Project
Inflammatory response time course, HUVEC (Wada, 2009)
Navigation
Downloads
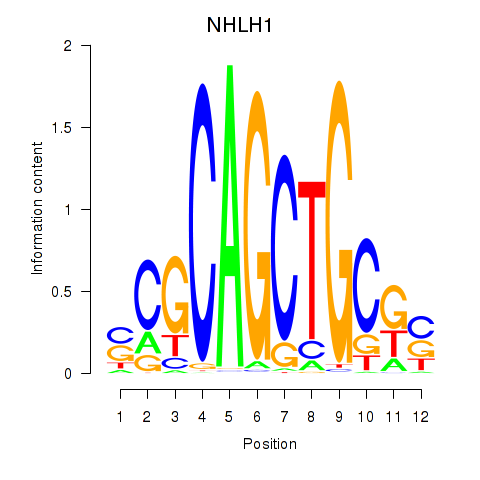
Results for NHLH1
Z-value: 0.67
Transcription factors associated with NHLH1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NHLH1
|
ENSG00000171786.6 | NHLH1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NHLH1 | hg38_v1_chr1_+_160367061_160367078 | -0.28 | 1.8e-01 | Click! |
Activity profile of NHLH1 motif
Sorted Z-values of NHLH1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of NHLH1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_+_41226648 | 3.41 |
ENST00000377721.3
|
KRTAP9-2
|
keratin associated protein 9-2 |
| chr17_+_41237998 | 2.22 |
ENST00000254072.7
|
KRTAP9-8
|
keratin associated protein 9-8 |
| chr17_-_41140487 | 1.02 |
ENST00000345847.4
|
KRTAP4-6
|
keratin associated protein 4-6 |
| chr6_-_154510114 | 0.98 |
ENST00000673182.1
|
ENSG00000288520.1
|
novel protein |
| chr13_-_71866769 | 0.92 |
ENST00000619232.1
|
DACH1
|
dachshund family transcription factor 1 |
| chr4_-_185775890 | 0.76 |
ENST00000437304.6
|
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr4_-_185775271 | 0.75 |
ENST00000430503.5
ENST00000319454.10 ENST00000450341.5 |
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr6_-_31714062 | 0.66 |
ENST00000409239.5
|
LY6G6E
|
lymphocyte antigen 6 family member G6E |
| chr5_+_56815534 | 0.64 |
ENST00000399503.4
|
MAP3K1
|
mitogen-activated protein kinase kinase kinase 1 |
| chr6_-_154510675 | 0.55 |
ENST00000607772.6
|
CNKSR3
|
CNKSR family member 3 |
| chr14_+_63204859 | 0.53 |
ENST00000555125.1
|
RHOJ
|
ras homolog family member J |
| chr11_-_1622093 | 0.53 |
ENST00000616115.1
ENST00000399682.1 |
KRTAP5-4
|
keratin associated protein 5-4 |
| chr4_+_41360759 | 0.50 |
ENST00000508501.5
ENST00000512946.5 ENST00000313860.11 ENST00000512632.5 ENST00000512820.5 |
LIMCH1
|
LIM and calponin homology domains 1 |
| chr19_+_1407731 | 0.46 |
ENST00000592453.2
|
DAZAP1
|
DAZ associated protein 1 |
| chr8_+_141128581 | 0.46 |
ENST00000519811.6
|
DENND3
|
DENN domain containing 3 |
| chr8_+_141128612 | 0.45 |
ENST00000518347.5
ENST00000262585.6 ENST00000520986.5 ENST00000523058.5 ENST00000518668.5 |
DENND3
|
DENN domain containing 3 |
| chr2_-_11670186 | 0.43 |
ENST00000306928.6
|
NTSR2
|
neurotensin receptor 2 |
| chr15_-_68820861 | 0.41 |
ENST00000560303.1
ENST00000465139.6 |
ANP32A
|
acidic nuclear phosphoprotein 32 family member A |
| chr14_-_53956811 | 0.41 |
ENST00000559087.5
ENST00000245451.9 |
BMP4
|
bone morphogenetic protein 4 |
| chr11_-_10808304 | 0.40 |
ENST00000532082.6
|
EIF4G2
|
eukaryotic translation initiation factor 4 gamma 2 |
| chr22_-_37486357 | 0.39 |
ENST00000356998.8
ENST00000416983.7 ENST00000424765.2 |
MFNG
|
MFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase |
| chr15_+_67125707 | 0.38 |
ENST00000540846.6
|
SMAD3
|
SMAD family member 3 |
| chr17_+_82237134 | 0.38 |
ENST00000583025.1
|
SLC16A3
|
solute carrier family 16 member 3 |
| chr19_+_45340774 | 0.38 |
ENST00000589837.5
|
KLC3
|
kinesin light chain 3 |
| chr6_+_125790922 | 0.37 |
ENST00000453302.5
ENST00000417494.5 ENST00000392477.7 ENST00000229634.13 |
NCOA7
|
nuclear receptor coactivator 7 |
| chr2_-_288056 | 0.37 |
ENST00000403610.9
|
ALKAL2
|
ALK and LTK ligand 2 |
| chr5_+_114362286 | 0.37 |
ENST00000610748.4
ENST00000264773.7 |
KCNN2
|
potassium calcium-activated channel subfamily N member 2 |
| chr2_-_229714478 | 0.37 |
ENST00000341772.5
|
DNER
|
delta/notch like EGF repeat containing |
| chr3_-_64268161 | 0.36 |
ENST00000564377.6
|
PRICKLE2
|
prickle planar cell polarity protein 2 |
| chr12_+_130162456 | 0.36 |
ENST00000539839.1
ENST00000229030.5 |
FZD10
|
frizzled class receptor 10 |
| chr3_-_31981228 | 0.35 |
ENST00000396556.7
ENST00000438237.6 |
OSBPL10
|
oxysterol binding protein like 10 |
| chr1_-_46668317 | 0.35 |
ENST00000574428.5
|
ATPAF1
|
ATP synthase mitochondrial F1 complex assembly factor 1 |
| chr1_+_78045956 | 0.35 |
ENST00000370759.4
|
GIPC2
|
GIPC PDZ domain containing family member 2 |
| chr5_+_76819022 | 0.34 |
ENST00000296677.5
|
F2RL1
|
F2R like trypsin receptor 1 |
| chr6_-_75205843 | 0.34 |
ENST00000345356.10
|
COL12A1
|
collagen type XII alpha 1 chain |
| chr8_+_97775829 | 0.34 |
ENST00000517924.5
|
LAPTM4B
|
lysosomal protein transmembrane 4 beta |
| chr1_-_46668394 | 0.33 |
ENST00000371937.8
ENST00000329231.8 |
ATPAF1
|
ATP synthase mitochondrial F1 complex assembly factor 1 |
| chr6_+_142301926 | 0.33 |
ENST00000296932.13
ENST00000367609.8 |
ADGRG6
|
adhesion G protein-coupled receptor G6 |
| chr5_+_139648338 | 0.33 |
ENST00000302517.8
|
CXXC5
|
CXXC finger protein 5 |
| chr7_-_11832190 | 0.33 |
ENST00000423059.9
ENST00000617773.1 |
THSD7A
|
thrombospondin type 1 domain containing 7A |
| chr22_-_30387078 | 0.32 |
ENST00000215798.10
|
RNF215
|
ring finger protein 215 |
| chr20_+_35455141 | 0.32 |
ENST00000446710.5
ENST00000420564.5 |
CEP250
|
centrosomal protein 250 |
| chr2_-_287320 | 0.32 |
ENST00000401503.5
|
ALKAL2
|
ALK and LTK ligand 2 |
| chr19_+_4304632 | 0.31 |
ENST00000597590.5
|
FSD1
|
fibronectin type III and SPRY domain containing 1 |
| chr12_+_19439469 | 0.31 |
ENST00000266508.14
|
AEBP2
|
AE binding protein 2 |
| chr12_-_6606642 | 0.31 |
ENST00000545584.2
ENST00000545942.6 |
CHD4
|
chromodomain helicase DNA binding protein 4 |
| chr20_+_38805686 | 0.30 |
ENST00000299824.6
ENST00000373331.2 |
PPP1R16B
|
protein phosphatase 1 regulatory subunit 16B |
| chr1_-_182391783 | 0.30 |
ENST00000331872.11
ENST00000339526.8 |
GLUL
|
glutamate-ammonia ligase |
| chr14_+_63204436 | 0.30 |
ENST00000316754.8
|
RHOJ
|
ras homolog family member J |
| chr5_+_167754918 | 0.30 |
ENST00000519204.5
|
TENM2
|
teneurin transmembrane protein 2 |
| chr1_-_209806124 | 0.30 |
ENST00000367021.8
ENST00000542854.5 |
IRF6
|
interferon regulatory factor 6 |
| chr12_+_19439669 | 0.30 |
ENST00000398864.7
|
AEBP2
|
AE binding protein 2 |
| chr10_-_87863533 | 0.30 |
ENST00000445946.5
|
KLLN
|
killin, p53 regulated DNA replication inhibitor |
| chr3_-_133895753 | 0.30 |
ENST00000460865.3
|
RAB6B
|
RAB6B, member RAS oncogene family |
| chr2_+_15940537 | 0.29 |
ENST00000281043.4
ENST00000638417.1 |
MYCN
|
MYCN proto-oncogene, bHLH transcription factor |
| chr2_-_19358612 | 0.29 |
ENST00000272223.3
|
OSR1
|
odd-skipped related transcription factor 1 |
| chr8_+_37796906 | 0.29 |
ENST00000315215.11
|
ADGRA2
|
adhesion G protein-coupled receptor A2 |
| chr2_+_33134620 | 0.29 |
ENST00000402934.5
ENST00000404525.5 ENST00000407925.5 |
LTBP1
|
latent transforming growth factor beta binding protein 1 |
| chr1_-_182391363 | 0.29 |
ENST00000417584.6
|
GLUL
|
glutamate-ammonia ligase |
| chr11_+_71538025 | 0.28 |
ENST00000398534.4
|
KRTAP5-8
|
keratin associated protein 5-8 |
| chr20_-_17682060 | 0.28 |
ENST00000455029.3
|
RRBP1
|
ribosome binding protein 1 |
| chr9_+_2017383 | 0.28 |
ENST00000382194.6
|
SMARCA2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr2_-_37672178 | 0.27 |
ENST00000457889.1
|
CDC42EP3
|
CDC42 effector protein 3 |
| chr3_+_141402322 | 0.27 |
ENST00000510338.5
ENST00000504673.5 |
ZBTB38
|
zinc finger and BTB domain containing 38 |
| chr16_+_25216943 | 0.27 |
ENST00000219660.6
|
AQP8
|
aquaporin 8 |
| chrX_+_85244075 | 0.27 |
ENST00000276123.7
|
ZNF711
|
zinc finger protein 711 |
| chr6_-_159727324 | 0.27 |
ENST00000401980.3
ENST00000545162.5 |
SOD2
|
superoxide dismutase 2 |
| chr6_+_36027773 | 0.26 |
ENST00000468133.5
|
MAPK14
|
mitogen-activated protein kinase 14 |
| chr2_+_33134579 | 0.26 |
ENST00000418533.6
|
LTBP1
|
latent transforming growth factor beta binding protein 1 |
| chr1_-_21279520 | 0.26 |
ENST00000357071.8
|
ECE1
|
endothelin converting enzyme 1 |
| chr2_+_70258088 | 0.26 |
ENST00000433351.7
ENST00000264441.9 |
PCYOX1
|
prenylcysteine oxidase 1 |
| chr18_-_51197671 | 0.26 |
ENST00000406189.4
|
MEX3C
|
mex-3 RNA binding family member C |
| chr11_+_33257265 | 0.26 |
ENST00000303296.9
ENST00000379016.7 |
HIPK3
|
homeodomain interacting protein kinase 3 |
| chr12_-_79690814 | 0.25 |
ENST00000548426.1
|
PAWR
|
pro-apoptotic WT1 regulator |
| chr2_+_202376301 | 0.25 |
ENST00000374580.10
|
BMPR2
|
bone morphogenetic protein receptor type 2 |
| chr19_-_18940289 | 0.25 |
ENST00000542541.6
ENST00000433218.6 |
HOMER3
|
homer scaffold protein 3 |
| chr1_-_6602859 | 0.25 |
ENST00000377658.8
|
KLHL21
|
kelch like family member 21 |
| chr8_+_17497078 | 0.25 |
ENST00000494857.6
ENST00000522656.5 |
SLC7A2
|
solute carrier family 7 member 2 |
| chrX_+_9786420 | 0.25 |
ENST00000380913.8
|
SHROOM2
|
shroom family member 2 |
| chr7_+_150368790 | 0.24 |
ENST00000397281.6
ENST00000444957.3 ENST00000466559.1 ENST00000475514.5 ENST00000482680.1 ENST00000488943.1 ENST00000489432.7 ENST00000518514.1 ENST00000478789.5 |
REPIN1
ZNF775
|
replication initiator 1 zinc finger protein 775 |
| chr1_-_6602885 | 0.24 |
ENST00000377663.3
|
KLHL21
|
kelch like family member 21 |
| chr2_+_65056382 | 0.24 |
ENST00000377990.7
ENST00000537589.1 ENST00000260569.4 |
CEP68
|
centrosomal protein 68 |
| chr9_+_128460559 | 0.24 |
ENST00000372807.9
ENST00000444119.6 |
ODF2
|
outer dense fiber of sperm tails 2 |
| chr8_+_17497108 | 0.23 |
ENST00000470360.5
|
SLC7A2
|
solute carrier family 7 member 2 |
| chr8_+_96493803 | 0.23 |
ENST00000518385.5
ENST00000302190.9 |
SDC2
|
syndecan 2 |
| chr6_-_30744537 | 0.23 |
ENST00000259874.6
ENST00000376377.2 |
IER3
|
immediate early response 3 |
| chr3_-_165837412 | 0.23 |
ENST00000479451.5
ENST00000488954.1 ENST00000264381.8 |
BCHE
|
butyrylcholinesterase |
| chr16_-_23149378 | 0.23 |
ENST00000219689.12
|
USP31
|
ubiquitin specific peptidase 31 |
| chr6_+_30880780 | 0.23 |
ENST00000460944.6
ENST00000324771.12 |
DDR1
|
discoidin domain receptor tyrosine kinase 1 |
| chr13_+_30906263 | 0.23 |
ENST00000380482.9
|
MEDAG
|
mesenteric estrogen dependent adipogenesis |
| chr15_-_74906872 | 0.23 |
ENST00000562698.5
|
FAM219B
|
family with sequence similarity 219 member B |
| chr18_+_3448456 | 0.23 |
ENST00000549780.5
|
TGIF1
|
TGFB induced factor homeobox 1 |
| chr15_-_34338033 | 0.22 |
ENST00000558667.5
ENST00000561120.5 ENST00000559236.5 ENST00000397702.6 |
SLC12A6
|
solute carrier family 12 member 6 |
| chr16_+_165966 | 0.21 |
ENST00000356815.4
|
HBM
|
hemoglobin subunit mu |
| chr7_+_131110087 | 0.21 |
ENST00000421797.6
|
MKLN1
|
muskelin 1 |
| chr1_+_81800906 | 0.21 |
ENST00000674393.1
ENST00000674208.1 |
ADGRL2
|
adhesion G protein-coupled receptor L2 |
| chr7_-_93226449 | 0.21 |
ENST00000394468.7
ENST00000453812.2 |
HEPACAM2
|
HEPACAM family member 2 |
| chrX_+_85243983 | 0.21 |
ENST00000674551.1
|
ZNF711
|
zinc finger protein 711 |
| chr14_+_73058591 | 0.21 |
ENST00000525161.5
|
RBM25
|
RNA binding motif protein 25 |
| chr5_+_114362043 | 0.21 |
ENST00000673685.1
|
KCNN2
|
potassium calcium-activated channel subfamily N member 2 |
| chr8_+_89902529 | 0.21 |
ENST00000451899.7
|
OSGIN2
|
oxidative stress induced growth inhibitor family member 2 |
| chr14_-_100587404 | 0.21 |
ENST00000554140.2
|
BEGAIN
|
brain enriched guanylate kinase associated |
| chr14_-_23352741 | 0.21 |
ENST00000354772.9
|
SLC22A17
|
solute carrier family 22 member 17 |
| chr1_-_91886144 | 0.20 |
ENST00000212355.9
|
TGFBR3
|
transforming growth factor beta receptor 3 |
| chr17_+_28042839 | 0.20 |
ENST00000582037.2
|
NLK
|
nemo like kinase |
| chr6_+_36027677 | 0.20 |
ENST00000622903.4
ENST00000229795.7 |
MAPK14
|
mitogen-activated protein kinase 14 |
| chr2_+_190408324 | 0.20 |
ENST00000417958.5
ENST00000432036.5 ENST00000392328.6 |
MFSD6
|
major facilitator superfamily domain containing 6 |
| chr17_-_33293247 | 0.20 |
ENST00000225823.7
|
ASIC2
|
acid sensing ion channel subunit 2 |
| chr6_-_83709019 | 0.20 |
ENST00000519779.5
ENST00000369694.6 ENST00000195649.10 |
SNAP91
|
synaptosome associated protein 91 |
| chr5_+_131264043 | 0.20 |
ENST00000505065.2
ENST00000503291.5 ENST00000360515.7 |
CDC42SE2
|
CDC42 small effector 2 |
| chrX_+_85244032 | 0.20 |
ENST00000373165.7
|
ZNF711
|
zinc finger protein 711 |
| chr10_-_132307935 | 0.20 |
ENST00000298630.8
|
STK32C
|
serine/threonine kinase 32C |
| chr9_-_76692181 | 0.19 |
ENST00000376717.6
ENST00000223609.10 |
PRUNE2
|
prune homolog 2 with BCH domain |
| chr22_-_30387334 | 0.19 |
ENST00000382363.8
|
RNF215
|
ring finger protein 215 |
| chr6_+_71886703 | 0.19 |
ENST00000491071.6
|
RIMS1
|
regulating synaptic membrane exocytosis 1 |
| chr16_-_30096170 | 0.19 |
ENST00000566134.5
ENST00000565110.5 ENST00000398841.6 ENST00000398838.8 |
YPEL3
|
yippee like 3 |
| chr16_-_88977196 | 0.19 |
ENST00000268679.9
|
CBFA2T3
|
CBFA2/RUNX1 partner transcriptional co-repressor 3 |
| chr15_-_34337719 | 0.19 |
ENST00000559484.1
ENST00000558589.5 ENST00000458406.6 |
SLC12A6
|
solute carrier family 12 member 6 |
| chr13_+_48976069 | 0.19 |
ENST00000378383.5
|
FNDC3A
|
fibronectin type III domain containing 3A |
| chr4_-_88823165 | 0.19 |
ENST00000508369.5
|
FAM13A
|
family with sequence similarity 13 member A |
| chr1_+_151721508 | 0.19 |
ENST00000479191.2
|
RIIAD1
|
regulatory subunit of type II PKA R-subunit domain containing 1 |
| chr11_+_62789124 | 0.18 |
ENST00000526546.1
|
TMEM179B
|
transmembrane protein 179B |
| chr19_-_46661132 | 0.18 |
ENST00000410105.2
ENST00000391916.7 |
DACT3
|
dishevelled binding antagonist of beta catenin 3 |
| chr1_+_10210562 | 0.18 |
ENST00000377093.9
ENST00000676179.1 |
KIF1B
|
kinesin family member 1B |
| chr6_+_36027796 | 0.18 |
ENST00000229794.9
|
MAPK14
|
mitogen-activated protein kinase 14 |
| chr4_-_101347327 | 0.18 |
ENST00000394853.8
|
PPP3CA
|
protein phosphatase 3 catalytic subunit alpha |
| chr7_+_102912983 | 0.18 |
ENST00000339431.9
ENST00000249377.4 |
LRRC17
|
leucine rich repeat containing 17 |
| chr12_-_118103998 | 0.18 |
ENST00000359236.10
|
VSIG10
|
V-set and immunoglobulin domain containing 10 |
| chr1_-_85465147 | 0.18 |
ENST00000284031.13
|
DDAH1
|
dimethylarginine dimethylaminohydrolase 1 |
| chrX_-_134915203 | 0.18 |
ENST00000370779.8
|
MOSPD1
|
motile sperm domain containing 1 |
| chr12_-_120116730 | 0.17 |
ENST00000538903.1
ENST00000229340.10 ENST00000534951.5 |
RAB35
|
RAB35, member RAS oncogene family |
| chr6_-_83709141 | 0.17 |
ENST00000521743.5
|
SNAP91
|
synaptosome associated protein 91 |
| chr3_+_42685535 | 0.17 |
ENST00000287777.5
|
KLHL40
|
kelch like family member 40 |
| chrX_+_150983350 | 0.17 |
ENST00000455596.5
ENST00000448905.6 |
HMGB3
|
high mobility group box 3 |
| chr6_-_75206044 | 0.17 |
ENST00000322507.13
|
COL12A1
|
collagen type XII alpha 1 chain |
| chr20_-_57525123 | 0.17 |
ENST00000243914.8
|
CTCFL
|
CCCTC-binding factor like |
| chr4_-_101347492 | 0.17 |
ENST00000394854.8
|
PPP3CA
|
protein phosphatase 3 catalytic subunit alpha |
| chr9_-_23821275 | 0.17 |
ENST00000380110.8
|
ELAVL2
|
ELAV like RNA binding protein 2 |
| chr11_+_61766073 | 0.17 |
ENST00000675319.1
|
MYRF
|
myelin regulatory factor |
| chr12_-_6606320 | 0.17 |
ENST00000642594.1
ENST00000644289.1 ENST00000645095.1 |
CHD4
|
chromodomain helicase DNA binding protein 4 |
| chr17_+_41105332 | 0.17 |
ENST00000391415.1
ENST00000617453.1 |
KRTAP4-9
|
keratin associated protein 4-9 |
| chr6_+_14117764 | 0.17 |
ENST00000379153.4
|
CD83
|
CD83 molecule |
| chr17_+_82735551 | 0.17 |
ENST00000300784.8
|
FN3K
|
fructosamine 3 kinase |
| chr4_-_101347471 | 0.17 |
ENST00000323055.10
ENST00000512215.5 |
PPP3CA
|
protein phosphatase 3 catalytic subunit alpha |
| chr17_-_8799365 | 0.16 |
ENST00000329805.6
|
MFSD6L
|
major facilitator superfamily domain containing 6 like |
| chr1_-_32222322 | 0.16 |
ENST00000344461.7
ENST00000373593.5 ENST00000309777.11 |
TMEM234
|
transmembrane protein 234 |
| chr19_-_45914775 | 0.16 |
ENST00000341294.4
|
NANOS2
|
nanos C2HC-type zinc finger 2 |
| chr16_-_2720217 | 0.16 |
ENST00000302641.8
|
PRSS27
|
serine protease 27 |
| chr1_-_53142577 | 0.16 |
ENST00000620347.5
ENST00000611397.5 |
SLC1A7
|
solute carrier family 1 member 7 |
| chr1_+_89524871 | 0.16 |
ENST00000639264.1
|
LRRC8B
|
leucine rich repeat containing 8 VRAC subunit B |
| chr1_+_89524819 | 0.16 |
ENST00000439853.6
ENST00000330947.7 ENST00000449440.5 ENST00000640258.1 |
LRRC8B
|
leucine rich repeat containing 8 VRAC subunit B |
| chr8_+_97775775 | 0.16 |
ENST00000521545.7
|
LAPTM4B
|
lysosomal protein transmembrane 4 beta |
| chr22_+_41381923 | 0.16 |
ENST00000266304.9
|
TEF
|
TEF transcription factor, PAR bZIP family member |
| chr3_-_71753582 | 0.16 |
ENST00000295612.7
|
EIF4E3
|
eukaryotic translation initiation factor 4E family member 3 |
| chr5_-_134411847 | 0.16 |
ENST00000458198.3
ENST00000395009.3 |
CDKN2AIPNL
|
CDKN2A interacting protein N-terminal like |
| chr15_-_74906613 | 0.16 |
ENST00000565772.5
|
FAM219B
|
family with sequence similarity 219 member B |
| chr2_+_115162107 | 0.16 |
ENST00000310323.12
|
DPP10
|
dipeptidyl peptidase like 10 |
| chrX_-_40177213 | 0.16 |
ENST00000672922.2
ENST00000378455.8 ENST00000342274.8 |
BCOR
|
BCL6 corepressor |
| chr9_+_112750722 | 0.16 |
ENST00000374232.8
|
SNX30
|
sorting nexin family member 30 |
| chr17_+_83079595 | 0.16 |
ENST00000320095.12
|
METRNL
|
meteorin like, glial cell differentiation regulator |
| chr11_-_35419899 | 0.15 |
ENST00000646847.1
ENST00000449068.2 ENST00000643401.1 ENST00000645966.1 ENST00000647104.1 |
SLC1A2
|
solute carrier family 1 member 2 |
| chr10_-_100185993 | 0.15 |
ENST00000421367.7
ENST00000370408.2 ENST00000407654.7 |
ERLIN1
|
ER lipid raft associated 1 |
| chr2_+_233354474 | 0.15 |
ENST00000264057.7
ENST00000427930.5 ENST00000442524.4 |
DGKD
|
diacylglycerol kinase delta |
| chr15_+_24954912 | 0.15 |
ENST00000584968.5
ENST00000346403.10 ENST00000554227.6 ENST00000390687.9 ENST00000579070.5 ENST00000577565.1 ENST00000577949.5 ENST00000338327.4 |
SNRPN
SNURF
|
small nuclear ribonucleoprotein polypeptide N SNRPN upstream reading frame |
| chrX_-_136780925 | 0.15 |
ENST00000250617.7
|
ARHGEF6
|
Rac/Cdc42 guanine nucleotide exchange factor 6 |
| chr1_+_161722659 | 0.15 |
ENST00000336830.9
ENST00000367944.3 ENST00000367945.5 ENST00000367946.7 |
FCRLB
|
Fc receptor like B |
| chr4_-_138242325 | 0.15 |
ENST00000280612.9
|
SLC7A11
|
solute carrier family 7 member 11 |
| chr22_+_22030934 | 0.15 |
ENST00000390282.2
|
IGLV4-69
|
immunoglobulin lambda variable 4-69 |
| chr17_+_68259164 | 0.15 |
ENST00000448504.6
|
ARSG
|
arylsulfatase G |
| chr8_+_85463997 | 0.15 |
ENST00000285379.10
|
CA2
|
carbonic anhydrase 2 |
| chr7_-_139036017 | 0.15 |
ENST00000275766.2
|
ZC3HAV1L
|
zinc finger CCCH-type containing, antiviral 1 like |
| chr16_+_22206255 | 0.15 |
ENST00000263026.10
|
EEF2K
|
eukaryotic elongation factor 2 kinase |
| chr10_-_107164692 | 0.15 |
ENST00000263054.11
|
SORCS1
|
sortilin related VPS10 domain containing receptor 1 |
| chr20_-_17682234 | 0.15 |
ENST00000377813.6
ENST00000377807.6 ENST00000360807.8 ENST00000398782.2 |
RRBP1
|
ribosome binding protein 1 |
| chr16_+_12901757 | 0.15 |
ENST00000423335.2
|
SHISA9
|
shisa family member 9 |
| chr21_+_33070133 | 0.15 |
ENST00000382348.2
|
OLIG1
|
oligodendrocyte transcription factor 1 |
| chr2_-_168913277 | 0.14 |
ENST00000451987.5
|
SPC25
|
SPC25 component of NDC80 kinetochore complex |
| chr12_-_29783798 | 0.14 |
ENST00000552618.5
ENST00000551659.5 ENST00000539277.6 |
TMTC1
|
transmembrane O-mannosyltransferase targeting cadherins 1 |
| chr19_+_14073556 | 0.14 |
ENST00000590772.1
|
MISP3
|
MISP family member 3 |
| chr11_-_35526024 | 0.14 |
ENST00000615849.4
|
PAMR1
|
peptidase domain containing associated with muscle regeneration 1 |
| chr1_-_32870775 | 0.14 |
ENST00000649537.2
ENST00000373471.9 |
FNDC5
|
fibronectin type III domain containing 5 |
| chr7_-_33109363 | 0.14 |
ENST00000684207.1
ENST00000297157.8 |
RP9
|
RP9 pre-mRNA splicing factor |
| chrX_+_119236245 | 0.14 |
ENST00000535419.2
|
PGRMC1
|
progesterone receptor membrane component 1 |
| chr15_-_52179881 | 0.14 |
ENST00000396335.8
ENST00000560116.1 ENST00000358784.11 |
GNB5
|
G protein subunit beta 5 |
| chr12_-_70609788 | 0.14 |
ENST00000547715.1
ENST00000538708.5 ENST00000550857.5 ENST00000261266.9 |
PTPRB
|
protein tyrosine phosphatase receptor type B |
| chr20_-_36646146 | 0.14 |
ENST00000262866.9
|
SLA2
|
Src like adaptor 2 |
| chr6_-_83709382 | 0.14 |
ENST00000520302.5
ENST00000520213.5 ENST00000439399.6 |
SNAP91
|
synaptosome associated protein 91 |
| chr4_-_88823214 | 0.14 |
ENST00000513837.5
ENST00000503556.5 |
FAM13A
|
family with sequence similarity 13 member A |
| chr2_-_234497035 | 0.14 |
ENST00000390645.2
ENST00000339728.6 |
ARL4C
|
ADP ribosylation factor like GTPase 4C |
| chr2_+_171922442 | 0.14 |
ENST00000264108.5
|
HAT1
|
histone acetyltransferase 1 |
| chr16_+_70646536 | 0.14 |
ENST00000288098.6
|
IL34
|
interleukin 34 |
| chrX_+_119236274 | 0.14 |
ENST00000217971.8
|
PGRMC1
|
progesterone receptor membrane component 1 |
| chr16_+_11345429 | 0.14 |
ENST00000576027.1
ENST00000312499.6 ENST00000648619.1 |
RMI2
|
RecQ mediated genome instability 2 |
| chr13_+_33818122 | 0.14 |
ENST00000380071.8
|
RFC3
|
replication factor C subunit 3 |
| chr16_+_81779279 | 0.14 |
ENST00000564138.6
|
PLCG2
|
phospholipase C gamma 2 |
| chr10_-_101588126 | 0.14 |
ENST00000339310.7
ENST00000299206.8 ENST00000413344.5 ENST00000429502.1 ENST00000430045.1 ENST00000370172.5 ENST00000370162.8 ENST00000628479.2 |
POLL
|
DNA polymerase lambda |
| chr15_+_90884496 | 0.13 |
ENST00000452243.5
ENST00000328850.8 |
FES
|
FES proto-oncogene, tyrosine kinase |
| chr17_+_42536510 | 0.13 |
ENST00000585572.1
ENST00000586516.5 ENST00000591587.1 |
ENSG00000266929.1
NAGLU
|
novel transcript N-acetyl-alpha-glucosaminidase |
| chr19_-_14206168 | 0.13 |
ENST00000361434.7
ENST00000340736.10 |
ADGRL1
|
adhesion G protein-coupled receptor L1 |
| chr3_+_122795039 | 0.13 |
ENST00000261038.6
|
SLC49A4
|
solute carrier family 49 member 4 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.3 | GO:2000137 | negative regulation of cell proliferation involved in heart morphogenesis(GO:2000137) |
| 0.2 | 0.7 | GO:0072098 | intermediate mesoderm development(GO:0048389) pattern specification involved in mesonephros development(GO:0061227) anterior/posterior pattern specification involved in kidney development(GO:0072098) |
| 0.2 | 0.7 | GO:0070378 | positive regulation of ERK5 cascade(GO:0070378) |
| 0.2 | 0.5 | GO:1903400 | L-arginine transmembrane transport(GO:1903400) |
| 0.1 | 0.6 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 0.1 | 0.5 | GO:1905205 | positive regulation of connective tissue replacement(GO:1905205) |
| 0.1 | 0.8 | GO:0033615 | mitochondrial proton-transporting ATP synthase complex assembly(GO:0033615) |
| 0.1 | 0.6 | GO:0014835 | myoblast differentiation involved in skeletal muscle regeneration(GO:0014835) positive regulation of interleukin-12 secretion(GO:2001184) |
| 0.1 | 0.9 | GO:0060244 | negative regulation of cell proliferation involved in contact inhibition(GO:0060244) |
| 0.1 | 0.3 | GO:1902309 | negative regulation of peptidyl-serine dephosphorylation(GO:1902309) |
| 0.1 | 0.3 | GO:0043311 | positive regulation of eosinophil degranulation(GO:0043311) positive regulation of eosinophil activation(GO:1902568) |
| 0.1 | 0.4 | GO:0001306 | age-dependent response to oxidative stress(GO:0001306) age-dependent response to reactive oxygen species(GO:0001315) regulation of systemic arterial blood pressure by acetylcholine(GO:0003068) vasodilation by acetylcholine involved in regulation of systemic arterial blood pressure(GO:0003069) regulation of systemic arterial blood pressure by neurotransmitter(GO:0003070) age-dependent general metabolic decline(GO:0007571) |
| 0.1 | 0.6 | GO:0035583 | sequestering of TGFbeta in extracellular matrix(GO:0035583) |
| 0.1 | 0.2 | GO:0014016 | neuroblast differentiation(GO:0014016) |
| 0.1 | 0.2 | GO:0070676 | intralumenal vesicle formation(GO:0070676) |
| 0.1 | 0.3 | GO:0015688 | iron chelate transport(GO:0015688) siderophore transport(GO:0015891) |
| 0.1 | 0.3 | GO:0034699 | response to luteinizing hormone(GO:0034699) |
| 0.1 | 0.4 | GO:0061767 | negative regulation of lung blood pressure(GO:0061767) |
| 0.1 | 0.3 | GO:0010816 | substance P catabolic process(GO:0010814) calcitonin catabolic process(GO:0010816) endothelin maturation(GO:0034959) |
| 0.1 | 0.5 | GO:0035879 | plasma membrane lactate transport(GO:0035879) |
| 0.1 | 0.4 | GO:0042539 | hypotonic salinity response(GO:0042539) cellular hypotonic salinity response(GO:0071477) |
| 0.1 | 0.2 | GO:0050915 | sensory perception of sour taste(GO:0050915) |
| 0.1 | 0.2 | GO:0030718 | germ-line stem cell population maintenance(GO:0030718) |
| 0.1 | 0.2 | GO:0030327 | prenylated protein catabolic process(GO:0030327) |
| 0.1 | 0.3 | GO:0042986 | positive regulation of amyloid precursor protein biosynthetic process(GO:0042986) |
| 0.1 | 0.6 | GO:0072553 | terminal button organization(GO:0072553) |
| 0.1 | 0.2 | GO:0097089 | methyl-branched fatty acid metabolic process(GO:0097089) |
| 0.0 | 0.1 | GO:0042938 | dipeptide transport(GO:0042938) |
| 0.0 | 0.1 | GO:0002316 | follicular B cell differentiation(GO:0002316) |
| 0.0 | 0.4 | GO:0038031 | non-canonical Wnt signaling pathway via JNK cascade(GO:0038031) |
| 0.0 | 1.6 | GO:0003301 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.0 | 0.6 | GO:0098914 | membrane repolarization during atrial cardiac muscle cell action potential(GO:0098914) |
| 0.0 | 0.3 | GO:0003415 | chondrocyte hypertrophy(GO:0003415) |
| 0.0 | 0.1 | GO:1900194 | negative regulation of oocyte development(GO:0060283) negative regulation of oocyte maturation(GO:1900194) |
| 0.0 | 0.2 | GO:0000415 | negative regulation of histone H3-K36 methylation(GO:0000415) |
| 0.0 | 0.2 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.0 | 0.6 | GO:0061299 | retina vasculature morphogenesis in camera-type eye(GO:0061299) |
| 0.0 | 0.1 | GO:1904397 | negative regulation of neuromuscular junction development(GO:1904397) |
| 0.0 | 0.1 | GO:1990637 | response to prolactin(GO:1990637) |
| 0.0 | 0.3 | GO:0030997 | regulation of centriole-centriole cohesion(GO:0030997) positive regulation of protein localization to centrosome(GO:1904781) |
| 0.0 | 0.1 | GO:0060739 | mesenchymal-epithelial cell signaling involved in prostate gland development(GO:0060739) |
| 0.0 | 0.1 | GO:0006408 | snRNA export from nucleus(GO:0006408) positive regulation of RNA export from nucleus(GO:0046833) |
| 0.0 | 0.1 | GO:0043376 | regulation of CD8-positive, alpha-beta T cell differentiation(GO:0043376) |
| 0.0 | 0.3 | GO:0090336 | positive regulation of brown fat cell differentiation(GO:0090336) |
| 0.0 | 0.1 | GO:0036111 | very long-chain fatty-acyl-CoA metabolic process(GO:0036111) |
| 0.0 | 0.1 | GO:0071288 | cellular response to mercury ion(GO:0071288) |
| 0.0 | 0.2 | GO:0090210 | regulation of establishment of blood-brain barrier(GO:0090210) |
| 0.0 | 0.3 | GO:2000334 | response to linoleic acid(GO:0070543) blood microparticle formation(GO:0072564) regulation of blood microparticle formation(GO:2000332) positive regulation of blood microparticle formation(GO:2000334) |
| 0.0 | 0.1 | GO:1904381 | Golgi apparatus mannose trimming(GO:1904381) |
| 0.0 | 0.1 | GO:0060744 | thelarche(GO:0042695) mammary gland branching involved in thelarche(GO:0060744) |
| 0.0 | 0.2 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.0 | 0.1 | GO:0006059 | hexitol metabolic process(GO:0006059) |
| 0.0 | 0.2 | GO:1900262 | regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.0 | 0.2 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.0 | 0.1 | GO:0006287 | base-excision repair, gap-filling(GO:0006287) |
| 0.0 | 0.4 | GO:0002315 | marginal zone B cell differentiation(GO:0002315) |
| 0.0 | 0.3 | GO:1903800 | positive regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903800) |
| 0.0 | 0.2 | GO:0048539 | bone marrow development(GO:0048539) |
| 0.0 | 0.0 | GO:0001794 | type IIa hypersensitivity(GO:0001794) regulation of type IIa hypersensitivity(GO:0001796) type II hypersensitivity(GO:0002445) regulation of type II hypersensitivity(GO:0002892) |
| 0.0 | 0.3 | GO:0010578 | regulation of adenylate cyclase activity involved in G-protein coupled receptor signaling pathway(GO:0010578) positive regulation of adenylate cyclase activity involved in G-protein coupled receptor signaling pathway(GO:0010579) |
| 0.0 | 0.2 | GO:0045541 | negative regulation of cholesterol biosynthetic process(GO:0045541) negative regulation of cholesterol metabolic process(GO:0090206) |
| 0.0 | 0.1 | GO:0098528 | skeletal muscle fiber differentiation(GO:0098528) |
| 0.0 | 0.1 | GO:0010760 | negative regulation of macrophage chemotaxis(GO:0010760) |
| 0.0 | 0.1 | GO:0071344 | diphosphate metabolic process(GO:0071344) |
| 0.0 | 0.1 | GO:0033693 | neurofilament bundle assembly(GO:0033693) |
| 0.0 | 0.3 | GO:0031274 | positive regulation of pseudopodium assembly(GO:0031274) |
| 0.0 | 0.1 | GO:0002159 | desmosome assembly(GO:0002159) |
| 0.0 | 0.4 | GO:0007220 | Notch receptor processing(GO:0007220) |
| 0.0 | 0.1 | GO:1900242 | regulation of synaptic vesicle endocytosis(GO:1900242) |
| 0.0 | 0.6 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.0 | 0.2 | GO:0032713 | negative regulation of interleukin-4 production(GO:0032713) |
| 0.0 | 0.1 | GO:1903862 | positive regulation of oxidative phosphorylation(GO:1903862) |
| 0.0 | 0.3 | GO:0045176 | apical protein localization(GO:0045176) |
| 0.0 | 0.4 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.0 | 0.1 | GO:0014876 | negative regulation of muscle atrophy(GO:0014736) response to injury involved in regulation of muscle adaptation(GO:0014876) |
| 0.0 | 0.1 | GO:0044789 | modulation by host of viral release from host cell(GO:0044789) positive regulation by host of viral release from host cell(GO:0044791) |
| 0.0 | 0.4 | GO:2000651 | positive regulation of sodium ion transmembrane transporter activity(GO:2000651) |
| 0.0 | 0.1 | GO:0033563 | dorsal/ventral axon guidance(GO:0033563) |
| 0.0 | 0.1 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.0 | 0.1 | GO:0044313 | protein K6-linked deubiquitination(GO:0044313) |
| 0.0 | 0.1 | GO:0046098 | guanine metabolic process(GO:0046098) |
| 0.0 | 0.1 | GO:0030311 | poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.0 | 0.1 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.0 | 0.1 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.0 | 0.0 | GO:0010931 | macrophage tolerance induction(GO:0010931) regulation of macrophage tolerance induction(GO:0010932) positive regulation of macrophage tolerance induction(GO:0010933) |
| 0.0 | 0.3 | GO:1900038 | negative regulation of cellular response to hypoxia(GO:1900038) |
| 0.0 | 0.1 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 | 0.2 | GO:1903715 | regulation of aerobic respiration(GO:1903715) |
| 0.0 | 0.1 | GO:1902202 | regulation of hepatocyte growth factor receptor signaling pathway(GO:1902202) |
| 0.0 | 0.2 | GO:0050703 | interleukin-1 alpha secretion(GO:0050703) |
| 0.0 | 0.1 | GO:0080009 | mRNA methylation(GO:0080009) |
| 0.0 | 0.1 | GO:0006121 | mitochondrial electron transport, succinate to ubiquinone(GO:0006121) |
| 0.0 | 0.1 | GO:0045079 | negative regulation of chemokine biosynthetic process(GO:0045079) |
| 0.0 | 0.0 | GO:1902512 | positive regulation of apoptotic DNA fragmentation(GO:1902512) |
| 0.0 | 0.2 | GO:0048227 | plasma membrane to endosome transport(GO:0048227) |
| 0.0 | 0.5 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.0 | 0.1 | GO:2001137 | positive regulation of endocytic recycling(GO:2001137) |
| 0.0 | 0.2 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.0 | 0.1 | GO:0008295 | spermidine biosynthetic process(GO:0008295) |
| 0.0 | 0.2 | GO:2000774 | positive regulation of cellular senescence(GO:2000774) |
| 0.0 | 0.4 | GO:0007216 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.0 | 0.1 | GO:0061101 | neuroendocrine cell differentiation(GO:0061101) |
| 0.0 | 0.0 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.0 | 0.1 | GO:1903566 | positive regulation of protein localization to cilium(GO:1903566) |
| 0.0 | 0.8 | GO:0008333 | endosome to lysosome transport(GO:0008333) |
| 0.0 | 0.0 | GO:0097476 | spinal cord motor neuron migration(GO:0097476) |
| 0.0 | 0.0 | GO:0006663 | platelet activating factor biosynthetic process(GO:0006663) |
| 0.0 | 0.4 | GO:0036150 | phosphatidylserine acyl-chain remodeling(GO:0036150) |
| 0.0 | 0.3 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.0 | 0.1 | GO:0038170 | somatostatin receptor signaling pathway(GO:0038169) somatostatin signaling pathway(GO:0038170) |
| 0.0 | 0.0 | GO:0060743 | epithelial cell maturation involved in prostate gland development(GO:0060743) |
| 0.0 | 0.2 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.0 | 0.1 | GO:0019375 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.0 | 0.2 | GO:1903608 | protein localization to cytoplasmic stress granule(GO:1903608) |
| 0.0 | 0.6 | GO:0002755 | MyD88-dependent toll-like receptor signaling pathway(GO:0002755) |
| 0.0 | 0.0 | GO:0033152 | immunoglobulin V(D)J recombination(GO:0033152) |
| 0.0 | 0.0 | GO:0090119 | vesicle-mediated cholesterol transport(GO:0090119) |
| 0.0 | 0.0 | GO:0006113 | fermentation(GO:0006113) regulation of fermentation(GO:0043465) |
| 0.0 | 0.1 | GO:0070673 | response to interleukin-18(GO:0070673) |
| 0.0 | 0.1 | GO:0008354 | germ cell migration(GO:0008354) |
| 0.0 | 0.0 | GO:0098937 | dendritic transport(GO:0098935) anterograde dendritic transport(GO:0098937) |
| 0.0 | 0.1 | GO:0021633 | optic nerve structural organization(GO:0021633) |
| 0.0 | 0.5 | GO:0034629 | cellular protein complex localization(GO:0034629) |
| 0.0 | 0.1 | GO:0015798 | myo-inositol transport(GO:0015798) |
| 0.0 | 0.0 | GO:0002581 | negative regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002581) |
| 0.0 | 0.1 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.0 | 0.1 | GO:2000048 | negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 0.0 | 0.2 | GO:0060009 | Sertoli cell development(GO:0060009) |
| 0.0 | 0.2 | GO:0086046 | membrane depolarization during SA node cell action potential(GO:0086046) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0005595 | collagen type XII trimer(GO:0005595) |
| 0.1 | 0.3 | GO:0031302 | intrinsic component of endosome membrane(GO:0031302) |
| 0.1 | 0.5 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.0 | 0.5 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.0 | 0.1 | GO:0034686 | integrin alphav-beta8 complex(GO:0034686) |
| 0.0 | 0.4 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.0 | 0.3 | GO:0044214 | spanning component of plasma membrane(GO:0044214) spanning component of membrane(GO:0089717) |
| 0.0 | 0.1 | GO:0005846 | nuclear cap binding complex(GO:0005846) |
| 0.0 | 0.5 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.0 | 0.1 | GO:0000839 | Hrd1p ubiquitin ligase ERAD-L complex(GO:0000839) |
| 0.0 | 2.5 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 0.1 | GO:0045293 | MIS complex(GO:0036396) mRNA editing complex(GO:0045293) |
| 0.0 | 0.6 | GO:0001527 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.0 | 0.2 | GO:0097165 | nuclear stress granule(GO:0097165) |
| 0.0 | 0.1 | GO:0035841 | new growing cell tip(GO:0035841) |
| 0.0 | 0.1 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 0.0 | 0.4 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.1 | GO:0097224 | sperm connecting piece(GO:0097224) |
| 0.0 | 0.7 | GO:0090545 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 0.2 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.0 | 0.2 | GO:0000808 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.0 | 0.6 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 0.1 | GO:0020018 | ciliary pocket(GO:0020016) ciliary pocket membrane(GO:0020018) |
| 0.0 | 0.1 | GO:0070381 | endosome to plasma membrane transport vesicle(GO:0070381) |
| 0.0 | 0.1 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.0 | 0.5 | GO:0097386 | glial cell projection(GO:0097386) |
| 0.0 | 0.1 | GO:0000110 | nucleotide-excision repair factor 1 complex(GO:0000110) |
| 0.0 | 0.3 | GO:0001520 | outer dense fiber(GO:0001520) |
| 0.0 | 0.2 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.0 | 0.2 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.0 | 0.2 | GO:0005845 | mRNA cap binding complex(GO:0005845) |
| 0.0 | 0.1 | GO:0016035 | zeta DNA polymerase complex(GO:0016035) |
| 0.0 | 0.1 | GO:0005749 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.0 | 0.1 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.0 | 0.0 | GO:0071756 | IgM immunoglobulin complex(GO:0071753) IgM immunoglobulin complex, circulating(GO:0071754) pentameric IgM immunoglobulin complex(GO:0071756) |
| 0.0 | 0.3 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.0 | 0.1 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.0 | 0.2 | GO:0005687 | U4 snRNP(GO:0005687) |
| 0.0 | 0.2 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0005289 | high-affinity basic amino acid transmembrane transporter activity(GO:0005287) high-affinity arginine transmembrane transporter activity(GO:0005289) high-affinity lysine transmembrane transporter activity(GO:0005292) |
| 0.1 | 0.6 | GO:0016211 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.1 | 0.7 | GO:0030298 | receptor signaling protein tyrosine kinase activator activity(GO:0030298) |
| 0.1 | 0.5 | GO:0050436 | microfibril binding(GO:0050436) |
| 0.1 | 0.4 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.1 | 0.3 | GO:0005519 | cytoskeletal regulatory protein binding(GO:0005519) |
| 0.1 | 0.3 | GO:0016524 | latrotoxin receptor activity(GO:0016524) |
| 0.1 | 0.4 | GO:0031962 | mineralocorticoid receptor binding(GO:0031962) |
| 0.1 | 0.6 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.1 | 0.6 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.1 | 0.2 | GO:0003990 | acetylcholinesterase activity(GO:0003990) choline binding(GO:0033265) |
| 0.1 | 0.5 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.1 | 1.7 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.1 | 0.9 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.1 | 0.2 | GO:0086059 | voltage-gated calcium channel activity involved SA node cell action potential(GO:0086059) |
| 0.0 | 0.3 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.0 | 0.5 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.0 | 0.2 | GO:0044736 | acid-sensing ion channel activity(GO:0044736) |
| 0.0 | 0.4 | GO:0016721 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.0 | 0.2 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.0 | 0.1 | GO:0032093 | SAM domain binding(GO:0032093) |
| 0.0 | 0.4 | GO:0030020 | extracellular matrix structural constituent conferring tensile strength(GO:0030020) |
| 0.0 | 0.3 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 0.0 | 0.3 | GO:0098821 | BMP receptor activity(GO:0098821) |
| 0.0 | 0.4 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.0 | 0.2 | GO:0005294 | neutral L-amino acid secondary active transmembrane transporter activity(GO:0005294) |
| 0.0 | 0.1 | GO:0033989 | 3alpha,7alpha,12alpha-trihydroxy-5beta-cholest-24-enoyl-CoA hydratase activity(GO:0033989) 17-beta-hydroxysteroid dehydrogenase (NAD+) activity(GO:0044594) |
| 0.0 | 0.4 | GO:0022820 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.0 | 0.4 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.0 | 0.1 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.0 | 0.1 | GO:0016838 | carbon-oxygen lyase activity, acting on phosphates(GO:0016838) |
| 0.0 | 0.3 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.0 | 0.1 | GO:0051575 | 5'-deoxyribose-5-phosphate lyase activity(GO:0051575) |
| 0.0 | 0.1 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.0 | 0.2 | GO:0016403 | dimethylargininase activity(GO:0016403) |
| 0.0 | 0.5 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.0 | 0.4 | GO:0015250 | water channel activity(GO:0015250) |
| 0.0 | 0.1 | GO:0030060 | L-malate dehydrogenase activity(GO:0030060) |
| 0.0 | 0.8 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.2 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.0 | 0.1 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 0.0 | 0.1 | GO:0032038 | myosin II heavy chain binding(GO:0032038) |
| 0.0 | 0.1 | GO:0005157 | macrophage colony-stimulating factor receptor binding(GO:0005157) |
| 0.0 | 0.2 | GO:0033170 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.0 | 0.1 | GO:0004905 | type I interferon receptor activity(GO:0004905) |
| 0.0 | 0.1 | GO:0004335 | galactokinase activity(GO:0004335) |
| 0.0 | 0.2 | GO:0016670 | oxidoreductase activity, acting on a sulfur group of donors, oxygen as acceptor(GO:0016670) |
| 0.0 | 0.1 | GO:0098988 | adenylate cyclase inhibiting G-protein coupled glutamate receptor activity(GO:0001640) G-protein coupled glutamate receptor activity(GO:0098988) |
| 0.0 | 0.6 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.2 | GO:0005114 | type II transforming growth factor beta receptor binding(GO:0005114) |
| 0.0 | 0.5 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.1 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 0.0 | 0.1 | GO:0016900 | oxidoreductase activity, acting on the CH-OH group of donors, disulfide as acceptor(GO:0016900) vitamin-K-epoxide reductase (warfarin-sensitive) activity(GO:0047057) |
| 0.0 | 0.1 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.0 | 0.1 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.0 | 0.1 | GO:0008177 | succinate dehydrogenase (ubiquinone) activity(GO:0008177) |
| 0.0 | 0.7 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.0 | 0.6 | GO:0008188 | neuropeptide receptor activity(GO:0008188) |
| 0.0 | 0.3 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 0.0 | 0.1 | GO:0097677 | STAT family protein binding(GO:0097677) |
| 0.0 | 0.1 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.0 | 0.3 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.0 | 0.1 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) |
| 0.0 | 0.2 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.0 | 0.1 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.1 | GO:0034618 | arginine binding(GO:0034618) |
| 0.0 | 0.0 | GO:0016794 | guanosine-3',5'-bis(diphosphate) 3'-diphosphatase activity(GO:0008893) diphosphoric monoester hydrolase activity(GO:0016794) |
| 0.0 | 0.1 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.0 | 0.0 | GO:0004698 | calcium-dependent protein kinase C activity(GO:0004698) |
| 0.0 | 0.8 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.1 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 0.9 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 0.1 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.0 | 0.2 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.0 | 0.4 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.2 | GO:0015245 | fatty acid transporter activity(GO:0015245) |
| 0.0 | 0.3 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.1 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.4 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 0.5 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.1 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.5 | PID BMP PATHWAY | BMP receptor signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.0 | 0.1 | REACTOME TRANSPORT OF MATURE MRNA DERIVED FROM AN INTRONLESS TRANSCRIPT | Genes involved in Transport of Mature mRNA Derived from an Intronless Transcript |
| 0.0 | 0.5 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.3 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 0.6 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 0.7 | REACTOME TRAF6 MEDIATED NFKB ACTIVATION | Genes involved in TRAF6 mediated NF-kB activation |
| 0.0 | 0.3 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.0 | 0.7 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 7ALPHA HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 7alpha-hydroxycholesterol |
| 0.0 | 0.4 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.0 | 0.4 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.0 | 0.1 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.0 | 0.6 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 0.2 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.0 | 0.3 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 0.2 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 0.2 | REACTOME POL SWITCHING | Genes involved in Polymerase switching |