|
chr1_+_81800906
Show fit
|
5.07 |
ENST00000674393.1
ENST00000674208.1
|
ADGRL2
|
adhesion G protein-coupled receptor L2
|
|
chr4_+_41612892
Show fit
|
5.05 |
ENST00000509454.5
ENST00000396595.7
ENST00000381753.8
|
LIMCH1
|
LIM and calponin homology domains 1
|
|
chr14_-_91946989
Show fit
|
4.72 |
ENST00000556154.5
|
FBLN5
|
fibulin 5
|
|
chr10_+_92831153
Show fit
|
4.36 |
ENST00000672817.1
|
EXOC6
|
exocyst complex component 6
|
|
chr4_+_41613476
Show fit
|
4.22 |
ENST00000508466.1
|
LIMCH1
|
LIM and calponin homology domains 1
|
|
chr14_-_91947383
Show fit
|
3.55 |
ENST00000267620.14
|
FBLN5
|
fibulin 5
|
|
chr4_-_185812209
Show fit
|
3.44 |
ENST00000393523.6
ENST00000393528.7
ENST00000449407.6
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr1_+_61082553
Show fit
|
3.41 |
ENST00000403491.8
ENST00000371187.7
|
NFIA
|
nuclear factor I A
|
|
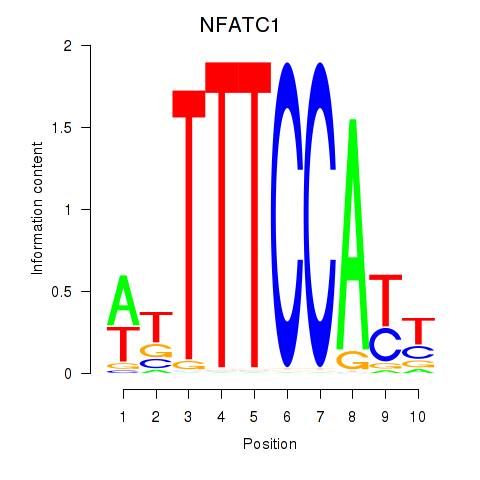
chr18_+_79395856
Show fit
|
3.27 |
ENST00000253506.9
ENST00000591814.5
ENST00000427363.7
|
NFATC1
|
nuclear factor of activated T cells 1
|
|
chr18_+_79395942
Show fit
|
3.05 |
ENST00000397790.6
|
NFATC1
|
nuclear factor of activated T cells 1
|
|
chr1_+_61081728
Show fit
|
3.02 |
ENST00000371189.8
|
NFIA
|
nuclear factor I A
|
|
chr18_-_25352116
Show fit
|
2.82 |
ENST00000584787.5
ENST00000538137.6
ENST00000361524.8
|
ZNF521
|
zinc finger protein 521
|
|
chr1_+_61082702
Show fit
|
2.81 |
ENST00000485903.6
ENST00000371185.6
ENST00000371184.6
|
NFIA
|
nuclear factor I A
|
|
chr9_-_14314519
Show fit
|
2.70 |
ENST00000397581.6
|
NFIB
|
nuclear factor I B
|
|
chr9_-_14314567
Show fit
|
2.69 |
ENST00000397579.6
|
NFIB
|
nuclear factor I B
|
|
chr9_-_16870702
Show fit
|
2.62 |
ENST00000380667.6
ENST00000545497.5
ENST00000486514.5
|
BNC2
|
basonuclin 2
|
|
chrX_-_63785510
Show fit
|
2.56 |
ENST00000437457.6
ENST00000374878.5
ENST00000623517.3
|
ARHGEF9
|
Cdc42 guanine nucleotide exchange factor 9
|
|
chr5_-_88883701
Show fit
|
2.38 |
ENST00000636998.1
|
MEF2C
|
myocyte enhancer factor 2C
|
|
chr21_-_34526815
Show fit
|
2.34 |
ENST00000492600.1
|
RCAN1
|
regulator of calcineurin 1
|
|
chr21_-_34526850
Show fit
|
2.33 |
ENST00000481448.5
ENST00000381132.6
|
RCAN1
|
regulator of calcineurin 1
|
|
chr21_-_26573211
Show fit
|
2.25 |
ENST00000299340.9
ENST00000652641.2
|
CYYR1
|
cysteine and tyrosine rich 1
|
|
chr1_+_81800368
Show fit
|
2.06 |
ENST00000674489.1
ENST00000674442.1
ENST00000674419.1
ENST00000674407.1
ENST00000674168.1
ENST00000674307.1
ENST00000674209.1
ENST00000370715.5
ENST00000370713.5
ENST00000319517.10
ENST00000627151.2
ENST00000370717.6
|
ADGRL2
|
adhesion G protein-coupled receptor L2
|
|
chr7_-_120858066
Show fit
|
2.01 |
ENST00000222747.8
|
TSPAN12
|
tetraspanin 12
|
|
chr20_+_43945677
Show fit
|
1.96 |
ENST00000358131.5
|
TOX2
|
TOX high mobility group box family member 2
|
|
chr2_-_156342348
Show fit
|
1.93 |
ENST00000409572.5
|
NR4A2
|
nuclear receptor subfamily 4 group A member 2
|
|
chr12_+_93572664
Show fit
|
1.88 |
ENST00000551556.2
|
SOCS2
|
suppressor of cytokine signaling 2
|
|
chr19_+_39406831
Show fit
|
1.85 |
ENST00000597629.3
ENST00000594442.2
ENST00000594045.2
|
ZFP36
|
ZFP36 ring finger protein
|
|
chr3_-_142028597
Show fit
|
1.81 |
ENST00000467667.5
|
TFDP2
|
transcription factor Dp-2
|
|
chr5_-_88883420
Show fit
|
1.77 |
ENST00000437473.6
|
MEF2C
|
myocyte enhancer factor 2C
|
|
chr3_-_15798184
Show fit
|
1.76 |
ENST00000624145.3
|
ANKRD28
|
ankyrin repeat domain 28
|
|
chr12_-_7695752
Show fit
|
1.75 |
ENST00000329913.4
|
GDF3
|
growth differentiation factor 3
|
|
chr3_-_15797930
Show fit
|
1.74 |
ENST00000683139.1
|
ANKRD28
|
ankyrin repeat domain 28
|
|
chr18_+_44700796
Show fit
|
1.74 |
ENST00000677130.1
|
SETBP1
|
SET binding protein 1
|
|
chr12_+_78863962
Show fit
|
1.71 |
ENST00000393240.7
|
SYT1
|
synaptotagmin 1
|
|
chr3_-_142028617
Show fit
|
1.69 |
ENST00000477292.5
ENST00000478006.5
ENST00000495310.5
ENST00000486111.5
|
TFDP2
|
transcription factor Dp-2
|
|
chr6_+_15246054
Show fit
|
1.64 |
ENST00000341776.7
|
JARID2
|
jumonji and AT-rich interaction domain containing 2
|
|
chr10_-_103452384
Show fit
|
1.58 |
ENST00000369788.7
|
CALHM2
|
calcium homeostasis modulator family member 2
|
|
chr8_-_3409528
Show fit
|
1.52 |
ENST00000335551.11
|
CSMD1
|
CUB and Sushi multiple domains 1
|
|
chr10_-_103452356
Show fit
|
1.52 |
ENST00000260743.10
|
CALHM2
|
calcium homeostasis modulator family member 2
|
|
chr4_-_158173042
Show fit
|
1.52 |
ENST00000592057.1
ENST00000393807.9
|
GASK1B
|
golgi associated kinase 1B
|
|
chr4_-_89838289
Show fit
|
1.48 |
ENST00000336904.7
|
SNCA
|
synuclein alpha
|
|
chr8_-_17697654
Show fit
|
1.41 |
ENST00000297488.10
|
MTUS1
|
microtubule associated scaffold protein 1
|
|
chr4_-_158173004
Show fit
|
1.25 |
ENST00000585682.6
|
GASK1B
|
golgi associated kinase 1B
|
|
chr20_+_32010429
Show fit
|
1.24 |
ENST00000452892.3
ENST00000262659.12
|
CCM2L
|
CCM2 like scaffold protein
|
|
chr18_-_55510753
Show fit
|
1.22 |
ENST00000543082.5
|
TCF4
|
transcription factor 4
|
|
chr3_-_158672612
Show fit
|
1.16 |
ENST00000264265.4
|
LXN
|
latexin
|
|
chr14_+_63204859
Show fit
|
1.15 |
ENST00000555125.1
|
RHOJ
|
ras homolog family member J
|
|
chr14_+_63204436
Show fit
|
1.13 |
ENST00000316754.8
|
RHOJ
|
ras homolog family member J
|
|
chr2_+_33476640
Show fit
|
1.12 |
ENST00000425210.5
ENST00000444784.5
ENST00000423159.5
ENST00000403687.8
|
RASGRP3
|
RAS guanyl releasing protein 3
|
|
chr2_-_55296361
Show fit
|
1.09 |
ENST00000647547.1
|
CCDC88A
|
coiled-coil domain containing 88A
|
|
chr4_+_185209577
Show fit
|
1.08 |
ENST00000652585.1
|
SNX25
|
sorting nexin 25
|
|
chr3_+_132597260
Show fit
|
1.07 |
ENST00000249887.3
|
ACKR4
|
atypical chemokine receptor 4
|
|
chr12_+_96194501
Show fit
|
1.05 |
ENST00000552142.5
|
ELK3
|
ETS transcription factor ELK3
|
|
chr2_-_55334529
Show fit
|
1.04 |
ENST00000645860.1
ENST00000642563.1
ENST00000647396.1
|
CCDC88A
|
coiled-coil domain containing 88A
|
|
chr12_-_58920465
Show fit
|
1.04 |
ENST00000320743.8
|
LRIG3
|
leucine rich repeats and immunoglobulin like domains 3
|
|
chr11_-_102530738
Show fit
|
1.04 |
ENST00000260227.5
|
MMP7
|
matrix metallopeptidase 7
|
|
chr8_+_39913881
Show fit
|
1.03 |
ENST00000518237.6
|
IDO1
|
indoleamine 2,3-dioxygenase 1
|
|
chr8_+_97887903
Show fit
|
1.02 |
ENST00000520016.5
|
MATN2
|
matrilin 2
|
|
chr5_-_111758061
Show fit
|
1.01 |
ENST00000509979.5
ENST00000513100.5
ENST00000508161.5
ENST00000455559.6
|
NREP
|
neuronal regeneration related protein
|
|
chr17_-_29176752
Show fit
|
1.00 |
ENST00000533112.5
|
MYO18A
|
myosin XVIIIA
|
|
chr12_-_50222348
Show fit
|
1.00 |
ENST00000552823.5
ENST00000552909.5
|
LIMA1
|
LIM domain and actin binding 1
|
|
chr18_-_55422306
Show fit
|
0.97 |
ENST00000566777.5
ENST00000626584.2
|
TCF4
|
transcription factor 4
|
|
chr2_+_28395511
Show fit
|
0.97 |
ENST00000436647.1
|
FOSL2
|
FOS like 2, AP-1 transcription factor subunit
|
|
chr18_-_55589770
Show fit
|
0.97 |
ENST00000565018.6
ENST00000636400.2
|
TCF4
|
transcription factor 4
|
|
chr7_-_120858303
Show fit
|
0.96 |
ENST00000415871.5
ENST00000430985.1
|
TSPAN12
|
tetraspanin 12
|
|
chr18_-_55321640
Show fit
|
0.96 |
ENST00000637169.2
|
TCF4
|
transcription factor 4
|
|
chr3_+_25428233
Show fit
|
0.96 |
ENST00000437042.6
ENST00000330688.9
|
RARB
|
retinoic acid receptor beta
|
|
chr2_+_188974364
Show fit
|
0.94 |
ENST00000304636.9
ENST00000317840.9
|
COL3A1
|
collagen type III alpha 1 chain
|
|
chr18_-_55589836
Show fit
|
0.93 |
ENST00000537578.5
ENST00000564403.6
|
TCF4
|
transcription factor 4
|
|
chr14_-_89417148
Show fit
|
0.91 |
ENST00000557258.6
|
FOXN3
|
forkhead box N3
|
|
chrX_+_153494970
Show fit
|
0.91 |
ENST00000331595.9
ENST00000431891.1
|
BGN
|
biglycan
|
|
chr12_+_93377883
Show fit
|
0.90 |
ENST00000337179.9
ENST00000415493.7
|
NUDT4
|
nudix hydrolase 4
|
|
chr3_-_183099508
Show fit
|
0.90 |
ENST00000476176.5
|
MCCC1
|
methylcrotonoyl-CoA carboxylase 1
|
|
chr20_-_17558811
Show fit
|
0.89 |
ENST00000536626.7
ENST00000377868.6
|
BFSP1
|
beaded filament structural protein 1
|
|
chr3_+_69084929
Show fit
|
0.88 |
ENST00000273258.4
|
ARL6IP5
|
ADP ribosylation factor like GTPase 6 interacting protein 5
|
|
chr18_-_55587335
Show fit
|
0.87 |
ENST00000638154.3
|
TCF4
|
transcription factor 4
|
|
chr18_-_55589795
Show fit
|
0.87 |
ENST00000568740.5
ENST00000629387.2
|
TCF4
|
transcription factor 4
|
|
chr14_+_24429932
Show fit
|
0.83 |
ENST00000556842.5
ENST00000553935.6
|
KHNYN
|
KH and NYN domain containing
|
|
chr3_+_69084973
Show fit
|
0.83 |
ENST00000478935.1
|
ARL6IP5
|
ADP ribosylation factor like GTPase 6 interacting protein 5
|
|
chr17_-_51260032
Show fit
|
0.83 |
ENST00000586178.6
|
MBTD1
|
mbt domain containing 1
|
|
chr4_-_177442427
Show fit
|
0.82 |
ENST00000264595.7
|
AGA
|
aspartylglucosaminidase
|
|
chr14_+_104581141
Show fit
|
0.81 |
ENST00000410013.1
|
C14orf180
|
chromosome 14 open reading frame 180
|
|
chr13_-_67230377
Show fit
|
0.81 |
ENST00000544246.5
ENST00000377861.4
|
PCDH9
|
protocadherin 9
|
|
chr10_+_99732211
Show fit
|
0.81 |
ENST00000370476.10
ENST00000370472.4
|
CUTC
|
cutC copper transporter
|
|
chr18_-_55422492
Show fit
|
0.81 |
ENST00000561992.5
ENST00000630712.2
|
TCF4
|
transcription factor 4
|
|
chr6_-_110815408
Show fit
|
0.80 |
ENST00000368911.8
|
CDK19
|
cyclin dependent kinase 19
|
|
chr1_-_160954801
Show fit
|
0.79 |
ENST00000368029.4
|
ITLN2
|
intelectin 2
|
|
chr8_-_80874771
Show fit
|
0.79 |
ENST00000327835.7
|
ZNF704
|
zinc finger protein 704
|
|
chr1_+_85580751
Show fit
|
0.77 |
ENST00000451137.7
|
CCN1
|
cellular communication network factor 1
|
|
chr6_-_79234619
Show fit
|
0.75 |
ENST00000344726.9
ENST00000275036.11
|
HMGN3
|
high mobility group nucleosomal binding domain 3
|
|
chr18_-_55322215
Show fit
|
0.72 |
ENST00000457482.7
|
TCF4
|
transcription factor 4
|
|
chr6_-_79234713
Show fit
|
0.72 |
ENST00000620514.1
|
HMGN3
|
high mobility group nucleosomal binding domain 3
|
|
chr2_+_1413456
Show fit
|
0.69 |
ENST00000539820.5
ENST00000382269.7
ENST00000345913.8
ENST00000329066.9
ENST00000382201.7
|
TPO
|
thyroid peroxidase
|
|
chr5_-_74641419
Show fit
|
0.69 |
ENST00000618628.4
ENST00000510316.5
ENST00000508331.1
|
ENC1
|
ectodermal-neural cortex 1
|
|
chrX_-_10576901
Show fit
|
0.68 |
ENST00000380779.5
|
MID1
|
midline 1
|
|
chr10_+_75431605
Show fit
|
0.68 |
ENST00000611255.5
|
LRMDA
|
leucine rich melanocyte differentiation associated
|
|
chr9_-_70869076
Show fit
|
0.64 |
ENST00000677594.1
|
TRPM3
|
transient receptor potential cation channel subfamily M member 3
|
|
chr2_-_157874976
Show fit
|
0.62 |
ENST00000682025.1
ENST00000683487.1
ENST00000682300.1
ENST00000683441.1
ENST00000684595.1
ENST00000683426.1
ENST00000683820.1
ENST00000263640.7
|
ACVR1
|
activin A receptor type 1
|
|
chr18_-_55321986
Show fit
|
0.62 |
ENST00000570287.6
|
TCF4
|
transcription factor 4
|
|
chr17_-_48613468
Show fit
|
0.60 |
ENST00000498634.2
|
HOXB8
|
homeobox B8
|
|
chr3_+_152300135
Show fit
|
0.60 |
ENST00000465907.6
ENST00000492948.5
ENST00000485509.5
ENST00000464596.5
|
MBNL1
|
muscleblind like splicing regulator 1
|
|
chr9_+_121299793
Show fit
|
0.60 |
ENST00000373818.8
|
GSN
|
gelsolin
|
|
chr4_+_122339221
Show fit
|
0.60 |
ENST00000442707.1
|
KIAA1109
|
KIAA1109
|
|
chr13_-_99016034
Show fit
|
0.58 |
ENST00000448493.7
|
DOCK9
|
dedicator of cytokinesis 9
|
|
chr3_-_183099577
Show fit
|
0.58 |
ENST00000610757.4
ENST00000629669.2
|
MCCC1
|
methylcrotonoyl-CoA carboxylase 1
|
|
chr13_+_73054969
Show fit
|
0.56 |
ENST00000539231.5
|
KLF5
|
Kruppel like factor 5
|
|
chr1_+_226063466
Show fit
|
0.56 |
ENST00000666609.1
ENST00000661429.1
|
H3-3A
|
H3.3 histone A
|
|
chr4_-_103099811
Show fit
|
0.56 |
ENST00000504285.5
ENST00000296424.9
|
BDH2
|
3-hydroxybutyrate dehydrogenase 2
|
|
chr16_+_30000944
Show fit
|
0.54 |
ENST00000562291.2
|
INO80E
|
INO80 complex subunit E
|
|
chr8_-_167024
Show fit
|
0.53 |
ENST00000320901.4
|
OR4F21
|
olfactory receptor family 4 subfamily F member 21
|
|
chr16_+_15434577
Show fit
|
0.53 |
ENST00000300006.9
|
BMERB1
|
bMERB domain containing 1
|
|
chr8_+_85209213
Show fit
|
0.53 |
ENST00000520225.1
|
E2F5
|
E2F transcription factor 5
|
|
chr1_-_94237562
Show fit
|
0.53 |
ENST00000260526.11
ENST00000370217.3
|
ARHGAP29
|
Rho GTPase activating protein 29
|
|
chr10_-_72626053
Show fit
|
0.52 |
ENST00000603011.5
ENST00000361114.10
|
MICU1
|
mitochondrial calcium uptake 1
|
|
chr13_-_75482114
Show fit
|
0.52 |
ENST00000377625.6
ENST00000431480.6
|
TBC1D4
|
TBC1 domain family member 4
|
|
chr3_-_65597886
Show fit
|
0.51 |
ENST00000460329.6
|
MAGI1
|
membrane associated guanylate kinase, WW and PDZ domain containing 1
|
|
chr15_+_40807590
Show fit
|
0.51 |
ENST00000299173.14
ENST00000566407.5
|
ZFYVE19
|
zinc finger FYVE-type containing 19
|
|
chr9_+_35161951
Show fit
|
0.50 |
ENST00000617908.4
ENST00000619578.4
|
UNC13B
|
unc-13 homolog B
|
|
chr1_-_226739271
Show fit
|
0.50 |
ENST00000429204.6
ENST00000366784.1
|
ITPKB
|
inositol-trisphosphate 3-kinase B
|
|
chr16_+_15434475
Show fit
|
0.50 |
ENST00000566490.5
|
BMERB1
|
bMERB domain containing 1
|
|
chr9_+_35162000
Show fit
|
0.48 |
ENST00000396787.5
ENST00000378495.7
ENST00000635942.1
ENST00000378496.8
|
UNC13B
|
unc-13 homolog B
|
|
chr12_+_4909895
Show fit
|
0.48 |
ENST00000638821.1
ENST00000382545.5
|
ENSG00000256654.4
KCNA1
|
novel transcript, sense overlapping KCNA1
potassium voltage-gated channel subfamily A member 1
|
|
chr5_-_95081482
Show fit
|
0.48 |
ENST00000312216.12
ENST00000512425.5
ENST00000505208.5
ENST00000429576.6
ENST00000508509.5
ENST00000510732.5
|
MCTP1
|
multiple C2 and transmembrane domain containing 1
|
|
chr3_-_183099464
Show fit
|
0.48 |
ENST00000265594.9
|
MCCC1
|
methylcrotonoyl-CoA carboxylase 1
|
|
chr2_-_197675578
Show fit
|
0.48 |
ENST00000295049.9
|
RFTN2
|
raftlin family member 2
|
|
chr13_-_75482151
Show fit
|
0.48 |
ENST00000377636.8
|
TBC1D4
|
TBC1 domain family member 4
|
|
chr8_-_94262308
Show fit
|
0.47 |
ENST00000297596.3
ENST00000396194.6
|
GEM
|
GTP binding protein overexpressed in skeletal muscle
|
|
chr13_-_67230313
Show fit
|
0.47 |
ENST00000377865.7
|
PCDH9
|
protocadherin 9
|
|
chr9_+_22446808
Show fit
|
0.47 |
ENST00000325870.3
|
DMRTA1
|
DMRT like family A1
|
|
chr2_-_229921316
Show fit
|
0.47 |
ENST00000428959.5
ENST00000675423.1
|
TRIP12
|
thyroid hormone receptor interactor 12
|
|
chr1_+_162632454
Show fit
|
0.46 |
ENST00000367921.8
ENST00000367922.7
|
DDR2
|
discoidin domain receptor tyrosine kinase 2
|
|
chrX_-_120629936
Show fit
|
0.46 |
ENST00000371313.2
ENST00000304661.6
|
C1GALT1C1
|
C1GALT1 specific chaperone 1
|
|
chr22_+_40177917
Show fit
|
0.45 |
ENST00000454349.7
ENST00000335727.13
|
TNRC6B
|
trinucleotide repeat containing adaptor 6B
|
|
chr12_-_91179472
Show fit
|
0.45 |
ENST00000550099.5
ENST00000546391.5
|
DCN
|
decorin
|
|
chr11_+_36594369
Show fit
|
0.45 |
ENST00000678060.1
ENST00000446510.6
ENST00000676979.1
ENST00000677808.1
ENST00000617650.5
ENST00000334307.10
ENST00000531554.6
ENST00000679022.1
ENST00000347206.8
ENST00000534635.5
ENST00000676921.1
ENST00000678950.1
ENST00000530697.6
ENST00000527108.6
ENST00000532470.3
|
IFTAP
|
intraflagellar transport associated protein
|
|
chr17_+_47522931
Show fit
|
0.45 |
ENST00000525007.5
ENST00000530173.6
|
NPEPPS
|
aminopeptidase puromycin sensitive
|
|
chr8_-_140764386
Show fit
|
0.45 |
ENST00000520151.5
ENST00000519024.5
ENST00000519465.5
|
PTK2
|
protein tyrosine kinase 2
|
|
chr7_+_30852273
Show fit
|
0.44 |
ENST00000509504.2
|
ENSG00000250424.4
|
novel protein, MINDY4 and AQP1 readthrough
|
|
chr17_+_47941562
Show fit
|
0.44 |
ENST00000225573.5
ENST00000434554.7
ENST00000642017.2
|
PNPO
|
pyridoxamine 5'-phosphate oxidase
|
|
chr10_-_63269057
Show fit
|
0.44 |
ENST00000542921.5
|
JMJD1C
|
jumonji domain containing 1C
|
|
chr1_-_77219399
Show fit
|
0.43 |
ENST00000359130.1
ENST00000370812.8
ENST00000445065.5
|
PIGK
|
phosphatidylinositol glycan anchor biosynthesis class K
|
|
chr12_-_50283472
Show fit
|
0.43 |
ENST00000551691.5
ENST00000394943.7
ENST00000341247.8
|
LIMA1
|
LIM domain and actin binding 1
|
|
chr9_-_122159742
Show fit
|
0.43 |
ENST00000373768.4
|
NDUFA8
|
NADH:ubiquinone oxidoreductase subunit A8
|
|
chr7_+_1086800
Show fit
|
0.42 |
ENST00000413368.5
ENST00000397092.5
ENST00000297469.3
|
GPER1
|
G protein-coupled estrogen receptor 1
|
|
chr1_-_156082412
Show fit
|
0.42 |
ENST00000532414.3
|
MEX3A
|
mex-3 RNA binding family member A
|
|
chr22_+_24011397
Show fit
|
0.42 |
ENST00000445422.5
ENST00000398319.6
|
CABIN1
|
calcineurin binding protein 1
|
|
chr9_-_20622479
Show fit
|
0.41 |
ENST00000380338.9
|
MLLT3
|
MLLT3 super elongation complex subunit
|
|
chr14_+_35122660
Show fit
|
0.41 |
ENST00000603544.5
|
PRORP
|
protein only RNase P catalytic subunit
|
|
chr13_-_61427849
Show fit
|
0.40 |
ENST00000409186.1
ENST00000472649.2
|
ENSG00000197991.11
LINC02339
|
novel protein
long intergenic non-protein coding RNA 2339
|
|
chr12_+_96194365
Show fit
|
0.40 |
ENST00000228741.8
ENST00000547249.1
|
ELK3
|
ETS transcription factor ELK3
|
|
chr15_-_32455634
Show fit
|
0.40 |
ENST00000509311.7
|
GOLGA8O
|
golgin A8 family member O
|
|
chrX_+_155026835
Show fit
|
0.40 |
ENST00000369498.8
|
FUNDC2
|
FUN14 domain containing 2
|
|
chr4_+_158210479
Show fit
|
0.40 |
ENST00000504569.5
ENST00000509278.5
ENST00000514558.5
ENST00000503200.5
ENST00000296529.11
|
TMEM144
|
transmembrane protein 144
|
|
chr22_+_24011192
Show fit
|
0.40 |
ENST00000454754.5
ENST00000263119.10
ENST00000617531.4
|
CABIN1
|
calcineurin binding protein 1
|
|
chr4_+_26584064
Show fit
|
0.39 |
ENST00000264866.9
ENST00000505206.5
ENST00000511789.5
|
TBC1D19
|
TBC1 domain family member 19
|
|
chr22_+_24011325
Show fit
|
0.39 |
ENST00000405822.6
|
CABIN1
|
calcineurin binding protein 1
|
|
chr6_-_159045010
Show fit
|
0.38 |
ENST00000338313.5
|
TAGAP
|
T cell activation RhoGTPase activating protein
|
|
chr15_-_82349437
Show fit
|
0.38 |
ENST00000621197.4
ENST00000610657.2
ENST00000619556.4
|
GOLGA6L10
|
golgin A6 family like 10
|
|
chr15_+_84235773
Show fit
|
0.37 |
ENST00000510439.7
ENST00000422563.6
|
GOLGA6L4
|
golgin A6 family like 4
|
|
chr1_+_161721563
Show fit
|
0.37 |
ENST00000367948.6
|
FCRLB
|
Fc receptor like B
|
|
chrX_+_41334154
Show fit
|
0.37 |
ENST00000441189.4
ENST00000644513.1
ENST00000644109.1
ENST00000646122.1
ENST00000644074.1
ENST00000644876.2
ENST00000399959.7
ENST00000646319.1
|
DDX3X
|
DEAD-box helicase 3 X-linked
|
|
chr12_-_123436664
Show fit
|
0.37 |
ENST00000280571.10
|
RILPL2
|
Rab interacting lysosomal protein like 2
|
|
chr15_+_30604028
Show fit
|
0.37 |
ENST00000566740.2
|
GOLGA8H
|
golgin A8 family member H
|
|
chr14_+_58427970
Show fit
|
0.36 |
ENST00000261244.9
|
KIAA0586
|
KIAA0586
|
|
chr1_-_26360050
Show fit
|
0.35 |
ENST00000475866.3
|
CRYBG2
|
crystallin beta-gamma domain containing 2
|
|
chr1_+_162790702
Show fit
|
0.35 |
ENST00000254521.8
ENST00000367915.1
|
HSD17B7
|
hydroxysteroid 17-beta dehydrogenase 7
|
|
chr12_+_109477368
Show fit
|
0.35 |
ENST00000434735.6
|
UBE3B
|
ubiquitin protein ligase E3B
|
|
chr19_-_43883964
Show fit
|
0.35 |
ENST00000587539.2
|
ZNF404
|
zinc finger protein 404
|
|
chr8_-_144529048
Show fit
|
0.35 |
ENST00000527462.1
ENST00000313465.5
ENST00000524821.6
|
C8orf82
|
chromosome 8 open reading frame 82
|
|
chr16_-_21425278
Show fit
|
0.34 |
ENST00000504841.6
ENST00000419180.6
|
NPIPB3
|
nuclear pore complex interacting protein family member B3
|
|
chr3_-_115147237
Show fit
|
0.33 |
ENST00000357258.8
|
ZBTB20
|
zinc finger and BTB domain containing 20
|
|
chr15_+_32593456
Show fit
|
0.33 |
ENST00000448387.6
ENST00000569659.5
|
GOLGA8N
|
golgin A8 family member N
|
|
chr17_-_41521719
Show fit
|
0.32 |
ENST00000393976.6
|
KRT15
|
keratin 15
|
|
chr3_-_115147277
Show fit
|
0.32 |
ENST00000675478.1
|
ZBTB20
|
zinc finger and BTB domain containing 20
|
|
chr10_-_92573910
Show fit
|
0.32 |
ENST00000678715.1
|
IDE
|
insulin degrading enzyme
|
|
chr15_+_58410543
Show fit
|
0.31 |
ENST00000356113.10
ENST00000414170.7
|
LIPC
|
lipase C, hepatic type
|
|
chr6_-_110415539
Show fit
|
0.31 |
ENST00000368923.8
ENST00000368924.9
|
DDO
|
D-aspartate oxidase
|
|
chr12_-_7503744
Show fit
|
0.31 |
ENST00000396620.7
ENST00000432237.3
|
CD163
|
CD163 molecule
|
|
chr11_+_118883884
Show fit
|
0.30 |
ENST00000292174.5
|
CXCR5
|
C-X-C motif chemokine receptor 5
|
|
chr16_-_66918876
Show fit
|
0.30 |
ENST00000570262.5
ENST00000299752.9
ENST00000394055.7
|
CDH16
|
cadherin 16
|
|
chrX_-_19887585
Show fit
|
0.30 |
ENST00000397821.8
|
SH3KBP1
|
SH3 domain containing kinase binding protein 1
|
|
chr11_-_14337074
Show fit
|
0.30 |
ENST00000531421.5
|
RRAS2
|
RAS related 2
|
|
chr1_+_46175079
Show fit
|
0.30 |
ENST00000372003.6
|
TSPAN1
|
tetraspanin 1
|
|
chr16_+_28638065
Show fit
|
0.29 |
ENST00000683297.1
|
NPIPB8
|
nuclear pore complex interacting protein family member B8
|
|
chr16_-_66918839
Show fit
|
0.29 |
ENST00000565235.2
ENST00000568632.5
ENST00000565796.5
|
CDH16
|
cadherin 16
|
|
chr16_-_21857418
Show fit
|
0.28 |
ENST00000415645.6
|
NPIPB4
|
nuclear pore complex interacting protein family member B4
|
|
chr17_+_40287861
Show fit
|
0.28 |
ENST00000209728.9
ENST00000580824.5
ENST00000577249.1
ENST00000649662.1
|
CDC6
|
cell division cycle 6
|
|
chr16_+_22513523
Show fit
|
0.28 |
ENST00000538606.5
ENST00000451409.5
ENST00000424340.5
ENST00000517539.5
ENST00000528249.5
|
NPIPB5
|
nuclear pore complex interacting protein family member B5
|
|
chr14_+_71598229
Show fit
|
0.28 |
ENST00000537413.5
|
SIPA1L1
|
signal induced proliferation associated 1 like 1
|
|
chr1_+_180632001
Show fit
|
0.27 |
ENST00000367590.9
ENST00000367589.3
|
XPR1
|
xenotropic and polytropic retrovirus receptor 1
|
|
chr16_-_29404029
Show fit
|
0.27 |
ENST00000524087.5
|
NPIPB11
|
nuclear pore complex interacting protein family member B11
|
|
chr15_-_74082550
Show fit
|
0.27 |
ENST00000290438.3
|
GOLGA6A
|
golgin A6 family member A
|
|
chr19_-_18281612
Show fit
|
0.27 |
ENST00000252818.5
|
JUND
|
JunD proto-oncogene, AP-1 transcription factor subunit
|
|
chr5_+_136059151
Show fit
|
0.26 |
ENST00000503087.1
|
TGFBI
|
transforming growth factor beta induced
|
|
chr9_+_27109393
Show fit
|
0.26 |
ENST00000406359.8
|
TEK
|
TEK receptor tyrosine kinase
|
|
chr19_+_14941489
Show fit
|
0.25 |
ENST00000248072.3
|
OR7C2
|
olfactory receptor family 7 subfamily C member 2
|
|
chr12_-_123272234
Show fit
|
0.25 |
ENST00000544658.5
|
CDK2AP1
|
cyclin dependent kinase 2 associated protein 1
|
|
chr12_+_20815672
Show fit
|
0.25 |
ENST00000261196.6
ENST00000381541.7
ENST00000540229.1
|
SLCO1B3
SLCO1B3-SLCO1B7
|
solute carrier organic anion transporter family member 1B3
SLCO1B3-SLCO1B7 readthrough
|
|
chr16_-_21857657
Show fit
|
0.25 |
ENST00000341400.11
ENST00000518761.8
ENST00000682606.1
|
NPIPB4
|
nuclear pore complex interacting protein family member B4
|
|
chr8_+_80485611
Show fit
|
0.25 |
ENST00000379091.8
|
ZBTB10
|
zinc finger and BTB domain containing 10
|