Project
Inflammatory response time course, HUVEC (Wada, 2009)
Navigation
Downloads
Results for NKX2-2
Z-value: 0.87
Transcription factors associated with NKX2-2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NKX2-2
|
ENSG00000125820.6 | NKX2-2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NKX2-2 | hg38_v1_chr20_-_21514046_21514070 | 0.03 | 8.9e-01 | Click! |
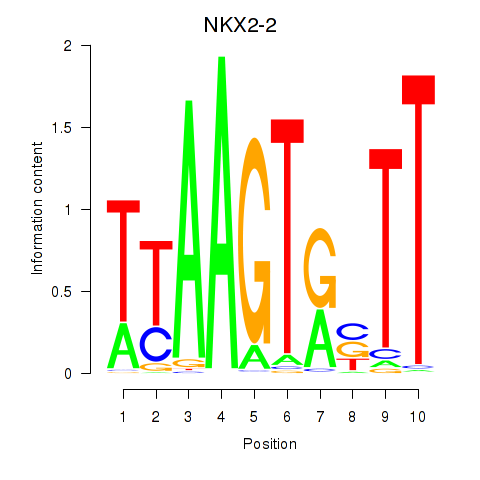
Activity profile of NKX2-2 motif
Sorted Z-values of NKX2-2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of NKX2-2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_-_69141878 | 2.41 |
ENST00000590645.1
ENST00000284425.7 |
ABCA6
|
ATP binding cassette subfamily A member 6 |
| chr1_+_84164370 | 2.15 |
ENST00000446538.5
ENST00000610703.4 ENST00000370682.7 ENST00000394838.6 ENST00000432111.5 |
PRKACB
|
protein kinase cAMP-activated catalytic subunit beta |
| chr4_-_98657635 | 1.76 |
ENST00000515287.5
ENST00000511651.5 ENST00000505184.5 |
TSPAN5
|
tetraspanin 5 |
| chr8_+_96584920 | 1.73 |
ENST00000521590.5
|
SDC2
|
syndecan 2 |
| chr2_+_172860038 | 1.65 |
ENST00000538974.5
ENST00000540783.5 |
RAPGEF4
|
Rap guanine nucleotide exchange factor 4 |
| chr10_+_35167516 | 1.40 |
ENST00000361599.8
|
CREM
|
cAMP responsive element modulator |
| chrX_+_136205982 | 1.36 |
ENST00000628568.1
|
FHL1
|
four and a half LIM domains 1 |
| chr6_+_143677935 | 1.27 |
ENST00000440869.6
ENST00000367582.7 ENST00000451827.6 |
PHACTR2
|
phosphatase and actin regulator 2 |
| chr13_+_102656933 | 1.04 |
ENST00000650757.1
|
TPP2
|
tripeptidyl peptidase 2 |
| chr4_+_87006988 | 0.97 |
ENST00000307808.10
|
AFF1
|
AF4/FMR2 family member 1 |
| chr18_+_44700796 | 0.95 |
ENST00000677130.1
|
SETBP1
|
SET binding protein 1 |
| chr7_-_22194709 | 0.95 |
ENST00000458533.5
|
RAPGEF5
|
Rap guanine nucleotide exchange factor 5 |
| chr5_+_141421064 | 0.91 |
ENST00000518882.2
|
PCDHGA11
|
protocadherin gamma subfamily A, 11 |
| chr14_+_22495890 | 0.89 |
ENST00000390494.1
|
TRAJ43
|
T cell receptor alpha joining 43 |
| chr3_+_41200080 | 0.87 |
ENST00000644524.1
|
CTNNB1
|
catenin beta 1 |
| chr5_-_11588842 | 0.87 |
ENST00000503622.5
|
CTNND2
|
catenin delta 2 |
| chr9_+_2159672 | 0.86 |
ENST00000634343.1
|
SMARCA2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr14_-_100375333 | 0.85 |
ENST00000557297.5
ENST00000555813.5 ENST00000392882.7 ENST00000557135.5 ENST00000556698.5 ENST00000554509.5 ENST00000555410.5 |
WARS1
|
tryptophanyl-tRNA synthetase 1 |
| chr7_+_116222804 | 0.84 |
ENST00000393481.6
|
TES
|
testin LIM domain protein |
| chr2_+_201129826 | 0.83 |
ENST00000457277.5
|
CFLAR
|
CASP8 and FADD like apoptosis regulator |
| chr1_+_84164684 | 0.81 |
ENST00000370680.5
|
PRKACB
|
protein kinase cAMP-activated catalytic subunit beta |
| chr3_+_41200104 | 0.78 |
ENST00000643297.1
ENST00000450969.6 |
CTNNB1
|
catenin beta 1 |
| chr18_-_55321640 | 0.76 |
ENST00000637169.2
|
TCF4
|
transcription factor 4 |
| chr4_+_143381939 | 0.73 |
ENST00000505913.5
|
GAB1
|
GRB2 associated binding protein 1 |
| chr6_+_28281555 | 0.71 |
ENST00000259883.3
ENST00000682144.1 |
PGBD1
|
piggyBac transposable element derived 1 |
| chr1_-_243843226 | 0.69 |
ENST00000336199.9
|
AKT3
|
AKT serine/threonine kinase 3 |
| chrX_-_71255060 | 0.69 |
ENST00000373988.5
ENST00000373998.5 |
ZMYM3
|
zinc finger MYM-type containing 3 |
| chr13_-_44474296 | 0.68 |
ENST00000611198.4
|
TSC22D1
|
TSC22 domain family member 1 |
| chr14_-_100375602 | 0.68 |
ENST00000553395.5
ENST00000553545.5 ENST00000344102.9 ENST00000556338.5 ENST00000553934.1 |
WARS1
|
tryptophanyl-tRNA synthetase 1 |
| chr15_-_52652031 | 0.67 |
ENST00000546305.6
|
FAM214A
|
family with sequence similarity 214 member A |
| chr7_-_120858066 | 0.67 |
ENST00000222747.8
|
TSPAN12
|
tetraspanin 12 |
| chr12_-_102197827 | 0.67 |
ENST00000329406.5
|
PMCH
|
pro-melanin concentrating hormone |
| chr13_-_19782923 | 0.66 |
ENST00000338910.9
|
PSPC1
|
paraspeckle component 1 |
| chr5_-_78985288 | 0.64 |
ENST00000264914.10
|
ARSB
|
arylsulfatase B |
| chr13_-_19782970 | 0.63 |
ENST00000427943.1
ENST00000619300.4 |
PSPC1
|
paraspeckle component 1 |
| chr17_-_3592877 | 0.61 |
ENST00000399756.8
|
TRPV1
|
transient receptor potential cation channel subfamily V member 1 |
| chr4_+_94455245 | 0.60 |
ENST00000508216.5
ENST00000514743.5 |
PDLIM5
|
PDZ and LIM domain 5 |
| chr7_-_120857124 | 0.59 |
ENST00000441017.5
ENST00000424710.5 ENST00000433758.5 |
TSPAN12
|
tetraspanin 12 |
| chrX_+_11759678 | 0.58 |
ENST00000649797.1
ENST00000648614.1 |
MSL3
|
MSL complex subunit 3 |
| chr12_-_9869345 | 0.58 |
ENST00000228438.3
|
CLEC2B
|
C-type lectin domain family 2 member B |
| chr2_-_182522703 | 0.58 |
ENST00000410103.1
|
PDE1A
|
phosphodiesterase 1A |
| chr5_-_11589019 | 0.58 |
ENST00000511377.5
|
CTNND2
|
catenin delta 2 |
| chr7_-_22193824 | 0.57 |
ENST00000401957.6
|
RAPGEF5
|
Rap guanine nucleotide exchange factor 5 |
| chr18_-_55321986 | 0.54 |
ENST00000570287.6
|
TCF4
|
transcription factor 4 |
| chrX_+_105822531 | 0.53 |
ENST00000243300.14
ENST00000536164.5 |
NRK
|
Nik related kinase |
| chr11_-_125111708 | 0.53 |
ENST00000531909.5
ENST00000529530.1 |
TMEM218
|
transmembrane protein 218 |
| chr17_+_68525795 | 0.51 |
ENST00000592800.5
|
PRKAR1A
|
protein kinase cAMP-dependent type I regulatory subunit alpha |
| chr3_+_159273235 | 0.50 |
ENST00000638749.1
|
IQCJ-SCHIP1
|
IQCJ-SCHIP1 readthrough |
| chr2_+_161231078 | 0.50 |
ENST00000439442.1
|
TANK
|
TRAF family member associated NFKB activator |
| chr14_+_23185316 | 0.49 |
ENST00000399910.5
ENST00000492621.5 |
RNF212B
|
ring finger protein 212B |
| chrX_-_154805386 | 0.49 |
ENST00000393531.5
ENST00000369534.8 ENST00000453245.5 ENST00000428488.1 ENST00000369531.1 |
MPP1
|
membrane palmitoylated protein 1 |
| chr13_+_57631735 | 0.49 |
ENST00000377918.8
|
PCDH17
|
protocadherin 17 |
| chr12_-_8650529 | 0.48 |
ENST00000543467.5
|
MFAP5
|
microfibril associated protein 5 |
| chrX_+_71254781 | 0.48 |
ENST00000677446.1
|
NONO
|
non-POU domain containing octamer binding |
| chr8_-_100950549 | 0.48 |
ENST00000395951.7
|
YWHAZ
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein zeta |
| chr15_+_65550819 | 0.47 |
ENST00000569894.5
|
HACD3
|
3-hydroxyacyl-CoA dehydratase 3 |
| chr7_+_80135694 | 0.46 |
ENST00000457358.7
|
GNAI1
|
G protein subunit alpha i1 |
| chr13_-_98978022 | 0.46 |
ENST00000682017.1
ENST00000442173.5 ENST00000627024.2 |
DOCK9
|
dedicator of cytokinesis 9 |
| chr16_+_22490337 | 0.45 |
ENST00000415833.6
|
NPIPB5
|
nuclear pore complex interacting protein family member B5 |
| chr22_-_35824373 | 0.45 |
ENST00000473487.6
|
RBFOX2
|
RNA binding fox-1 homolog 2 |
| chr1_-_246507237 | 0.45 |
ENST00000490107.6
|
SMYD3
|
SET and MYND domain containing 3 |
| chr8_-_140800535 | 0.44 |
ENST00000521986.5
ENST00000523539.5 |
PTK2
|
protein tyrosine kinase 2 |
| chr9_+_96928310 | 0.44 |
ENST00000354649.7
|
NUTM2G
|
NUT family member 2G |
| chr9_+_113501359 | 0.44 |
ENST00000343817.9
ENST00000394646.7 |
RGS3
|
regulator of G protein signaling 3 |
| chr5_+_151025343 | 0.44 |
ENST00000521632.1
|
GPX3
|
glutathione peroxidase 3 |
| chr9_+_121207774 | 0.44 |
ENST00000373823.7
|
GSN
|
gelsolin |
| chr8_+_11808417 | 0.43 |
ENST00000525954.5
|
FDFT1
|
farnesyl-diphosphate farnesyltransferase 1 |
| chr1_-_243843164 | 0.43 |
ENST00000491219.6
ENST00000680056.1 ENST00000492957.2 |
AKT3
|
AKT serine/threonine kinase 3 |
| chr2_-_219571241 | 0.42 |
ENST00000373876.5
ENST00000603926.5 ENST00000373873.8 ENST00000289656.3 |
OBSL1
|
obscurin like cytoskeletal adaptor 1 |
| chr5_+_141223332 | 0.42 |
ENST00000239449.7
ENST00000624896.1 ENST00000624396.1 |
PCDHB14
ENSG00000279983.1
|
protocadherin beta 14 novel protein |
| chr8_+_11802667 | 0.42 |
ENST00000443614.6
ENST00000220584.9 ENST00000525900.5 |
FDFT1
|
farnesyl-diphosphate farnesyltransferase 1 |
| chr3_-_15798184 | 0.42 |
ENST00000624145.3
|
ANKRD28
|
ankyrin repeat domain 28 |
| chr6_+_32969345 | 0.42 |
ENST00000678250.1
|
BRD2
|
bromodomain containing 2 |
| chr8_+_11802611 | 0.41 |
ENST00000623368.3
|
FDFT1
|
farnesyl-diphosphate farnesyltransferase 1 |
| chr8_+_11803012 | 0.41 |
ENST00000528812.5
ENST00000622850.2 |
FDFT1
|
farnesyl-diphosphate farnesyltransferase 1 |
| chr1_-_154502402 | 0.41 |
ENST00000304760.3
|
SHE
|
Src homology 2 domain containing E |
| chr19_-_2427538 | 0.41 |
ENST00000591871.1
ENST00000215570.8 |
TIMM13
|
translocase of inner mitochondrial membrane 13 |
| chr10_-_70376779 | 0.40 |
ENST00000395011.5
|
LRRC20
|
leucine rich repeat containing 20 |
| chr9_-_94593810 | 0.40 |
ENST00000375337.4
|
FBP2
|
fructose-bisphosphatase 2 |
| chr6_+_32153441 | 0.39 |
ENST00000414204.5
ENST00000361568.6 ENST00000395523.5 |
PPT2
|
palmitoyl-protein thioesterase 2 |
| chr8_+_11809135 | 0.39 |
ENST00000528643.5
ENST00000525777.5 |
FDFT1
|
farnesyl-diphosphate farnesyltransferase 1 |
| chr12_+_80099535 | 0.38 |
ENST00000646859.1
ENST00000547103.7 |
OTOGL
|
otogelin like |
| chr6_+_32969165 | 0.38 |
ENST00000496118.2
ENST00000449085.4 |
BRD2
|
bromodomain containing 2 |
| chr3_+_98763331 | 0.38 |
ENST00000485391.5
ENST00000492254.5 |
ST3GAL6
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 6 |
| chr13_-_77919390 | 0.38 |
ENST00000475537.2
ENST00000646605.1 |
EDNRB
|
endothelin receptor type B |
| chr2_+_188292771 | 0.38 |
ENST00000359135.7
|
GULP1
|
GULP PTB domain containing engulfment adaptor 1 |
| chrX_+_123961696 | 0.36 |
ENST00000371145.8
ENST00000371157.7 ENST00000371144.7 |
STAG2
|
stromal antigen 2 |
| chrX_+_47232866 | 0.36 |
ENST00000218348.7
ENST00000377107.7 |
USP11
|
ubiquitin specific peptidase 11 |
| chr6_-_109690500 | 0.36 |
ENST00000448084.6
|
AK9
|
adenylate kinase 9 |
| chr6_+_32968557 | 0.36 |
ENST00000374825.9
|
BRD2
|
bromodomain containing 2 |
| chr7_+_150368189 | 0.36 |
ENST00000519397.1
ENST00000479668.5 |
REPIN1
|
replication initiator 1 |
| chr11_+_34632464 | 0.34 |
ENST00000531794.5
|
EHF
|
ETS homologous factor |
| chr14_-_81533800 | 0.34 |
ENST00000555824.5
ENST00000557372.1 ENST00000336735.9 |
SEL1L
|
SEL1L adaptor subunit of ERAD E3 ubiquitin ligase |
| chr6_-_109690515 | 0.33 |
ENST00000532976.1
|
AK9
|
adenylate kinase 9 |
| chr10_+_5094405 | 0.33 |
ENST00000380554.5
|
AKR1C3
|
aldo-keto reductase family 1 member C3 |
| chrX_+_101488044 | 0.33 |
ENST00000423738.4
|
ARMCX4
|
armadillo repeat containing X-linked 4 |
| chr4_-_155866277 | 0.33 |
ENST00000537611.3
|
ASIC5
|
acid sensing ion channel subunit family member 5 |
| chr20_-_61998132 | 0.33 |
ENST00000474089.5
|
TAF4
|
TATA-box binding protein associated factor 4 |
| chr11_-_119196769 | 0.33 |
ENST00000415318.2
|
CCDC153
|
coiled-coil domain containing 153 |
| chrX_+_108439866 | 0.32 |
ENST00000361603.7
|
COL4A5
|
collagen type IV alpha 5 chain |
| chr1_-_44788168 | 0.32 |
ENST00000372207.4
|
BEST4
|
bestrophin 4 |
| chr5_+_160229499 | 0.32 |
ENST00000402432.4
|
FABP6
|
fatty acid binding protein 6 |
| chr1_+_174700413 | 0.32 |
ENST00000529145.6
ENST00000325589.9 |
RABGAP1L
|
RAB GTPase activating protein 1 like |
| chr19_-_39391029 | 0.32 |
ENST00000221265.8
|
PAF1
|
PAF1 homolog, Paf1/RNA polymerase II complex component |
| chr20_+_58651785 | 0.31 |
ENST00000358029.8
|
STX16
|
syntaxin 16 |
| chr19_-_17255448 | 0.31 |
ENST00000594059.1
|
ENSG00000269095.1
|
novel protein |
| chr14_-_89412025 | 0.31 |
ENST00000553840.5
ENST00000556916.5 |
FOXN3
|
forkhead box N3 |
| chr1_-_216805367 | 0.30 |
ENST00000360012.7
|
ESRRG
|
estrogen related receptor gamma |
| chr1_+_241532121 | 0.30 |
ENST00000366558.7
|
KMO
|
kynurenine 3-monooxygenase |
| chr12_-_57846686 | 0.30 |
ENST00000548823.1
ENST00000398073.7 |
CTDSP2
|
CTD small phosphatase 2 |
| chr8_-_140764386 | 0.30 |
ENST00000520151.5
ENST00000519024.5 ENST00000519465.5 |
PTK2
|
protein tyrosine kinase 2 |
| chr19_+_57682019 | 0.29 |
ENST00000596085.1
ENST00000594684.1 |
ZNF551
ENSG00000269026.2
|
zinc finger protein 551 novel protein |
| chr2_-_73233206 | 0.29 |
ENST00000258083.3
|
PRADC1
|
protease associated domain containing 1 |
| chr20_+_11890785 | 0.29 |
ENST00000399006.6
ENST00000405977.5 |
BTBD3
|
BTB domain containing 3 |
| chr3_+_184053047 | 0.29 |
ENST00000318351.2
|
HTR3C
|
5-hydroxytryptamine receptor 3C |
| chr1_-_74733253 | 0.29 |
ENST00000417775.5
|
CRYZ
|
crystallin zeta |
| chr22_-_50085331 | 0.29 |
ENST00000395876.6
|
MLC1
|
modulator of VRAC current 1 |
| chr2_-_27335403 | 0.28 |
ENST00000454704.5
|
GTF3C2
|
general transcription factor IIIC subunit 2 |
| chr13_+_101489940 | 0.28 |
ENST00000376162.7
|
ITGBL1
|
integrin subunit beta like 1 |
| chr15_-_42548763 | 0.28 |
ENST00000563454.5
ENST00000397130.8 ENST00000570160.1 ENST00000323443.6 |
LRRC57
|
leucine rich repeat containing 57 |
| chr9_+_128685062 | 0.28 |
ENST00000409104.7
|
SET
|
SET nuclear proto-oncogene |
| chr2_-_32039776 | 0.28 |
ENST00000342166.10
ENST00000295066.3 |
DPY30
|
dpy-30 histone methyltransferase complex regulatory subunit |
| chr1_-_53940100 | 0.27 |
ENST00000371376.1
|
HSPB11
|
heat shock protein family B (small) member 11 |
| chr7_+_32979445 | 0.27 |
ENST00000418354.5
ENST00000490776.3 |
FKBP9
|
FKBP prolyl isomerase 9 |
| chr5_+_141412979 | 0.27 |
ENST00000612503.1
ENST00000398610.3 |
PCDHGA10
|
protocadherin gamma subfamily A, 10 |
| chrX_-_71254106 | 0.27 |
ENST00000373984.7
ENST00000314425.9 ENST00000373982.5 |
ZMYM3
|
zinc finger MYM-type containing 3 |
| chr10_+_13161543 | 0.27 |
ENST00000378714.8
ENST00000479669.5 ENST00000484800.6 |
MCM10
|
minichromosome maintenance 10 replication initiation factor |
| chr4_+_122339221 | 0.27 |
ENST00000442707.1
|
KIAA1109
|
KIAA1109 |
| chr2_+_201129483 | 0.26 |
ENST00000440180.5
|
CFLAR
|
CASP8 and FADD like apoptosis regulator |
| chr5_+_141172637 | 0.26 |
ENST00000231137.6
|
PCDHB7
|
protocadherin beta 7 |
| chr9_+_89311187 | 0.26 |
ENST00000314355.7
|
CKS2
|
CDC28 protein kinase regulatory subunit 2 |
| chrX_+_34942790 | 0.26 |
ENST00000329357.6
|
FAM47B
|
family with sequence similarity 47 member B |
| chr8_-_123737378 | 0.25 |
ENST00000419625.6
ENST00000262219.10 |
ANXA13
|
annexin A13 |
| chr9_-_20622479 | 0.25 |
ENST00000380338.9
|
MLLT3
|
MLLT3 super elongation complex subunit |
| chr20_+_58651228 | 0.25 |
ENST00000361830.7
ENST00000458280.5 ENST00000355957.9 ENST00000312283.12 ENST00000412911.5 ENST00000359617.8 ENST00000371141.8 |
STX16
|
syntaxin 16 |
| chr6_+_32154010 | 0.25 |
ENST00000375137.6
|
PPT2
|
palmitoyl-protein thioesterase 2 |
| chr12_+_10505602 | 0.25 |
ENST00000322446.3
|
EIF2S3B
|
eukaryotic translation initiation factor 2 subunit gamma B |
| chr2_+_188292814 | 0.25 |
ENST00000409580.5
ENST00000409637.7 |
GULP1
|
GULP PTB domain containing engulfment adaptor 1 |
| chr5_+_141421020 | 0.25 |
ENST00000622044.1
ENST00000398587.7 |
PCDHGA11
|
protocadherin gamma subfamily A, 11 |
| chr9_-_92404559 | 0.24 |
ENST00000262551.8
ENST00000375561.10 |
OGN
|
osteoglycin |
| chr6_+_158536398 | 0.24 |
ENST00000367090.4
|
TMEM181
|
transmembrane protein 181 |
| chr15_+_67067780 | 0.24 |
ENST00000679624.1
|
SMAD3
|
SMAD family member 3 |
| chrX_+_11759604 | 0.23 |
ENST00000649602.1
ENST00000650215.1 ENST00000649685.1 ENST00000649649.1 ENST00000380693.8 ENST00000380692.7 ENST00000650628.1 ENST00000649851.1 ENST00000421368.3 |
MSL3
|
MSL complex subunit 3 |
| chr8_-_132760548 | 0.23 |
ENST00000519187.5
ENST00000523829.5 ENST00000677595.1 ENST00000356838.7 ENST00000377901.8 ENST00000519304.1 |
TMEM71
|
transmembrane protein 71 |
| chrM_+_12329 | 0.23 |
ENST00000361567.2
|
MT-ND5
|
mitochondrially encoded NADH:ubiquinone oxidoreductase core subunit 5 |
| chr9_+_37667997 | 0.23 |
ENST00000539465.5
|
FRMPD1
|
FERM and PDZ domain containing 1 |
| chr11_-_11353241 | 0.23 |
ENST00000528848.3
|
CSNK2A3
|
casein kinase 2 alpha 3 |
| chr7_+_92057602 | 0.23 |
ENST00000491695.2
|
AKAP9
|
A-kinase anchoring protein 9 |
| chr8_-_120445092 | 0.23 |
ENST00000518918.1
|
MRPL13
|
mitochondrial ribosomal protein L13 |
| chr1_-_226739271 | 0.22 |
ENST00000429204.6
ENST00000366784.1 |
ITPKB
|
inositol-trisphosphate 3-kinase B |
| chr15_+_58410543 | 0.22 |
ENST00000356113.10
ENST00000414170.7 |
LIPC
|
lipase C, hepatic type |
| chr10_+_79841358 | 0.22 |
ENST00000602967.5
|
NUTM2E
|
NUT family member 2E |
| chr12_+_56152579 | 0.22 |
ENST00000551834.5
ENST00000552568.5 |
MYL6B
|
myosin light chain 6B |
| chr5_-_78985951 | 0.22 |
ENST00000396151.7
|
ARSB
|
arylsulfatase B |
| chr5_+_126423363 | 0.22 |
ENST00000285689.8
|
GRAMD2B
|
GRAM domain containing 2B |
| chr20_+_58888779 | 0.21 |
ENST00000488546.6
ENST00000667293.2 ENST00000481039.6 ENST00000467321.6 |
GNAS
|
GNAS complex locus |
| chr6_-_73225465 | 0.21 |
ENST00000370388.4
|
KHDC1L
|
KH domain containing 1 like |
| chrX_+_11760087 | 0.21 |
ENST00000649271.1
|
MSL3
|
MSL complex subunit 3 |
| chr1_+_119507203 | 0.21 |
ENST00000369413.8
ENST00000528909.1 |
HSD3B1
|
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 1 |
| chr5_+_126423122 | 0.21 |
ENST00000515200.5
|
GRAMD2B
|
GRAM domain containing 2B |
| chr19_-_3971051 | 0.21 |
ENST00000545797.7
ENST00000596311.5 |
DAPK3
|
death associated protein kinase 3 |
| chr5_+_126423176 | 0.20 |
ENST00000542322.5
ENST00000544396.5 |
GRAMD2B
|
GRAM domain containing 2B |
| chr13_-_77919459 | 0.20 |
ENST00000643890.1
|
EDNRB
|
endothelin receptor type B |
| chr3_+_152299392 | 0.20 |
ENST00000498502.5
ENST00000545754.5 ENST00000357472.7 ENST00000324196.9 |
MBNL1
|
muscleblind like splicing regulator 1 |
| chr2_-_219571529 | 0.20 |
ENST00000404537.6
|
OBSL1
|
obscurin like cytoskeletal adaptor 1 |
| chrX_+_11760035 | 0.19 |
ENST00000482871.6
ENST00000648013.1 |
MSL3
|
MSL complex subunit 3 |
| chrX_-_19799751 | 0.18 |
ENST00000379698.8
|
SH3KBP1
|
SH3 domain containing kinase binding protein 1 |
| chr2_-_144517663 | 0.18 |
ENST00000427902.5
ENST00000462355.2 ENST00000470879.5 ENST00000409487.7 ENST00000435831.5 ENST00000630572.2 |
ZEB2
|
zinc finger E-box binding homeobox 2 |
| chr11_-_129024157 | 0.18 |
ENST00000392657.7
|
ARHGAP32
|
Rho GTPase activating protein 32 |
| chr9_-_15510289 | 0.18 |
ENST00000397519.6
|
PSIP1
|
PC4 and SFRS1 interacting protein 1 |
| chr13_+_101452569 | 0.18 |
ENST00000618057.4
|
ITGBL1
|
integrin subunit beta like 1 |
| chr2_+_10368645 | 0.18 |
ENST00000613496.4
|
HPCAL1
|
hippocalcin like 1 |
| chr12_+_56152439 | 0.18 |
ENST00000550443.5
|
MYL6B
|
myosin light chain 6B |
| chr14_+_56117702 | 0.18 |
ENST00000559044.5
|
PELI2
|
pellino E3 ubiquitin protein ligase family member 2 |
| chr6_-_27472681 | 0.18 |
ENST00000377419.1
|
ZNF184
|
zinc finger protein 184 |
| chr1_-_109613070 | 0.17 |
ENST00000351050.8
|
GNAT2
|
G protein subunit alpha transducin 2 |
| chr7_-_130441136 | 0.17 |
ENST00000675596.1
ENST00000676312.1 |
CEP41
|
centrosomal protein 41 |
| chr11_-_124673708 | 0.17 |
ENST00000263593.8
|
SIAE
|
sialic acid acetylesterase |
| chr5_+_136059151 | 0.17 |
ENST00000503087.1
|
TGFBI
|
transforming growth factor beta induced |
| chr12_-_71157872 | 0.17 |
ENST00000546561.2
|
TSPAN8
|
tetraspanin 8 |
| chrM_+_4467 | 0.17 |
ENST00000361453.3
|
MT-ND2
|
mitochondrially encoded NADH:ubiquinone oxidoreductase core subunit 2 |
| chr5_+_119476530 | 0.16 |
ENST00000645099.1
ENST00000513628.5 |
HSD17B4
|
hydroxysteroid 17-beta dehydrogenase 4 |
| chr10_+_72893734 | 0.16 |
ENST00000334011.10
|
OIT3
|
oncoprotein induced transcript 3 |
| chr12_-_10826358 | 0.16 |
ENST00000240619.2
|
TAS2R10
|
taste 2 receptor member 10 |
| chr17_+_50746534 | 0.16 |
ENST00000511974.5
|
LUC7L3
|
LUC7 like 3 pre-mRNA splicing factor |
| chr2_+_10368764 | 0.16 |
ENST00000620771.4
|
HPCAL1
|
hippocalcin like 1 |
| chr9_-_39239174 | 0.16 |
ENST00000358144.6
|
CNTNAP3
|
contactin associated protein family member 3 |
| chr2_+_151410090 | 0.16 |
ENST00000430328.6
|
RIF1
|
replication timing regulatory factor 1 |
| chr1_-_202967229 | 0.16 |
ENST00000367249.9
|
CYB5R1
|
cytochrome b5 reductase 1 |
| chr16_+_71808439 | 0.15 |
ENST00000683775.1
|
ATXN1L
|
ataxin 1 like |
| chr13_-_106568107 | 0.15 |
ENST00000400198.8
|
ARGLU1
|
arginine and glutamate rich 1 |
| chr1_+_180632001 | 0.15 |
ENST00000367590.9
ENST00000367589.3 |
XPR1
|
xenotropic and polytropic retrovirus receptor 1 |
| chr1_+_241532370 | 0.15 |
ENST00000366559.9
ENST00000366557.8 |
KMO
|
kynurenine 3-monooxygenase |
| chr11_+_22668101 | 0.15 |
ENST00000630668.2
ENST00000278187.7 |
GAS2
|
growth arrest specific 2 |
| chr1_+_53014926 | 0.15 |
ENST00000430330.6
ENST00000408941.7 ENST00000478274.6 ENST00000484100.5 ENST00000435345.6 ENST00000488965.1 |
SCP2
|
sterol carrier protein 2 |
| chr13_-_75366973 | 0.15 |
ENST00000648194.1
|
TBC1D4
|
TBC1 domain family member 4 |
| chr8_-_120445140 | 0.15 |
ENST00000306185.8
|
MRPL13
|
mitochondrial ribosomal protein L13 |
| chr4_-_87529092 | 0.14 |
ENST00000503414.5
|
SPARCL1
|
SPARC like 1 |
| chrX_-_135764444 | 0.14 |
ENST00000597510.6
|
CT45A3
|
cancer/testis antigen family 45 member A3 |
| chr2_+_69013379 | 0.14 |
ENST00000409349.7
|
ANTXR1
|
ANTXR cell adhesion molecule 1 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 3.0 | GO:0097338 | response to clozapine(GO:0097338) |
| 0.3 | 2.1 | GO:0008204 | ergosterol biosynthetic process(GO:0006696) ergosterol metabolic process(GO:0008204) |
| 0.3 | 1.7 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.3 | 1.6 | GO:1904499 | glial cell fate determination(GO:0007403) canonical Wnt signaling pathway involved in positive regulation of cardiac outflow tract cell proliferation(GO:0061324) regulation of chromatin-mediated maintenance of transcription(GO:1904499) positive regulation of chromatin-mediated maintenance of transcription(GO:1904501) regulation of euchromatin binding(GO:1904793) |
| 0.3 | 0.8 | GO:0061580 | colon epithelial cell migration(GO:0061580) |
| 0.2 | 0.7 | GO:0061569 | dADP phosphorylation(GO:0006174) dGDP phosphorylation(GO:0006186) AMP phosphorylation(GO:0006756) CDP phosphorylation(GO:0061508) dAMP phosphorylation(GO:0061565) CMP phosphorylation(GO:0061566) dCMP phosphorylation(GO:0061567) GDP phosphorylation(GO:0061568) UDP phosphorylation(GO:0061569) dCDP phosphorylation(GO:0061570) TDP phosphorylation(GO:0061571) |
| 0.2 | 0.5 | GO:0060720 | spongiotrophoblast cell proliferation(GO:0060720) regulation of spongiotrophoblast cell proliferation(GO:0060721) cell proliferation involved in embryonic placenta development(GO:0060722) regulation of cell proliferation involved in embryonic placenta development(GO:0060723) |
| 0.2 | 0.6 | GO:0050960 | detection of temperature stimulus involved in thermoception(GO:0050960) response to capsazepine(GO:1901594) |
| 0.1 | 0.6 | GO:0035645 | enteric smooth muscle cell differentiation(GO:0035645) |
| 0.1 | 0.3 | GO:0016107 | sesquiterpenoid metabolic process(GO:0006714) sesquiterpenoid catabolic process(GO:0016107) farnesol metabolic process(GO:0016487) farnesol catabolic process(GO:0016488) regulation of testosterone biosynthetic process(GO:2000224) |
| 0.1 | 0.4 | GO:0005986 | sucrose biosynthetic process(GO:0005986) |
| 0.1 | 1.1 | GO:1903943 | regulation of hepatocyte apoptotic process(GO:1903943) negative regulation of hepatocyte apoptotic process(GO:1903944) |
| 0.1 | 0.5 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.1 | 0.4 | GO:0009405 | pathogenesis(GO:0009405) |
| 0.1 | 0.4 | GO:1903906 | plasma membrane raft distribution(GO:0044855) plasma membrane raft localization(GO:0044856) plasma membrane raft polarization(GO:0044858) regulation of plasma membrane raft polarization(GO:1903906) |
| 0.1 | 0.5 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 0.1 | 0.4 | GO:0006982 | response to lipid hydroperoxide(GO:0006982) |
| 0.1 | 1.1 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.1 | 0.5 | GO:0046726 | positive regulation by virus of viral protein levels in host cell(GO:0046726) |
| 0.1 | 0.2 | GO:0036111 | very long-chain fatty-acyl-CoA metabolic process(GO:0036111) |
| 0.1 | 0.6 | GO:0098734 | macromolecule depalmitoylation(GO:0098734) |
| 0.0 | 0.5 | GO:1903377 | negative regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903377) |
| 0.0 | 0.4 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.0 | 0.3 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.0 | 0.4 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.0 | 0.7 | GO:0051964 | negative regulation of synapse assembly(GO:0051964) |
| 0.0 | 0.2 | GO:2001034 | positive regulation of double-strand break repair via nonhomologous end joining(GO:2001034) |
| 0.0 | 0.2 | GO:1901202 | negative regulation of extracellular matrix assembly(GO:1901202) |
| 0.0 | 0.1 | GO:2000523 | regulation of T cell costimulation(GO:2000523) positive regulation of T cell costimulation(GO:2000525) regulation of dendritic cell dendrite assembly(GO:2000547) |
| 0.0 | 1.3 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 0.2 | GO:0061767 | negative regulation of lung blood pressure(GO:0061767) |
| 0.0 | 0.8 | GO:0031033 | myosin filament organization(GO:0031033) |
| 0.0 | 0.5 | GO:0090168 | Golgi reassembly(GO:0090168) |
| 0.0 | 0.3 | GO:0042791 | 5S class rRNA transcription from RNA polymerase III type 1 promoter(GO:0042791) tRNA transcription from RNA polymerase III promoter(GO:0042797) |
| 0.0 | 0.1 | GO:0042640 | anagen(GO:0042640) |
| 0.0 | 0.1 | GO:0003245 | cardiac muscle tissue growth involved in heart morphogenesis(GO:0003245) |
| 0.0 | 0.5 | GO:1904321 | response to forskolin(GO:1904321) cellular response to forskolin(GO:1904322) |
| 0.0 | 0.1 | GO:1990637 | response to prolactin(GO:1990637) |
| 0.0 | 0.6 | GO:2000807 | regulation of synaptic vesicle clustering(GO:2000807) |
| 0.0 | 0.2 | GO:0033029 | regulation of neutrophil apoptotic process(GO:0033029) |
| 0.0 | 0.6 | GO:0090161 | Golgi ribbon formation(GO:0090161) |
| 0.0 | 0.3 | GO:0006348 | chromatin silencing at telomere(GO:0006348) |
| 0.0 | 0.4 | GO:0032876 | negative regulation of DNA endoreduplication(GO:0032876) |
| 0.0 | 1.3 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.0 | 0.1 | GO:2000969 | positive regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000969) |
| 0.0 | 0.5 | GO:2000480 | negative regulation of meiotic nuclear division(GO:0045835) negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.0 | 0.2 | GO:0048549 | positive regulation of pinocytosis(GO:0048549) |
| 0.0 | 0.6 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.0 | 0.1 | GO:0021699 | cerebellum maturation(GO:0021590) cerebellar cortex maturation(GO:0021699) |
| 0.0 | 1.7 | GO:0048384 | retinoic acid receptor signaling pathway(GO:0048384) |
| 0.0 | 0.1 | GO:0060448 | dichotomous subdivision of terminal units involved in lung branching(GO:0060448) |
| 0.0 | 0.1 | GO:0036215 | response to stem cell factor(GO:0036215) cellular response to stem cell factor stimulus(GO:0036216) Kit signaling pathway(GO:0038109) |
| 0.0 | 0.2 | GO:1902748 | positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.0 | 0.7 | GO:0035728 | response to hepatocyte growth factor(GO:0035728) |
| 0.0 | 1.1 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.0 | 0.2 | GO:0032383 | regulation of intracellular lipid transport(GO:0032377) regulation of intracellular sterol transport(GO:0032380) regulation of intracellular cholesterol transport(GO:0032383) |
| 0.0 | 0.1 | GO:0000964 | mitochondrial RNA 5'-end processing(GO:0000964) |
| 0.0 | 0.2 | GO:0051661 | maintenance of centrosome location(GO:0051661) |
| 0.0 | 0.1 | GO:1904253 | positive regulation of bile acid biosynthetic process(GO:0070859) positive regulation of bile acid metabolic process(GO:1904253) |
| 0.0 | 0.2 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.0 | 2.1 | GO:0060997 | dendritic spine morphogenesis(GO:0060997) |
| 0.0 | 0.4 | GO:0019321 | pentose metabolic process(GO:0019321) |
| 0.0 | 0.3 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) |
| 0.0 | 0.3 | GO:0070986 | left/right axis specification(GO:0070986) |
| 0.0 | 0.5 | GO:1904380 | endoplasmic reticulum mannose trimming(GO:1904380) |
| 0.0 | 0.3 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.0 | 0.3 | GO:0042178 | xenobiotic catabolic process(GO:0042178) |
| 0.0 | 0.2 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.0 | 0.1 | GO:0046671 | negative regulation of cellular pH reduction(GO:0032848) CD8-positive, alpha-beta T cell lineage commitment(GO:0043375) negative regulation of retinal cell programmed cell death(GO:0046671) |
| 0.0 | 0.1 | GO:2000342 | negative regulation of chemokine (C-X-C motif) ligand 2 production(GO:2000342) |
| 0.0 | 0.1 | GO:0090290 | positive regulation of osteoclast proliferation(GO:0090290) |
| 0.0 | 1.5 | GO:0043268 | positive regulation of potassium ion transport(GO:0043268) |
| 0.0 | 0.6 | GO:0018146 | keratan sulfate biosynthetic process(GO:0018146) |
| 0.0 | 0.3 | GO:0071397 | cellular response to cholesterol(GO:0071397) |
| 0.0 | 0.2 | GO:0046549 | response to light intensity(GO:0009642) retinal cone cell differentiation(GO:0042670) retinal cone cell development(GO:0046549) |
| 0.0 | 1.8 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.0 | 0.2 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.0 | 0.0 | GO:0071676 | negative regulation of mononuclear cell migration(GO:0071676) |
| 0.0 | 0.5 | GO:0060216 | definitive hemopoiesis(GO:0060216) |
| 0.0 | 0.2 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.0 | 0.2 | GO:0034372 | very-low-density lipoprotein particle remodeling(GO:0034372) phosphatidylcholine catabolic process(GO:0034638) |
| 0.0 | 0.0 | GO:1902283 | negative regulation of primary amine oxidase activity(GO:1902283) |
| 0.0 | 0.2 | GO:0040032 | post-embryonic body morphogenesis(GO:0040032) |
| 0.0 | 0.5 | GO:0090022 | regulation of neutrophil chemotaxis(GO:0090022) |
| 0.0 | 0.1 | GO:0035524 | proline transmembrane transport(GO:0035524) |
| 0.0 | 0.4 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 0.3 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.6 | GO:0034750 | Scrib-APC-beta-catenin complex(GO:0034750) |
| 0.2 | 1.8 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.2 | 3.5 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.1 | 1.2 | GO:0072487 | MSL complex(GO:0072487) |
| 0.1 | 0.6 | GO:1990393 | 3M complex(GO:1990393) |
| 0.1 | 0.3 | GO:0000839 | Hrd1p ubiquitin ligase ERAD-L complex(GO:0000839) |
| 0.1 | 0.2 | GO:0044307 | dendritic branch(GO:0044307) |
| 0.0 | 1.1 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) |
| 0.0 | 0.4 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.0 | 0.3 | GO:0005587 | collagen type IV trimer(GO:0005587) |
| 0.0 | 0.3 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.0 | 1.4 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 0.4 | GO:0000808 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.0 | 0.4 | GO:0030478 | actin cap(GO:0030478) |
| 0.0 | 0.2 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.0 | 0.3 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.0 | 0.6 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 0.2 | GO:0097425 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 0.0 | 0.2 | GO:0001940 | female pronucleus(GO:0001939) male pronucleus(GO:0001940) |
| 0.0 | 0.9 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.3 | GO:0031298 | replication fork protection complex(GO:0031298) |
| 0.0 | 0.1 | GO:0030678 | mitochondrial ribonuclease P complex(GO:0030678) |
| 0.0 | 0.3 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 0.5 | GO:0001527 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.0 | 0.1 | GO:0097209 | epidermal lamellar body(GO:0097209) |
| 0.0 | 0.4 | GO:0016461 | unconventional myosin complex(GO:0016461) |
| 0.0 | 0.1 | GO:1990667 | PCSK9-AnxA2 complex(GO:1990667) |
| 0.0 | 0.3 | GO:0033276 | transcription factor TFTC complex(GO:0033276) |
| 0.0 | 1.1 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.0 | 2.3 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.0 | 0.0 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.0 | 0.1 | GO:0071458 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) |
| 0.0 | 0.1 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 0.0 | 0.3 | GO:0071004 | U2-type prespliceosome(GO:0071004) |
| 0.0 | 0.9 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.1 | GO:0070652 | HAUS complex(GO:0070652) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.1 | GO:0004310 | farnesyl-diphosphate farnesyltransferase activity(GO:0004310) squalene synthase activity(GO:0051996) |
| 0.2 | 1.0 | GO:0008240 | tripeptidyl-peptidase activity(GO:0008240) |
| 0.2 | 0.8 | GO:0003943 | N-acetylgalactosamine-4-sulfatase activity(GO:0003943) |
| 0.2 | 3.0 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.1 | 0.4 | GO:0052798 | beta-galactoside alpha-2,3-sialyltransferase activity(GO:0052798) |
| 0.1 | 0.3 | GO:0047787 | geranylgeranyl reductase activity(GO:0045550) delta4-3-oxosteroid 5beta-reductase activity(GO:0047787) |
| 0.1 | 1.5 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.1 | 0.6 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.1 | 0.4 | GO:0042132 | fructose 1,6-bisphosphate 1-phosphatase activity(GO:0042132) |
| 0.1 | 1.6 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.1 | 0.6 | GO:0097603 | temperature-gated ion channel activity(GO:0097603) |
| 0.1 | 0.6 | GO:0004962 | endothelin receptor activity(GO:0004962) |
| 0.1 | 0.5 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.1 | 1.6 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.1 | 0.3 | GO:0070404 | NADH binding(GO:0070404) |
| 0.1 | 0.5 | GO:0102344 | 3-hydroxy-behenoyl-CoA dehydratase activity(GO:0102344) 3-hydroxy-lignoceroyl-CoA dehydratase activity(GO:0102345) |
| 0.1 | 0.3 | GO:0044736 | acid-sensing ion channel activity(GO:0044736) |
| 0.1 | 1.3 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.1 | 0.2 | GO:0044594 | 3alpha,7alpha,12alpha-trihydroxy-5beta-cholest-24-enoyl-CoA hydratase activity(GO:0033989) 17-beta-hydroxysteroid dehydrogenase (NAD+) activity(GO:0044594) |
| 0.1 | 0.7 | GO:0050145 | nucleoside phosphate kinase activity(GO:0050145) |
| 0.1 | 0.6 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.1 | 0.2 | GO:0004769 | steroid delta-isomerase activity(GO:0004769) |
| 0.1 | 0.2 | GO:0070538 | oleic acid binding(GO:0070538) |
| 0.0 | 0.2 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.0 | 0.3 | GO:0022850 | serotonin-gated cation channel activity(GO:0022850) |
| 0.0 | 1.3 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.4 | GO:0001162 | RNA polymerase II intronic transcription regulatory region sequence-specific DNA binding(GO:0001162) |
| 0.0 | 0.3 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.0 | 0.2 | GO:0031962 | mineralocorticoid receptor binding(GO:0031962) |
| 0.0 | 0.2 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) |
| 0.0 | 0.5 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.0 | 1.1 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.0 | 0.3 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 0.0 | 0.1 | GO:0071987 | WD40-repeat domain binding(GO:0071987) |
| 0.0 | 0.7 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.0 | 0.3 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.0 | 0.3 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.0 | 0.4 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.0 | 0.5 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.0 | 0.3 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.0 | 1.2 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 0.5 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.0 | 0.2 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.0 | 0.1 | GO:0005298 | proline:sodium symporter activity(GO:0005298) |
| 0.0 | 0.5 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 0.6 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.0 | 1.5 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 0.1 | GO:0004939 | beta-adrenergic receptor activity(GO:0004939) |
| 0.0 | 0.3 | GO:0032052 | bile acid binding(GO:0032052) |
| 0.0 | 0.6 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.0 | 0.2 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.0 | 0.3 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 2.4 | GO:0042626 | ATPase activity, coupled to transmembrane movement of substances(GO:0042626) |
| 0.0 | 0.1 | GO:0009019 | tRNA (guanine-N1-)-methyltransferase activity(GO:0009019) |
| 0.0 | 0.3 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 0.0 | GO:0004556 | alpha-amylase activity(GO:0004556) |
| 0.0 | 0.2 | GO:0097100 | supercoiled DNA binding(GO:0097100) |
| 0.0 | 0.4 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 1.2 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 0.1 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.0 | 0.2 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 0.1 | GO:0010385 | double-stranded methylated DNA binding(GO:0010385) |
| 0.0 | 0.2 | GO:0051430 | corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.0 | 0.8 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.0 | 0.3 | GO:0019789 | SUMO transferase activity(GO:0019789) |
| 0.0 | 0.2 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.0 | 0.2 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.0 | 0.1 | GO:0099580 | ion antiporter activity involved in regulation of postsynaptic membrane potential(GO:0099580) |
| 0.0 | 0.6 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.0 | GO:0052596 | tryptamine:oxygen oxidoreductase (deaminating) activity(GO:0052593) aminoacetone:oxygen oxidoreductase(deaminating) activity(GO:0052594) aliphatic-amine oxidase activity(GO:0052595) phenethylamine:oxygen oxidoreductase (deaminating) activity(GO:0052596) |
| 0.0 | 0.1 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.4 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.1 | 2.4 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.0 | 1.1 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 1.7 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.0 | 1.2 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 1.6 | PID ERBB1 RECEPTOR PROXIMAL PATHWAY | EGF receptor (ErbB1) signaling pathway |
| 0.0 | 1.4 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 0.5 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 0.2 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.0 | 0.5 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 0.6 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.0 | 0.6 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 0.5 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 5.1 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.1 | 2.4 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.1 | 1.1 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.1 | 1.7 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.1 | 1.7 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.1 | 2.1 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.8 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 0.5 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 1.1 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 0.7 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.0 | 1.8 | REACTOME PLC BETA MEDIATED EVENTS | Genes involved in PLC beta mediated events |
| 0.0 | 0.7 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.7 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 0.3 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 0.6 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 0.6 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 0.2 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 0.3 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 1.3 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.2 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.0 | 0.2 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.0 | 0.2 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 0.5 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.0 | 0.2 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |