Project
Inflammatory response time course, HUVEC (Wada, 2009)
Navigation
Downloads
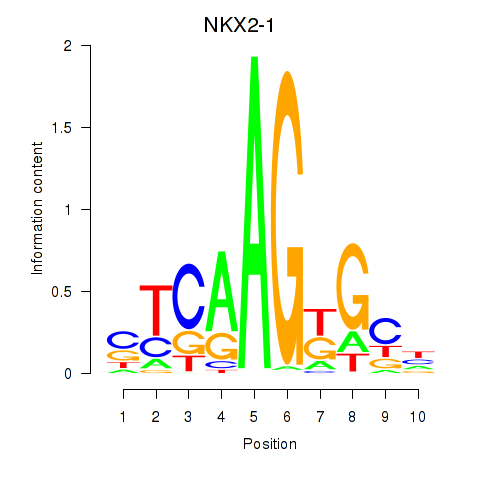
Results for NKX2-1
Z-value: 0.79
Transcription factors associated with NKX2-1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NKX2-1
|
ENSG00000136352.19 | NKX2-1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NKX2-1 | hg38_v1_chr14_-_36520222_36520237, hg38_v1_chr14_-_36521149_36521191, hg38_v1_chr14_-_36519679_36519693 | 0.19 | 3.6e-01 | Click! |
Activity profile of NKX2-1 motif
Sorted Z-values of NKX2-1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of NKX2-1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.0 | GO:1904582 | regulation of intracellular mRNA localization(GO:1904580) positive regulation of intracellular mRNA localization(GO:1904582) |
| 0.5 | 1.5 | GO:0070662 | erythropoietin-mediated signaling pathway(GO:0038162) mast cell proliferation(GO:0070662) |
| 0.5 | 1.4 | GO:0021538 | epithalamus development(GO:0021538) habenula development(GO:0021986) |
| 0.2 | 0.7 | GO:0006667 | sphinganine metabolic process(GO:0006667) |
| 0.2 | 1.4 | GO:0021773 | striatal medium spiny neuron differentiation(GO:0021773) |
| 0.2 | 2.1 | GO:2000795 | negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.2 | 1.0 | GO:1901907 | diadenosine polyphosphate catabolic process(GO:0015961) diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 0.2 | 0.7 | GO:0005986 | sucrose biosynthetic process(GO:0005986) |
| 0.2 | 0.5 | GO:1903465 | vacuolar phosphate transport(GO:0007037) positive regulation of mitotic cell cycle DNA replication(GO:1903465) positive regulation of parathyroid hormone secretion(GO:2000830) |
| 0.2 | 0.5 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) |
| 0.2 | 0.6 | GO:0018199 | peptidyl-glutamine modification(GO:0018199) |
| 0.1 | 1.7 | GO:0098734 | macromolecule depalmitoylation(GO:0098734) |
| 0.1 | 0.4 | GO:0035674 | tricarboxylic acid transmembrane transport(GO:0035674) |
| 0.1 | 0.4 | GO:2000777 | positive regulation of oocyte maturation(GO:1900195) positive regulation of proteasomal ubiquitin-dependent protein catabolic process involved in cellular response to hypoxia(GO:2000777) |
| 0.1 | 0.5 | GO:0002032 | desensitization of G-protein coupled receptor protein signaling pathway by arrestin(GO:0002032) |
| 0.1 | 0.3 | GO:0010808 | positive regulation of synaptic vesicle priming(GO:0010808) |
| 0.1 | 1.1 | GO:1904338 | regulation of dopaminergic neuron differentiation(GO:1904338) |
| 0.1 | 0.3 | GO:0009085 | lysine biosynthetic process(GO:0009085) lysine biosynthetic process via aminoadipic acid(GO:0019878) |
| 0.1 | 0.3 | GO:0070682 | proteasome regulatory particle assembly(GO:0070682) |
| 0.1 | 3.4 | GO:0061049 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.1 | 1.1 | GO:0061299 | retina vasculature morphogenesis in camera-type eye(GO:0061299) |
| 0.1 | 1.0 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) |
| 0.1 | 0.8 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 0.1 | 1.9 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.1 | 0.2 | GO:0043049 | hepatocyte cell migration(GO:0002194) otic placode formation(GO:0043049) branching involved in pancreas morphogenesis(GO:0061114) acinar cell differentiation(GO:0090425) positive regulation of forebrain neuron differentiation(GO:2000979) |
| 0.1 | 0.5 | GO:0031339 | negative regulation of vesicle fusion(GO:0031339) |
| 0.1 | 1.2 | GO:0070914 | UV-damage excision repair(GO:0070914) |
| 0.1 | 0.2 | GO:0019483 | beta-alanine biosynthetic process(GO:0019483) |
| 0.1 | 0.2 | GO:1901594 | detection of temperature stimulus involved in thermoception(GO:0050960) response to capsazepine(GO:1901594) |
| 0.1 | 0.3 | GO:0061181 | regulation of chondrocyte development(GO:0061181) |
| 0.0 | 0.2 | GO:0072092 | ureteric bud invasion(GO:0072092) |
| 0.0 | 0.9 | GO:0007501 | mesodermal cell fate specification(GO:0007501) |
| 0.0 | 0.9 | GO:2001135 | regulation of endocytic recycling(GO:2001135) |
| 0.0 | 0.6 | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.0 | 0.3 | GO:1903385 | regulation of homophilic cell adhesion(GO:1903385) |
| 0.0 | 0.1 | GO:0006500 | N-terminal protein palmitoylation(GO:0006500) |
| 0.0 | 0.4 | GO:0009249 | protein lipoylation(GO:0009249) |
| 0.0 | 0.3 | GO:0035811 | regulation of systemic arterial blood pressure by vasopressin(GO:0001992) negative regulation of urine volume(GO:0035811) |
| 0.0 | 0.2 | GO:0010286 | heat acclimation(GO:0010286) cellular heat acclimation(GO:0070370) |
| 0.0 | 0.5 | GO:0050703 | interleukin-1 alpha secretion(GO:0050703) |
| 0.0 | 1.2 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.0 | 0.7 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.0 | 1.1 | GO:0035728 | response to hepatocyte growth factor(GO:0035728) |
| 0.0 | 0.4 | GO:0030210 | heparin metabolic process(GO:0030202) heparin biosynthetic process(GO:0030210) |
| 0.0 | 0.2 | GO:1905123 | regulation of glucosylceramidase activity(GO:1905123) |
| 0.0 | 0.2 | GO:1903377 | negative regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903377) |
| 0.0 | 0.5 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.0 | 1.1 | GO:1904659 | hexose transmembrane transport(GO:0035428) glucose transmembrane transport(GO:1904659) |
| 0.0 | 0.5 | GO:0010991 | negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.0 | 0.5 | GO:0006089 | lactate metabolic process(GO:0006089) |
| 0.0 | 1.1 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.0 | 0.2 | GO:0006011 | UDP-glucose metabolic process(GO:0006011) |
| 0.0 | 0.2 | GO:2000381 | negative regulation of mesoderm development(GO:2000381) |
| 0.0 | 0.2 | GO:0045959 | regulation of complement activation, classical pathway(GO:0030450) negative regulation of complement activation, classical pathway(GO:0045959) regulation of opsonization(GO:1903027) |
| 0.0 | 0.2 | GO:0048368 | lateral mesoderm development(GO:0048368) |
| 0.0 | 0.3 | GO:0061158 | 3'-UTR-mediated mRNA destabilization(GO:0061158) |
| 0.0 | 0.1 | GO:1902938 | regulation of intracellular calcium activated chloride channel activity(GO:1902938) |
| 0.0 | 0.2 | GO:0015986 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 0.1 | GO:1901475 | pyruvate transport(GO:0006848) pyruvate transmembrane transport(GO:1901475) |
| 0.0 | 0.2 | GO:1990035 | calcium ion import across plasma membrane(GO:0098703) calcium ion import into cell(GO:1990035) |
| 0.0 | 0.2 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.0 | 0.2 | GO:0036010 | protein localization to endosome(GO:0036010) |
| 0.0 | 0.2 | GO:1901409 | positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.0 | 0.1 | GO:0060136 | embryonic process involved in female pregnancy(GO:0060136) |
| 0.0 | 0.4 | GO:0019372 | lipoxygenase pathway(GO:0019372) |
| 0.0 | 0.4 | GO:0097320 | membrane tubulation(GO:0097320) |
| 0.0 | 0.2 | GO:0090385 | phagosome-lysosome fusion(GO:0090385) |
| 0.0 | 0.7 | GO:1901661 | quinone metabolic process(GO:1901661) |
| 0.0 | 0.4 | GO:0042994 | cytoplasmic sequestering of transcription factor(GO:0042994) |
| 0.0 | 0.2 | GO:0060033 | anatomical structure regression(GO:0060033) |
| 0.0 | 0.1 | GO:0030070 | insulin processing(GO:0030070) |
| 0.0 | 0.8 | GO:0048384 | retinoic acid receptor signaling pathway(GO:0048384) |
| 0.0 | 0.7 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 1.4 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.0 | 0.2 | GO:0051964 | negative regulation of synapse assembly(GO:0051964) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.9 | GO:0032039 | integrator complex(GO:0032039) |
| 0.1 | 2.0 | GO:0070578 | RISC-loading complex(GO:0070578) |
| 0.1 | 1.2 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.1 | 1.7 | GO:0012510 | trans-Golgi network transport vesicle membrane(GO:0012510) |
| 0.1 | 0.7 | GO:0031211 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.1 | 0.2 | GO:1905103 | integral component of lysosomal membrane(GO:1905103) |
| 0.1 | 1.9 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.1 | 0.4 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.1 | 0.2 | GO:0071149 | TEAD-2-YAP complex(GO:0071149) |
| 0.1 | 0.7 | GO:0097486 | multivesicular body lumen(GO:0097486) |
| 0.1 | 2.0 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.0 | 0.3 | GO:0044305 | calyx of Held(GO:0044305) |
| 0.0 | 0.6 | GO:0016013 | syntrophin complex(GO:0016013) |
| 0.0 | 1.0 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.0 | 0.4 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.0 | 0.8 | GO:0001931 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.0 | 0.2 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.0 | 2.2 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.2 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.0 | 0.6 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 0.3 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.0 | 0.2 | GO:0031588 | cAMP-dependent protein kinase complex(GO:0005952) nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.2 | GO:0005686 | U2 snRNP(GO:0005686) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.0 | GO:0000298 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) inositol diphosphate tetrakisphosphate diphosphatase activity(GO:0052840) inositol bisdiphosphate tetrakisphosphate diphosphatase activity(GO:0052841) inositol diphosphate pentakisphosphate diphosphatase activity(GO:0052842) inositol-1-diphosphate-2,3,4,5,6-pentakisphosphate diphosphatase activity(GO:0052843) inositol-3-diphosphate-1,2,4,5,6-pentakisphosphate diphosphatase activity(GO:0052844) inositol-5-diphosphate-1,2,3,4,6-pentakisphosphate diphosphatase activity(GO:0052845) inositol-1,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 1-diphosphatase activity(GO:0052846) inositol-1,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 5-diphosphatase activity(GO:0052847) inositol-3,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 5-diphosphatase activity(GO:0052848) |
| 0.2 | 0.7 | GO:0042132 | fructose 1,6-bisphosphate 1-phosphatase activity(GO:0042132) |
| 0.2 | 1.4 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.1 | 0.7 | GO:0016454 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.1 | 1.7 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.1 | 2.0 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.1 | 0.4 | GO:0034739 | histone deacetylase activity (H4-K16 specific)(GO:0034739) |
| 0.1 | 0.5 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.1 | 3.4 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.1 | 0.4 | GO:0102007 | lactonohydrolase activity(GO:0046573) acyl-L-homoserine-lactone lactonohydrolase activity(GO:0102007) |
| 0.1 | 0.5 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.1 | 0.7 | GO:0003960 | NADPH:quinone reductase activity(GO:0003960) |
| 0.1 | 1.5 | GO:0004716 | receptor signaling protein tyrosine kinase activity(GO:0004716) |
| 0.1 | 1.1 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.1 | 0.5 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.1 | 0.4 | GO:0015137 | citrate transmembrane transporter activity(GO:0015137) tricarboxylic acid transmembrane transporter activity(GO:0015142) |
| 0.1 | 1.4 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.1 | 0.7 | GO:0070679 | inositol 1,4,5 trisphosphate binding(GO:0070679) |
| 0.1 | 0.9 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 0.4 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.2 | GO:1903135 | cupric ion binding(GO:1903135) |
| 0.0 | 0.1 | GO:0005477 | pyruvate secondary active transmembrane transporter activity(GO:0005477) |
| 0.0 | 1.1 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.7 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.0 | 2.7 | GO:0016303 | 1-phosphatidylinositol-3-kinase activity(GO:0016303) |
| 0.0 | 0.1 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 0.0 | 0.4 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.0 | 0.7 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 0.2 | GO:0097603 | temperature-gated ion channel activity(GO:0097603) |
| 0.0 | 0.3 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 0.0 | 0.3 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.0 | 0.2 | GO:0035251 | UDP-glucosyltransferase activity(GO:0035251) |
| 0.0 | 0.4 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.0 | 0.6 | GO:0017166 | vinculin binding(GO:0017166) nitric-oxide synthase binding(GO:0050998) |
| 0.0 | 0.5 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.0 | 0.2 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.0 | 0.1 | GO:0030345 | extracellular matrix structural constituent conferring compression resistance(GO:0030021) structural constituent of tooth enamel(GO:0030345) |
| 0.0 | 0.5 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 0.3 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.0 | 0.3 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.0 | 0.7 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.3 | GO:0016780 | phosphotransferase activity, for other substituted phosphate groups(GO:0016780) |
| 0.0 | 0.2 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.0 | 0.5 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.2 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.0 | 0.2 | GO:0001134 | transcription factor activity, transcription factor recruiting(GO:0001134) |
| 0.0 | 1.2 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.0 | 0.1 | GO:0052658 | inositol-1,4,5-trisphosphate 5-phosphatase activity(GO:0052658) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 2.6 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.0 | 0.7 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 2.1 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 1.5 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 0.5 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 1.4 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.2 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 0.6 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 0.7 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 0.5 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 0.3 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.0 | REACTOME DESTABILIZATION OF MRNA BY TRISTETRAPROLIN TTP | Genes involved in Destabilization of mRNA by Tristetraprolin (TTP) |
| 0.1 | 1.6 | REACTOME SIGNALING BY ACTIVATED POINT MUTANTS OF FGFR1 | Genes involved in Signaling by activated point mutants of FGFR1 |
| 0.1 | 1.5 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.1 | 2.1 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.1 | 1.1 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 0.8 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 1.4 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 1.1 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.0 | 0.7 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.8 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.0 | 0.3 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 0.5 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.0 | 0.5 | REACTOME PYRUVATE METABOLISM | Genes involved in Pyruvate metabolism |
| 0.0 | 1.1 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 0.2 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 1.4 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.1 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.5 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.0 | 0.3 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |