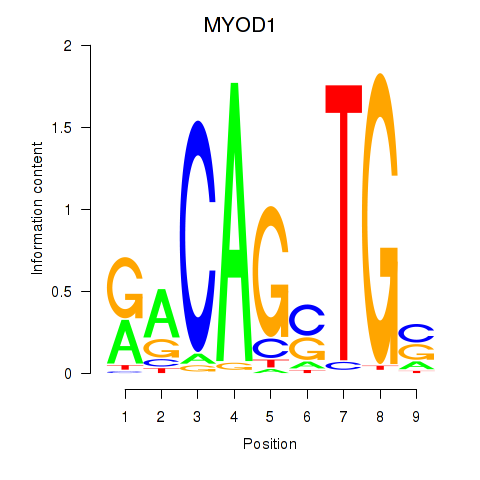
Results for MYOD1
Z-value: 0.64
Transcription factors associated with MYOD1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
MYOD1
|
ENSG00000129152.4 | MYOD1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| MYOD1 | hg38_v1_chr11_+_17719564_17719577 | 0.16 | 4.4e-01 | Click! |
Activity profile of MYOD1 motif
Sorted Z-values of MYOD1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of MYOD1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_+_70169516 | 1.22 |
ENST00000243457.4
|
KCNJ2
|
potassium inwardly rectifying channel subfamily J member 2 |
| chr1_+_25616780 | 1.19 |
ENST00000374332.9
|
MAN1C1
|
mannosidase alpha class 1C member 1 |
| chr14_-_22815421 | 1.10 |
ENST00000674313.1
ENST00000555959.1 |
SLC7A7
|
solute carrier family 7 member 7 |
| chr3_-_64268161 | 1.05 |
ENST00000564377.6
|
PRICKLE2
|
prickle planar cell polarity protein 2 |
| chr1_-_32702736 | 1.03 |
ENST00000373484.4
ENST00000409190.8 |
SYNC
|
syncoilin, intermediate filament protein |
| chr2_-_174764407 | 0.94 |
ENST00000409219.5
ENST00000409542.5 |
CHRNA1
|
cholinergic receptor nicotinic alpha 1 subunit |
| chr17_+_41226648 | 0.94 |
ENST00000377721.3
|
KRTAP9-2
|
keratin associated protein 9-2 |
| chr4_+_155758990 | 0.91 |
ENST00000505154.5
ENST00000652626.1 ENST00000502959.5 ENST00000264424.13 ENST00000505764.5 ENST00000507146.5 ENST00000503520.5 |
GUCY1B1
|
guanylate cyclase 1 soluble subunit beta 1 |
| chr4_+_155759365 | 0.88 |
ENST00000513437.1
|
GUCY1B1
|
guanylate cyclase 1 soluble subunit beta 1 |
| chr6_+_30884063 | 0.85 |
ENST00000511510.5
ENST00000376569.7 ENST00000376570.8 ENST00000504927.5 |
DDR1
|
discoidin domain receptor tyrosine kinase 1 |
| chr22_-_37486357 | 0.82 |
ENST00000356998.8
ENST00000416983.7 ENST00000424765.2 |
MFNG
|
MFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase |
| chr7_+_155298561 | 0.80 |
ENST00000476756.1
|
INSIG1
|
insulin induced gene 1 |
| chr6_+_30884353 | 0.79 |
ENST00000428153.6
ENST00000376568.8 ENST00000452441.5 ENST00000515219.5 |
DDR1
|
discoidin domain receptor tyrosine kinase 1 |
| chr17_+_4498866 | 0.77 |
ENST00000329078.8
|
SPNS2
|
sphingolipid transporter 2 |
| chr16_-_46831043 | 0.75 |
ENST00000565112.1
|
C16orf87
|
chromosome 16 open reading frame 87 |
| chr2_-_219571241 | 0.73 |
ENST00000373876.5
ENST00000603926.5 ENST00000373873.8 ENST00000289656.3 |
OBSL1
|
obscurin like cytoskeletal adaptor 1 |
| chr11_-_33869816 | 0.72 |
ENST00000395833.7
|
LMO2
|
LIM domain only 2 |
| chrX_+_9912434 | 0.68 |
ENST00000418909.6
|
SHROOM2
|
shroom family member 2 |
| chr12_+_93572664 | 0.68 |
ENST00000551556.2
|
SOCS2
|
suppressor of cytokine signaling 2 |
| chr15_-_70702273 | 0.67 |
ENST00000558758.5
ENST00000379983.6 ENST00000560441.5 |
UACA
|
uveal autoantigen with coiled-coil domains and ankyrin repeats |
| chr6_-_75493629 | 0.66 |
ENST00000393004.6
|
FILIP1
|
filamin A interacting protein 1 |
| chr19_-_18940289 | 0.66 |
ENST00000542541.6
ENST00000433218.6 |
HOMER3
|
homer scaffold protein 3 |
| chr7_-_120857124 | 0.65 |
ENST00000441017.5
ENST00000424710.5 ENST00000433758.5 |
TSPAN12
|
tetraspanin 12 |
| chr6_-_42048648 | 0.65 |
ENST00000502771.1
ENST00000508143.5 ENST00000514588.1 ENST00000510503.5 |
CCND3
|
cyclin D3 |
| chr1_+_89524871 | 0.63 |
ENST00000639264.1
|
LRRC8B
|
leucine rich repeat containing 8 VRAC subunit B |
| chr2_-_207624551 | 0.63 |
ENST00000272839.7
ENST00000426075.5 |
METTL21A
|
methyltransferase like 21A |
| chr1_-_54887161 | 0.62 |
ENST00000535035.6
ENST00000371269.9 ENST00000436604.2 |
DHCR24
|
24-dehydrocholesterol reductase |
| chr5_+_69492767 | 0.61 |
ENST00000681041.1
ENST00000680098.1 ENST00000680784.1 ENST00000396442.7 ENST00000681895.1 |
OCLN
|
occludin |
| chr1_+_89524819 | 0.60 |
ENST00000439853.6
ENST00000330947.7 ENST00000449440.5 ENST00000640258.1 |
LRRC8B
|
leucine rich repeat containing 8 VRAC subunit B |
| chr11_-_2149603 | 0.59 |
ENST00000643349.1
|
ENSG00000284779.2
|
novel protein |
| chr3_-_66038537 | 0.59 |
ENST00000483466.5
|
MAGI1
|
membrane associated guanylate kinase, WW and PDZ domain containing 1 |
| chr12_-_56957587 | 0.58 |
ENST00000398138.5
|
RDH16
|
retinol dehydrogenase 16 |
| chr6_+_30880780 | 0.57 |
ENST00000460944.6
ENST00000324771.12 |
DDR1
|
discoidin domain receptor tyrosine kinase 1 |
| chr8_+_119208322 | 0.57 |
ENST00000614891.5
|
MAL2
|
mal, T cell differentiation protein 2 |
| chr7_-_11832190 | 0.56 |
ENST00000423059.9
ENST00000617773.1 |
THSD7A
|
thrombospondin type 1 domain containing 7A |
| chrX_+_151694967 | 0.56 |
ENST00000448726.5
ENST00000538575.5 |
PRRG3
|
proline rich and Gla domain 3 |
| chr1_+_61404076 | 0.55 |
ENST00000357977.5
|
NFIA
|
nuclear factor I A |
| chr9_-_16728165 | 0.55 |
ENST00000603713.5
ENST00000603313.5 |
BNC2
|
basonuclin 2 |
| chr3_+_9933805 | 0.54 |
ENST00000684493.1
ENST00000673935.2 ENST00000684181.1 ENST00000683189.1 ENST00000383811.8 ENST00000452070.6 ENST00000682642.1 ENST00000684659.1 ENST00000491527.2 ENST00000326434.9 ENST00000682783.1 ENST00000683835.1 ENST00000682570.1 |
CRELD1
|
cysteine rich with EGF like domains 1 |
| chr2_-_174764436 | 0.54 |
ENST00000409323.1
ENST00000261007.9 ENST00000348749.9 ENST00000672640.1 |
CHRNA1
|
cholinergic receptor nicotinic alpha 1 subunit |
| chrX_-_63785510 | 0.54 |
ENST00000437457.6
ENST00000374878.5 ENST00000623517.3 |
ARHGEF9
|
Cdc42 guanine nucleotide exchange factor 9 |
| chr6_+_17281341 | 0.54 |
ENST00000379052.10
|
RBM24
|
RNA binding motif protein 24 |
| chr6_-_75493773 | 0.54 |
ENST00000237172.12
|
FILIP1
|
filamin A interacting protein 1 |
| chr2_-_177264686 | 0.53 |
ENST00000397062.8
ENST00000430047.1 |
NFE2L2
|
nuclear factor, erythroid 2 like 2 |
| chr17_+_28042839 | 0.53 |
ENST00000582037.2
|
NLK
|
nemo like kinase |
| chr5_+_66828762 | 0.52 |
ENST00000490016.6
ENST00000403666.5 ENST00000450827.5 |
MAST4
|
microtubule associated serine/threonine kinase family member 4 |
| chr11_+_44726811 | 0.51 |
ENST00000533202.5
ENST00000520358.7 ENST00000533080.5 ENST00000520999.6 |
TSPAN18
|
tetraspanin 18 |
| chr21_-_34526850 | 0.50 |
ENST00000481448.5
ENST00000381132.6 |
RCAN1
|
regulator of calcineurin 1 |
| chr12_+_78864768 | 0.49 |
ENST00000261205.9
ENST00000457153.6 |
SYT1
|
synaptotagmin 1 |
| chr1_-_154956086 | 0.49 |
ENST00000368463.8
ENST00000368460.7 ENST00000368465.5 |
PBXIP1
|
PBX homeobox interacting protein 1 |
| chr3_+_32106612 | 0.49 |
ENST00000282541.10
ENST00000425459.5 ENST00000431009.1 |
GPD1L
|
glycerol-3-phosphate dehydrogenase 1 like |
| chr17_+_41237998 | 0.48 |
ENST00000254072.7
|
KRTAP9-8
|
keratin associated protein 9-8 |
| chr5_+_56815534 | 0.48 |
ENST00000399503.4
|
MAP3K1
|
mitogen-activated protein kinase kinase kinase 1 |
| chr4_+_128811311 | 0.47 |
ENST00000413543.6
|
JADE1
|
jade family PHD finger 1 |
| chr14_+_103121457 | 0.47 |
ENST00000333007.8
|
TNFAIP2
|
TNF alpha induced protein 2 |
| chr3_+_52794768 | 0.46 |
ENST00000621946.4
ENST00000416872.6 ENST00000449956.2 |
ITIH3
|
inter-alpha-trypsin inhibitor heavy chain 3 |
| chr7_-_132576493 | 0.46 |
ENST00000321063.8
|
PLXNA4
|
plexin A4 |
| chr9_-_76692181 | 0.46 |
ENST00000376717.6
ENST00000223609.10 |
PRUNE2
|
prune homolog 2 with BCH domain |
| chr19_-_49155130 | 0.45 |
ENST00000595625.1
|
HRC
|
histidine rich calcium binding protein |
| chrX_-_136780925 | 0.44 |
ENST00000250617.7
|
ARHGEF6
|
Rac/Cdc42 guanine nucleotide exchange factor 6 |
| chr19_-_49155384 | 0.44 |
ENST00000252825.9
|
HRC
|
histidine rich calcium binding protein |
| chr2_+_232697362 | 0.44 |
ENST00000482666.5
ENST00000483164.5 ENST00000490229.5 ENST00000464805.5 ENST00000489328.1 |
GIGYF2
|
GRB10 interacting GYF protein 2 |
| chr8_+_80486209 | 0.43 |
ENST00000426744.5
ENST00000455036.8 |
ZBTB10
|
zinc finger and BTB domain containing 10 |
| chr16_+_30395400 | 0.42 |
ENST00000320159.2
ENST00000613509.2 |
ZNF48
|
zinc finger protein 48 |
| chr1_+_77888490 | 0.42 |
ENST00000401035.7
ENST00000330010.12 |
NEXN
|
nexilin F-actin binding protein |
| chr21_-_34526815 | 0.41 |
ENST00000492600.1
|
RCAN1
|
regulator of calcineurin 1 |
| chr7_+_140404034 | 0.41 |
ENST00000537763.6
|
RAB19
|
RAB19, member RAS oncogene family |
| chr17_-_8630749 | 0.41 |
ENST00000379980.8
ENST00000269243.8 |
MYH10
|
myosin heavy chain 10 |
| chr12_+_93574965 | 0.40 |
ENST00000551883.1
ENST00000549510.1 |
SOCS2
|
suppressor of cytokine signaling 2 |
| chr22_+_40679273 | 0.40 |
ENST00000381433.2
|
MCHR1
|
melanin concentrating hormone receptor 1 |
| chr3_+_52420955 | 0.39 |
ENST00000465863.1
|
PHF7
|
PHD finger protein 7 |
| chr17_-_8630713 | 0.39 |
ENST00000411957.1
ENST00000360416.8 |
MYH10
|
myosin heavy chain 10 |
| chr12_-_108827384 | 0.39 |
ENST00000326470.9
|
SSH1
|
slingshot protein phosphatase 1 |
| chr19_-_18280806 | 0.38 |
ENST00000600972.1
|
JUND
|
JunD proto-oncogene, AP-1 transcription factor subunit |
| chr21_+_44573724 | 0.38 |
ENST00000622352.3
ENST00000400374.4 ENST00000616689.2 |
KRTAP10-4
|
keratin associated protein 10-4 |
| chr16_-_31202733 | 0.38 |
ENST00000350605.4
ENST00000247470.10 |
PYCARD
|
PYD and CARD domain containing |
| chr2_-_182522703 | 0.37 |
ENST00000410103.1
|
PDE1A
|
phosphodiesterase 1A |
| chr9_+_89605004 | 0.37 |
ENST00000252506.11
ENST00000375769.1 |
GADD45G
|
growth arrest and DNA damage inducible gamma |
| chr2_-_196176467 | 0.37 |
ENST00000409228.5
|
STK17B
|
serine/threonine kinase 17b |
| chr17_-_47957824 | 0.37 |
ENST00000300557.3
|
PRR15L
|
proline rich 15 like |
| chr13_-_98977975 | 0.35 |
ENST00000376460.5
|
DOCK9
|
dedicator of cytokinesis 9 |
| chr8_+_80485641 | 0.35 |
ENST00000430430.5
|
ZBTB10
|
zinc finger and BTB domain containing 10 |
| chr17_+_49219503 | 0.35 |
ENST00000573347.5
|
ABI3
|
ABI family member 3 |
| chr1_-_16978276 | 0.34 |
ENST00000375534.7
|
MFAP2
|
microfibril associated protein 2 |
| chr11_+_62789124 | 0.34 |
ENST00000526546.1
|
TMEM179B
|
transmembrane protein 179B |
| chr12_-_12350222 | 0.34 |
ENST00000543314.1
ENST00000396349.3 ENST00000535902.6 |
MANSC1
|
MANSC domain containing 1 |
| chr2_+_176188658 | 0.34 |
ENST00000331462.6
|
HOXD1
|
homeobox D1 |
| chr1_+_77888612 | 0.33 |
ENST00000334785.12
|
NEXN
|
nexilin F-actin binding protein |
| chr10_+_123135938 | 0.33 |
ENST00000357878.7
|
HMX3
|
H6 family homeobox 3 |
| chr2_-_156342348 | 0.33 |
ENST00000409572.5
|
NR4A2
|
nuclear receptor subfamily 4 group A member 2 |
| chr7_-_120858303 | 0.33 |
ENST00000415871.5
ENST00000430985.1 |
TSPAN12
|
tetraspanin 12 |
| chr20_+_36091409 | 0.33 |
ENST00000202028.9
|
EPB41L1
|
erythrocyte membrane protein band 4.1 like 1 |
| chr19_+_35248879 | 0.32 |
ENST00000347609.8
|
LSR
|
lipolysis stimulated lipoprotein receptor |
| chr9_-_13165442 | 0.32 |
ENST00000542239.1
ENST00000538841.5 ENST00000433359.6 |
MPDZ
|
multiple PDZ domain crumbs cell polarity complex component |
| chr14_-_106511856 | 0.32 |
ENST00000390622.2
|
IGHV1-46
|
immunoglobulin heavy variable 1-46 |
| chr2_+_190649062 | 0.32 |
ENST00000409581.5
ENST00000337386.10 |
NAB1
|
NGFI-A binding protein 1 |
| chr17_+_60677822 | 0.32 |
ENST00000407086.8
ENST00000589222.5 ENST00000626960.2 ENST00000390652.9 |
BCAS3
|
BCAS3 microtubule associated cell migration factor |
| chr15_+_32641665 | 0.32 |
ENST00000300175.8
ENST00000413748.6 ENST00000494364.5 ENST00000497208.5 |
SCG5
|
secretogranin V |
| chr14_+_75279637 | 0.31 |
ENST00000555686.1
ENST00000555672.1 |
FOS
|
Fos proto-oncogene, AP-1 transcription factor subunit |
| chr3_+_141324208 | 0.31 |
ENST00000509842.5
|
ZBTB38
|
zinc finger and BTB domain containing 38 |
| chr17_+_32927910 | 0.31 |
ENST00000394642.7
|
TMEM98
|
transmembrane protein 98 |
| chr1_+_37474572 | 0.30 |
ENST00000373087.7
|
ZC3H12A
|
zinc finger CCCH-type containing 12A |
| chr19_-_39335999 | 0.30 |
ENST00000602185.5
ENST00000598034.5 ENST00000601387.5 ENST00000595636.1 ENST00000253054.12 ENST00000594700.5 ENST00000597595.6 |
GMFG
|
glia maturation factor gamma |
| chrX_+_150983350 | 0.30 |
ENST00000455596.5
ENST00000448905.6 |
HMGB3
|
high mobility group box 3 |
| chr19_+_55485176 | 0.29 |
ENST00000205194.5
ENST00000591590.1 ENST00000587400.1 |
NAT14
|
N-acetyltransferase 14 (putative) |
| chr2_+_172735838 | 0.29 |
ENST00000397081.8
|
RAPGEF4
|
Rap guanine nucleotide exchange factor 4 |
| chr1_+_162381703 | 0.29 |
ENST00000458626.4
|
C1orf226
|
chromosome 1 open reading frame 226 |
| chr17_+_32928126 | 0.29 |
ENST00000579849.6
ENST00000578289.5 ENST00000439138.5 |
TMEM98
|
transmembrane protein 98 |
| chr3_-_49813880 | 0.29 |
ENST00000333486.4
|
UBA7
|
ubiquitin like modifier activating enzyme 7 |
| chr2_+_172735912 | 0.29 |
ENST00000409036.5
|
RAPGEF4
|
Rap guanine nucleotide exchange factor 4 |
| chr21_+_44697427 | 0.29 |
ENST00000618832.1
|
KRTAP10-12
|
keratin associated protein 10-12 |
| chr13_-_98978022 | 0.29 |
ENST00000682017.1
ENST00000442173.5 ENST00000627024.2 |
DOCK9
|
dedicator of cytokinesis 9 |
| chr17_-_7262343 | 0.28 |
ENST00000571881.2
ENST00000360325.11 |
CLDN7
|
claudin 7 |
| chr4_-_177442427 | 0.28 |
ENST00000264595.7
|
AGA
|
aspartylglucosaminidase |
| chr1_-_12616762 | 0.28 |
ENST00000464917.5
|
DHRS3
|
dehydrogenase/reductase 3 |
| chr22_-_42720813 | 0.28 |
ENST00000381278.4
|
A4GALT
|
alpha 1,4-galactosyltransferase (P blood group) |
| chrX_-_63785149 | 0.28 |
ENST00000671741.2
ENST00000625116.3 ENST00000624355.1 |
ARHGEF9
|
Cdc42 guanine nucleotide exchange factor 9 |
| chr22_-_42720861 | 0.28 |
ENST00000642412.2
|
A4GALT
|
alpha 1,4-galactosyltransferase (P blood group) |
| chr11_-_66958366 | 0.28 |
ENST00000651036.1
ENST00000652125.1 ENST00000531614.6 ENST00000524491.6 ENST00000529047.6 ENST00000393960.7 ENST00000393958.7 ENST00000528403.6 ENST00000651854.1 |
PC
|
pyruvate carboxylase |
| chr2_+_232539720 | 0.27 |
ENST00000389492.3
|
CHRNG
|
cholinergic receptor nicotinic gamma subunit |
| chrX_+_87517784 | 0.27 |
ENST00000373119.9
ENST00000373114.4 |
KLHL4
|
kelch like family member 4 |
| chr2_+_94871419 | 0.27 |
ENST00000295201.5
|
TEKT4
|
tektin 4 |
| chr12_-_27014300 | 0.27 |
ENST00000535819.1
ENST00000543803.5 ENST00000535423.5 ENST00000539741.5 ENST00000343028.9 ENST00000545600.1 ENST00000543088.5 |
TM7SF3
|
transmembrane 7 superfamily member 3 |
| chr3_-_122793772 | 0.27 |
ENST00000306103.3
|
HSPBAP1
|
HSPB1 associated protein 1 |
| chr4_-_42657085 | 0.27 |
ENST00000264449.14
ENST00000510289.1 ENST00000381668.9 |
ATP8A1
|
ATPase phospholipid transporting 8A1 |
| chr17_-_41811930 | 0.27 |
ENST00000393928.6
|
P3H4
|
prolyl 3-hydroxylase family member 4 (inactive) |
| chr16_-_46831134 | 0.27 |
ENST00000394806.6
ENST00000285697.9 |
C16orf87
|
chromosome 16 open reading frame 87 |
| chr8_-_90645512 | 0.26 |
ENST00000422900.1
|
TMEM64
|
transmembrane protein 64 |
| chr1_-_93847150 | 0.26 |
ENST00000370244.5
|
BCAR3
|
BCAR3 adaptor protein, NSP family member |
| chr11_+_842807 | 0.26 |
ENST00000397411.6
|
TSPAN4
|
tetraspanin 4 |
| chr2_+_64454145 | 0.26 |
ENST00000238875.10
|
LGALSL
|
galectin like |
| chr9_+_87497852 | 0.26 |
ENST00000408954.8
|
DAPK1
|
death associated protein kinase 1 |
| chr2_-_73233206 | 0.25 |
ENST00000258083.3
|
PRADC1
|
protease associated domain containing 1 |
| chr15_-_75455767 | 0.25 |
ENST00000360439.8
|
SIN3A
|
SIN3 transcription regulator family member A |
| chr2_-_219571529 | 0.25 |
ENST00000404537.6
|
OBSL1
|
obscurin like cytoskeletal adaptor 1 |
| chr5_-_160312756 | 0.25 |
ENST00000644313.1
|
CCNJL
|
cyclin J like |
| chr3_-_15798184 | 0.25 |
ENST00000624145.3
|
ANKRD28
|
ankyrin repeat domain 28 |
| chr5_-_160312524 | 0.25 |
ENST00000520748.1
ENST00000257536.13 ENST00000393977.7 |
CCNJL
|
cyclin J like |
| chr8_-_29263063 | 0.25 |
ENST00000524189.6
|
KIF13B
|
kinesin family member 13B |
| chr6_+_135181323 | 0.25 |
ENST00000367814.8
|
MYB
|
MYB proto-oncogene, transcription factor |
| chr1_+_100538131 | 0.24 |
ENST00000315033.5
|
GPR88
|
G protein-coupled receptor 88 |
| chrX_+_101078861 | 0.24 |
ENST00000372930.5
|
TMEM35A
|
transmembrane protein 35A |
| chr19_+_35248728 | 0.24 |
ENST00000602003.1
ENST00000360798.7 ENST00000354900.7 |
LSR
|
lipolysis stimulated lipoprotein receptor |
| chr17_-_41140487 | 0.24 |
ENST00000345847.4
|
KRTAP4-6
|
keratin associated protein 4-6 |
| chr6_+_144583198 | 0.24 |
ENST00000367526.8
|
UTRN
|
utrophin |
| chr7_-_130441136 | 0.24 |
ENST00000675596.1
ENST00000676312.1 |
CEP41
|
centrosomal protein 41 |
| chr18_-_55422492 | 0.23 |
ENST00000561992.5
ENST00000630712.2 |
TCF4
|
transcription factor 4 |
| chr6_+_135181268 | 0.23 |
ENST00000341911.10
ENST00000442647.7 ENST00000618728.4 ENST00000316528.12 ENST00000616088.4 |
MYB
|
MYB proto-oncogene, transcription factor |
| chr13_+_75760362 | 0.23 |
ENST00000534657.5
|
LMO7
|
LIM domain 7 |
| chr3_+_137764296 | 0.23 |
ENST00000306087.3
|
SOX14
|
SRY-box transcription factor 14 |
| chr19_+_35248694 | 0.23 |
ENST00000361790.7
|
LSR
|
lipolysis stimulated lipoprotein receptor |
| chr2_+_74002685 | 0.23 |
ENST00000305799.8
|
TET3
|
tet methylcytosine dioxygenase 3 |
| chr19_+_35248375 | 0.23 |
ENST00000602122.5
|
LSR
|
lipolysis stimulated lipoprotein receptor |
| chr9_+_128552558 | 0.23 |
ENST00000372731.8
ENST00000630804.2 ENST00000372739.7 ENST00000627441.2 ENST00000358161.9 ENST00000636257.1 |
SPTAN1
|
spectrin alpha, non-erythrocytic 1 |
| chr11_-_76669985 | 0.23 |
ENST00000407242.6
ENST00000421973.1 |
LRRC32
|
leucine rich repeat containing 32 |
| chr9_-_16727980 | 0.23 |
ENST00000418777.5
ENST00000468187.6 |
BNC2
|
basonuclin 2 |
| chr11_-_119381629 | 0.23 |
ENST00000260187.7
ENST00000455332.6 |
USP2
|
ubiquitin specific peptidase 2 |
| chr4_-_151015263 | 0.23 |
ENST00000510413.5
ENST00000507224.5 ENST00000651943.2 |
LRBA
|
LPS responsive beige-like anchor protein |
| chr11_-_417385 | 0.23 |
ENST00000332725.7
|
SIGIRR
|
single Ig and TIR domain containing |
| chr11_+_92224801 | 0.22 |
ENST00000525166.6
|
FAT3
|
FAT atypical cadherin 3 |
| chr12_-_123268244 | 0.22 |
ENST00000618072.4
|
CDK2AP1
|
cyclin dependent kinase 2 associated protein 1 |
| chr11_-_417304 | 0.22 |
ENST00000397632.7
|
SIGIRR
|
single Ig and TIR domain containing |
| chr19_+_35248656 | 0.22 |
ENST00000621372.4
|
LSR
|
lipolysis stimulated lipoprotein receptor |
| chr5_+_73626158 | 0.22 |
ENST00000296794.10
ENST00000545377.5 ENST00000509848.5 ENST00000513042.7 |
ARHGEF28
|
Rho guanine nucleotide exchange factor 28 |
| chr21_+_10521536 | 0.22 |
ENST00000622113.4
|
TPTE
|
transmembrane phosphatase with tensin homology |
| chr1_+_13892791 | 0.22 |
ENST00000636564.1
ENST00000636203.1 |
KAZN
|
kazrin, periplakin interacting protein |
| chr3_-_139539577 | 0.22 |
ENST00000619087.4
|
RBP1
|
retinol binding protein 1 |
| chr11_-_76670737 | 0.22 |
ENST00000260061.9
ENST00000404995.5 |
LRRC32
|
leucine rich repeat containing 32 |
| chrX_-_112679919 | 0.21 |
ENST00000371968.8
|
LHFPL1
|
LHFPL tetraspan subfamily member 1 |
| chr20_+_45306834 | 0.21 |
ENST00000343694.8
ENST00000372741.7 ENST00000372743.5 |
RBPJL
|
recombination signal binding protein for immunoglobulin kappa J region like |
| chr3_+_52779916 | 0.21 |
ENST00000537050.5
|
ITIH1
|
inter-alpha-trypsin inhibitor heavy chain 1 |
| chr9_-_69672341 | 0.21 |
ENST00000265381.7
|
APBA1
|
amyloid beta precursor protein binding family A member 1 |
| chr5_-_95284535 | 0.21 |
ENST00000515393.5
|
MCTP1
|
multiple C2 and transmembrane domain containing 1 |
| chr16_-_636253 | 0.21 |
ENST00000565163.5
|
METTL26
|
methyltransferase like 26 |
| chr21_+_10521569 | 0.21 |
ENST00000612957.4
ENST00000427445.6 ENST00000612746.1 ENST00000618007.5 |
TPTE
|
transmembrane phosphatase with tensin homology |
| chr11_+_842928 | 0.21 |
ENST00000397408.5
|
TSPAN4
|
tetraspanin 4 |
| chr17_+_15999815 | 0.21 |
ENST00000261647.10
|
TTC19
|
tetratricopeptide repeat domain 19 |
| chr2_-_37672178 | 0.21 |
ENST00000457889.1
|
CDC42EP3
|
CDC42 effector protein 3 |
| chr1_+_99850485 | 0.21 |
ENST00000370165.7
ENST00000370163.7 ENST00000294724.8 |
AGL
|
amylo-alpha-1, 6-glucosidase, 4-alpha-glucanotransferase |
| chr11_+_842824 | 0.20 |
ENST00000397396.5
ENST00000397397.7 |
TSPAN4
|
tetraspanin 4 |
| chr7_-_28958321 | 0.20 |
ENST00000539664.3
|
TRIL
|
TLR4 interactor with leucine rich repeats |
| chr1_+_2073986 | 0.20 |
ENST00000461106.6
|
PRKCZ
|
protein kinase C zeta |
| chr12_-_70609788 | 0.20 |
ENST00000547715.1
ENST00000538708.5 ENST00000550857.5 ENST00000261266.9 |
PTPRB
|
protein tyrosine phosphatase receptor type B |
| chr6_-_31728877 | 0.20 |
ENST00000437288.5
|
DDAH2
|
dimethylarginine dimethylaminohydrolase 2 |
| chr19_+_35248994 | 0.19 |
ENST00000427250.5
ENST00000605618.6 |
LSR
|
lipolysis stimulated lipoprotein receptor |
| chr20_+_43945677 | 0.19 |
ENST00000358131.5
|
TOX2
|
TOX high mobility group box family member 2 |
| chrX_+_129738942 | 0.19 |
ENST00000371106.4
|
XPNPEP2
|
X-prolyl aminopeptidase 2 |
| chr8_-_17002327 | 0.19 |
ENST00000180166.6
|
FGF20
|
fibroblast growth factor 20 |
| chr17_+_76079182 | 0.19 |
ENST00000334586.10
|
ZACN
|
zinc activated ion channel |
| chr14_+_67619911 | 0.19 |
ENST00000261783.4
|
ARG2
|
arginase 2 |
| chr15_+_75043263 | 0.19 |
ENST00000563393.1
|
PPCDC
|
phosphopantothenoylcysteine decarboxylase |
| chr16_+_30000944 | 0.19 |
ENST00000562291.2
|
INO80E
|
INO80 complex subunit E |
| chr19_-_38253238 | 0.19 |
ENST00000587515.5
|
PPP1R14A
|
protein phosphatase 1 regulatory inhibitor subunit 14A |
| chr11_-_111912871 | 0.19 |
ENST00000528628.5
|
CRYAB
|
crystallin alpha B |
| chr17_+_47209338 | 0.19 |
ENST00000393450.5
|
MYL4
|
myosin light chain 4 |
| chr1_+_99850348 | 0.19 |
ENST00000361915.8
|
AGL
|
amylo-alpha-1, 6-glucosidase, 4-alpha-glucanotransferase |
| chrX_-_100731504 | 0.19 |
ENST00000372989.5
ENST00000276141.10 |
SYTL4
|
synaptotagmin like 4 |
| chr15_+_76995118 | 0.19 |
ENST00000558012.6
ENST00000379595.7 |
PSTPIP1
|
proline-serine-threonine phosphatase interacting protein 1 |
| chrX_+_71301742 | 0.19 |
ENST00000373829.8
ENST00000538820.1 |
ITGB1BP2
|
integrin subunit beta 1 binding protein 2 |
| chr6_-_31729260 | 0.19 |
ENST00000375789.7
|
DDAH2
|
dimethylarginine dimethylaminohydrolase 2 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.2 | GO:0090076 | relaxation of skeletal muscle(GO:0090076) |
| 0.3 | 1.2 | GO:1904381 | Golgi apparatus mannose trimming(GO:1904381) |
| 0.3 | 0.9 | GO:1901899 | positive regulation of relaxation of cardiac muscle(GO:1901899) |
| 0.3 | 0.8 | GO:1901301 | regulation of cargo loading into COPII-coated vesicle(GO:1901301) |
| 0.2 | 2.2 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.2 | 1.4 | GO:1904274 | tricellular tight junction assembly(GO:1904274) |
| 0.2 | 0.6 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 0.2 | 0.8 | GO:0021592 | fourth ventricle development(GO:0021592) |
| 0.2 | 0.8 | GO:0048073 | regulation of eye pigmentation(GO:0048073) |
| 0.2 | 1.6 | GO:0048630 | skeletal muscle tissue growth(GO:0048630) |
| 0.2 | 0.5 | GO:0098746 | fast, calcium ion-dependent exocytosis of neurotransmitter(GO:0098746) |
| 0.2 | 1.1 | GO:0000821 | regulation of arginine metabolic process(GO:0000821) |
| 0.2 | 0.5 | GO:1904897 | regulation of hepatic stellate cell proliferation(GO:1904897) positive regulation of hepatic stellate cell proliferation(GO:1904899) hepatic stellate cell proliferation(GO:1990922) |
| 0.1 | 0.4 | GO:0021793 | chemorepulsion of branchiomotor axon(GO:0021793) |
| 0.1 | 0.5 | GO:1903786 | mycotoxin catabolic process(GO:0043387) aflatoxin catabolic process(GO:0046223) organic heteropentacyclic compound catabolic process(GO:1901377) regulation of glutathione biosynthetic process(GO:1903786) positive regulation of glutathione biosynthetic process(GO:1903788) |
| 0.1 | 0.4 | GO:1904717 | excitatory chemical synaptic transmission(GO:0098976) regulation of AMPA glutamate receptor clustering(GO:1904717) positive regulation of AMPA glutamate receptor clustering(GO:1904719) |
| 0.1 | 0.4 | GO:0002582 | positive regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002582) positive regulation of antigen processing and presentation of peptide antigen(GO:0002585) positive regulation of antigen processing and presentation of peptide antigen via MHC class II(GO:0002588) |
| 0.1 | 0.6 | GO:0001575 | globoside metabolic process(GO:0001575) |
| 0.1 | 0.3 | GO:0021986 | epithalamus development(GO:0021538) habenula development(GO:0021986) |
| 0.1 | 0.6 | GO:0033490 | cholesterol biosynthetic process via desmosterol(GO:0033489) cholesterol biosynthetic process via lathosterol(GO:0033490) |
| 0.1 | 0.3 | GO:1901675 | negative regulation of histone H3-K27 acetylation(GO:1901675) |
| 0.1 | 0.2 | GO:0044725 | chromatin reprogramming in the zygote(GO:0044725) |
| 0.1 | 0.3 | GO:0019074 | viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 0.1 | 0.4 | GO:0045079 | negative regulation of chemokine biosynthetic process(GO:0045079) |
| 0.1 | 0.7 | GO:0070673 | response to interleukin-18(GO:0070673) |
| 0.1 | 0.4 | GO:0044339 | canonical Wnt signaling pathway involved in osteoblast differentiation(GO:0044339) |
| 0.1 | 0.3 | GO:0010587 | miRNA catabolic process(GO:0010587) negative regulation by host of viral genome replication(GO:0044828) |
| 0.1 | 0.1 | GO:0006113 | fermentation(GO:0006113) regulation of fermentation(GO:0043465) |
| 0.1 | 1.0 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.1 | 0.5 | GO:0001661 | conditioned taste aversion(GO:0001661) |
| 0.1 | 0.8 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.1 | 0.2 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.1 | 0.2 | GO:0060723 | spongiotrophoblast cell proliferation(GO:0060720) regulation of spongiotrophoblast cell proliferation(GO:0060721) cell proliferation involved in embryonic placenta development(GO:0060722) regulation of cell proliferation involved in embryonic placenta development(GO:0060723) |
| 0.0 | 0.1 | GO:0010933 | macrophage tolerance induction(GO:0010931) regulation of macrophage tolerance induction(GO:0010932) positive regulation of macrophage tolerance induction(GO:0010933) |
| 0.0 | 0.3 | GO:0048050 | post-embryonic eye morphogenesis(GO:0048050) |
| 0.0 | 0.7 | GO:0045176 | apical protein localization(GO:0045176) |
| 0.0 | 0.6 | GO:0001766 | membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) |
| 0.0 | 1.1 | GO:0040015 | negative regulation of multicellular organism growth(GO:0040015) |
| 0.0 | 0.3 | GO:0051697 | protein delipidation(GO:0051697) |
| 0.0 | 0.2 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.0 | 0.2 | GO:0098957 | anterograde axonal transport of mitochondrion(GO:0098957) |
| 0.0 | 0.3 | GO:0061091 | regulation of phospholipid translocation(GO:0061091) positive regulation of phospholipid translocation(GO:0061092) |
| 0.0 | 0.1 | GO:0003032 | detection of oxygen(GO:0003032) |
| 0.0 | 0.8 | GO:0000052 | citrulline metabolic process(GO:0000052) |
| 0.0 | 0.1 | GO:0003064 | regulation of heart rate by hormone(GO:0003064) negative regulation of potassium ion export(GO:1902303) |
| 0.0 | 0.2 | GO:0061743 | motor learning(GO:0061743) |
| 0.0 | 0.2 | GO:2001181 | positive regulation of interleukin-10 secretion(GO:2001181) |
| 0.0 | 0.1 | GO:1904772 | hepatocyte homeostasis(GO:0036333) response to tetrachloromethane(GO:1904772) |
| 0.0 | 0.1 | GO:0033082 | regulation of extrathymic T cell differentiation(GO:0033082) sebum secreting cell proliferation(GO:1990654) |
| 0.0 | 0.2 | GO:1903435 | positive regulation of constitutive secretory pathway(GO:1903435) |
| 0.0 | 0.3 | GO:0034316 | negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) |
| 0.0 | 0.5 | GO:0043983 | histone H4-K12 acetylation(GO:0043983) |
| 0.0 | 0.9 | GO:0042789 | mRNA transcription from RNA polymerase II promoter(GO:0042789) |
| 0.0 | 0.4 | GO:0014877 | response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.0 | 0.5 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.0 | 0.1 | GO:1904247 | positive regulation of polynucleotide adenylyltransferase activity(GO:1904247) |
| 0.0 | 0.4 | GO:0098914 | membrane repolarization during atrial cardiac muscle cell action potential(GO:0098914) |
| 0.0 | 0.1 | GO:0016333 | morphogenesis of follicular epithelium(GO:0016333) establishment or maintenance of polarity of follicular epithelium(GO:0016334) establishment of planar polarity of follicular epithelium(GO:0042247) |
| 0.0 | 0.1 | GO:0018032 | protein amidation(GO:0018032) |
| 0.0 | 0.2 | GO:1901341 | activation of store-operated calcium channel activity(GO:0032237) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.0 | 0.1 | GO:1903778 | protein localization to vacuolar membrane(GO:1903778) |
| 0.0 | 0.1 | GO:0015688 | iron chelate transport(GO:0015688) siderophore transport(GO:0015891) |
| 0.0 | 0.1 | GO:0009405 | pathogenesis(GO:0009405) |
| 0.0 | 0.5 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.0 | 0.3 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.0 | 0.1 | GO:0032765 | positive regulation of mast cell cytokine production(GO:0032765) |
| 0.0 | 0.1 | GO:1903061 | positive regulation of protein lipidation(GO:1903061) |
| 0.0 | 0.2 | GO:0032252 | secretory granule localization(GO:0032252) |
| 0.0 | 0.1 | GO:0097195 | pilomotor reflex(GO:0097195) positive regulation of transcription from RNA polymerase II promoter involved in smooth muscle cell differentiation(GO:2000721) |
| 0.0 | 0.3 | GO:0002138 | retinoic acid biosynthetic process(GO:0002138) |
| 0.0 | 0.2 | GO:0072383 | plus-end-directed vesicle transport along microtubule(GO:0072383) |
| 0.0 | 0.3 | GO:2000271 | positive regulation of fibroblast apoptotic process(GO:2000271) |
| 0.0 | 0.0 | GO:0090244 | Wnt signaling pathway involved in somitogenesis(GO:0090244) |
| 0.0 | 0.1 | GO:2000813 | negative regulation of barbed-end actin filament capping(GO:2000813) |
| 0.0 | 0.2 | GO:1904338 | regulation of dopaminergic neuron differentiation(GO:1904338) |
| 0.0 | 0.2 | GO:0036484 | trunk segmentation(GO:0035290) trunk neural crest cell migration(GO:0036484) ventral trunk neural crest cell migration(GO:0036486) |
| 0.0 | 0.2 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.0 | 0.1 | GO:0021633 | optic nerve structural organization(GO:0021633) |
| 0.0 | 0.9 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.0 | 0.3 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.0 | 0.8 | GO:0003416 | endochondral bone growth(GO:0003416) |
| 0.0 | 0.1 | GO:0023021 | termination of signal transduction(GO:0023021) |
| 0.0 | 0.6 | GO:0042501 | serine phosphorylation of STAT protein(GO:0042501) |
| 0.0 | 0.6 | GO:0072189 | ureter development(GO:0072189) |
| 0.0 | 0.1 | GO:1903026 | regulation of transcription from RNA polymerase II promoter in response to UV-induced DNA damage(GO:0010767) negative regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903026) |
| 0.0 | 0.1 | GO:0060754 | positive regulation of mast cell chemotaxis(GO:0060754) |
| 0.0 | 0.1 | GO:0097032 | mitochondrial electron transport, succinate to ubiquinone(GO:0006121) respiratory chain complex II assembly(GO:0034552) mitochondrial respiratory chain complex II assembly(GO:0034553) mitochondrial respiratory chain complex II biogenesis(GO:0097032) |
| 0.0 | 0.2 | GO:0006729 | tetrahydrobiopterin biosynthetic process(GO:0006729) regulation of lung blood pressure(GO:0014916) tetrahydrobiopterin metabolic process(GO:0046146) |
| 0.0 | 0.1 | GO:2000035 | regulation of stem cell division(GO:2000035) |
| 0.0 | 0.2 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.0 | 0.2 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.0 | 0.2 | GO:0034551 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) |
| 0.0 | 0.1 | GO:0090070 | positive regulation of ribosome biogenesis(GO:0090070) positive regulation of rRNA processing(GO:2000234) |
| 0.0 | 0.3 | GO:0007220 | Notch receptor processing(GO:0007220) |
| 0.0 | 0.5 | GO:0032463 | negative regulation of protein homooligomerization(GO:0032463) |
| 0.0 | 0.1 | GO:0044691 | tooth eruption(GO:0044691) |
| 0.0 | 0.2 | GO:0000050 | urea cycle(GO:0000050) |
| 0.0 | 0.1 | GO:0070682 | proteasome regulatory particle assembly(GO:0070682) |
| 0.0 | 0.2 | GO:1903416 | response to glycoside(GO:1903416) |
| 0.0 | 0.4 | GO:1900745 | positive regulation of p38MAPK cascade(GO:1900745) |
| 0.0 | 0.7 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.0 | 0.0 | GO:0090187 | positive regulation of pancreatic juice secretion(GO:0090187) regulation of somatostatin secretion(GO:0090273) positive regulation of somatostatin secretion(GO:0090274) |
| 0.0 | 0.5 | GO:0007216 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.0 | 0.3 | GO:0038092 | nodal signaling pathway(GO:0038092) |
| 0.0 | 0.1 | GO:0060671 | epithelial cell differentiation involved in embryonic placenta development(GO:0060671) epithelial cell morphogenesis involved in placental branching(GO:0060672) |
| 0.0 | 0.3 | GO:0045475 | locomotor rhythm(GO:0045475) |
| 0.0 | 0.3 | GO:0060294 | cilium movement involved in cell motility(GO:0060294) |
| 0.0 | 0.1 | GO:0045586 | regulation of gamma-delta T cell differentiation(GO:0045586) |
| 0.0 | 0.3 | GO:0038007 | netrin-activated signaling pathway(GO:0038007) |
| 0.0 | 0.1 | GO:0060373 | regulation of ventricular cardiac muscle cell membrane depolarization(GO:0060373) |
| 0.0 | 0.1 | GO:0019375 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.0 | 0.1 | GO:0006850 | mitochondrial pyruvate transport(GO:0006850) mitochondrial pyruvate transmembrane transport(GO:1902361) |
| 0.0 | 0.4 | GO:0034391 | smooth muscle cell apoptotic process(GO:0034390) regulation of smooth muscle cell apoptotic process(GO:0034391) |
| 0.0 | 0.3 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.0 | 0.2 | GO:0014051 | gamma-aminobutyric acid secretion(GO:0014051) |
| 0.0 | 0.0 | GO:0035625 | epidermal growth factor-activated receptor transactivation by G-protein coupled receptor signaling pathway(GO:0035625) |
| 0.0 | 0.1 | GO:1903385 | regulation of homophilic cell adhesion(GO:1903385) |
| 0.0 | 0.5 | GO:0045737 | positive regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045737) |
| 0.0 | 0.1 | GO:0097475 | motor neuron migration(GO:0097475) |
| 0.0 | 0.0 | GO:0001868 | regulation of complement activation, lectin pathway(GO:0001868) negative regulation of complement activation, lectin pathway(GO:0001869) |
| 0.0 | 0.1 | GO:2000676 | positive regulation of type B pancreatic cell apoptotic process(GO:2000676) |
| 0.0 | 0.7 | GO:0045103 | intermediate filament-based process(GO:0045103) |
| 0.0 | 0.0 | GO:0038018 | Wnt receptor catabolic process(GO:0038018) |
| 0.0 | 0.1 | GO:0046959 | habituation(GO:0046959) |
| 0.0 | 0.3 | GO:0002076 | osteoblast development(GO:0002076) |
| 0.0 | 0.0 | GO:0060596 | mammary placode formation(GO:0060596) |
| 0.0 | 0.3 | GO:0007130 | synaptonemal complex assembly(GO:0007130) |
| 0.0 | 0.1 | GO:0006348 | chromatin silencing at telomere(GO:0006348) |
| 0.0 | 0.3 | GO:0043162 | ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
| 0.0 | 0.1 | GO:0009732 | detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) detection of monosaccharide stimulus(GO:0034287) detection of glucose(GO:0051594) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.8 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.2 | 1.0 | GO:1990393 | 3M complex(GO:1990393) |
| 0.2 | 0.8 | GO:0097513 | myosin II filament(GO:0097513) |
| 0.1 | 1.5 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.1 | 0.9 | GO:0035976 | AP1 complex(GO:0035976) |
| 0.1 | 0.5 | GO:0060200 | clathrin-sculpted acetylcholine transport vesicle(GO:0060200) clathrin-sculpted acetylcholine transport vesicle membrane(GO:0060201) |
| 0.1 | 0.5 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.1 | 0.9 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.1 | 0.4 | GO:0072558 | NLRP1 inflammasome complex(GO:0072558) |
| 0.1 | 2.0 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.1 | 0.3 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.0 | 2.6 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 0.2 | GO:0044326 | dendritic spine neck(GO:0044326) |
| 0.0 | 0.4 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.0 | 0.3 | GO:0042584 | chromaffin granule membrane(GO:0042584) |
| 0.0 | 0.3 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.0 | 0.3 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.0 | 1.1 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 1.3 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.0 | 0.3 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.0 | 0.2 | GO:0001940 | male pronucleus(GO:0001940) |
| 0.0 | 0.1 | GO:1902937 | inward rectifier potassium channel complex(GO:1902937) |
| 0.0 | 0.3 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.0 | 0.5 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 0.0 | 0.2 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.0 | 0.2 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.0 | 0.1 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.0 | 0.1 | GO:0042643 | actomyosin, actin portion(GO:0042643) |
| 0.0 | 0.2 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.0 | 0.5 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 0.2 | GO:0043203 | axon hillock(GO:0043203) |
| 0.0 | 0.2 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.0 | 0.3 | GO:0001527 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.0 | 0.2 | GO:0070938 | contractile ring(GO:0070938) |
| 0.0 | 0.3 | GO:0001931 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.0 | 0.1 | GO:0070436 | Grb2-EGFR complex(GO:0070436) |
| 0.0 | 0.1 | GO:0043291 | RAVE complex(GO:0043291) |
| 0.0 | 0.1 | GO:0045120 | pronucleus(GO:0045120) |
| 0.0 | 0.1 | GO:0033010 | paranodal junction(GO:0033010) |
| 0.0 | 0.2 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.0 | 0.0 | GO:1990851 | Wnt-Frizzled-LRP5/6 complex(GO:1990851) |
| 0.0 | 0.1 | GO:0000808 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.0 | 0.3 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.0 | 0.1 | GO:0035061 | interchromatin granule(GO:0035061) |
| 0.0 | 0.3 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.0 | 0.1 | GO:0044354 | pinosome(GO:0044352) macropinosome(GO:0044354) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 2.2 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.2 | 0.8 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.2 | 0.8 | GO:0046624 | sphingolipid transporter activity(GO:0046624) |
| 0.1 | 1.1 | GO:0008269 | JAK pathway signal transduction adaptor activity(GO:0008269) |
| 0.1 | 1.2 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.1 | 0.4 | GO:0032090 | Pyrin domain binding(GO:0032090) |
| 0.1 | 0.3 | GO:0010698 | acetyltransferase activator activity(GO:0010698) |
| 0.1 | 0.9 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.1 | 0.7 | GO:0016403 | dimethylargininase activity(GO:0016403) |
| 0.1 | 1.5 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.1 | 0.5 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.1 | 0.5 | GO:0016807 | cysteine-type carboxypeptidase activity(GO:0016807) cysteine-type exopeptidase activity(GO:0070004) |
| 0.1 | 0.2 | GO:0004874 | aryl hydrocarbon receptor activity(GO:0004874) |
| 0.1 | 2.0 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.1 | 0.2 | GO:0008336 | gamma-butyrobetaine dioxygenase activity(GO:0008336) |
| 0.1 | 0.2 | GO:0034416 | bisphosphoglycerate phosphatase activity(GO:0034416) inositol-1,3,4,5-tetrakisphosphate 3-phosphatase activity(GO:0051717) |
| 0.1 | 1.1 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.1 | 1.2 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.1 | 0.4 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.1 | 0.3 | GO:0032038 | myosin II heavy chain binding(GO:0032038) |
| 0.0 | 0.4 | GO:0090599 | alpha-glucosidase activity(GO:0090599) |
| 0.0 | 0.7 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.0 | 0.2 | GO:0022850 | serotonin-gated cation channel activity(GO:0022850) |
| 0.0 | 0.3 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.0 | 0.2 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.0 | 0.1 | GO:0052810 | 1-phosphatidylinositol-5-kinase activity(GO:0052810) |
| 0.0 | 0.7 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.0 | 0.2 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 1.0 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.5 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.0 | 0.4 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.0 | 0.6 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 1.1 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.7 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 0.0 | 0.9 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.0 | 0.3 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.0 | 0.1 | GO:0004598 | peptidylglycine monooxygenase activity(GO:0004504) peptidylamidoglycolate lyase activity(GO:0004598) |
| 0.0 | 0.7 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.1 | GO:0001132 | RNA polymerase II transcription factor activity, TBP-class protein binding, involved in preinitiation complex assembly(GO:0001129) RNA polymerase II transcription factor activity, TBP-class protein binding(GO:0001132) |
| 0.0 | 0.1 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.0 | 0.3 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.3 | GO:0004075 | biotin carboxylase activity(GO:0004075) |
| 0.0 | 0.5 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.1 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.0 | 0.4 | GO:0019841 | retinol binding(GO:0019841) |
| 0.0 | 0.0 | GO:0005026 | transforming growth factor beta receptor activity, type II(GO:0005026) |
| 0.0 | 0.1 | GO:0035514 | DNA demethylase activity(GO:0035514) DNA-N1-methyladenine dioxygenase activity(GO:0043734) |
| 0.0 | 0.2 | GO:1903763 | gap junction channel activity involved in cell communication by electrical coupling(GO:1903763) |
| 0.0 | 0.5 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.2 | GO:0019238 | cyclohydrolase activity(GO:0019238) |
| 0.0 | 0.6 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 0.6 | GO:0016628 | oxidoreductase activity, acting on the CH-CH group of donors, NAD or NADP as acceptor(GO:0016628) |
| 0.0 | 0.4 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.0 | 0.3 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.0 | 1.7 | GO:0098811 | transcriptional activator activity, RNA polymerase II transcription factor binding(GO:0001190) transcriptional repressor activity, RNA polymerase II activating transcription factor binding(GO:0098811) |
| 0.0 | 0.4 | GO:0000400 | four-way junction DNA binding(GO:0000400) |
| 0.0 | 0.3 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.0 | 0.1 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.0 | 0.1 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 0.0 | 0.8 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.0 | 0.3 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.1 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.0 | 0.1 | GO:0030144 | alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase activity(GO:0030144) |
| 0.0 | 0.2 | GO:1990239 | steroid hormone binding(GO:1990239) |
| 0.0 | 1.0 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 0.6 | GO:0001221 | transcription cofactor binding(GO:0001221) |
| 0.0 | 0.4 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.1 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) C3HC4-type RING finger domain binding(GO:0055131) |
| 0.0 | 0.0 | GO:0004938 | alpha2-adrenergic receptor activity(GO:0004938) |
| 0.0 | 0.3 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 0.0 | GO:0001588 | dopamine neurotransmitter receptor activity, coupled via Gs(GO:0001588) |
| 0.0 | 0.1 | GO:0061513 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 0.0 | 0.0 | GO:0030158 | protein xylosyltransferase activity(GO:0030158) |
| 0.0 | 1.9 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 0.2 | GO:0001083 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, RNA polymerase II basal transcription factor binding(GO:0001083) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.0 | 0.1 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.0 | 0.3 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.1 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 1.8 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 1.0 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.0 | 1.6 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 0.8 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.0 | 0.4 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 0.8 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.0 | 0.4 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 0.2 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.0 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.0 | 0.6 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 0.8 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 1.1 | REACTOME GROWTH HORMONE RECEPTOR SIGNALING | Genes involved in Growth hormone receptor signaling |
| 0.0 | 1.0 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 0.5 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.0 | 0.5 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 1.1 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.0 | 0.3 | REACTOME SIGNALING BY NOTCH2 | Genes involved in Signaling by NOTCH2 |
| 0.0 | 0.2 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.0 | 0.3 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.0 | 0.5 | REACTOME TRAF6 MEDIATED NFKB ACTIVATION | Genes involved in TRAF6 mediated NF-kB activation |
| 0.0 | 0.3 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 0.8 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 0.6 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 1.3 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.6 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 0.4 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 0.3 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 0.0 | 0.4 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 0.7 | REACTOME INTEGRIN ALPHAIIB BETA3 SIGNALING | Genes involved in Integrin alphaIIb beta3 signaling |
| 0.0 | 0.2 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 0.2 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.0 | 0.2 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 0.3 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |