Project
Inflammatory response time course, HUVEC (Wada, 2009)
Navigation
Downloads
Results for NFAT5
Z-value: 0.52
Transcription factors associated with NFAT5
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NFAT5
|
ENSG00000102908.22 | NFAT5 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NFAT5 | hg38_v1_chr16_+_69565958_69565984 | -0.41 | 4.1e-02 | Click! |
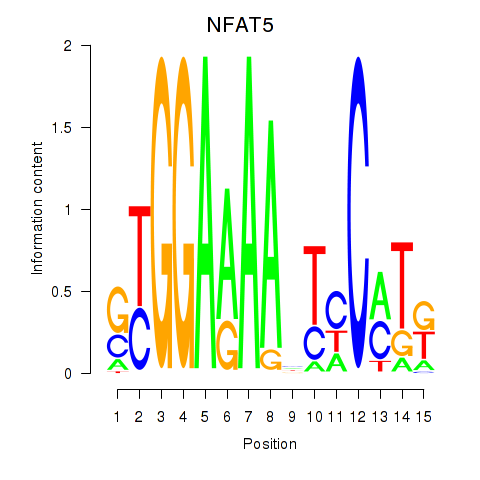
Activity profile of NFAT5 motif
Sorted Z-values of NFAT5 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of NFAT5
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr20_-_45547420 | 2.42 |
ENST00000504988.1
|
EPPIN-WFDC6
|
EPPIN-WFDC6 readthrough |
| chr20_-_45547648 | 2.12 |
ENST00000651288.1
|
EPPIN-WFDC6
|
EPPIN-WFDC6 readthrough |
| chr14_+_103123452 | 1.77 |
ENST00000558056.1
ENST00000560869.6 |
TNFAIP2
|
TNF alpha induced protein 2 |
| chr12_-_214146 | 1.30 |
ENST00000684302.1
|
SLC6A12
|
solute carrier family 6 member 12 |
| chr19_+_44751251 | 1.16 |
ENST00000444487.1
|
BCL3
|
BCL3 transcription coactivator |
| chr18_-_36122110 | 0.87 |
ENST00000586829.1
|
SLC39A6
|
solute carrier family 39 member 6 |
| chr1_-_56579555 | 0.85 |
ENST00000371250.4
|
PLPP3
|
phospholipid phosphatase 3 |
| chr11_-_8810635 | 0.72 |
ENST00000527510.5
ENST00000528527.5 ENST00000313726.11 ENST00000528523.5 |
DENND2B
|
DENN domain containing 2B |
| chr19_+_43533384 | 0.71 |
ENST00000601282.1
|
ZNF575
|
zinc finger protein 575 |
| chr3_+_122680802 | 0.66 |
ENST00000474629.7
|
PARP14
|
poly(ADP-ribose) polymerase family member 14 |
| chr7_+_143222037 | 0.61 |
ENST00000408947.4
|
TAS2R40
|
taste 2 receptor member 40 |
| chr11_+_8911280 | 0.55 |
ENST00000530281.5
ENST00000396648.6 ENST00000534147.5 ENST00000529942.1 |
AKIP1
|
A-kinase interacting protein 1 |
| chr19_-_49155130 | 0.55 |
ENST00000595625.1
|
HRC
|
histidine rich calcium binding protein |
| chr2_+_108377947 | 0.53 |
ENST00000272452.7
|
SULT1C4
|
sulfotransferase family 1C member 4 |
| chr7_-_19145306 | 0.51 |
ENST00000275461.3
|
FERD3L
|
Fer3 like bHLH transcription factor |
| chr16_-_75464364 | 0.45 |
ENST00000569540.5
ENST00000566594.1 |
TMEM170A
ENSG00000261717.5
|
transmembrane protein 170A novel TMEM170A-CFDP1 readthrough protein |
| chr14_+_22052503 | 0.44 |
ENST00000390449.3
|
TRAV21
|
T cell receptor alpha variable 21 |
| chr6_+_4889992 | 0.44 |
ENST00000343762.5
|
CDYL
|
chromodomain Y like |
| chrX_+_30235894 | 0.42 |
ENST00000620842.1
|
MAGEB3
|
MAGE family member B3 |
| chr1_-_150765735 | 0.40 |
ENST00000679898.1
ENST00000448301.7 ENST00000680664.1 ENST00000679512.1 ENST00000368985.8 ENST00000679582.1 |
CTSS
|
cathepsin S |
| chr19_+_48325522 | 0.40 |
ENST00000594198.1
ENST00000270221.11 ENST00000597279.5 ENST00000593437.1 |
EMP3
|
epithelial membrane protein 3 |
| chr12_+_22625357 | 0.39 |
ENST00000545979.2
|
ETNK1
|
ethanolamine kinase 1 |
| chr19_+_48325323 | 0.38 |
ENST00000596315.5
|
EMP3
|
epithelial membrane protein 3 |
| chr20_-_45547752 | 0.36 |
ENST00000409554.1
|
EPPIN
|
epididymal peptidase inhibitor |
| chr6_-_30684744 | 0.35 |
ENST00000615892.4
|
PPP1R18
|
protein phosphatase 1 regulatory subunit 18 |
| chr11_+_57181945 | 0.34 |
ENST00000497933.3
|
LRRC55
|
leucine rich repeat containing 55 |
| chr16_-_75464655 | 0.32 |
ENST00000569276.1
ENST00000357613.8 ENST00000561878.2 ENST00000566980.1 ENST00000567194.5 |
TMEM170A
ENSG00000261717.5
|
transmembrane protein 170A novel TMEM170A-CFDP1 readthrough protein |
| chr1_+_159204860 | 0.31 |
ENST00000368122.4
ENST00000368121.6 |
ACKR1
|
atypical chemokine receptor 1 (Duffy blood group) |
| chr3_+_98168700 | 0.31 |
ENST00000383696.4
|
OR5H15
|
olfactory receptor family 5 subfamily H member 15 |
| chr11_+_128693887 | 0.31 |
ENST00000281428.12
|
FLI1
|
Fli-1 proto-oncogene, ETS transcription factor |
| chr7_+_66205325 | 0.29 |
ENST00000304842.6
ENST00000649664.1 |
TPST1
|
tyrosylprotein sulfotransferase 1 |
| chr9_-_37465402 | 0.29 |
ENST00000307750.5
|
ZBTB5
|
zinc finger and BTB domain containing 5 |
| chrX_+_83861126 | 0.28 |
ENST00000621735.4
ENST00000329312.5 |
CYLC1
|
cylicin 1 |
| chr2_+_26346086 | 0.27 |
ENST00000613142.4
ENST00000260585.12 ENST00000447170.1 |
SELENOI
|
selenoprotein I |
| chr10_-_67696115 | 0.26 |
ENST00000433211.7
|
CTNNA3
|
catenin alpha 3 |
| chr3_+_128051610 | 0.26 |
ENST00000464451.5
|
SEC61A1
|
SEC61 translocon subunit alpha 1 |
| chr17_+_7012417 | 0.26 |
ENST00000548577.5
|
RNASEK
|
ribonuclease K |
| chr14_+_64113054 | 0.25 |
ENST00000673869.1
|
SYNE2
|
spectrin repeat containing nuclear envelope protein 2 |
| chr17_-_41140487 | 0.25 |
ENST00000345847.4
|
KRTAP4-6
|
keratin associated protein 4-6 |
| chr17_+_34356472 | 0.24 |
ENST00000225844.7
|
CCL13
|
C-C motif chemokine ligand 13 |
| chr6_+_101393699 | 0.23 |
ENST00000369134.9
ENST00000684068.1 ENST00000683903.1 ENST00000681975.1 |
GRIK2
|
glutamate ionotropic receptor kainate type subunit 2 |
| chr19_-_43596123 | 0.22 |
ENST00000422989.6
ENST00000598324.1 |
IRGQ
|
immunity related GTPase Q |
| chr7_+_112423137 | 0.22 |
ENST00000005558.8
ENST00000621379.4 |
IFRD1
|
interferon related developmental regulator 1 |
| chr14_+_94612383 | 0.22 |
ENST00000393080.8
ENST00000555820.1 ENST00000393078.5 ENST00000467132.5 |
SERPINA3
|
serpin family A member 3 |
| chr22_-_37984534 | 0.21 |
ENST00000396884.8
|
SOX10
|
SRY-box transcription factor 10 |
| chr17_-_81961177 | 0.21 |
ENST00000409678.8
|
NOTUM
|
notum, palmitoleoyl-protein carboxylesterase |
| chr19_-_54364807 | 0.21 |
ENST00000474878.5
|
LAIR1
|
leukocyte associated immunoglobulin like receptor 1 |
| chr14_+_64113084 | 0.21 |
ENST00000673797.1
|
SYNE2
|
spectrin repeat containing nuclear envelope protein 2 |
| chr18_+_31447732 | 0.20 |
ENST00000257189.5
|
DSG3
|
desmoglein 3 |
| chr10_+_70052806 | 0.20 |
ENST00000676609.1
ENST00000679349.1 ENST00000373255.9 ENST00000676608.1 ENST00000455786.1 |
MACROH2A2
|
macroH2A.2 histone |
| chr1_-_159925496 | 0.20 |
ENST00000368097.9
|
TAGLN2
|
transgelin 2 |
| chr18_-_77127935 | 0.20 |
ENST00000581878.5
|
MBP
|
myelin basic protein |
| chr1_-_8879170 | 0.19 |
ENST00000489867.2
|
ENO1
|
enolase 1 |
| chr19_+_7534004 | 0.19 |
ENST00000221249.10
ENST00000601668.5 ENST00000601001.5 |
PNPLA6
|
patatin like phospholipase domain containing 6 |
| chr19_+_17309531 | 0.19 |
ENST00000359866.9
|
DDA1
|
DET1 and DDB1 associated 1 |
| chr3_+_9792495 | 0.19 |
ENST00000498623.6
|
ARPC4
|
actin related protein 2/3 complex subunit 4 |
| chr11_-_58844695 | 0.19 |
ENST00000287275.6
|
GLYATL2
|
glycine-N-acyltransferase like 2 |
| chr3_+_98149326 | 0.19 |
ENST00000437310.1
|
OR5H14
|
olfactory receptor family 5 subfamily H member 14 |
| chr6_-_31958431 | 0.17 |
ENST00000625905.1
ENST00000454913.5 ENST00000436289.6 |
NELFE
|
negative elongation factor complex member E |
| chr6_+_131199979 | 0.17 |
ENST00000537868.5
|
AKAP7
|
A-kinase anchoring protein 7 |
| chr11_+_131911396 | 0.17 |
ENST00000425719.6
ENST00000374784.5 |
NTM
|
neurotrimin |
| chr14_-_25010604 | 0.16 |
ENST00000550887.5
|
STXBP6
|
syntaxin binding protein 6 |
| chr8_+_49911396 | 0.16 |
ENST00000642720.2
|
SNTG1
|
syntrophin gamma 1 |
| chr9_-_34589716 | 0.15 |
ENST00000378980.8
|
CNTFR
|
ciliary neurotrophic factor receptor |
| chr8_+_49911604 | 0.15 |
ENST00000642164.1
ENST00000644093.1 ENST00000643999.1 ENST00000647073.1 ENST00000646880.1 |
SNTG1
|
syntrophin gamma 1 |
| chr9_+_136807911 | 0.15 |
ENST00000371671.9
ENST00000311502.12 ENST00000371663.10 |
RABL6
|
RAB, member RAS oncogene family like 6 |
| chr9_-_34589701 | 0.15 |
ENST00000351266.8
|
CNTFR
|
ciliary neurotrophic factor receptor |
| chr17_+_42798881 | 0.15 |
ENST00000588527.5
|
CNTD1
|
cyclin N-terminal domain containing 1 |
| chr7_-_100100716 | 0.15 |
ENST00000354230.7
ENST00000425308.5 |
MCM7
|
minichromosome maintenance complex component 7 |
| chr22_-_29315656 | 0.14 |
ENST00000401450.3
ENST00000216101.7 |
RASL10A
|
RAS like family 10 member A |
| chr17_+_42798857 | 0.14 |
ENST00000588408.6
|
CNTD1
|
cyclin N-terminal domain containing 1 |
| chr7_+_1088106 | 0.14 |
ENST00000397088.4
|
GPER1
|
G protein-coupled estrogen receptor 1 |
| chr17_+_70075317 | 0.14 |
ENST00000589377.1
|
KCNJ16
|
potassium inwardly rectifying channel subfamily J member 16 |
| chr19_+_17795129 | 0.14 |
ENST00000600777.1
ENST00000318683.7 ENST00000595387.1 |
B3GNT3
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 3 |
| chr9_-_95317671 | 0.13 |
ENST00000490972.7
ENST00000647778.1 ENST00000649611.1 ENST00000289081.8 |
FANCC
|
FA complementation group C |
| chr16_-_1981463 | 0.13 |
ENST00000356120.9
ENST00000354249.8 |
NOXO1
|
NADPH oxidase organizer 1 |
| chr3_-_44510602 | 0.13 |
ENST00000436261.6
|
ZNF852
|
zinc finger protein 852 |
| chr8_-_51408974 | 0.13 |
ENST00000522933.5
|
PXDNL
|
peroxidasin like |
| chr3_+_9793074 | 0.13 |
ENST00000397261.8
ENST00000485273.1 ENST00000433034.1 ENST00000397256.5 |
ARPC4
ARPC4-TTLL3
|
actin related protein 2/3 complex subunit 4 ARPC4-TTLL3 readthrough |
| chr6_-_27912396 | 0.13 |
ENST00000303324.4
|
OR2B2
|
olfactory receptor family 2 subfamily B member 2 |
| chr17_+_70075215 | 0.12 |
ENST00000283936.5
ENST00000615244.4 ENST00000392671.6 |
KCNJ16
|
potassium inwardly rectifying channel subfamily J member 16 |
| chr9_+_133376334 | 0.12 |
ENST00000371957.4
|
STKLD1
|
serine/threonine kinase like domain containing 1 |
| chr1_-_53738024 | 0.12 |
ENST00000628545.1
|
GLIS1
|
GLIS family zinc finger 1 |
| chr9_-_133376111 | 0.12 |
ENST00000545297.5
ENST00000613129.4 ENST00000618229.4 |
SURF4
|
surfeit 4 |
| chr10_-_133308844 | 0.11 |
ENST00000682905.1
ENST00000682515.1 ENST00000683612.1 ENST00000252936.8 ENST00000417178.7 ENST00000682161.1 ENST00000683014.1 ENST00000683060.1 ENST00000683383.1 |
TUBGCP2
|
tubulin gamma complex associated protein 2 |
| chr2_+_68157877 | 0.11 |
ENST00000263657.7
|
PNO1
|
partner of NOB1 homolog |
| chr1_-_85404494 | 0.11 |
ENST00000633113.1
|
DDAH1
|
dimethylarginine dimethylaminohydrolase 1 |
| chr19_-_55831987 | 0.11 |
ENST00000592953.5
ENST00000589093.5 |
NLRP11
|
NLR family pyrin domain containing 11 |
| chr7_+_149872955 | 0.11 |
ENST00000421974.7
ENST00000456496.7 |
ATP6V0E2
|
ATPase H+ transporting V0 subunit e2 |
| chr17_+_42798779 | 0.11 |
ENST00000585355.5
|
CNTD1
|
cyclin N-terminal domain containing 1 |
| chr7_+_128758947 | 0.11 |
ENST00000493278.1
|
CALU
|
calumenin |
| chr19_-_54364863 | 0.10 |
ENST00000348231.8
|
LAIR1
|
leukocyte associated immunoglobulin like receptor 1 |
| chrX_-_45200828 | 0.10 |
ENST00000398000.7
|
DIPK2B
|
divergent protein kinase domain 2B |
| chr1_-_60073750 | 0.10 |
ENST00000371201.3
|
C1orf87
|
chromosome 1 open reading frame 87 |
| chr7_-_93226449 | 0.10 |
ENST00000394468.7
ENST00000453812.2 |
HEPACAM2
|
HEPACAM family member 2 |
| chrX_-_70931089 | 0.10 |
ENST00000374299.8
ENST00000298085.4 |
SLC7A3
|
solute carrier family 7 member 3 |
| chr14_-_106803221 | 0.10 |
ENST00000390636.2
|
IGHV3-73
|
immunoglobulin heavy variable 3-73 |
| chr9_-_27005659 | 0.10 |
ENST00000380055.6
|
LRRC19
|
leucine rich repeat containing 19 |
| chr2_+_184598520 | 0.10 |
ENST00000302277.7
|
ZNF804A
|
zinc finger protein 804A |
| chr19_-_45782479 | 0.09 |
ENST00000447742.6
ENST00000354227.9 ENST00000291270.9 ENST00000683086.1 ENST00000343373.10 |
DMPK
|
DM1 protein kinase |
| chr16_+_2029636 | 0.09 |
ENST00000561844.5
|
SLC9A3R2
|
SLC9A3 regulator 2 |
| chr11_-_82846128 | 0.09 |
ENST00000679809.1
ENST00000680186.1 ENST00000681592.1 |
PRCP
|
prolylcarboxypeptidase |
| chr9_+_122614738 | 0.09 |
ENST00000297913.3
|
OR1Q1
|
olfactory receptor family 1 subfamily Q member 1 |
| chr19_-_54364983 | 0.09 |
ENST00000434277.6
|
LAIR1
|
leukocyte associated immunoglobulin like receptor 1 |
| chr20_+_34516394 | 0.09 |
ENST00000357156.7
ENST00000300469.13 ENST00000374846.3 |
DYNLRB1
|
dynein light chain roadblock-type 1 |
| chr18_+_10454584 | 0.09 |
ENST00000355285.10
|
APCDD1
|
APC down-regulated 1 |
| chr1_-_60073733 | 0.09 |
ENST00000450089.6
|
C1orf87
|
chromosome 1 open reading frame 87 |
| chr1_+_155033824 | 0.09 |
ENST00000295542.6
ENST00000423025.6 ENST00000368419.2 |
DCST1
|
DC-STAMP domain containing 1 |
| chr11_-_111923722 | 0.09 |
ENST00000527950.5
|
CRYAB
|
crystallin alpha B |
| chr12_+_119334722 | 0.09 |
ENST00000327554.3
|
CCDC60
|
coiled-coil domain containing 60 |
| chr17_+_4950147 | 0.08 |
ENST00000522301.5
|
ENO3
|
enolase 3 |
| chrX_+_48897900 | 0.08 |
ENST00000376566.8
ENST00000376563.5 |
PQBP1
|
polyglutamine binding protein 1 |
| chr1_+_93345893 | 0.08 |
ENST00000370272.9
ENST00000370267.1 |
DR1
|
down-regulator of transcription 1 |
| chr13_+_112376341 | 0.08 |
ENST00000414180.5
ENST00000283550.8 ENST00000443541.5 |
SPACA7
|
sperm acrosome associated 7 |
| chr10_-_56361235 | 0.08 |
ENST00000373944.8
ENST00000361148.6 ENST00000395405.5 |
ZWINT
|
ZW10 interacting kinetochore protein |
| chr10_-_62236112 | 0.07 |
ENST00000315289.6
|
RTKN2
|
rhotekin 2 |
| chr19_-_46784905 | 0.07 |
ENST00000594991.5
|
SLC1A5
|
solute carrier family 1 member 5 |
| chr6_+_89080739 | 0.07 |
ENST00000369472.1
ENST00000336032.4 |
PNRC1
|
proline rich nuclear receptor coactivator 1 |
| chr7_-_102478895 | 0.07 |
ENST00000393794.4
ENST00000292614.9 |
POLR2J
|
RNA polymerase II subunit J |
| chr4_-_154590735 | 0.07 |
ENST00000403106.8
ENST00000622532.1 ENST00000651975.1 |
FGA
|
fibrinogen alpha chain |
| chr7_+_1086800 | 0.07 |
ENST00000413368.5
ENST00000397092.5 ENST00000297469.3 |
GPER1
|
G protein-coupled estrogen receptor 1 |
| chr1_-_155033778 | 0.06 |
ENST00000368424.4
|
DCST2
|
DC-STAMP domain containing 2 |
| chr1_+_161707244 | 0.06 |
ENST00000349527.8
ENST00000294796.8 ENST00000309691.10 ENST00000367953.7 ENST00000367950.2 |
FCRLA
|
Fc receptor like A |
| chr3_-_9792404 | 0.06 |
ENST00000301964.7
|
TADA3
|
transcriptional adaptor 3 |
| chr3_-_116445458 | 0.06 |
ENST00000490035.7
|
LSAMP
|
limbic system associated membrane protein |
| chr19_+_49496424 | 0.05 |
ENST00000596873.1
ENST00000594493.1 ENST00000270625.7 ENST00000599561.1 |
RPS11
|
ribosomal protein S11 |
| chr3_-_116444983 | 0.05 |
ENST00000333617.8
|
LSAMP
|
limbic system associated membrane protein |
| chr12_-_75209814 | 0.05 |
ENST00000549446.6
|
KCNC2
|
potassium voltage-gated channel subfamily C member 2 |
| chr5_+_140882116 | 0.05 |
ENST00000289272.3
ENST00000409494.5 ENST00000617769.1 |
PCDHA13
|
protocadherin alpha 13 |
| chr5_-_170267285 | 0.05 |
ENST00000521416.5
ENST00000520344.1 |
LCP2
|
lymphocyte cytosolic protein 2 |
| chr4_+_154743993 | 0.05 |
ENST00000336356.4
|
LRAT
|
lecithin retinol acyltransferase |
| chr7_+_33904911 | 0.05 |
ENST00000297161.6
|
BMPER
|
BMP binding endothelial regulator |
| chr11_+_131370478 | 0.05 |
ENST00000374791.7
ENST00000683400.1 ENST00000436745.5 |
NTM
|
neurotrimin |
| chr3_-_149576203 | 0.05 |
ENST00000472417.1
|
WWTR1
|
WW domain containing transcription regulator 1 |
| chr16_+_89921851 | 0.05 |
ENST00000554444.5
ENST00000556565.5 |
TUBB3
|
tubulin beta 3 class III |
| chr12_-_75209701 | 0.04 |
ENST00000350228.6
ENST00000298972.5 |
KCNC2
|
potassium voltage-gated channel subfamily C member 2 |
| chr15_-_65792283 | 0.04 |
ENST00000443035.8
ENST00000564674.5 |
DENND4A
|
DENN domain containing 4A |
| chr19_-_54364908 | 0.04 |
ENST00000391742.7
|
LAIR1
|
leukocyte associated immunoglobulin like receptor 1 |
| chr12_+_53454819 | 0.04 |
ENST00000562264.5
|
PCBP2
|
poly(rC) binding protein 2 |
| chr12_+_53454764 | 0.04 |
ENST00000439930.7
ENST00000548933.5 |
PCBP2
|
poly(rC) binding protein 2 |
| chr7_+_143183419 | 0.04 |
ENST00000446620.1
|
TAS2R39
|
taste 2 receptor member 39 |
| chr14_-_106389858 | 0.03 |
ENST00000390617.2
|
IGHV3-35
|
immunoglobulin heavy variable 3-35 (non-functional) |
| chr20_-_23751256 | 0.03 |
ENST00000398402.1
|
CST1
|
cystatin SN |
| chr12_+_69348372 | 0.03 |
ENST00000261267.7
ENST00000549690.1 ENST00000548839.1 |
LYZ
|
lysozyme |
| chr1_-_153544997 | 0.03 |
ENST00000368715.5
|
S100A4
|
S100 calcium binding protein A4 |
| chr17_+_18781155 | 0.03 |
ENST00000574226.5
ENST00000575261.5 ENST00000307767.13 |
TVP23B
|
trans-golgi network vesicle protein 23 homolog B |
| chr10_+_97849824 | 0.03 |
ENST00000370602.6
|
GOLGA7B
|
golgin A7 family member B |
| chr5_-_151141631 | 0.03 |
ENST00000523714.5
ENST00000521749.5 |
ANXA6
|
annexin A6 |
| chr3_-_41961989 | 0.03 |
ENST00000420927.5
ENST00000301831.9 ENST00000414606.1 |
ULK4
|
unc-51 like kinase 4 |
| chr4_+_119027335 | 0.03 |
ENST00000627783.2
|
SYNPO2
|
synaptopodin 2 |
| chr17_-_15563428 | 0.02 |
ENST00000584811.5
ENST00000419890.3 ENST00000518321.6 ENST00000438826.7 ENST00000225576.7 ENST00000428082.6 ENST00000522212.6 |
TVP23C
TVP23C-CDRT4
|
trans-golgi network vesicle protein 23 homolog C TVP23C-CDRT4 readthrough |
| chr16_+_33009175 | 0.02 |
ENST00000565407.2
|
IGHV3OR16-8
|
immunoglobulin heavy variable 3/OR16-8 (non-functional) |
| chr15_+_90872162 | 0.02 |
ENST00000680053.1
|
FURIN
|
furin, paired basic amino acid cleaving enzyme |
| chr14_-_59630582 | 0.02 |
ENST00000395090.5
|
RTN1
|
reticulon 1 |
| chr13_+_30422487 | 0.02 |
ENST00000638137.1
ENST00000635918.2 |
UBE2L5
|
ubiquitin conjugating enzyme E2 L5 |
| chr11_+_65386611 | 0.02 |
ENST00000531296.1
ENST00000533782.5 ENST00000355991.9 ENST00000317568.10 ENST00000416776.6 ENST00000526201.1 |
FRMD8
|
FERM domain containing 8 |
| chr7_-_101245056 | 0.02 |
ENST00000223136.5
|
FIS1
|
fission, mitochondrial 1 |
| chr12_+_2053311 | 0.02 |
ENST00000399617.6
ENST00000683482.1 |
CACNA1C
|
calcium voltage-gated channel subunit alpha1 C |
| chr6_+_150143018 | 0.02 |
ENST00000361131.5
|
PPP1R14C
|
protein phosphatase 1 regulatory inhibitor subunit 14C |
| chr15_+_44736556 | 0.02 |
ENST00000338264.8
|
TRIM69
|
tripartite motif containing 69 |
| chr2_+_233712905 | 0.02 |
ENST00000373414.4
|
UGT1A5
|
UDP glucuronosyltransferase family 1 member A5 |
| chr1_-_155241220 | 0.02 |
ENST00000368373.8
ENST00000427500.7 |
GBA
|
glucosylceramidase beta |
| chr15_+_44736522 | 0.02 |
ENST00000329464.9
ENST00000558329.5 ENST00000560442.5 ENST00000561043.5 |
TRIM69
|
tripartite motif containing 69 |
| chr1_-_44141631 | 0.02 |
ENST00000634670.1
|
KLF18
|
Kruppel like factor 18 |
| chr3_+_40505992 | 0.02 |
ENST00000420891.5
ENST00000314529.10 ENST00000418905.1 |
ZNF620
|
zinc finger protein 620 |
| chr17_+_18781250 | 0.02 |
ENST00000476139.5
|
TVP23B
|
trans-golgi network vesicle protein 23 homolog B |
| chr20_-_23750929 | 0.01 |
ENST00000304749.7
|
CST1
|
cystatin SN |
| chr9_+_68356603 | 0.01 |
ENST00000396396.6
|
PGM5
|
phosphoglucomutase 5 |
| chr6_-_46954922 | 0.01 |
ENST00000265417.7
|
ADGRF5
|
adhesion G protein-coupled receptor F5 |
| chr1_-_114757971 | 0.01 |
ENST00000261443.9
ENST00000534699.5 ENST00000339438.10 ENST00000358528.9 ENST00000529046.5 ENST00000525970.5 ENST00000369530.5 ENST00000530886.5 ENST00000610726.4 |
CSDE1
|
cold shock domain containing E1 |
| chr4_-_139556199 | 0.01 |
ENST00000274031.8
ENST00000506866.6 |
SETD7
|
SET domain containing 7, histone lysine methyltransferase |
| chr21_-_32279012 | 0.01 |
ENST00000290130.4
|
MIS18A
|
MIS18 kinetochore protein A |
| chr19_+_859654 | 0.00 |
ENST00000592860.2
ENST00000327726.11 |
CFD
|
complement factor D |
| chr22_+_37849372 | 0.00 |
ENST00000624234.3
ENST00000381683.10 ENST00000652021.1 ENST00000414316.5 |
EIF3L
|
eukaryotic translation initiation factor 3 subunit L |
| chr4_+_17577487 | 0.00 |
ENST00000606142.5
|
LAP3
|
leucine aminopeptidase 3 |
| chr22_+_37849437 | 0.00 |
ENST00000406934.5
ENST00000451427.1 |
EIF3L
|
eukaryotic translation initiation factor 3 subunit L |
| chr17_-_352784 | 0.00 |
ENST00000577079.5
ENST00000331302.12 ENST00000618002.4 ENST00000536489.6 |
RPH3AL
|
rabphilin 3A like (without C2 domains) |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.2 | GO:0045082 | positive regulation of interleukin-10 biosynthetic process(GO:0045082) |
| 0.2 | 0.5 | GO:1901899 | positive regulation of relaxation of cardiac muscle(GO:1901899) |
| 0.1 | 0.4 | GO:0034769 | basement membrane disassembly(GO:0034769) |
| 0.1 | 1.9 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.1 | 0.3 | GO:0006478 | peptidyl-tyrosine sulfation(GO:0006478) |
| 0.1 | 0.2 | GO:0002434 | immune complex clearance(GO:0002434) immune complex clearance by monocytes and macrophages(GO:0002436) regulation of immune complex clearance by monocytes and macrophages(GO:0090264) response to cyclosporin A(GO:1905237) |
| 0.1 | 0.2 | GO:0007518 | myoblast fate determination(GO:0007518) |
| 0.1 | 0.2 | GO:0031959 | mineralocorticoid receptor signaling pathway(GO:0031959) positive regulation of cardiac vascular smooth muscle cell differentiation(GO:2000724) |
| 0.1 | 0.8 | GO:0008354 | germ cell migration(GO:0008354) |
| 0.1 | 0.5 | GO:0033504 | floor plate development(GO:0033504) |
| 0.1 | 1.3 | GO:0015812 | gamma-aminobutyric acid transport(GO:0015812) |
| 0.1 | 0.5 | GO:0031022 | nuclear migration along microfilament(GO:0031022) |
| 0.1 | 0.9 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.0 | 0.3 | GO:0039019 | pronephric nephron development(GO:0039019) |
| 0.0 | 0.2 | GO:0071393 | cellular response to progesterone stimulus(GO:0071393) |
| 0.0 | 0.3 | GO:1901318 | negative regulation of sperm motility(GO:1901318) |
| 0.0 | 0.2 | GO:1904207 | maintenance of blood-brain barrier(GO:0035633) regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904207) positive regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904209) |
| 0.0 | 0.3 | GO:0086042 | cardiac muscle cell-cardiac muscle cell adhesion(GO:0086042) bundle of His cell-Purkinje myocyte adhesion involved in cell communication(GO:0086073) |
| 0.0 | 0.8 | GO:0071786 | nuclear pore complex assembly(GO:0051292) endoplasmic reticulum tubular network organization(GO:0071786) |
| 0.0 | 0.8 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.0 | 0.3 | GO:0070120 | brainstem development(GO:0003360) ciliary neurotrophic factor-mediated signaling pathway(GO:0070120) |
| 0.0 | 0.4 | GO:1903818 | positive regulation of voltage-gated potassium channel activity(GO:1903818) |
| 0.0 | 0.7 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.0 | 0.1 | GO:0018094 | protein polyglycylation(GO:0018094) |
| 0.0 | 0.1 | GO:1903401 | L-lysine transmembrane transport(GO:1903401) |
| 0.0 | 0.1 | GO:0030311 | poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.0 | 0.5 | GO:0051923 | sulfation(GO:0051923) |
| 0.0 | 0.2 | GO:0010756 | positive regulation of plasminogen activation(GO:0010756) |
| 0.0 | 0.1 | GO:0010585 | glutamine secretion(GO:0010585) L-glutamine import(GO:0036229) L-glutamine import into cell(GO:1903803) |
| 0.0 | 0.2 | GO:0051967 | negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.0 | 0.1 | GO:0051418 | interphase microtubule nucleation by interphase microtubule organizing center(GO:0051415) microtubule nucleation by microtubule organizing center(GO:0051418) |
| 0.0 | 0.2 | GO:0042159 | lipoprotein catabolic process(GO:0042159) |
| 0.0 | 0.1 | GO:0002353 | kinin cascade(GO:0002254) plasma kallikrein-kinin cascade(GO:0002353) |
| 0.0 | 0.1 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.0 | 0.3 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.0 | 0.2 | GO:1900364 | negative regulation of mRNA polyadenylation(GO:1900364) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.2 | GO:0033257 | Bcl3/NF-kappaB2 complex(GO:0033257) |
| 0.1 | 0.3 | GO:0043159 | cytoskeletal calyx(GO:0033150) acrosomal matrix(GO:0043159) |
| 0.1 | 0.3 | GO:0097059 | CNTFR-CLCF1 complex(GO:0097059) |
| 0.1 | 1.9 | GO:0000145 | exocyst(GO:0000145) |
| 0.1 | 0.7 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.1 | 0.4 | GO:0036021 | endolysosome lumen(GO:0036021) |
| 0.0 | 0.3 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.0 | 0.3 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.0 | 0.5 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.2 | GO:0032021 | NELF complex(GO:0032021) |
| 0.0 | 0.1 | GO:0008275 | gamma-tubulin small complex(GO:0008275) |
| 0.0 | 0.3 | GO:0016013 | syntrophin complex(GO:0016013) |
| 0.0 | 0.2 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.0 | 0.2 | GO:0044327 | dendritic spine head(GO:0044327) |
| 0.0 | 0.3 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 0.7 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.0 | 0.1 | GO:0071598 | neuronal ribonucleoprotein granule(GO:0071598) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.3 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.1 | 0.8 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 0.1 | 0.3 | GO:0008476 | protein-tyrosine sulfotransferase activity(GO:0008476) |
| 0.1 | 0.3 | GO:0004307 | ethanolaminephosphotransferase activity(GO:0004307) |
| 0.1 | 0.2 | GO:0031727 | CCR2 chemokine receptor binding(GO:0031727) |
| 0.1 | 0.4 | GO:0004305 | ethanolamine kinase activity(GO:0004305) |
| 0.0 | 0.1 | GO:0047223 | beta-1,3-galactosyl-O-glycosyl-glycoprotein beta-1,3-N-acetylglucosaminyltransferase activity(GO:0047223) |
| 0.0 | 0.3 | GO:0086083 | cell adhesive protein binding involved in bundle of His cell-Purkinje myocyte communication(GO:0086083) |
| 0.0 | 0.2 | GO:0047961 | glycine N-acyltransferase activity(GO:0047961) |
| 0.0 | 0.3 | GO:0004897 | ciliary neurotrophic factor receptor activity(GO:0004897) |
| 0.0 | 0.3 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.0 | 0.5 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.0 | 0.2 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.0 | 0.1 | GO:0070735 | protein-glycine ligase activity(GO:0070735) |
| 0.0 | 0.9 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.3 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.0 | 0.1 | GO:0015189 | L-lysine transmembrane transporter activity(GO:0015189) |
| 0.0 | 0.2 | GO:1990239 | steroid hormone binding(GO:1990239) |
| 0.0 | 0.1 | GO:0016715 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced ascorbate as one donor, and incorporation of one atom of oxygen(GO:0016715) |
| 0.0 | 0.5 | GO:0005549 | odorant binding(GO:0005549) |
| 0.0 | 0.7 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.1 | GO:0016403 | dimethylargininase activity(GO:0016403) |
| 0.0 | 0.3 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.0 | 0.2 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.1 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.2 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.3 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 0.4 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.0 | 0.9 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.5 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |