Project
Inflammatory response time course, HUVEC (Wada, 2009)
Navigation
Downloads
Results for NFKB1
Z-value: 3.40
Transcription factors associated with NFKB1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NFKB1
|
ENSG00000109320.13 | NFKB1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NFKB1 | hg38_v1_chr4_+_102501298_102501474, hg38_v1_chr4_+_102501885_102501957 | 0.90 | 1.2e-09 | Click! |
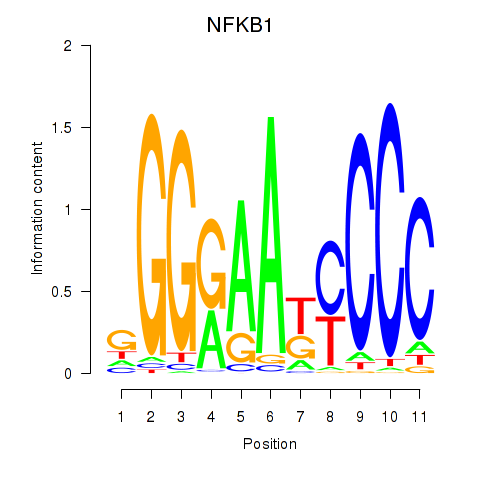
Activity profile of NFKB1 motif
Sorted Z-values of NFKB1 motif
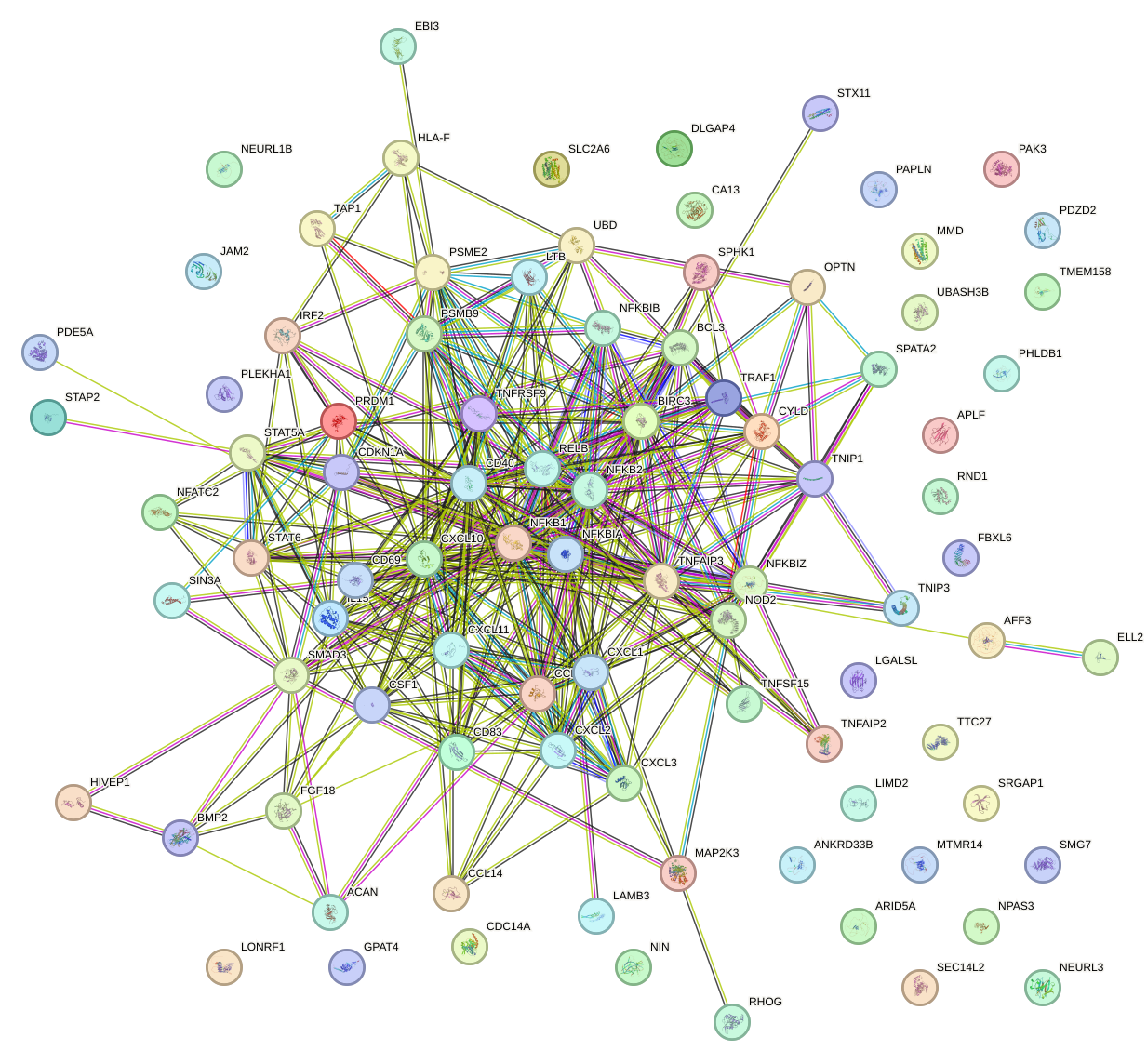
Network of associatons between targets according to the STRING database.
First level regulatory network of NFKB1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 11.7 | 35.2 | GO:0045062 | extrathymic T cell selection(GO:0045062) |
| 11.6 | 34.8 | GO:0052250 | modulation of signal transduction in other organism(GO:0044501) modulation by symbiont of host signal transduction pathway(GO:0052027) modulation of signal transduction in other organism involved in symbiotic interaction(GO:0052250) modulation by symbiont of host I-kappaB kinase/NF-kappaB cascade(GO:0085032) |
| 9.5 | 38.1 | GO:1990108 | protein linear deubiquitination(GO:1990108) |
| 9.1 | 36.4 | GO:1904141 | positive regulation of odontogenesis of dentin-containing tooth(GO:0042488) mammary gland fat development(GO:0060611) positive regulation of macrophage colony-stimulating factor signaling pathway(GO:1902228) positive regulation of response to macrophage colony-stimulating factor(GO:1903971) positive regulation of cellular response to macrophage colony-stimulating factor stimulus(GO:1903974) positive regulation of microglial cell migration(GO:1904141) |
| 9.0 | 26.9 | GO:0070429 | regulation of toll-like receptor 5 signaling pathway(GO:0034147) negative regulation of toll-like receptor 5 signaling pathway(GO:0034148) negative regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070429) |
| 8.6 | 25.7 | GO:1900161 | regulation of phospholipid scramblase activity(GO:1900161) positive regulation of phospholipid scramblase activity(GO:1900163) regulation of glucosylceramide catabolic process(GO:2000752) positive regulation of glucosylceramide catabolic process(GO:2000753) regulation of sphingomyelin catabolic process(GO:2000754) positive regulation of sphingomyelin catabolic process(GO:2000755) |
| 8.1 | 24.2 | GO:0045362 | regulation of interleukin-1 biosynthetic process(GO:0045360) positive regulation of interleukin-1 biosynthetic process(GO:0045362) |
| 7.5 | 29.9 | GO:0002268 | follicular dendritic cell differentiation(GO:0002268) |
| 6.9 | 20.8 | GO:0046967 | cytosol to ER transport(GO:0046967) |
| 6.6 | 46.4 | GO:0000255 | allantoin metabolic process(GO:0000255) |
| 5.3 | 32.1 | GO:0061767 | negative regulation of lung blood pressure(GO:0061767) |
| 5.2 | 57.5 | GO:0043950 | positive regulation of cAMP-mediated signaling(GO:0043950) |
| 4.6 | 32.1 | GO:0045084 | positive regulation of interleukin-12 biosynthetic process(GO:0045084) |
| 4.6 | 36.7 | GO:0070842 | aggresome assembly(GO:0070842) |
| 4.4 | 17.7 | GO:0002296 | T-helper 1 cell lineage commitment(GO:0002296) |
| 4.3 | 21.3 | GO:1900127 | positive regulation of hyaluronan biosynthetic process(GO:1900127) |
| 4.2 | 58.4 | GO:0070424 | regulation of nucleotide-binding oligomerization domain containing signaling pathway(GO:0070424) |
| 3.7 | 7.4 | GO:1902523 | positive regulation of protein K63-linked ubiquitination(GO:1902523) |
| 3.6 | 10.8 | GO:0060128 | regulation of calcium-independent cell-cell adhesion(GO:0051040) corticotropin hormone secreting cell differentiation(GO:0060128) |
| 3.5 | 14.1 | GO:0033590 | response to cobalamin(GO:0033590) |
| 3.5 | 24.2 | GO:0060282 | positive regulation of oocyte development(GO:0060282) |
| 3.4 | 30.2 | GO:0032713 | negative regulation of interleukin-4 production(GO:0032713) |
| 3.2 | 16.2 | GO:0034344 | type III interferon production(GO:0034343) regulation of type III interferon production(GO:0034344) |
| 2.9 | 8.6 | GO:0090222 | centrosome-templated microtubule nucleation(GO:0090222) |
| 2.6 | 5.1 | GO:0002730 | regulation of dendritic cell cytokine production(GO:0002730) |
| 2.4 | 14.3 | GO:2001180 | negative regulation of interleukin-10 secretion(GO:2001180) |
| 2.3 | 6.8 | GO:1901675 | negative regulation of histone H3-K27 acetylation(GO:1901675) |
| 2.2 | 6.5 | GO:0046521 | sphingoid catabolic process(GO:0046521) |
| 1.8 | 12.3 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 1.7 | 3.3 | GO:0070427 | nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070427) |
| 1.6 | 4.9 | GO:1904897 | regulation of hepatic stellate cell proliferation(GO:1904897) positive regulation of hepatic stellate cell proliferation(GO:1904899) hepatic stellate cell proliferation(GO:1990922) |
| 1.5 | 13.5 | GO:0060574 | intestinal epithelial cell maturation(GO:0060574) |
| 1.3 | 19.3 | GO:0032688 | negative regulation of interferon-beta production(GO:0032688) |
| 1.2 | 18.6 | GO:0045078 | positive regulation of interferon-gamma biosynthetic process(GO:0045078) |
| 1.2 | 3.6 | GO:0033078 | extrathymic T cell differentiation(GO:0033078) |
| 1.2 | 3.5 | GO:1900169 | regulation of glucocorticoid mediated signaling pathway(GO:1900169) |
| 1.2 | 3.5 | GO:0002071 | glandular epithelial cell maturation(GO:0002071) |
| 1.2 | 20.8 | GO:0016322 | neuron remodeling(GO:0016322) |
| 1.1 | 5.6 | GO:0033031 | positive regulation of neutrophil apoptotic process(GO:0033031) |
| 1.0 | 10.4 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 1.0 | 2.0 | GO:0097252 | oligodendrocyte apoptotic process(GO:0097252) |
| 0.8 | 7.9 | GO:0002480 | antigen processing and presentation of exogenous peptide antigen via MHC class I, TAP-independent(GO:0002480) |
| 0.8 | 2.3 | GO:1904616 | regulation of actin filament binding(GO:1904529) regulation of actin binding(GO:1904616) |
| 0.7 | 25.1 | GO:0090023 | positive regulation of neutrophil chemotaxis(GO:0090023) |
| 0.7 | 2.1 | GO:0006579 | amino-acid betaine catabolic process(GO:0006579) |
| 0.7 | 2.1 | GO:1903422 | negative regulation of synaptic vesicle recycling(GO:1903422) |
| 0.7 | 5.4 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.6 | 1.9 | GO:1903015 | regulation of exo-alpha-sialidase activity(GO:1903015) |
| 0.6 | 5.8 | GO:2000286 | receptor internalization involved in canonical Wnt signaling pathway(GO:2000286) |
| 0.6 | 1.9 | GO:0002876 | positive regulation of chronic inflammatory response to antigenic stimulus(GO:0002876) |
| 0.6 | 5.7 | GO:0038110 | interleukin-2-mediated signaling pathway(GO:0038110) |
| 0.6 | 5.6 | GO:0035897 | proteolysis in other organism(GO:0035897) |
| 0.6 | 2.5 | GO:1901842 | negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.6 | 9.3 | GO:0045779 | negative regulation of bone resorption(GO:0045779) |
| 0.6 | 4.0 | GO:1904849 | positive regulation of cell chemotaxis to fibroblast growth factor(GO:1904849) positive regulation of endothelial cell chemotaxis to fibroblast growth factor(GO:2000546) |
| 0.5 | 1.6 | GO:0009386 | translational attenuation(GO:0009386) |
| 0.5 | 1.6 | GO:0060166 | olfactory pit development(GO:0060166) |
| 0.5 | 8.2 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.5 | 8.1 | GO:0007253 | cytoplasmic sequestering of NF-kappaB(GO:0007253) |
| 0.5 | 2.5 | GO:0071393 | cellular response to progesterone stimulus(GO:0071393) |
| 0.5 | 1.5 | GO:2000630 | positive regulation of miRNA metabolic process(GO:2000630) |
| 0.5 | 2.3 | GO:0097039 | protein linear polyubiquitination(GO:0097039) |
| 0.5 | 1.8 | GO:0035407 | histone H3-T11 phosphorylation(GO:0035407) |
| 0.4 | 2.6 | GO:0019264 | glycine biosynthetic process from serine(GO:0019264) |
| 0.4 | 3.4 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.4 | 3.7 | GO:0061052 | negative regulation of cell growth involved in cardiac muscle cell development(GO:0061052) |
| 0.4 | 3.8 | GO:0051106 | positive regulation of DNA ligation(GO:0051106) |
| 0.4 | 1.5 | GO:0000415 | negative regulation of histone H3-K36 methylation(GO:0000415) |
| 0.4 | 5.8 | GO:0042340 | keratan sulfate catabolic process(GO:0042340) |
| 0.4 | 2.1 | GO:2000234 | positive regulation of ribosome biogenesis(GO:0090070) positive regulation of rRNA processing(GO:2000234) |
| 0.3 | 2.4 | GO:0007406 | negative regulation of neuroblast proliferation(GO:0007406) |
| 0.3 | 1.0 | GO:0044027 | hypermethylation of CpG island(GO:0044027) |
| 0.3 | 4.9 | GO:0033327 | Leydig cell differentiation(GO:0033327) |
| 0.3 | 24.5 | GO:0010803 | regulation of tumor necrosis factor-mediated signaling pathway(GO:0010803) |
| 0.3 | 31.4 | GO:0002479 | antigen processing and presentation of exogenous peptide antigen via MHC class I, TAP-dependent(GO:0002479) |
| 0.3 | 7.2 | GO:0031629 | synaptic vesicle fusion to presynaptic active zone membrane(GO:0031629) vesicle fusion to plasma membrane(GO:0099500) |
| 0.3 | 4.9 | GO:0051256 | mitotic spindle midzone assembly(GO:0051256) |
| 0.3 | 10.4 | GO:0035428 | hexose transmembrane transport(GO:0035428) glucose transmembrane transport(GO:1904659) |
| 0.3 | 10.9 | GO:0002756 | MyD88-independent toll-like receptor signaling pathway(GO:0002756) |
| 0.3 | 7.5 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.3 | 2.9 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 0.2 | 3.4 | GO:0070777 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.2 | 7.8 | GO:0060334 | regulation of interferon-gamma-mediated signaling pathway(GO:0060334) |
| 0.2 | 2.7 | GO:0001766 | membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) |
| 0.2 | 0.6 | GO:0044725 | chromatin reprogramming in the zygote(GO:0044725) |
| 0.2 | 0.6 | GO:0003245 | cardiac muscle tissue growth involved in heart morphogenesis(GO:0003245) regulation of chromatin silencing at telomere(GO:0031938) |
| 0.2 | 10.1 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.2 | 2.0 | GO:0072553 | terminal button organization(GO:0072553) |
| 0.2 | 1.7 | GO:0006477 | protein sulfation(GO:0006477) |
| 0.2 | 1.1 | GO:0002483 | antigen processing and presentation of endogenous peptide antigen(GO:0002483) antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 0.2 | 3.5 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.2 | 1.2 | GO:1902261 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) |
| 0.2 | 2.3 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.2 | 0.8 | GO:1902766 | skeletal muscle satellite cell migration(GO:1902766) |
| 0.2 | 3.2 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.2 | 3.6 | GO:1901741 | positive regulation of myoblast fusion(GO:1901741) |
| 0.1 | 1.0 | GO:2000324 | positive regulation of glucocorticoid receptor signaling pathway(GO:2000324) |
| 0.1 | 2.8 | GO:1904259 | regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904259) positive regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904261) basement membrane assembly involved in embryonic body morphogenesis(GO:2001197) |
| 0.1 | 5.7 | GO:0071577 | zinc II ion transmembrane transport(GO:0071577) |
| 0.1 | 0.8 | GO:0035105 | sterol regulatory element binding protein import into nucleus(GO:0035105) |
| 0.1 | 9.7 | GO:0071391 | cellular response to estrogen stimulus(GO:0071391) |
| 0.1 | 9.4 | GO:0042531 | positive regulation of tyrosine phosphorylation of STAT protein(GO:0042531) |
| 0.1 | 0.4 | GO:0043449 | cellular alkene metabolic process(GO:0043449) |
| 0.1 | 0.8 | GO:0098814 | spontaneous neurotransmitter secretion(GO:0061669) spontaneous synaptic transmission(GO:0098814) |
| 0.1 | 4.3 | GO:1900027 | regulation of ruffle assembly(GO:1900027) |
| 0.1 | 1.6 | GO:0060371 | regulation of atrial cardiac muscle cell membrane depolarization(GO:0060371) |
| 0.1 | 0.3 | GO:1904204 | regulation of skeletal muscle hypertrophy(GO:1904204) |
| 0.1 | 2.4 | GO:0045717 | negative regulation of fatty acid biosynthetic process(GO:0045717) |
| 0.1 | 0.3 | GO:1902595 | regulation of DNA replication origin binding(GO:1902595) |
| 0.1 | 5.7 | GO:0019835 | cytolysis(GO:0019835) |
| 0.1 | 2.4 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.1 | 1.8 | GO:0051531 | NFAT protein import into nucleus(GO:0051531) |
| 0.1 | 1.2 | GO:0060088 | auditory receptor cell stereocilium organization(GO:0060088) |
| 0.1 | 1.3 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.1 | 1.6 | GO:0035563 | positive regulation of chromatin binding(GO:0035563) |
| 0.1 | 0.2 | GO:0035262 | gonad morphogenesis(GO:0035262) |
| 0.1 | 1.8 | GO:0010832 | negative regulation of myotube differentiation(GO:0010832) |
| 0.1 | 8.7 | GO:0032436 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) |
| 0.1 | 0.2 | GO:0090271 | regulation of prostaglandin biosynthetic process(GO:0031392) positive regulation of prostaglandin biosynthetic process(GO:0031394) positive regulation of fibroblast growth factor production(GO:0090271) regulation of unsaturated fatty acid biosynthetic process(GO:2001279) positive regulation of unsaturated fatty acid biosynthetic process(GO:2001280) |
| 0.1 | 1.2 | GO:0051770 | positive regulation of nitric-oxide synthase biosynthetic process(GO:0051770) |
| 0.1 | 5.8 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.1 | 1.3 | GO:0008334 | histone mRNA metabolic process(GO:0008334) |
| 0.1 | 6.2 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.1 | 0.5 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.1 | 0.2 | GO:0044376 | RNA polymerase II complex import to nucleus(GO:0044376) RNA polymerase III complex localization to nucleus(GO:1990022) |
| 0.1 | 1.5 | GO:0098743 | cell aggregation(GO:0098743) |
| 0.1 | 1.4 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.1 | 1.3 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.1 | 6.2 | GO:0071357 | type I interferon signaling pathway(GO:0060337) cellular response to type I interferon(GO:0071357) |
| 0.1 | 1.4 | GO:0046827 | positive regulation of protein export from nucleus(GO:0046827) |
| 0.1 | 4.4 | GO:0043966 | histone H3 acetylation(GO:0043966) |
| 0.1 | 1.4 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.1 | 4.9 | GO:0009301 | snRNA transcription(GO:0009301) snRNA transcription from RNA polymerase II promoter(GO:0042795) |
| 0.1 | 0.6 | GO:0042487 | regulation of odontogenesis of dentin-containing tooth(GO:0042487) |
| 0.0 | 8.4 | GO:0090501 | RNA phosphodiester bond hydrolysis(GO:0090501) |
| 0.0 | 0.4 | GO:2000543 | positive regulation of gastrulation(GO:2000543) |
| 0.0 | 1.0 | GO:0021794 | thalamus development(GO:0021794) |
| 0.0 | 2.1 | GO:0030513 | positive regulation of BMP signaling pathway(GO:0030513) |
| 0.0 | 6.7 | GO:0051092 | positive regulation of NF-kappaB transcription factor activity(GO:0051092) |
| 0.0 | 0.6 | GO:1990035 | calcium ion import across plasma membrane(GO:0098703) calcium ion import into cell(GO:1990035) |
| 0.0 | 1.2 | GO:0030511 | positive regulation of transforming growth factor beta receptor signaling pathway(GO:0030511) positive regulation of cellular response to transforming growth factor beta stimulus(GO:1903846) |
| 0.0 | 3.2 | GO:0007528 | neuromuscular junction development(GO:0007528) |
| 0.0 | 0.2 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 0.9 | GO:0034629 | cellular protein complex localization(GO:0034629) |
| 0.0 | 1.5 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
| 0.0 | 1.1 | GO:0072600 | establishment of protein localization to Golgi(GO:0072600) |
| 0.0 | 0.3 | GO:0021914 | negative regulation of smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021914) |
| 0.0 | 0.5 | GO:0010991 | negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.0 | 0.4 | GO:0032486 | Rap protein signal transduction(GO:0032486) |
| 0.0 | 0.9 | GO:0050901 | leukocyte tethering or rolling(GO:0050901) |
| 0.0 | 0.6 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.0 | 4.5 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.0 | 0.8 | GO:0051560 | mitochondrial calcium ion homeostasis(GO:0051560) |
| 0.0 | 1.5 | GO:0009948 | anterior/posterior axis specification(GO:0009948) |
| 0.0 | 1.0 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.0 | 0.2 | GO:0031022 | nuclear migration along microfilament(GO:0031022) |
| 0.0 | 2.8 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.0 | 0.4 | GO:0072619 | interleukin-21 production(GO:0032625) interleukin-21 secretion(GO:0072619) |
| 0.0 | 6.1 | GO:0030336 | negative regulation of cell migration(GO:0030336) |
| 0.0 | 0.5 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.0 | 0.9 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.0 | 2.0 | GO:0006501 | C-terminal protein lipidation(GO:0006501) |
| 0.0 | 0.1 | GO:0098828 | positive regulation of inhibitory postsynaptic potential(GO:0097151) modulation of inhibitory postsynaptic potential(GO:0098828) |
| 0.0 | 0.5 | GO:0035116 | embryonic hindlimb morphogenesis(GO:0035116) |
| 0.0 | 1.6 | GO:0001578 | microtubule bundle formation(GO:0001578) |
| 0.0 | 0.9 | GO:0070830 | bicellular tight junction assembly(GO:0070830) |
| 0.0 | 0.6 | GO:0001756 | somitogenesis(GO:0001756) |
| 0.0 | 0.2 | GO:0090331 | negative regulation of platelet aggregation(GO:0090331) |
| 0.0 | 0.1 | GO:0000056 | ribosomal large subunit export from nucleus(GO:0000055) ribosomal small subunit export from nucleus(GO:0000056) |
| 0.0 | 2.3 | GO:0007269 | neurotransmitter secretion(GO:0007269) signal release from synapse(GO:0099643) |
| 0.0 | 0.5 | GO:1903959 | regulation of anion transmembrane transport(GO:1903959) |
| 0.0 | 0.5 | GO:0051058 | negative regulation of Ras protein signal transduction(GO:0046580) negative regulation of small GTPase mediated signal transduction(GO:0051058) |
| 0.0 | 1.2 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 0.0 | 2.0 | GO:0007162 | negative regulation of cell adhesion(GO:0007162) |
| 0.0 | 0.7 | GO:0035914 | skeletal muscle cell differentiation(GO:0035914) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.5 | 29.9 | GO:0033257 | Bcl3/NF-kappaB2 complex(GO:0033257) |
| 6.1 | 36.4 | GO:1990682 | CSF1-CSF1R complex(GO:1990682) |
| 4.9 | 43.9 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 4.0 | 16.1 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 3.6 | 32.1 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 3.0 | 20.8 | GO:0042825 | TAP complex(GO:0042825) |
| 2.7 | 13.5 | GO:0070557 | PCNA-p21 complex(GO:0070557) |
| 1.8 | 14.1 | GO:0043196 | varicosity(GO:0043196) |
| 1.7 | 13.6 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 1.1 | 5.4 | GO:0045160 | myosin I complex(GO:0045160) |
| 1.1 | 7.5 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 0.8 | 12.7 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.8 | 9.0 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.7 | 7.9 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.6 | 38.1 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.6 | 36.7 | GO:0016235 | aggresome(GO:0016235) |
| 0.6 | 6.1 | GO:0060091 | kinocilium(GO:0060091) |
| 0.4 | 1.8 | GO:1990578 | perinuclear endoplasmic reticulum membrane(GO:1990578) |
| 0.3 | 5.4 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.3 | 4.4 | GO:0070775 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.3 | 5.0 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.3 | 2.6 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.3 | 8.2 | GO:0000145 | exocyst(GO:0000145) |
| 0.3 | 6.8 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.3 | 0.8 | GO:0016938 | kinesin I complex(GO:0016938) |
| 0.2 | 2.1 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.2 | 5.6 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.2 | 3.2 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 0.2 | 6.6 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.2 | 72.8 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.2 | 2.1 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.2 | 2.4 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.2 | 1.5 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.2 | 2.8 | GO:0045180 | basal cortex(GO:0045180) |
| 0.2 | 3.2 | GO:0071437 | invadopodium(GO:0071437) |
| 0.1 | 11.6 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.1 | 27.2 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.1 | 68.8 | GO:0045121 | membrane raft(GO:0045121) |
| 0.1 | 0.6 | GO:0031933 | telomeric heterochromatin(GO:0031933) |
| 0.1 | 1.0 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.1 | 5.8 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.1 | 1.3 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.1 | 0.2 | GO:0072536 | interleukin-23 receptor complex(GO:0072536) |
| 0.1 | 7.2 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.1 | 2.0 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.1 | 2.8 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.1 | 3.4 | GO:0030673 | axolemma(GO:0030673) |
| 0.1 | 0.8 | GO:0005638 | lamin filament(GO:0005638) |
| 0.1 | 1.8 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.1 | 1.4 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.1 | 7.0 | GO:1904724 | tertiary granule lumen(GO:1904724) |
| 0.1 | 3.5 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.1 | 2.5 | GO:0031312 | extrinsic component of organelle membrane(GO:0031312) |
| 0.1 | 4.7 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.1 | 0.9 | GO:0016942 | insulin-like growth factor binding protein complex(GO:0016942) growth factor complex(GO:0036454) |
| 0.1 | 4.6 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.1 | 3.1 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.1 | 0.6 | GO:0001940 | male pronucleus(GO:0001940) |
| 0.1 | 1.3 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.1 | 4.8 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.1 | 1.0 | GO:0044613 | nuclear pore central transport channel(GO:0044613) nuclear pore nuclear basket(GO:0044615) |
| 0.1 | 1.3 | GO:0031011 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 0.1 | 109.5 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 5.6 | GO:0030136 | clathrin-coated vesicle(GO:0030136) |
| 0.0 | 1.2 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 1.8 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 12.5 | GO:0005911 | cell-cell junction(GO:0005911) |
| 0.0 | 6.2 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 1.5 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.0 | 1.2 | GO:0000314 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 4.1 | GO:0030496 | midbody(GO:0030496) |
| 0.0 | 4.2 | GO:0001726 | ruffle(GO:0001726) |
| 0.0 | 1.4 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 17.0 | GO:0005912 | adherens junction(GO:0005912) |
| 0.0 | 4.3 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 1.5 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.0 | 0.5 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 1.4 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.8 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.2 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 9.9 | GO:0005764 | lytic vacuole(GO:0000323) lysosome(GO:0005764) |
| 0.0 | 1.1 | GO:0043679 | axon terminus(GO:0043679) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 11.5 | 57.5 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 8.1 | 24.2 | GO:0031731 | CCR6 chemokine receptor binding(GO:0031731) |
| 5.3 | 32.1 | GO:0031962 | mineralocorticoid receptor binding(GO:0031962) |
| 5.2 | 36.4 | GO:0005157 | macrophage colony-stimulating factor receptor binding(GO:0005157) |
| 4.3 | 25.7 | GO:0004699 | calcium-independent protein kinase C activity(GO:0004699) |
| 4.0 | 31.6 | GO:0046979 | TAP2 binding(GO:0046979) |
| 3.4 | 13.7 | GO:0004906 | interferon-gamma receptor activity(GO:0004906) |
| 3.4 | 64.9 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 2.7 | 13.5 | GO:0019912 | cyclin-dependent protein kinase activating kinase activity(GO:0019912) |
| 2.3 | 16.1 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 2.1 | 38.2 | GO:0031996 | thioesterase binding(GO:0031996) |
| 2.0 | 20.3 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 1.6 | 38.4 | GO:0070628 | proteasome binding(GO:0070628) |
| 1.1 | 55.5 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 1.0 | 4.0 | GO:0005111 | type 2 fibroblast growth factor receptor binding(GO:0005111) |
| 0.9 | 6.5 | GO:0017050 | sphinganine kinase activity(GO:0008481) D-erythro-sphingosine kinase activity(GO:0017050) |
| 0.9 | 12.9 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.9 | 3.4 | GO:0070137 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 0.8 | 28.4 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.8 | 7.4 | GO:0031730 | CCR5 chemokine receptor binding(GO:0031730) |
| 0.8 | 27.1 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.8 | 52.0 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.7 | 10.4 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.6 | 3.5 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.6 | 5.6 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.6 | 21.1 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.5 | 1.6 | GO:0086059 | voltage-gated calcium channel activity involved SA node cell action potential(GO:0086059) |
| 0.5 | 10.3 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.5 | 2.1 | GO:0046997 | oxidoreductase activity, acting on the CH-NH group of donors, flavin as acceptor(GO:0046997) |
| 0.5 | 13.6 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.5 | 1.8 | GO:0035402 | histone kinase activity (H3-T11 specific)(GO:0035402) |
| 0.4 | 7.4 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.4 | 2.6 | GO:0008732 | glycine hydroxymethyltransferase activity(GO:0004372) threonine aldolase activity(GO:0004793) L-allo-threonine aldolase activity(GO:0008732) |
| 0.4 | 4.9 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.4 | 2.8 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.4 | 3.2 | GO:0015450 | P-P-bond-hydrolysis-driven protein transmembrane transporter activity(GO:0015450) |
| 0.3 | 11.3 | GO:0046965 | retinoid X receptor binding(GO:0046965) |
| 0.3 | 1.3 | GO:0047391 | alkylglycerophosphoethanolamine phosphodiesterase activity(GO:0047391) |
| 0.3 | 2.3 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) |
| 0.3 | 0.9 | GO:0098640 | integrin binding involved in cell-matrix adhesion(GO:0098640) |
| 0.3 | 2.9 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 0.3 | 1.1 | GO:0005151 | interleukin-1, Type II receptor binding(GO:0005151) |
| 0.3 | 1.3 | GO:0071208 | histone pre-mRNA DCP binding(GO:0071208) |
| 0.3 | 5.5 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.2 | 3.4 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.2 | 0.7 | GO:0015275 | stretch-activated, cation-selective, calcium channel activity(GO:0015275) |
| 0.2 | 1.2 | GO:0047280 | nicotinamide phosphoribosyltransferase activity(GO:0047280) |
| 0.2 | 3.5 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.2 | 5.4 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.2 | 12.2 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.2 | 9.2 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.2 | 1.7 | GO:0043426 | MRF binding(GO:0043426) |
| 0.2 | 18.0 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.2 | 2.0 | GO:0000990 | transcription factor activity, core RNA polymerase binding(GO:0000990) |
| 0.2 | 2.3 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.2 | 0.9 | GO:0004466 | long-chain-acyl-CoA dehydrogenase activity(GO:0004466) |
| 0.2 | 1.8 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.2 | 2.7 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.2 | 17.7 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.2 | 8.8 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.2 | 1.3 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.2 | 8.1 | GO:0030159 | receptor signaling complex scaffold activity(GO:0030159) |
| 0.2 | 3.8 | GO:0003906 | DNA-(apurinic or apyrimidinic site) lyase activity(GO:0003906) |
| 0.2 | 6.1 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.2 | 6.2 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.2 | 5.7 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.2 | 8.4 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.1 | 37.0 | GO:0005125 | cytokine activity(GO:0005125) |
| 0.1 | 5.1 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.1 | 0.6 | GO:0052595 | tryptamine:oxygen oxidoreductase (deaminating) activity(GO:0052593) aminoacetone:oxygen oxidoreductase(deaminating) activity(GO:0052594) aliphatic-amine oxidase activity(GO:0052595) phenethylamine:oxygen oxidoreductase (deaminating) activity(GO:0052596) |
| 0.1 | 0.6 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.1 | 4.9 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.1 | 7.2 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.1 | 18.5 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.1 | 36.3 | GO:0004713 | protein tyrosine kinase activity(GO:0004713) |
| 0.1 | 19.4 | GO:0001047 | core promoter binding(GO:0001047) |
| 0.1 | 3.3 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.1 | 0.3 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.1 | 9.3 | GO:0051219 | phosphoprotein binding(GO:0051219) |
| 0.1 | 1.5 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.1 | 3.2 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.1 | 2.0 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.1 | 0.9 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.1 | 0.4 | GO:0070576 | vitamin D 24-hydroxylase activity(GO:0070576) |
| 0.1 | 2.3 | GO:0031005 | filamin binding(GO:0031005) |
| 0.1 | 0.6 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.1 | 3.8 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.1 | 1.6 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.1 | 25.3 | GO:0003713 | transcription coactivator activity(GO:0003713) |
| 0.1 | 8.2 | GO:0000149 | SNARE binding(GO:0000149) |
| 0.1 | 26.8 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.1 | 1.2 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.1 | 1.0 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.1 | 1.5 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.1 | 0.2 | GO:0042019 | interleukin-12 receptor binding(GO:0005143) interleukin-23 binding(GO:0042019) interleukin-23 receptor activity(GO:0042020) |
| 0.1 | 1.2 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.0 | 1.3 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.0 | 0.6 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.0 | 0.2 | GO:0050473 | prostaglandin-endoperoxide synthase activity(GO:0004666) arachidonate 15-lipoxygenase activity(GO:0050473) |
| 0.0 | 1.0 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 6.4 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 1.3 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 1.1 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 2.5 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 1.1 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.0 | 1.0 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.0 | 0.4 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.0 | 0.8 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 0.5 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.4 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 0.4 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 0.3 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.0 | 0.5 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.0 | 1.3 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 0.4 | GO:0050681 | androgen receptor binding(GO:0050681) |
| 0.0 | 1.4 | GO:0005525 | GTP binding(GO:0005525) |
| 0.0 | 1.2 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.1 | 214.2 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 2.3 | 4.6 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 1.3 | 49.8 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 1.0 | 67.4 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 1.0 | 35.0 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 1.0 | 34.0 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 1.0 | 13.5 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.8 | 28.5 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.6 | 25.1 | SIG IL4RECEPTOR IN B LYPHOCYTES | Genes related to IL4 rceptor signaling in B lymphocytes |
| 0.6 | 31.1 | PID THROMBIN PAR1 PATHWAY | PAR1-mediated thrombin signaling events |
| 0.5 | 3.7 | PID S1P S1P2 PATHWAY | S1P2 pathway |
| 0.4 | 10.3 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.4 | 17.9 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.4 | 7.5 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.4 | 12.1 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.4 | 6.7 | PID S1P S1P1 PATHWAY | S1P1 pathway |
| 0.4 | 3.7 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.3 | 12.9 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.3 | 107.1 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.2 | 4.9 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.2 | 4.7 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.1 | 3.8 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.1 | 6.2 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 6.8 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.1 | 7.0 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.1 | 17.8 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.1 | 8.4 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.1 | 4.1 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.1 | 1.3 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.1 | 1.9 | ST ADRENERGIC | Adrenergic Pathway |
| 0.1 | 1.2 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.1 | 4.0 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.0 | 1.2 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.0 | 0.9 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 2.3 | PID AVB3 INTEGRIN PATHWAY | Integrins in angiogenesis |
| 0.0 | 0.8 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.0 | 1.0 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.0 | 0.9 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.9 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 0.8 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 0.5 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 1.0 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.9 | 60.7 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 2.6 | 130.8 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 1.7 | 52.2 | REACTOME TRAF6 MEDIATED NFKB ACTIVATION | Genes involved in TRAF6 mediated NF-kB activation |
| 1.5 | 100.1 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 1.1 | 5.6 | REACTOME ACTIVATED TAK1 MEDIATES P38 MAPK ACTIVATION | Genes involved in activated TAK1 mediates p38 MAPK activation |
| 1.0 | 20.8 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 1.0 | 29.8 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.9 | 32.1 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.7 | 10.4 | REACTOME TRAF3 DEPENDENT IRF ACTIVATION PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |
| 0.6 | 10.1 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.6 | 27.1 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.5 | 25.7 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.5 | 42.7 | REACTOME P53 DEPENDENT G1 DNA DAMAGE RESPONSE | Genes involved in p53-Dependent G1 DNA Damage Response |
| 0.5 | 10.4 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.4 | 14.9 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.4 | 7.8 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.4 | 5.8 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.4 | 4.6 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.3 | 6.2 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.3 | 14.2 | REACTOME GPVI MEDIATED ACTIVATION CASCADE | Genes involved in GPVI-mediated activation cascade |
| 0.3 | 7.2 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.2 | 12.9 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.2 | 5.7 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.2 | 4.0 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 0.2 | 6.5 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.2 | 1.7 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.2 | 2.3 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.1 | 4.5 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.1 | 14.1 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.1 | 2.8 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.1 | 1.3 | REACTOME CD28 DEPENDENT PI3K AKT SIGNALING | Genes involved in CD28 dependent PI3K/Akt signaling |
| 0.1 | 3.8 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.1 | 2.4 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.1 | 1.3 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.1 | 12.4 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.1 | 6.6 | REACTOME DOWNSTREAM SIGNAL TRANSDUCTION | Genes involved in Downstream signal transduction |
| 0.1 | 5.8 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.1 | 2.1 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 0.9 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 3.7 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.0 | 2.4 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.9 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 2.5 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 4.1 | REACTOME CELL JUNCTION ORGANIZATION | Genes involved in Cell junction organization |
| 0.0 | 1.9 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 4.2 | REACTOME NONSENSE MEDIATED DECAY ENHANCED BY THE EXON JUNCTION COMPLEX | Genes involved in Nonsense Mediated Decay Enhanced by the Exon Junction Complex |
| 0.0 | 1.2 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 0.8 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.8 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.2 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |