Project
Inflammatory response time course, HUVEC (Wada, 2009)
Navigation
Downloads
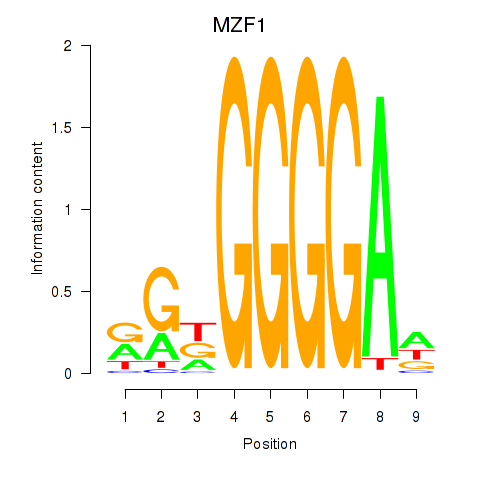
Results for MZF1
Z-value: 0.93
Transcription factors associated with MZF1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
MZF1
|
ENSG00000099326.9 | MZF1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| MZF1 | hg38_v1_chr19_-_58573280_58573354, hg38_v1_chr19_-_58573555_58573582 | 0.30 | 1.4e-01 | Click! |
Activity profile of MZF1 motif
Sorted Z-values of MZF1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of MZF1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 9.0 | GO:0060244 | negative regulation of cell proliferation involved in contact inhibition(GO:0060244) |
| 0.5 | 1.4 | GO:0044725 | chromatin reprogramming in the zygote(GO:0044725) |
| 0.4 | 6.7 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) |
| 0.4 | 0.4 | GO:2000137 | negative regulation of cell proliferation involved in heart morphogenesis(GO:2000137) |
| 0.3 | 2.4 | GO:0000821 | regulation of arginine metabolic process(GO:0000821) |
| 0.3 | 0.8 | GO:1904247 | positive regulation of polynucleotide adenylyltransferase activity(GO:1904247) |
| 0.2 | 1.2 | GO:0093001 | glycolysis from storage polysaccharide through glucose-1-phosphate(GO:0093001) |
| 0.2 | 0.9 | GO:0002032 | desensitization of G-protein coupled receptor protein signaling pathway by arrestin(GO:0002032) |
| 0.2 | 1.1 | GO:0014886 | transition between slow and fast fiber(GO:0014886) |
| 0.2 | 1.5 | GO:0045629 | negative regulation of T-helper 2 cell differentiation(GO:0045629) |
| 0.2 | 1.2 | GO:0051138 | positive regulation of NK T cell differentiation(GO:0051138) |
| 0.2 | 2.8 | GO:0015074 | DNA integration(GO:0015074) |
| 0.2 | 0.5 | GO:0090341 | negative regulation of secretion of lysosomal enzymes(GO:0090341) |
| 0.2 | 0.7 | GO:0060399 | positive regulation of growth hormone receptor signaling pathway(GO:0060399) |
| 0.2 | 0.9 | GO:0071461 | cellular response to redox state(GO:0071461) |
| 0.2 | 1.7 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.1 | 1.3 | GO:0045602 | negative regulation of endothelial cell differentiation(GO:0045602) |
| 0.1 | 3.1 | GO:0070914 | UV-damage excision repair(GO:0070914) |
| 0.1 | 0.4 | GO:0045715 | negative regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045715) |
| 0.1 | 4.2 | GO:0072189 | ureter development(GO:0072189) |
| 0.1 | 0.4 | GO:0052047 | interaction with other organism via secreted substance involved in symbiotic interaction(GO:0052047) |
| 0.1 | 0.4 | GO:0003192 | mitral valve formation(GO:0003192) |
| 0.1 | 0.6 | GO:0060748 | tertiary branching involved in mammary gland duct morphogenesis(GO:0060748) |
| 0.1 | 0.4 | GO:0042040 | molybdenum incorporation into molybdenum-molybdopterin complex(GO:0018315) metal incorporation into metallo-molybdopterin complex(GO:0042040) glycine receptor clustering(GO:0072579) |
| 0.1 | 1.0 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 0.1 | 0.6 | GO:1904637 | negative regulation by host of viral genome replication(GO:0044828) response to ionomycin(GO:1904636) cellular response to ionomycin(GO:1904637) |
| 0.1 | 1.0 | GO:0001661 | conditioned taste aversion(GO:0001661) |
| 0.1 | 0.6 | GO:0015891 | iron chelate transport(GO:0015688) siderophore transport(GO:0015891) |
| 0.1 | 0.3 | GO:1902460 | regulation of mesenchymal stem cell proliferation(GO:1902460) positive regulation of mesenchymal stem cell proliferation(GO:1902462) |
| 0.1 | 0.3 | GO:0061394 | regulation of transcription from RNA polymerase II promoter in response to arsenic-containing substance(GO:0061394) |
| 0.1 | 0.7 | GO:0071557 | histone H3-K27 demethylation(GO:0071557) |
| 0.1 | 0.3 | GO:0002572 | pro-T cell differentiation(GO:0002572) regulation of pro-T cell differentiation(GO:2000174) positive regulation of pro-T cell differentiation(GO:2000176) |
| 0.1 | 0.2 | GO:0060018 | astrocyte fate commitment(GO:0060018) |
| 0.1 | 0.6 | GO:0038016 | insulin receptor internalization(GO:0038016) |
| 0.1 | 0.3 | GO:0061445 | endocardial cell fate commitment(GO:0060957) endocardial cushion cell differentiation(GO:0061443) endocardial cushion cell fate commitment(GO:0061445) |
| 0.1 | 0.6 | GO:1903566 | positive regulation of protein localization to cilium(GO:1903566) |
| 0.1 | 0.4 | GO:0006344 | maintenance of chromatin silencing(GO:0006344) |
| 0.1 | 0.8 | GO:0032511 | late endosome to vacuole transport via multivesicular body sorting pathway(GO:0032511) |
| 0.1 | 0.7 | GO:0034316 | negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) |
| 0.1 | 0.1 | GO:0016539 | intein-mediated protein splicing(GO:0016539) protein splicing(GO:0030908) |
| 0.1 | 0.5 | GO:0090116 | C-5 methylation of cytosine(GO:0090116) |
| 0.1 | 0.3 | GO:0055014 | atrial cardiac muscle cell differentiation(GO:0055011) atrial cardiac muscle cell development(GO:0055014) |
| 0.1 | 0.3 | GO:1903750 | regulation of intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:1903750) negative regulation of intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:1903751) |
| 0.1 | 1.0 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.1 | 0.3 | GO:0032877 | positive regulation of DNA endoreduplication(GO:0032877) |
| 0.1 | 0.3 | GO:1904381 | Golgi apparatus mannose trimming(GO:1904381) |
| 0.1 | 0.7 | GO:0035093 | spermatogenesis, exchange of chromosomal proteins(GO:0035093) |
| 0.1 | 0.7 | GO:1903377 | negative regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903377) |
| 0.1 | 1.6 | GO:0017187 | peptidyl-glutamic acid carboxylation(GO:0017187) protein carboxylation(GO:0018214) |
| 0.1 | 0.3 | GO:0021691 | cerebellar Purkinje cell layer maturation(GO:0021691) |
| 0.1 | 0.4 | GO:0031630 | regulation of synaptic vesicle fusion to presynaptic membrane(GO:0031630) |
| 0.1 | 0.3 | GO:0042904 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.1 | 0.2 | GO:0060266 | negative regulation of respiratory burst involved in inflammatory response(GO:0060266) |
| 0.1 | 0.4 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 0.1 | 0.4 | GO:0034959 | substance P catabolic process(GO:0010814) calcitonin catabolic process(GO:0010816) endothelin maturation(GO:0034959) |
| 0.1 | 0.3 | GO:0010157 | response to chlorate(GO:0010157) |
| 0.1 | 0.3 | GO:1902498 | regulation of protein autoubiquitination(GO:1902498) |
| 0.1 | 2.1 | GO:0014850 | response to muscle activity(GO:0014850) |
| 0.1 | 0.3 | GO:0031117 | positive regulation of microtubule depolymerization(GO:0031117) |
| 0.1 | 0.5 | GO:2000507 | positive regulation of energy homeostasis(GO:2000507) |
| 0.1 | 0.3 | GO:0016480 | negative regulation of transcription from RNA polymerase III promoter(GO:0016480) |
| 0.1 | 0.2 | GO:0042938 | dipeptide transport(GO:0042938) |
| 0.1 | 0.3 | GO:0042335 | cuticle development(GO:0042335) |
| 0.1 | 1.0 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.1 | 0.4 | GO:0034499 | late endosome to Golgi transport(GO:0034499) |
| 0.1 | 0.3 | GO:0035261 | external genitalia morphogenesis(GO:0035261) |
| 0.1 | 0.2 | GO:0050915 | sensory perception of sour taste(GO:0050915) |
| 0.1 | 0.3 | GO:0061086 | negative regulation of histone H3-K27 methylation(GO:0061086) |
| 0.1 | 0.4 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.1 | 0.4 | GO:0030421 | defecation(GO:0030421) |
| 0.1 | 0.3 | GO:1903336 | negative regulation of vacuolar transport(GO:1903336) |
| 0.1 | 0.7 | GO:0000733 | DNA strand renaturation(GO:0000733) |
| 0.1 | 0.2 | GO:1904933 | regulation of cell proliferation in midbrain(GO:1904933) |
| 0.1 | 0.2 | GO:0032487 | regulation of Rap protein signal transduction(GO:0032487) |
| 0.1 | 1.5 | GO:0043153 | entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.1 | 0.1 | GO:0048850 | hypophysis morphogenesis(GO:0048850) |
| 0.1 | 2.3 | GO:0007202 | activation of phospholipase C activity(GO:0007202) |
| 0.1 | 0.4 | GO:0021615 | glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.1 | 1.0 | GO:0032688 | negative regulation of interferon-beta production(GO:0032688) |
| 0.1 | 0.2 | GO:0002378 | immunoglobulin biosynthetic process(GO:0002378) |
| 0.1 | 0.7 | GO:0015866 | ADP transport(GO:0015866) |
| 0.1 | 0.2 | GO:0002270 | plasmacytoid dendritic cell activation(GO:0002270) regulation of restriction endodeoxyribonuclease activity(GO:0032072) |
| 0.1 | 0.6 | GO:1902746 | regulation of lens fiber cell differentiation(GO:1902746) |
| 0.1 | 0.3 | GO:1902559 | 3'-phosphoadenosine 5'-phosphosulfate transport(GO:0046963) 3'-phospho-5'-adenylyl sulfate transmembrane transport(GO:1902559) |
| 0.1 | 0.2 | GO:1902952 | positive regulation of dendritic spine maintenance(GO:1902952) regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903756) negative regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903758) |
| 0.0 | 0.2 | GO:0060010 | Sertoli cell fate commitment(GO:0060010) |
| 0.0 | 0.2 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.0 | 0.6 | GO:1902414 | protein localization to cell junction(GO:1902414) |
| 0.0 | 0.2 | GO:1904327 | protein localization to cytosolic proteasome complex(GO:1904327) protein localization to cytosolic proteasome complex involved in ERAD pathway(GO:1904379) |
| 0.0 | 0.2 | GO:0071879 | positive regulation of adrenergic receptor signaling pathway(GO:0071879) |
| 0.0 | 0.4 | GO:1902966 | regulation of protein localization to early endosome(GO:1902965) positive regulation of protein localization to early endosome(GO:1902966) |
| 0.0 | 0.1 | GO:0031550 | positive regulation of brain-derived neurotrophic factor receptor signaling pathway(GO:0031550) |
| 0.0 | 0.2 | GO:0045204 | MAPK export from nucleus(GO:0045204) |
| 0.0 | 0.3 | GO:1903378 | positive regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903378) |
| 0.0 | 0.2 | GO:0014724 | regulation of twitch skeletal muscle contraction(GO:0014724) |
| 0.0 | 0.1 | GO:0050760 | negative regulation of thymidylate synthase biosynthetic process(GO:0050760) |
| 0.0 | 0.5 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.0 | 0.3 | GO:0051697 | protein delipidation(GO:0051697) |
| 0.0 | 0.3 | GO:0044806 | G-quadruplex DNA unwinding(GO:0044806) |
| 0.0 | 0.2 | GO:0060596 | mammary placode formation(GO:0060596) |
| 0.0 | 1.3 | GO:0070886 | positive regulation of calcineurin-NFAT signaling cascade(GO:0070886) |
| 0.0 | 0.3 | GO:1901187 | regulation of ephrin receptor signaling pathway(GO:1901187) |
| 0.0 | 0.3 | GO:0032876 | negative regulation of DNA endoreduplication(GO:0032876) |
| 0.0 | 0.2 | GO:0006196 | AMP catabolic process(GO:0006196) |
| 0.0 | 0.5 | GO:0098734 | macromolecule depalmitoylation(GO:0098734) |
| 0.0 | 0.6 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 | 0.5 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.0 | 1.1 | GO:0003416 | endochondral bone growth(GO:0003416) |
| 0.0 | 0.2 | GO:0010641 | positive regulation of platelet-derived growth factor receptor signaling pathway(GO:0010641) |
| 0.0 | 0.1 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.0 | 0.1 | GO:2000078 | glandular epithelial cell maturation(GO:0002071) regulation of transcription from RNA polymerase II promoter involved in ventral spinal cord interneuron specification(GO:0021913) positive regulation of type B pancreatic cell development(GO:2000078) |
| 0.0 | 0.0 | GO:0090273 | regulation of somatostatin secretion(GO:0090273) positive regulation of somatostatin secretion(GO:0090274) |
| 0.0 | 0.5 | GO:0015871 | choline transport(GO:0015871) |
| 0.0 | 0.4 | GO:0006657 | CDP-choline pathway(GO:0006657) |
| 0.0 | 0.2 | GO:1901341 | activation of store-operated calcium channel activity(GO:0032237) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.0 | 0.4 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.0 | 0.1 | GO:0060309 | elastin catabolic process(GO:0060309) |
| 0.0 | 0.2 | GO:0030167 | proteoglycan catabolic process(GO:0030167) regulation of hepatocyte growth factor receptor signaling pathway(GO:1902202) |
| 0.0 | 0.4 | GO:0045741 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) |
| 0.0 | 0.2 | GO:1902723 | negative regulation of skeletal muscle cell proliferation(GO:0014859) negative regulation of skeletal muscle satellite cell proliferation(GO:1902723) |
| 0.0 | 0.1 | GO:0098532 | histone H3-K27 trimethylation(GO:0098532) |
| 0.0 | 0.1 | GO:0035507 | regulation of myosin-light-chain-phosphatase activity(GO:0035507) |
| 0.0 | 0.2 | GO:0034154 | toll-like receptor 7 signaling pathway(GO:0034154) |
| 0.0 | 0.2 | GO:0008626 | granzyme-mediated apoptotic signaling pathway(GO:0008626) |
| 0.0 | 0.1 | GO:0007198 | adenylate cyclase-inhibiting serotonin receptor signaling pathway(GO:0007198) |
| 0.0 | 0.1 | GO:0030472 | mitotic spindle organization in nucleus(GO:0030472) |
| 0.0 | 0.0 | GO:0007210 | serotonin receptor signaling pathway(GO:0007210) |
| 0.0 | 0.7 | GO:0007176 | regulation of epidermal growth factor-activated receptor activity(GO:0007176) |
| 0.0 | 0.5 | GO:0002087 | regulation of respiratory gaseous exchange by neurological system process(GO:0002087) |
| 0.0 | 0.1 | GO:0001555 | oocyte growth(GO:0001555) |
| 0.0 | 0.7 | GO:0021516 | dorsal spinal cord development(GO:0021516) |
| 0.0 | 0.3 | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.0 | 0.1 | GO:2000653 | regulation of genetic imprinting(GO:2000653) |
| 0.0 | 0.2 | GO:0033029 | regulation of neutrophil apoptotic process(GO:0033029) |
| 0.0 | 0.3 | GO:1904153 | negative regulation of protein exit from endoplasmic reticulum(GO:0070862) negative regulation of retrograde protein transport, ER to cytosol(GO:1904153) |
| 0.0 | 0.3 | GO:0043983 | histone H4-K12 acetylation(GO:0043983) |
| 0.0 | 0.7 | GO:0016578 | histone deubiquitination(GO:0016578) |
| 0.0 | 0.4 | GO:0070734 | histone H3-K27 methylation(GO:0070734) |
| 0.0 | 0.1 | GO:1901675 | negative regulation of histone H3-K27 acetylation(GO:1901675) |
| 0.0 | 0.3 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 0.0 | 0.1 | GO:0060022 | hard palate development(GO:0060022) |
| 0.0 | 0.7 | GO:0035584 | calcium-mediated signaling using intracellular calcium source(GO:0035584) |
| 0.0 | 0.8 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.0 | 0.2 | GO:0043654 | recognition of apoptotic cell(GO:0043654) |
| 0.0 | 0.3 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.0 | 0.2 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.0 | 0.4 | GO:1904778 | regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.0 | 0.1 | GO:1905244 | regulation of modification of synaptic structure(GO:1905244) |
| 0.0 | 0.1 | GO:0038145 | macrophage colony-stimulating factor signaling pathway(GO:0038145) |
| 0.0 | 0.1 | GO:0060160 | negative regulation of dopamine receptor signaling pathway(GO:0060160) |
| 0.0 | 0.3 | GO:0035873 | lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) |
| 0.0 | 0.1 | GO:0072553 | terminal button organization(GO:0072553) |
| 0.0 | 0.2 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.0 | 0.2 | GO:0044375 | regulation of peroxisome size(GO:0044375) |
| 0.0 | 0.1 | GO:2000676 | positive regulation of type B pancreatic cell apoptotic process(GO:2000676) |
| 0.0 | 0.1 | GO:0045324 | late endosome to vacuole transport(GO:0045324) |
| 0.0 | 0.1 | GO:0048749 | compound eye development(GO:0048749) |
| 0.0 | 0.4 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.0 | 0.2 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.4 | GO:0007130 | synaptonemal complex assembly(GO:0007130) |
| 0.0 | 0.1 | GO:0090666 | scaRNA localization to Cajal body(GO:0090666) |
| 0.0 | 1.2 | GO:0050434 | positive regulation of viral transcription(GO:0050434) |
| 0.0 | 0.2 | GO:2000381 | negative regulation of mesoderm development(GO:2000381) |
| 0.0 | 1.7 | GO:1901222 | regulation of NIK/NF-kappaB signaling(GO:1901222) |
| 0.0 | 0.2 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.0 | 0.0 | GO:0060585 | regulation of prostaglandin-endoperoxide synthase activity(GO:0060584) positive regulation of prostaglandin-endoperoxide synthase activity(GO:0060585) |
| 0.0 | 0.2 | GO:0098909 | regulation of cardiac muscle cell action potential involved in regulation of contraction(GO:0098909) |
| 0.0 | 0.7 | GO:0030252 | growth hormone secretion(GO:0030252) |
| 0.0 | 1.2 | GO:0046626 | regulation of insulin receptor signaling pathway(GO:0046626) |
| 0.0 | 0.0 | GO:0090309 | positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.0 | 0.1 | GO:1904253 | circadian temperature homeostasis(GO:0060086) positive regulation of bile acid biosynthetic process(GO:0070859) positive regulation of bile acid metabolic process(GO:1904253) |
| 0.0 | 0.2 | GO:0046600 | negative regulation of centriole replication(GO:0046600) |
| 0.0 | 0.1 | GO:1902164 | mRNA localization resulting in posttranscriptional regulation of gene expression(GO:0010609) platelet alpha granule organization(GO:0070889) regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902162) positive regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902164) |
| 0.0 | 0.2 | GO:0060689 | cell differentiation involved in salivary gland development(GO:0060689) |
| 0.0 | 0.1 | GO:0030807 | positive regulation of cyclic nucleotide catabolic process(GO:0030807) positive regulation of cAMP catabolic process(GO:0030822) |
| 0.0 | 0.1 | GO:0043606 | histidine catabolic process to glutamate and formamide(GO:0019556) histidine catabolic process to glutamate and formate(GO:0019557) formamide metabolic process(GO:0043606) |
| 0.0 | 0.3 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.0 | 0.3 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.0 | 0.1 | GO:0031547 | brain-derived neurotrophic factor receptor signaling pathway(GO:0031547) |
| 0.0 | 0.1 | GO:0097026 | dendritic cell dendrite assembly(GO:0097026) |
| 0.0 | 0.2 | GO:0051256 | mitotic spindle midzone assembly(GO:0051256) |
| 0.0 | 0.2 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.0 | 0.2 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.0 | 0.4 | GO:0032897 | negative regulation of viral transcription(GO:0032897) |
| 0.0 | 0.0 | GO:1904017 | cellular response to Thyroglobulin triiodothyronine(GO:1904017) |
| 0.0 | 0.0 | GO:0061428 | negative regulation of transcription from RNA polymerase II promoter in response to hypoxia(GO:0061428) |
| 0.0 | 0.1 | GO:1900425 | negative regulation of defense response to bacterium(GO:1900425) negative regulation of interleukin-8 secretion(GO:2000483) |
| 0.0 | 0.3 | GO:0019511 | peptidyl-proline hydroxylation(GO:0019511) |
| 0.0 | 1.3 | GO:0006446 | regulation of translational initiation(GO:0006446) |
| 0.0 | 0.1 | GO:0061591 | calcium activated phospholipid scrambling(GO:0061588) calcium activated phosphatidylcholine scrambling(GO:0061590) calcium activated galactosylceramide scrambling(GO:0061591) |
| 0.0 | 0.1 | GO:2000786 | positive regulation of autophagosome assembly(GO:2000786) |
| 0.0 | 0.3 | GO:0033631 | cell-cell adhesion mediated by integrin(GO:0033631) |
| 0.0 | 0.2 | GO:0048368 | lateral mesoderm development(GO:0048368) |
| 0.0 | 0.1 | GO:0051151 | negative regulation of smooth muscle cell differentiation(GO:0051151) |
| 0.0 | 0.4 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
| 0.0 | 0.1 | GO:0099566 | regulation of postsynaptic cytosolic calcium ion concentration(GO:0099566) |
| 0.0 | 0.0 | GO:0061056 | sclerotome development(GO:0061056) |
| 0.0 | 0.0 | GO:0046603 | negative regulation of mitotic centrosome separation(GO:0046603) |
| 0.0 | 0.1 | GO:0010603 | regulation of cytoplasmic mRNA processing body assembly(GO:0010603) |
| 0.0 | 0.1 | GO:0061205 | alveolar primary septum development(GO:0061143) paramesonephric duct development(GO:0061205) |
| 0.0 | 0.0 | GO:0032848 | positive regulation of neuron maturation(GO:0014042) negative regulation of cellular pH reduction(GO:0032848) CD8-positive, alpha-beta T cell lineage commitment(GO:0043375) negative regulation of retinal cell programmed cell death(GO:0046671) |
| 0.0 | 0.6 | GO:0045672 | positive regulation of osteoclast differentiation(GO:0045672) |
| 0.0 | 0.3 | GO:0070932 | histone H3 deacetylation(GO:0070932) |
| 0.0 | 0.1 | GO:0006682 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.0 | 0.1 | GO:2000852 | regulation of corticosterone secretion(GO:2000852) |
| 0.0 | 0.1 | GO:0016198 | axon choice point recognition(GO:0016198) |
| 0.0 | 0.2 | GO:0035721 | intraciliary retrograde transport(GO:0035721) |
| 0.0 | 0.5 | GO:0006692 | prostanoid metabolic process(GO:0006692) prostaglandin metabolic process(GO:0006693) |
| 0.0 | 0.1 | GO:0035278 | miRNA mediated inhibition of translation(GO:0035278) negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) |
| 0.0 | 0.0 | GO:1903259 | exon-exon junction complex disassembly(GO:1903259) |
| 0.0 | 0.1 | GO:2000324 | positive regulation of glucocorticoid receptor signaling pathway(GO:2000324) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 3.1 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.2 | 6.8 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.2 | 0.7 | GO:0071062 | alphav-beta3 integrin-vitronectin complex(GO:0071062) |
| 0.2 | 1.2 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.2 | 1.5 | GO:0035976 | AP1 complex(GO:0035976) |
| 0.2 | 1.5 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.2 | 0.3 | GO:1990622 | CHOP-ATF3 complex(GO:1990622) |
| 0.2 | 1.4 | GO:0001940 | male pronucleus(GO:0001940) |
| 0.1 | 0.3 | GO:0070435 | Shc-EGFR complex(GO:0070435) |
| 0.1 | 1.8 | GO:0097470 | ribbon synapse(GO:0097470) |
| 0.1 | 0.3 | GO:0097135 | cyclin E2-CDK2 complex(GO:0097135) |
| 0.1 | 0.3 | GO:0032116 | SMC loading complex(GO:0032116) |
| 0.1 | 0.4 | GO:0031302 | intrinsic component of endosome membrane(GO:0031302) |
| 0.1 | 0.7 | GO:0016589 | NURF complex(GO:0016589) |
| 0.1 | 0.4 | GO:0044214 | spanning component of plasma membrane(GO:0044214) spanning component of membrane(GO:0089717) |
| 0.1 | 0.6 | GO:0072487 | MSL complex(GO:0072487) |
| 0.1 | 0.9 | GO:0070776 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.1 | 0.7 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.1 | 0.6 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.1 | 0.8 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.1 | 1.1 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.1 | 2.2 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.1 | 0.2 | GO:0044307 | dendritic branch(GO:0044307) |
| 0.1 | 0.3 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.1 | 0.8 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.0 | 0.7 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.0 | 0.2 | GO:0071149 | TEAD-2-YAP complex(GO:0071149) |
| 0.0 | 0.9 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.0 | 0.2 | GO:0044308 | axonal spine(GO:0044308) |
| 0.0 | 0.1 | GO:0034271 | phosphatidylinositol 3-kinase complex, class III, type I(GO:0034271) phosphatidylinositol 3-kinase complex, class III, type II(GO:0034272) |
| 0.0 | 0.4 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.3 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.0 | 1.1 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 0.3 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.0 | 0.3 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.0 | 0.6 | GO:0001741 | XY body(GO:0001741) |
| 0.0 | 0.1 | GO:0038039 | G-protein coupled receptor heterodimeric complex(GO:0038039) |
| 0.0 | 1.0 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 0.0 | GO:0098592 | cytoplasmic side of apical plasma membrane(GO:0098592) |
| 0.0 | 0.7 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 1.9 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 0.2 | GO:0060091 | kinocilium(GO:0060091) |
| 0.0 | 0.0 | GO:0048237 | rough endoplasmic reticulum lumen(GO:0048237) |
| 0.0 | 0.4 | GO:0031254 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.0 | 0.7 | GO:0032590 | dendrite membrane(GO:0032590) |
| 0.0 | 0.7 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 0.3 | GO:0010369 | chromocenter(GO:0010369) |
| 0.0 | 2.3 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.0 | 0.5 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 0.0 | GO:0034686 | integrin alphav-beta8 complex(GO:0034686) |
| 0.0 | 0.1 | GO:0098837 | postsynaptic recycling endosome(GO:0098837) |
| 0.0 | 0.1 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 0.0 | 1.1 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 0.1 | GO:1990682 | CSF1-CSF1R complex(GO:1990682) |
| 0.0 | 0.2 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.0 | 6.7 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 0.1 | GO:0031501 | mannosyltransferase complex(GO:0031501) |
| 0.0 | 0.2 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 0.1 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.0 | 0.2 | GO:0005845 | mRNA cap binding complex(GO:0005845) |
| 0.0 | 0.0 | GO:0097059 | CNTFR-CLCF1 complex(GO:0097059) |
| 0.0 | 0.3 | GO:0005849 | mRNA cleavage factor complex(GO:0005849) |
| 0.0 | 0.3 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.9 | GO:0016524 | latrotoxin receptor activity(GO:0016524) |
| 0.6 | 9.0 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.3 | 1.4 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.3 | 1.8 | GO:0016618 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 0.2 | 0.9 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.2 | 2.8 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.1 | 1.2 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.1 | 0.5 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.1 | 1.9 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.1 | 0.4 | GO:0061599 | nitrate reductase activity(GO:0008940) molybdopterin adenylyltransferase activity(GO:0061598) molybdopterin molybdotransferase activity(GO:0061599) |
| 0.1 | 0.3 | GO:0008267 | poly-glutamine tract binding(GO:0008267) |
| 0.1 | 0.5 | GO:0032810 | sterol response element binding(GO:0032810) |
| 0.1 | 0.7 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.1 | 3.2 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.1 | 0.7 | GO:0004305 | ethanolamine kinase activity(GO:0004305) |
| 0.1 | 1.9 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.1 | 0.7 | GO:0036132 | 13-prostaglandin reductase activity(GO:0036132) 15-oxoprostaglandin 13-oxidase activity(GO:0047522) |
| 0.1 | 0.5 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.1 | 0.3 | GO:0019797 | procollagen-proline 3-dioxygenase activity(GO:0019797) |
| 0.1 | 1.9 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.1 | 0.4 | GO:0005119 | smoothened binding(GO:0005119) |
| 0.1 | 0.6 | GO:0015235 | cobalamin transporter activity(GO:0015235) |
| 0.1 | 0.4 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.1 | 1.8 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.1 | 0.7 | GO:0036310 | annealing helicase activity(GO:0036310) |
| 0.1 | 0.6 | GO:0043184 | vascular endothelial growth factor receptor 2 binding(GO:0043184) |
| 0.1 | 0.4 | GO:0047057 | oxidoreductase activity, acting on the CH-OH group of donors, disulfide as acceptor(GO:0016900) vitamin-K-epoxide reductase (warfarin-sensitive) activity(GO:0047057) |
| 0.1 | 0.7 | GO:0017002 | activin-activated receptor activity(GO:0017002) |
| 0.1 | 0.8 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.1 | 0.5 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.1 | 0.7 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.1 | 0.5 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.1 | 0.3 | GO:0035276 | ethanol binding(GO:0035276) |
| 0.1 | 0.6 | GO:0008525 | phosphatidylcholine transporter activity(GO:0008525) |
| 0.1 | 0.2 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.1 | 0.5 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.1 | 0.7 | GO:0005347 | ATP transmembrane transporter activity(GO:0005347) ADP transmembrane transporter activity(GO:0015217) |
| 0.1 | 0.3 | GO:0046964 | 3'-phosphoadenosine 5'-phosphosulfate transmembrane transporter activity(GO:0046964) |
| 0.0 | 0.6 | GO:0003996 | acyl-CoA ligase activity(GO:0003996) |
| 0.0 | 0.4 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.0 | 0.1 | GO:0005477 | pyruvate secondary active transmembrane transporter activity(GO:0005477) |
| 0.0 | 0.4 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.0 | 0.1 | GO:0030272 | 5-formyltetrahydrofolate cyclo-ligase activity(GO:0030272) |
| 0.0 | 0.8 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.0 | 0.5 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 0.3 | GO:0048408 | epidermal growth factor binding(GO:0048408) |
| 0.0 | 0.3 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.0 | 0.2 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 0.0 | 0.3 | GO:0001025 | RNA polymerase III transcription factor binding(GO:0001025) |
| 0.0 | 0.6 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.0 | 0.3 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.0 | 0.2 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.0 | 0.5 | GO:0033592 | RNA strand annealing activity(GO:0033592) |
| 0.0 | 0.1 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 0.0 | 0.7 | GO:0005024 | transforming growth factor beta-activated receptor activity(GO:0005024) |
| 0.0 | 0.5 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.0 | 0.3 | GO:0004022 | alcohol dehydrogenase (NAD) activity(GO:0004022) |
| 0.0 | 1.4 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.0 | 0.1 | GO:0005011 | macrophage colony-stimulating factor receptor activity(GO:0005011) |
| 0.0 | 6.7 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 0.1 | GO:0004584 | dolichyl-phosphate-mannose-glycolipid alpha-mannosyltransferase activity(GO:0004584) |
| 0.0 | 0.1 | GO:0004768 | stearoyl-CoA 9-desaturase activity(GO:0004768) acyl-CoA desaturase activity(GO:0016215) |
| 0.0 | 3.3 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.0 | 0.2 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.1 | GO:0043208 | glycosphingolipid binding(GO:0043208) |
| 0.0 | 2.6 | GO:0008094 | DNA-dependent ATPase activity(GO:0008094) |
| 0.0 | 0.3 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 0.3 | GO:0097157 | pre-mRNA intronic binding(GO:0097157) |
| 0.0 | 0.2 | GO:0047696 | beta-adrenergic receptor kinase activity(GO:0047696) |
| 0.0 | 0.3 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.0 | 1.0 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.0 | 0.2 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.1 | GO:0001226 | RNA polymerase II transcription corepressor binding(GO:0001226) |
| 0.0 | 0.2 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.0 | 0.1 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.0 | 0.5 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 0.1 | GO:0008160 | protein tyrosine phosphatase activator activity(GO:0008160) |
| 0.0 | 0.3 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.0 | 0.1 | GO:0033981 | D-dopachrome decarboxylase activity(GO:0033981) |
| 0.0 | 0.1 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.0 | 0.5 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.0 | 0.1 | GO:0031750 | D3 dopamine receptor binding(GO:0031750) |
| 0.0 | 0.7 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 0.2 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.0 | 0.1 | GO:0003945 | N-acetyllactosamine synthase activity(GO:0003945) |
| 0.0 | 0.1 | GO:0044736 | acid-sensing ion channel activity(GO:0044736) |
| 0.0 | 0.1 | GO:0061649 | ubiquitinated histone binding(GO:0061649) |
| 0.0 | 0.1 | GO:0080023 | 3R-hydroxyacyl-CoA dehydratase activity(GO:0080023) |
| 0.0 | 0.3 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.0 | 8.3 | GO:0001077 | transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) |
| 0.0 | 1.4 | GO:0000980 | RNA polymerase II distal enhancer sequence-specific DNA binding(GO:0000980) |
| 0.0 | 0.2 | GO:0004727 | prenylated protein tyrosine phosphatase activity(GO:0004727) |
| 0.0 | 0.6 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 0.2 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.0 | 0.2 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.0 | 0.7 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.2 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.0 | 0.3 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 0.1 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.0 | 0.6 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 0.2 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.0 | 0.1 | GO:0000182 | rDNA binding(GO:0000182) |
| 0.0 | 1.0 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.2 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 0.1 | GO:1904315 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.0 | 0.1 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.0 | 0.1 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.0 | 0.3 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 0.4 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.0 | 0.2 | GO:0043176 | amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 0.0 | 2.4 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 0.3 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 0.4 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.1 | GO:0016841 | ammonia-lyase activity(GO:0016841) |
| 0.0 | 0.1 | GO:0016679 | oxidoreductase activity, acting on diphenols and related substances as donors(GO:0016679) |
| 0.0 | 0.2 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 0.1 | GO:0090599 | alpha-glucosidase activity(GO:0090599) |
| 0.0 | 0.1 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 0.0 | GO:0004803 | transposase activity(GO:0004803) |
| 0.0 | 0.0 | GO:0010385 | double-stranded methylated DNA binding(GO:0010385) |
| 0.0 | 0.3 | GO:0015299 | solute:proton antiporter activity(GO:0015299) |
| 0.0 | 0.2 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.0 | 0.2 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.2 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 0.1 | GO:0008048 | calcium sensitive guanylate cyclase activator activity(GO:0008048) |
| 0.0 | 0.3 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 0.0 | GO:0015056 | corticotrophin-releasing factor receptor activity(GO:0015056) |
| 0.0 | 0.2 | GO:0030297 | transmembrane receptor protein tyrosine kinase activator activity(GO:0030297) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.5 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.1 | 4.1 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 1.8 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.0 | 0.4 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.0 | 4.2 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 0.7 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.0 | 0.3 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 0.8 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 4.0 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 0.6 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 0.1 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.0 | 0.2 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.0 | 1.8 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 0.9 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 2.4 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 0.5 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 0.9 | PID ATM PATHWAY | ATM pathway |
| 0.0 | 2.0 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 0.9 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 0.1 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 0.4 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 0.3 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.0 | 0.3 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 0.3 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.0 | 0.6 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.4 | PID VEGFR1 PATHWAY | VEGFR1 specific signals |
| 0.0 | 0.3 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.0 | 0.9 | ST T CELL SIGNAL TRANSDUCTION | T Cell Signal Transduction |
| 0.0 | 0.6 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.0 | 0.9 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 0.4 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.3 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.1 | 1.1 | REACTOME SIGNALING BY NOTCH4 | Genes involved in Signaling by NOTCH4 |
| 0.1 | 1.2 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.1 | 1.5 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 2.5 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 1.8 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 0.9 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.0 | 0.9 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.0 | 0.8 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 0.7 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.6 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.0 | 0.8 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.0 | 0.3 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.0 | 0.3 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.2 | REACTOME P53 DEPENDENT G1 DNA DAMAGE RESPONSE | Genes involved in p53-Dependent G1 DNA Damage Response |
| 0.0 | 0.6 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.0 | 0.2 | REACTOME RAS ACTIVATION UOPN CA2 INFUX THROUGH NMDA RECEPTOR | Genes involved in Ras activation uopn Ca2+ infux through NMDA receptor |
| 0.0 | 1.2 | REACTOME CELL SURFACE INTERACTIONS AT THE VASCULAR WALL | Genes involved in Cell surface interactions at the vascular wall |
| 0.0 | 1.4 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.4 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.7 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 1.0 | REACTOME TRAFFICKING OF AMPA RECEPTORS | Genes involved in Trafficking of AMPA receptors |
| 0.0 | 0.2 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.0 | 0.5 | REACTOME RNA POL I TRANSCRIPTION INITIATION | Genes involved in RNA Polymerase I Transcription Initiation |
| 0.0 | 0.5 | REACTOME ACTIVATION OF GENES BY ATF4 | Genes involved in Activation of Genes by ATF4 |
| 0.0 | 0.2 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.0 | 0.2 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 0.1 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |