Project
Inflammatory response time course, HUVEC (Wada, 2009)
Navigation
Downloads
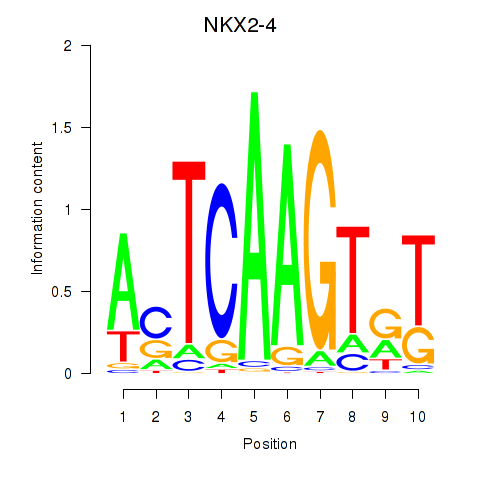
Results for NKX2-4
Z-value: 0.74
Transcription factors associated with NKX2-4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NKX2-4
|
ENSG00000125816.5 | NKX2-4 |
Activity profile of NKX2-4 motif
Sorted Z-values of NKX2-4 motif
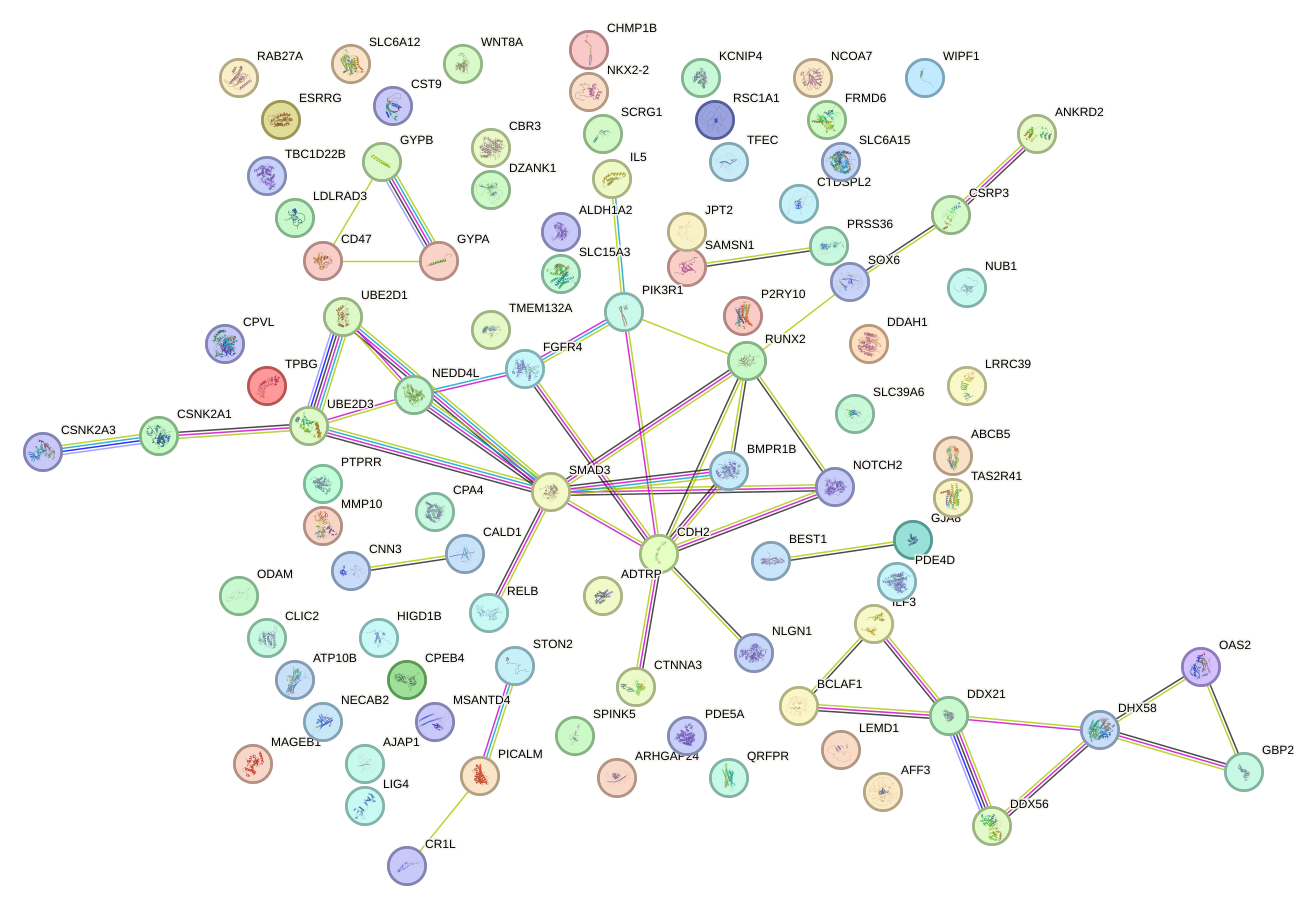
Network of associatons between targets according to the STRING database.
First level regulatory network of NKX2-4
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_-_102780620 | 2.01 |
ENST00000279441.9
ENST00000539681.1 |
MMP10
|
matrix metallopeptidase 10 |
| chr3_-_108058361 | 1.57 |
ENST00000398258.7
|
CD47
|
CD47 molecule |
| chr4_-_119628791 | 1.32 |
ENST00000354960.8
|
PDE5A
|
phosphodiesterase 5A |
| chr15_+_67138001 | 1.27 |
ENST00000439724.7
|
SMAD3
|
SMAD family member 3 |
| chr19_+_45001430 | 1.23 |
ENST00000625761.2
ENST00000505236.1 ENST00000221452.13 |
RELB
|
RELB proto-oncogene, NF-kB subunit |
| chr1_-_89126066 | 1.17 |
ENST00000370466.4
|
GBP2
|
guanylate binding protein 2 |
| chr7_+_134866831 | 1.13 |
ENST00000435928.1
|
CALD1
|
caldesmon 1 |
| chr6_-_11779606 | 1.13 |
ENST00000506810.1
|
ADTRP
|
androgen dependent TFPI regulating protein |
| chr1_-_85404494 | 1.12 |
ENST00000633113.1
|
DDAH1
|
dimethylarginine dimethylaminohydrolase 1 |
| chrX_-_155334580 | 1.11 |
ENST00000369449.7
ENST00000321926.4 |
CLIC2
|
chloride intracellular channel 2 |
| chr7_+_134843884 | 1.08 |
ENST00000445569.6
|
CALD1
|
caldesmon 1 |
| chr12_-_70788914 | 1.05 |
ENST00000342084.8
|
PTPRR
|
protein tyrosine phosphatase receptor type R |
| chr7_+_130293134 | 0.97 |
ENST00000445470.6
ENST00000492072.5 ENST00000222482.10 ENST00000473956.5 ENST00000493259.5 ENST00000486598.1 |
CPA4
|
carboxypeptidase A4 |
| chr15_+_44427793 | 0.92 |
ENST00000558966.5
ENST00000559793.5 ENST00000558968.1 |
CTDSPL2
|
CTD small phosphatase like 2 |
| chr1_-_120051714 | 0.89 |
ENST00000579475.7
|
NOTCH2
|
notch receptor 2 |
| chr7_-_115968302 | 0.86 |
ENST00000457268.5
|
TFEC
|
transcription factor EC |
| chr12_+_10505602 | 0.85 |
ENST00000322446.3
|
EIF2S3B
|
eukaryotic translation initiation factor 2 subunit gamma B |
| chr16_+_1680337 | 0.81 |
ENST00000566691.5
ENST00000382710.8 |
JPT2
|
Jupiter microtubule associated homolog 2 |
| chr21_+_36135071 | 0.79 |
ENST00000290354.6
|
CBR3
|
carbonyl reductase 3 |
| chr2_+_161136901 | 0.79 |
ENST00000259075.6
ENST00000432002.5 |
TANK
|
TRAF family member associated NFKB activator |
| chr18_-_36122110 | 0.77 |
ENST00000586829.1
|
SLC39A6
|
solute carrier family 39 member 6 |
| chr19_+_10655023 | 0.76 |
ENST00000590009.5
|
ILF3
|
interleukin enhancer binding factor 3 |
| chr20_-_18413216 | 0.76 |
ENST00000480488.2
|
DZANK1
|
double zinc ribbon and ankyrin repeat domains 1 |
| chr2_-_37672178 | 0.75 |
ENST00000457889.1
|
CDC42EP3
|
CDC42 effector protein 3 |
| chr4_+_95091462 | 0.74 |
ENST00000264568.8
|
BMPR1B
|
bone morphogenetic protein receptor type 1B |
| chr6_+_45422564 | 0.74 |
ENST00000625924.1
|
RUNX2
|
RUNX family transcription factor 2 |
| chr15_-_58014097 | 0.74 |
ENST00000559517.5
|
ALDH1A2
|
aldehyde dehydrogenase 1 family member A2 |
| chr7_+_151341764 | 0.73 |
ENST00000413040.7
ENST00000470229.6 ENST00000568733.6 |
NUB1
|
negative regulator of ubiquitin like proteins 1 |
| chr11_-_60952559 | 0.72 |
ENST00000538739.2
|
SLC15A3
|
solute carrier family 15 member 3 |
| chr5_+_68288346 | 0.71 |
ENST00000320694.12
|
PIK3R1
|
phosphoinositide-3-kinase regulatory subunit 1 |
| chr6_+_82363199 | 0.69 |
ENST00000535040.4
|
TPBG
|
trophoblast glycoprotein |
| chr2_-_37672448 | 0.67 |
ENST00000611976.1
|
CDC42EP3
|
CDC42 effector protein 3 |
| chr11_-_106022209 | 0.67 |
ENST00000301919.9
ENST00000534458.1 ENST00000530108.1 ENST00000530788.1 |
MSANTD4
|
Myb/SANT DNA binding domain containing 4 with coiled-coils |
| chrX_+_135309480 | 0.67 |
ENST00000635820.1
|
ETDC
|
embryonic testis differentiation homolog C |
| chr6_+_45422485 | 0.65 |
ENST00000359524.7
|
RUNX2
|
RUNX family transcription factor 2 |
| chr11_+_35943981 | 0.64 |
ENST00000528989.5
ENST00000524419.5 ENST00000315571.6 |
LDLRAD3
|
low density lipoprotein receptor class A domain containing 3 |
| chr5_-_59216826 | 0.63 |
ENST00000638939.1
|
PDE4D
|
phosphodiesterase 4D |
| chr10_+_97572771 | 0.62 |
ENST00000370655.6
ENST00000455090.1 |
ANKRD2
|
ankyrin repeat domain 2 |
| chr14_+_51489112 | 0.58 |
ENST00000356218.8
|
FRMD6
|
FERM domain containing 6 |
| chr7_-_29195186 | 0.58 |
ENST00000449801.5
ENST00000409850.5 |
CPVL
|
carboxypeptidase vitellogenic like |
| chr15_-_55249029 | 0.56 |
ENST00000566877.5
|
RAB27A
|
RAB27A, member RAS oncogene family |
| chr11_-_19202004 | 0.56 |
ENST00000648719.1
|
CSRP3
|
cysteine and glycine rich protein 3 |
| chr8_+_31639291 | 0.56 |
ENST00000651149.1
ENST00000650866.1 |
NRG1
|
neuregulin 1 |
| chrX_+_30243715 | 0.56 |
ENST00000378981.8
ENST00000397550.6 |
MAGEB1
|
MAGE family member B1 |
| chr5_+_173918186 | 0.55 |
ENST00000657000.1
|
CPEB4
|
cytoplasmic polyadenylation element binding protein 4 |
| chr2_-_174597728 | 0.54 |
ENST00000409891.5
ENST00000410117.5 |
WIPF1
|
WAS/WASL interacting protein family member 1 |
| chr5_+_177087412 | 0.53 |
ENST00000513166.1
|
FGFR4
|
fibroblast growth factor receptor 4 |
| chr4_-_102825767 | 0.53 |
ENST00000505207.5
ENST00000502404.5 ENST00000507845.5 |
UBE2D3
|
ubiquitin conjugating enzyme E2 D3 |
| chr6_+_125919210 | 0.52 |
ENST00000438495.6
|
NCOA7
|
nuclear receptor coactivator 7 |
| chr8_+_31639222 | 0.52 |
ENST00000519301.6
ENST00000652698.1 |
NRG1
|
neuregulin 1 |
| chr5_+_173918216 | 0.51 |
ENST00000519467.1
|
CPEB4
|
cytoplasmic polyadenylation element binding protein 4 |
| chr6_+_125919296 | 0.51 |
ENST00000444128.2
|
NCOA7
|
nuclear receptor coactivator 7 |
| chr13_-_108215498 | 0.50 |
ENST00000614526.1
ENST00000611712.4 ENST00000442234.6 |
LIG4
|
DNA ligase 4 |
| chr4_+_85827745 | 0.50 |
ENST00000509300.5
|
ARHGAP24
|
Rho GTPase activating protein 24 |
| chr4_+_85827891 | 0.50 |
ENST00000514229.5
|
ARHGAP24
|
Rho GTPase activating protein 24 |
| chr21_-_14546297 | 0.50 |
ENST00000400566.6
ENST00000400564.5 |
SAMSN1
|
SAM domain, SH3 domain and nuclear localization signals 1 |
| chr11_-_86069811 | 0.50 |
ENST00000531930.5
ENST00000528398.5 |
PICALM
|
phosphatidylinositol binding clathrin assembly protein |
| chr16_+_83998252 | 0.50 |
ENST00000564166.1
|
NECAB2
|
N-terminal EF-hand calcium binding protein 2 |
| chr21_-_14546351 | 0.50 |
ENST00000619120.4
|
SAMSN1
|
SAM domain, SH3 domain and nuclear localization signals 1 |
| chr4_-_102825526 | 0.50 |
ENST00000504211.5
ENST00000508476.5 |
UBE2D3
|
ubiquitin conjugating enzyme E2 D3 |
| chr4_-_102825854 | 0.49 |
ENST00000350435.11
|
UBE2D3
|
ubiquitin conjugating enzyme E2 D3 |
| chr11_+_61950343 | 0.49 |
ENST00000378043.9
|
BEST1
|
bestrophin 1 |
| chr12_+_112978562 | 0.47 |
ENST00000680122.1
|
OAS2
|
2'-5'-oligoadenylate synthetase 2 |
| chr2_+_161160420 | 0.47 |
ENST00000392749.7
ENST00000405852.5 |
TANK
|
TRAF family member associated NFKB activator |
| chr4_-_121381007 | 0.47 |
ENST00000394427.3
|
QRFPR
|
pyroglutamylated RFamide peptide receptor |
| chr1_-_94925759 | 0.47 |
ENST00000415017.1
ENST00000545882.5 |
CNN3
|
calponin 3 |
| chr5_+_138084015 | 0.46 |
ENST00000504809.5
ENST00000506684.6 ENST00000398754.1 |
WNT8A
|
Wnt family member 8A |
| chr18_-_27990256 | 0.46 |
ENST00000675173.1
|
CDH2
|
cadherin 2 |
| chr17_-_42112674 | 0.46 |
ENST00000251642.8
ENST00000591220.5 |
DHX58
|
DExH-box helicase 58 |
| chr6_-_36024635 | 0.45 |
ENST00000490799.6
|
SLC26A8
|
solute carrier family 26 member 8 |
| chr11_+_60931744 | 0.45 |
ENST00000536409.1
|
TMEM132A
|
transmembrane protein 132A |
| chr11_-_86068929 | 0.45 |
ENST00000630913.2
|
PICALM
|
phosphatidylinositol binding clathrin assembly protein |
| chr6_-_36024493 | 0.44 |
ENST00000394602.6
ENST00000355574.6 |
SLC26A8
|
solute carrier family 26 member 8 |
| chr10_+_58334998 | 0.44 |
ENST00000373910.9
|
UBE2D1
|
ubiquitin conjugating enzyme E2 D1 |
| chr6_+_37257762 | 0.44 |
ENST00000373491.3
|
TBC1D22B
|
TBC1 domain family member 22B |
| chr1_+_202462730 | 0.43 |
ENST00000290419.9
ENST00000491336.5 |
PPP1R12B
|
protein phosphatase 1 regulatory subunit 12B |
| chr20_-_21514046 | 0.43 |
ENST00000377142.5
|
NKX2-2
|
NK2 homeobox 2 |
| chr10_+_58334976 | 0.42 |
ENST00000615793.1
|
UBE2D1
|
ubiquitin conjugating enzyme E2 D1 |
| chr4_-_173399102 | 0.41 |
ENST00000296506.8
|
SCRG1
|
stimulator of chondrogenesis 1 |
| chr2_-_100142575 | 0.41 |
ENST00000317233.8
ENST00000672204.1 ENST00000416492.5 ENST00000672857.1 ENST00000672756.2 |
AFF3
|
AF4/FMR2 family member 3 |
| chr18_+_58149314 | 0.40 |
ENST00000435432.6
ENST00000357895.9 ENST00000586263.5 |
NEDD4L
|
NEDD4 like E3 ubiquitin protein ligase |
| chr5_-_160852200 | 0.38 |
ENST00000327245.10
|
ATP10B
|
ATPase phospholipid transporting 10B (putative) |
| chr11_+_61950370 | 0.38 |
ENST00000449131.6
|
BEST1
|
bestrophin 1 |
| chr11_-_86069043 | 0.37 |
ENST00000532317.5
ENST00000528256.1 ENST00000393346.8 ENST00000526033.5 |
PICALM
|
phosphatidylinositol binding clathrin assembly protein |
| chr1_+_4654601 | 0.36 |
ENST00000378191.5
|
AJAP1
|
adherens junctions associated protein 1 |
| chr4_-_21544650 | 0.35 |
ENST00000509207.1
|
KCNIP4
|
potassium voltage-gated channel interacting protein 4 |
| chr20_-_23605917 | 0.35 |
ENST00000376971.4
|
CST9
|
cystatin 9 |
| chr11_-_11353241 | 0.33 |
ENST00000528848.3
|
CSNK2A3
|
casein kinase 2 alpha 3 |
| chr6_-_136289778 | 0.33 |
ENST00000530767.5
ENST00000527759.5 |
BCLAF1
|
BCL2 associated transcription factor 1 |
| chr10_+_68956158 | 0.32 |
ENST00000354185.9
|
DDX21
|
DExD-box helicase 21 |
| chr1_-_205449924 | 0.32 |
ENST00000367154.5
|
LEMD1
|
LEM domain containing 1 |
| chr11_-_16356538 | 0.32 |
ENST00000683767.1
|
SOX6
|
SRY-box transcription factor 6 |
| chr12_-_213338 | 0.31 |
ENST00000424061.6
|
SLC6A12
|
solute carrier family 6 member 12 |
| chr12_-_21775045 | 0.31 |
ENST00000667884.1
|
KCNJ8
|
potassium inwardly rectifying channel subfamily J member 8 |
| chr7_+_143477873 | 0.31 |
ENST00000408916.1
|
TAS2R41
|
taste 2 receptor member 41 |
| chr4_+_70197924 | 0.31 |
ENST00000514097.5
|
ODAM
|
odontogenic, ameloblast associated |
| chr3_-_156555083 | 0.31 |
ENST00000265044.7
ENST00000476217.5 |
SSR3
|
signal sequence receptor subunit 3 |
| chr1_-_217076889 | 0.30 |
ENST00000493748.5
ENST00000463665.5 |
ESRRG
|
estrogen related receptor gamma |
| chr3_+_173584433 | 0.30 |
ENST00000361589.8
|
NLGN1
|
neuroligin 1 |
| chr5_+_148063971 | 0.30 |
ENST00000398454.5
ENST00000359874.7 ENST00000508733.5 ENST00000256084.8 |
SPINK5
|
serine peptidase inhibitor Kazal type 5 |
| chr10_-_67696115 | 0.29 |
ENST00000433211.7
|
CTNNA3
|
catenin alpha 3 |
| chr5_-_132543513 | 0.29 |
ENST00000231454.6
|
IL5
|
interleukin 5 |
| chr4_-_144140635 | 0.29 |
ENST00000512064.5
ENST00000512789.5 ENST00000504786.5 ENST00000503627.2 ENST00000642713.1 ENST00000643148.1 ENST00000642738.1 ENST00000642295.1 ENST00000646447.1 ENST00000535709.6 |
GYPA
|
glycophorin A (MNS blood group) |
| chr11_-_19201976 | 0.29 |
ENST00000647990.1
ENST00000649235.1 ENST00000265968.9 ENST00000649842.1 |
CSRP3
|
cysteine and glycine rich protein 3 |
| chr17_+_44846318 | 0.29 |
ENST00000591513.5
|
HIGD1B
|
HIG1 hypoxia inducible domain family member 1B |
| chr1_+_207645114 | 0.29 |
ENST00000508064.7
|
CR1L
|
complement C3b/C4b receptor 1 like |
| chr7_+_20647388 | 0.29 |
ENST00000258738.10
|
ABCB5
|
ATP binding cassette subfamily B member 5 |
| chr1_+_147902789 | 0.27 |
ENST00000369235.2
|
GJA8
|
gap junction protein alpha 8 |
| chr1_-_100178215 | 0.27 |
ENST00000370138.1
ENST00000370137.6 ENST00000342895.7 ENST00000620882.4 |
LRRC39
|
leucine rich repeat containing 39 |
| chr10_+_71396905 | 0.26 |
ENST00000224721.12
ENST00000398809.9 ENST00000461841.7 ENST00000299366.11 ENST00000644511.1 ENST00000616684.4 |
CDH23
|
cadherin related 23 |
| chr13_-_108218293 | 0.26 |
ENST00000405925.2
|
LIG4
|
DNA ligase 4 |
| chr1_+_15659869 | 0.26 |
ENST00000345034.1
|
RSC1A1
|
regulator of solute carriers 1 |
| chr16_-_31150058 | 0.26 |
ENST00000569305.1
ENST00000268281.9 ENST00000418068.6 |
PRSS36
|
serine protease 36 |
| chr12_-_84892120 | 0.26 |
ENST00000680379.1
|
SLC6A15
|
solute carrier family 6 member 15 |
| chr15_+_41286011 | 0.26 |
ENST00000661438.1
|
ENSG00000285920.2
|
novel protein |
| chr18_+_11851404 | 0.25 |
ENST00000526991.3
|
CHMP1B
|
charged multivesicular body protein 1B |
| chrX_+_78945332 | 0.25 |
ENST00000544091.1
ENST00000171757.3 |
P2RY10
|
P2Y receptor family member 10 |
| chr14_-_81427390 | 0.24 |
ENST00000555447.5
|
STON2
|
stonin 2 |
| chr12_+_49295138 | 0.24 |
ENST00000257860.9
|
PRPH
|
peripherin |
| chr16_+_10743786 | 0.24 |
ENST00000574334.5
ENST00000283027.10 ENST00000433392.6 |
NUBP1
|
nucleotide binding protein 1 |
| chr5_-_11588842 | 0.23 |
ENST00000503622.5
|
CTNND2
|
catenin delta 2 |
| chr19_+_10718114 | 0.23 |
ENST00000408974.8
|
DNM2
|
dynamin 2 |
| chr12_-_14938508 | 0.23 |
ENST00000266397.7
|
ERP27
|
endoplasmic reticulum protein 27 |
| chr1_-_241357085 | 0.23 |
ENST00000366564.5
|
RGS7
|
regulator of G protein signaling 7 |
| chr1_-_88992616 | 0.23 |
ENST00000260508.9
ENST00000652648.1 |
KYAT3
RBMXL1
|
kynurenine aminotransferase 3 RBMX like 1 |
| chr10_+_103245887 | 0.23 |
ENST00000441178.2
|
RPEL1
|
ribulose-5-phosphate-3-epimerase like 1 |
| chr12_+_1970809 | 0.23 |
ENST00000683781.1
ENST00000682462.1 |
CACNA1C
|
calcium voltage-gated channel subunit alpha1 C |
| chr19_+_48756067 | 0.23 |
ENST00000222157.5
|
FGF21
|
fibroblast growth factor 21 |
| chr4_-_99352730 | 0.23 |
ENST00000510055.5
ENST00000515683.6 ENST00000511397.3 |
ADH1C
|
alcohol dehydrogenase 1C (class I), gamma polypeptide |
| chr6_-_152637351 | 0.23 |
ENST00000367255.10
|
SYNE1
|
spectrin repeat containing nuclear envelope protein 1 |
| chrX_+_71223216 | 0.23 |
ENST00000361726.7
|
GJB1
|
gap junction protein beta 1 |
| chr22_-_50085331 | 0.23 |
ENST00000395876.6
|
MLC1
|
modulator of VRAC current 1 |
| chr1_-_241357225 | 0.22 |
ENST00000366565.5
|
RGS7
|
regulator of G protein signaling 7 |
| chr9_+_37667997 | 0.22 |
ENST00000539465.5
|
FRMPD1
|
FERM and PDZ domain containing 1 |
| chr1_+_109090726 | 0.22 |
ENST00000338272.9
ENST00000651489.1 |
TMEM167B
|
transmembrane protein 167B |
| chr12_+_1970772 | 0.22 |
ENST00000682544.1
|
CACNA1C
|
calcium voltage-gated channel subunit alpha1 C |
| chr22_-_50085414 | 0.22 |
ENST00000311597.10
|
MLC1
|
modulator of VRAC current 1 |
| chr19_+_37507129 | 0.22 |
ENST00000586138.5
ENST00000588578.5 ENST00000587986.5 |
ZNF793
|
zinc finger protein 793 |
| chr12_+_8822610 | 0.21 |
ENST00000299698.12
|
A2ML1
|
alpha-2-macroglobulin like 1 |
| chr1_-_241357171 | 0.21 |
ENST00000440928.6
|
RGS7
|
regulator of G protein signaling 7 |
| chr7_-_107803215 | 0.21 |
ENST00000340010.10
ENST00000453332.1 |
SLC26A3
|
solute carrier family 26 member 3 |
| chr1_-_116667668 | 0.21 |
ENST00000369486.8
ENST00000369483.5 |
IGSF3
|
immunoglobulin superfamily member 3 |
| chr21_-_33491386 | 0.20 |
ENST00000617313.4
ENST00000314399.3 |
DNAJC28
|
DnaJ heat shock protein family (Hsp40) member C28 |
| chr5_-_135452318 | 0.20 |
ENST00000537858.2
|
TIFAB
|
TIFA inhibitor |
| chr17_+_28357638 | 0.20 |
ENST00000292114.8
ENST00000591482.1 |
TMEM199
ENSG00000258924.4
|
transmembrane protein 199 novel transcript readthrough between TMEM199 and SARM1 |
| chrX_-_15854743 | 0.20 |
ENST00000450644.2
|
AP1S2
|
adaptor related protein complex 1 subunit sigma 2 |
| chr18_+_63542365 | 0.20 |
ENST00000269491.6
ENST00000382768.2 |
SERPINB12
|
serpin family B member 12 |
| chr10_-_119536533 | 0.20 |
ENST00000392865.5
|
RGS10
|
regulator of G protein signaling 10 |
| chr19_+_21082190 | 0.20 |
ENST00000618422.1
ENST00000618008.4 ENST00000425625.5 ENST00000456283.7 |
ZNF714
|
zinc finger protein 714 |
| chr19_-_9936501 | 0.19 |
ENST00000264833.9
|
OLFM2
|
olfactomedin 2 |
| chr19_-_17448664 | 0.19 |
ENST00000341130.6
|
TMEM221
|
transmembrane protein 221 |
| chr11_+_78063851 | 0.19 |
ENST00000281030.2
|
THRSP
|
thyroid hormone responsive |
| chr22_-_36817510 | 0.19 |
ENST00000443735.1
|
PVALB
|
parvalbumin |
| chr1_+_156338619 | 0.19 |
ENST00000481479.5
ENST00000368252.5 ENST00000466306.5 ENST00000368251.1 |
TSACC
|
TSSK6 activating cochaperone |
| chrX_-_11290478 | 0.19 |
ENST00000380717.7
|
ARHGAP6
|
Rho GTPase activating protein 6 |
| chr6_+_101398788 | 0.19 |
ENST00000369138.5
ENST00000413795.5 ENST00000358361.7 |
GRIK2
|
glutamate ionotropic receptor kainate type subunit 2 |
| chr1_-_204213943 | 0.19 |
ENST00000308302.4
|
GOLT1A
|
golgi transport 1A |
| chr3_+_46370854 | 0.18 |
ENST00000292303.4
|
CCR5
|
C-C motif chemokine receptor 5 |
| chr19_+_7669043 | 0.18 |
ENST00000221515.6
|
RETN
|
resistin |
| chr15_+_74174403 | 0.18 |
ENST00000560862.1
ENST00000395118.1 |
ISLR
|
immunoglobulin superfamily containing leucine rich repeat |
| chr6_-_30075767 | 0.18 |
ENST00000244360.8
ENST00000376751.8 |
RNF39
|
ring finger protein 39 |
| chr2_-_208190001 | 0.18 |
ENST00000451346.5
ENST00000341287.9 |
C2orf80
|
chromosome 2 open reading frame 80 |
| chr1_-_42335869 | 0.18 |
ENST00000372573.5
|
FOXJ3
|
forkhead box J3 |
| chr19_+_21082224 | 0.18 |
ENST00000620627.1
|
ZNF714
|
zinc finger protein 714 |
| chr3_-_185923893 | 0.18 |
ENST00000259043.11
|
TRA2B
|
transformer 2 beta homolog |
| chr14_-_70600643 | 0.18 |
ENST00000554963.5
ENST00000256379.10 ENST00000430055.6 ENST00000440435.2 ENST00000615788.4 |
MED6
|
mediator complex subunit 6 |
| chrX_-_139832235 | 0.18 |
ENST00000327569.7
ENST00000361648.6 |
ATP11C
|
ATPase phospholipid transporting 11C |
| chr1_+_175067831 | 0.18 |
ENST00000239462.9
|
TNN
|
tenascin N |
| chr14_-_22589157 | 0.18 |
ENST00000538631.1
ENST00000543337.1 ENST00000250498.9 |
DAD1
|
defender against cell death 1 |
| chr20_-_45101112 | 0.18 |
ENST00000306117.5
ENST00000537075.3 |
KCNS1
|
potassium voltage-gated channel modifier subfamily S member 1 |
| chr6_+_26156323 | 0.18 |
ENST00000304218.6
|
H1-4
|
H1.4 linker histone, cluster member |
| chr10_+_79512533 | 0.18 |
ENST00000520547.4
|
EIF5AL1
|
eukaryotic translation initiation factor 5A like 1 |
| chr4_-_174522791 | 0.17 |
ENST00000541923.5
ENST00000542498.5 |
HPGD
|
15-hydroxyprostaglandin dehydrogenase |
| chr2_-_201619144 | 0.17 |
ENST00000439802.5
ENST00000286195.7 ENST00000439140.6 ENST00000450242.1 |
C2CD6
|
C2 calcium dependent domain containing 6 |
| chr1_+_152878312 | 0.17 |
ENST00000368765.4
|
SMCP
|
sperm mitochondria associated cysteine rich protein |
| chr14_+_22226711 | 0.17 |
ENST00000390463.3
|
TRAV36DV7
|
T cell receptor alpha variable 36/delta variable 7 |
| chr11_-_33161502 | 0.17 |
ENST00000438862.6
|
CSTF3
|
cleavage stimulation factor subunit 3 |
| chr6_-_29487956 | 0.17 |
ENST00000377127.4
|
MAS1L
|
MAS1 proto-oncogene like, G protein-coupled receptor |
| chr1_-_212414815 | 0.17 |
ENST00000261455.9
ENST00000535273.2 |
PACC1
|
proton activated chloride channel 1 |
| chr20_-_44651683 | 0.17 |
ENST00000537820.1
ENST00000372874.9 |
ADA
|
adenosine deaminase |
| chr22_-_29061831 | 0.17 |
ENST00000216071.5
|
C22orf31
|
chromosome 22 open reading frame 31 |
| chr4_+_168497044 | 0.16 |
ENST00000505667.6
|
PALLD
|
palladin, cytoskeletal associated protein |
| chr2_-_151971750 | 0.16 |
ENST00000636598.1
|
CACNB4
|
calcium voltage-gated channel auxiliary subunit beta 4 |
| chr12_+_100503352 | 0.16 |
ENST00000551379.5
ENST00000188403.7 |
NR1H4
|
nuclear receptor subfamily 1 group H member 4 |
| chr16_+_77788554 | 0.16 |
ENST00000302536.3
|
VAT1L
|
vesicle amine transport 1 like |
| chr2_+_100974849 | 0.16 |
ENST00000450763.1
|
NPAS2
|
neuronal PAS domain protein 2 |
| chr5_+_162067764 | 0.16 |
ENST00000639213.2
ENST00000414552.6 |
GABRG2
|
gamma-aminobutyric acid type A receptor subunit gamma2 |
| chrX_-_111412162 | 0.16 |
ENST00000637570.1
ENST00000356220.8 ENST00000636035.2 ENST00000637453.1 ENST00000635795.1 |
DCX
|
doublecortin |
| chr11_-_33161461 | 0.16 |
ENST00000323959.9
ENST00000431742.2 ENST00000524827.6 |
CSTF3
|
cleavage stimulation factor subunit 3 |
| chr8_+_7848000 | 0.16 |
ENST00000326558.9
ENST00000351436.8 ENST00000528033.3 |
SPAG11A
|
sperm associated antigen 11A |
| chr11_-_104164361 | 0.16 |
ENST00000302251.9
|
PDGFD
|
platelet derived growth factor D |
| chr3_+_4493471 | 0.16 |
ENST00000544951.6
ENST00000650294.1 |
ITPR1
|
inositol 1,4,5-trisphosphate receptor type 1 |
| chr19_-_49813259 | 0.15 |
ENST00000313777.9
|
FUZ
|
fuzzy planar cell polarity protein |
| chr17_+_30477362 | 0.15 |
ENST00000225724.9
ENST00000451249.7 ENST00000467337.6 ENST00000581721.5 ENST00000414833.2 |
GOSR1
|
golgi SNAP receptor complex member 1 |
| chr15_-_85794902 | 0.15 |
ENST00000337975.6
|
KLHL25
|
kelch like family member 25 |
| chr2_+_190180835 | 0.15 |
ENST00000340623.4
|
C2orf88
|
chromosome 2 open reading frame 88 |
| chr4_+_168497066 | 0.15 |
ENST00000261509.10
|
PALLD
|
palladin, cytoskeletal associated protein |
| chr1_+_160127672 | 0.15 |
ENST00000447527.1
|
ATP1A2
|
ATPase Na+/K+ transporting subunit alpha 2 |
| chr16_-_67966793 | 0.15 |
ENST00000541864.6
|
SLC12A4
|
solute carrier family 12 member 4 |
| chr5_+_162067858 | 0.15 |
ENST00000361925.9
|
GABRG2
|
gamma-aminobutyric acid type A receptor subunit gamma2 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.3 | GO:0061767 | negative regulation of lung blood pressure(GO:0061767) |
| 0.2 | 1.3 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 0.2 | 1.3 | GO:0060282 | positive regulation of oocyte development(GO:0060282) |
| 0.2 | 1.3 | GO:1902963 | regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902962) negative regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902963) |
| 0.2 | 1.0 | GO:1903385 | regulation of homophilic cell adhesion(GO:1903385) |
| 0.2 | 0.8 | GO:0042376 | phylloquinone metabolic process(GO:0042374) phylloquinone catabolic process(GO:0042376) quinone catabolic process(GO:1901662) |
| 0.2 | 0.5 | GO:0014034 | neural crest cell fate commitment(GO:0014034) canonical Wnt signaling pathway involved in neural crest cell differentiation(GO:0044335) |
| 0.2 | 1.4 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 0.1 | 0.7 | GO:0021780 | oligodendrocyte cell fate specification(GO:0021778) oligodendrocyte cell fate commitment(GO:0021779) glial cell fate specification(GO:0021780) |
| 0.1 | 0.8 | GO:0033152 | immunoglobulin V(D)J recombination(GO:0033152) |
| 0.1 | 0.5 | GO:1904021 | negative regulation of G-protein coupled receptor internalization(GO:1904021) |
| 0.1 | 0.8 | GO:0035995 | detection of muscle stretch(GO:0035995) |
| 0.1 | 0.5 | GO:0039534 | negative regulation of MDA-5 signaling pathway(GO:0039534) |
| 0.1 | 0.4 | GO:2000314 | regulation of neural crest formation(GO:0090299) negative regulation of neural crest formation(GO:0090301) negative regulation of fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:2000314) |
| 0.1 | 0.7 | GO:0001550 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) negative regulation of chondrocyte proliferation(GO:1902731) |
| 0.1 | 0.6 | GO:0003383 | apical constriction(GO:0003383) |
| 0.1 | 0.3 | GO:0045643 | regulation of eosinophil differentiation(GO:0045643) positive regulation of eosinophil differentiation(GO:0045645) |
| 0.1 | 0.3 | GO:0048058 | compound eye corneal lens development(GO:0048058) |
| 0.1 | 0.4 | GO:1904640 | response to methionine(GO:1904640) |
| 0.1 | 0.6 | GO:1903435 | positive regulation of constitutive secretory pathway(GO:1903435) |
| 0.1 | 0.2 | GO:0002314 | germinal center B cell differentiation(GO:0002314) |
| 0.1 | 1.1 | GO:0021840 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) ERBB3 signaling pathway(GO:0038129) |
| 0.1 | 0.7 | GO:0042904 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.1 | 0.9 | GO:0030321 | transepithelial chloride transport(GO:0030321) |
| 0.1 | 1.1 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.1 | 1.4 | GO:0031274 | positive regulation of pseudopodium assembly(GO:0031274) |
| 0.1 | 0.2 | GO:1903526 | negative regulation of membrane tubulation(GO:1903526) |
| 0.1 | 0.5 | GO:1903412 | response to bile acid(GO:1903412) |
| 0.1 | 0.3 | GO:0098923 | retrograde trans-synaptic signaling by soluble gas(GO:0098923) trans-synaptic signaling by soluble gas(GO:0099543) |
| 0.1 | 0.2 | GO:1900111 | positive regulation of histone H3-K9 dimethylation(GO:1900111) |
| 0.1 | 1.6 | GO:0008228 | opsonization(GO:0008228) |
| 0.1 | 1.1 | GO:0060315 | negative regulation of ryanodine-sensitive calcium-release channel activity(GO:0060315) |
| 0.1 | 0.2 | GO:0048213 | Golgi vesicle prefusion complex stabilization(GO:0048213) |
| 0.1 | 0.2 | GO:0098583 | mastication(GO:0071626) learned vocalization behavior(GO:0098583) |
| 0.1 | 1.2 | GO:0032688 | negative regulation of interferon-beta production(GO:0032688) |
| 0.1 | 0.6 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.1 | 1.1 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.1 | 0.1 | GO:1903015 | regulation of exo-alpha-sialidase activity(GO:1903015) |
| 0.1 | 0.4 | GO:0030860 | regulation of polarized epithelial cell differentiation(GO:0030860) |
| 0.1 | 1.1 | GO:0006527 | arginine catabolic process(GO:0006527) |
| 0.1 | 0.2 | GO:0070681 | glutaminyl-tRNAGln biosynthesis via transamidation(GO:0070681) |
| 0.1 | 1.0 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.1 | 0.9 | GO:0002315 | marginal zone B cell differentiation(GO:0002315) |
| 0.1 | 0.9 | GO:1902916 | positive regulation of protein polyubiquitination(GO:1902916) |
| 0.1 | 0.2 | GO:2000439 | positive regulation of monocyte extravasation(GO:2000439) |
| 0.1 | 0.5 | GO:1902897 | regulation of postsynaptic density protein 95 clustering(GO:1902897) |
| 0.1 | 0.2 | GO:1900920 | regulation of amino acid uptake involved in synaptic transmission(GO:0051941) regulation of glutamate uptake involved in transmission of nerve impulse(GO:0051946) regulation of L-glutamate import(GO:1900920) |
| 0.0 | 0.3 | GO:1900425 | negative regulation of defense response to bacterium(GO:1900425) |
| 0.0 | 0.2 | GO:2000196 | positive regulation of female gonad development(GO:2000196) |
| 0.0 | 0.8 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.0 | 0.5 | GO:0018377 | protein myristoylation(GO:0018377) |
| 0.0 | 0.1 | GO:1904933 | regulation of cell proliferation in midbrain(GO:1904933) |
| 0.0 | 0.4 | GO:0035585 | calcium-mediated signaling using extracellular calcium source(GO:0035585) |
| 0.0 | 0.2 | GO:1903314 | nitrogen catabolite regulation of transcription from RNA polymerase II promoter(GO:0001079) nitrogen catabolite activation of transcription from RNA polymerase II promoter(GO:0001080) regulation of urea metabolic process(GO:0034255) intracellular bile acid receptor signaling pathway(GO:0038185) interleukin-17 secretion(GO:0072615) nitrogen catabolite regulation of transcription(GO:0090293) nitrogen catabolite activation of transcription(GO:0090294) regulation of nitrogen cycle metabolic process(GO:1903314) positive regulation of glutamate metabolic process(GO:2000213) regulation of ammonia assimilation cycle(GO:2001248) positive regulation of ammonia assimilation cycle(GO:2001250) |
| 0.0 | 0.1 | GO:0006288 | base-excision repair, DNA ligation(GO:0006288) |
| 0.0 | 0.1 | GO:1901420 | negative regulation of response to alcohol(GO:1901420) |
| 0.0 | 0.1 | GO:0034970 | histone H3-R2 methylation(GO:0034970) |
| 0.0 | 0.3 | GO:0060054 | positive regulation of epithelial cell proliferation involved in wound healing(GO:0060054) |
| 0.0 | 0.3 | GO:0086073 | bundle of His cell-Purkinje myocyte adhesion involved in cell communication(GO:0086073) |
| 0.0 | 0.1 | GO:1900147 | Schwann cell migration(GO:0036135) regulation of Schwann cell migration(GO:1900147) |
| 0.0 | 0.4 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.0 | 0.1 | GO:0070682 | proteasome regulatory particle assembly(GO:0070682) |
| 0.0 | 0.7 | GO:0051531 | NFAT protein import into nucleus(GO:0051531) |
| 0.0 | 0.6 | GO:2000291 | regulation of myoblast proliferation(GO:2000291) |
| 0.0 | 0.2 | GO:0007341 | penetration of zona pellucida(GO:0007341) |
| 0.0 | 0.1 | GO:1901355 | response to rapamycin(GO:1901355) |
| 0.0 | 0.2 | GO:0044339 | canonical Wnt signaling pathway involved in osteoblast differentiation(GO:0044339) |
| 0.0 | 0.1 | GO:0007499 | ectoderm and mesoderm interaction(GO:0007499) |
| 0.0 | 0.2 | GO:0097052 | L-kynurenine metabolic process(GO:0097052) |
| 0.0 | 0.2 | GO:0009052 | pentose-phosphate shunt, non-oxidative branch(GO:0009052) pentose catabolic process(GO:0019323) |
| 0.0 | 0.9 | GO:0046827 | positive regulation of protein export from nucleus(GO:0046827) |
| 0.0 | 0.4 | GO:0047484 | regulation of response to osmotic stress(GO:0047484) |
| 0.0 | 0.1 | GO:0003095 | pressure natriuresis(GO:0003095) |
| 0.0 | 0.7 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.0 | 0.3 | GO:0050957 | equilibrioception(GO:0050957) |
| 0.0 | 1.5 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.0 | 0.1 | GO:0035625 | negative regulation of epinephrine secretion(GO:0032811) epidermal growth factor-activated receptor transactivation by G-protein coupled receptor signaling pathway(GO:0035625) |
| 0.0 | 0.1 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.0 | 0.3 | GO:1990573 | potassium ion import across plasma membrane(GO:1990573) |
| 0.0 | 0.3 | GO:0035524 | proline transmembrane transport(GO:0035524) |
| 0.0 | 0.1 | GO:0003366 | cell-matrix adhesion involved in ameboidal cell migration(GO:0003366) |
| 0.0 | 0.2 | GO:0002329 | pre-B cell differentiation(GO:0002329) |
| 0.0 | 0.2 | GO:0045905 | translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 0.0 | 0.1 | GO:0070858 | negative regulation of bile acid biosynthetic process(GO:0070858) negative regulation of bile acid metabolic process(GO:1904252) |
| 0.0 | 0.2 | GO:0043578 | nuclear matrix organization(GO:0043578) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.0 | 0.2 | GO:0044800 | fusion of virus membrane with host plasma membrane(GO:0019064) membrane fusion involved in viral entry into host cell(GO:0039663) multi-organism membrane fusion(GO:0044800) |
| 0.0 | 0.1 | GO:0090579 | transcriptional activation by promoter-enhancer looping(GO:0071733) gene looping(GO:0090202) dsDNA loop formation(GO:0090579) |
| 0.0 | 0.1 | GO:0030961 | peptidyl-arginine hydroxylation(GO:0030961) |
| 0.0 | 0.3 | GO:1904903 | ESCRT complex disassembly(GO:1904896) ESCRT III complex disassembly(GO:1904903) |
| 0.0 | 0.1 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 0.0 | 0.4 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.0 | 0.1 | GO:0043589 | skin morphogenesis(GO:0043589) |
| 0.0 | 0.8 | GO:0050869 | negative regulation of B cell activation(GO:0050869) |
| 0.0 | 0.2 | GO:0050882 | voluntary musculoskeletal movement(GO:0050882) |
| 0.0 | 0.1 | GO:1902748 | melanocyte migration(GO:0097324) positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.0 | 0.3 | GO:0006069 | ethanol oxidation(GO:0006069) |
| 0.0 | 0.1 | GO:0030263 | apoptotic chromosome condensation(GO:0030263) |
| 0.0 | 0.1 | GO:0050859 | negative regulation of B cell receptor signaling pathway(GO:0050859) |
| 0.0 | 0.0 | GO:1900133 | regulation of renin secretion into blood stream(GO:1900133) |
| 0.0 | 0.4 | GO:0072697 | protein localization to cell cortex(GO:0072697) |
| 0.0 | 0.2 | GO:1905146 | lysosomal lumen acidification(GO:0007042) lysosomal protein catabolic process(GO:1905146) |
| 0.0 | 0.1 | GO:1990414 | replication-born double-strand break repair via sister chromatid exchange(GO:1990414) |
| 0.0 | 1.5 | GO:0030574 | collagen catabolic process(GO:0030574) |
| 0.0 | 0.2 | GO:0032808 | lacrimal gland development(GO:0032808) |
| 0.0 | 0.3 | GO:0035589 | G-protein coupled purinergic nucleotide receptor signaling pathway(GO:0035589) |
| 0.0 | 0.2 | GO:0075713 | establishment of integrated proviral latency(GO:0075713) |
| 0.0 | 0.3 | GO:0015812 | gamma-aminobutyric acid transport(GO:0015812) |
| 0.0 | 0.2 | GO:0051152 | positive regulation of smooth muscle cell differentiation(GO:0051152) |
| 0.0 | 0.4 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.0 | 0.2 | GO:0097070 | ductus arteriosus closure(GO:0097070) lipoxin metabolic process(GO:2001300) |
| 0.0 | 0.1 | GO:0016480 | negative regulation of transcription from RNA polymerase III promoter(GO:0016480) |
| 0.0 | 0.1 | GO:0060715 | syncytiotrophoblast cell differentiation involved in labyrinthine layer development(GO:0060715) |
| 0.0 | 0.2 | GO:0051967 | negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.0 | 0.1 | GO:0033088 | regulation of immature T cell proliferation in thymus(GO:0033084) negative regulation of immature T cell proliferation in thymus(GO:0033088) |
| 0.0 | 0.0 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.0 | 0.2 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.0 | 0.0 | GO:0015722 | canalicular bile acid transport(GO:0015722) |
| 0.0 | 0.2 | GO:0016264 | gap junction assembly(GO:0016264) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.3 | GO:0070381 | endosome to plasma membrane transport vesicle(GO:0070381) |
| 0.2 | 0.7 | GO:1990578 | perinuclear endoplasmic reticulum membrane(GO:1990578) |
| 0.1 | 2.2 | GO:0030478 | actin cap(GO:0030478) |
| 0.1 | 1.3 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.1 | 0.7 | GO:0032807 | DNA ligase IV complex(GO:0032807) |
| 0.1 | 1.2 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.1 | 0.4 | GO:0044214 | spanning component of plasma membrane(GO:0044214) spanning component of membrane(GO:0089717) |
| 0.1 | 0.2 | GO:0030956 | glutamyl-tRNA(Gln) amidotransferase complex(GO:0030956) |
| 0.1 | 0.7 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.0 | 0.3 | GO:0097209 | epidermal lamellar body(GO:0097209) |
| 0.0 | 0.1 | GO:0036195 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 0.0 | 0.6 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.0 | 0.3 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.0 | 1.1 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.8 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.0 | 0.2 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.0 | 2.7 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 0.1 | GO:0034669 | integrin alpha4-beta7 complex(GO:0034669) |
| 0.0 | 0.4 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.5 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 0.9 | GO:0000791 | euchromatin(GO:0000791) |
| 0.0 | 0.4 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.0 | 0.2 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.0 | 0.1 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 0.0 | 0.1 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.0 | 0.3 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 0.1 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.0 | 0.2 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.0 | 0.7 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 0.8 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 0.2 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 0.2 | GO:0001741 | XY body(GO:0001741) |
| 0.0 | 0.2 | GO:0030663 | COPI-coated vesicle membrane(GO:0030663) |
| 0.0 | 1.5 | GO:0070821 | tertiary granule membrane(GO:0070821) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.4 | GO:0005519 | cytoskeletal regulatory protein binding(GO:0005519) |
| 0.3 | 0.8 | GO:0000253 | 3-keto sterol reductase activity(GO:0000253) |
| 0.2 | 1.3 | GO:0031962 | mineralocorticoid receptor binding(GO:0031962) |
| 0.2 | 1.3 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.2 | 0.6 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.2 | 0.5 | GO:0031687 | A2A adenosine receptor binding(GO:0031687) |
| 0.1 | 1.1 | GO:0016403 | dimethylargininase activity(GO:0016403) |
| 0.1 | 1.6 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.1 | 1.0 | GO:0038049 | transcription factor activity, ligand-activated RNA polymerase II transcription factor binding(GO:0038049) |
| 0.1 | 0.7 | GO:0015333 | peptide:proton symporter activity(GO:0015333) proton-dependent peptide secondary active transmembrane transporter activity(GO:0022897) |
| 0.1 | 0.3 | GO:0005137 | interleukin-5 receptor binding(GO:0005137) |
| 0.1 | 0.8 | GO:0003910 | DNA ligase (ATP) activity(GO:0003910) |
| 0.1 | 1.3 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.1 | 1.9 | GO:0043125 | ErbB-3 class receptor binding(GO:0043125) |
| 0.1 | 1.1 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.1 | 0.1 | GO:0003909 | DNA ligase activity(GO:0003909) |
| 0.1 | 2.2 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.1 | 0.2 | GO:0047315 | kynurenine-glyoxylate transaminase activity(GO:0047315) |
| 0.1 | 0.2 | GO:0004750 | ribulose-phosphate 3-epimerase activity(GO:0004750) |
| 0.1 | 1.0 | GO:0031433 | telethonin binding(GO:0031433) |
| 0.1 | 0.7 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.1 | 0.7 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.1 | 0.2 | GO:0050567 | amidase activity(GO:0004040) glutaminyl-tRNA synthase (glutamine-hydrolyzing) activity(GO:0050567) |
| 0.1 | 0.5 | GO:0004983 | neuropeptide Y receptor activity(GO:0004983) |
| 0.1 | 0.3 | GO:0017098 | sulfonylurea receptor binding(GO:0017098) |
| 0.0 | 0.1 | GO:0061505 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 0.0 | 0.3 | GO:0015562 | efflux transmembrane transporter activity(GO:0015562) |
| 0.0 | 0.2 | GO:0031749 | D2 dopamine receptor binding(GO:0031749) |
| 0.0 | 2.4 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 0.3 | GO:0005298 | proline:sodium symporter activity(GO:0005298) |
| 0.0 | 0.2 | GO:1902122 | chenodeoxycholic acid binding(GO:1902122) |
| 0.0 | 0.7 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) |
| 0.0 | 0.3 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.0 | 0.7 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.0 | 0.3 | GO:0086083 | cell adhesive protein binding involved in bundle of His cell-Purkinje myocyte communication(GO:0086083) |
| 0.0 | 0.5 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.0 | 0.2 | GO:0016404 | 15-hydroxyprostaglandin dehydrogenase (NAD+) activity(GO:0016404) |
| 0.0 | 0.1 | GO:0004938 | alpha2-adrenergic receptor activity(GO:0004938) |
| 0.0 | 0.3 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 0.0 | 0.6 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 1.1 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 1.0 | GO:0019198 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 0.5 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.0 | 1.1 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.0 | 0.7 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.0 | 0.9 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.0 | 0.3 | GO:0004024 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) |
| 0.0 | 0.1 | GO:0035241 | protein-arginine omega-N monomethyltransferase activity(GO:0035241) |
| 0.0 | 0.4 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.0 | 0.3 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.0 | 0.8 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.0 | 0.1 | GO:0004348 | glucosylceramidase activity(GO:0004348) |
| 0.0 | 0.1 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.0 | 0.5 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.0 | 1.4 | GO:0043425 | bHLH transcription factor binding(GO:0043425) |
| 0.0 | 0.6 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.1 | GO:0000994 | RNA polymerase III core binding(GO:0000994) |
| 0.0 | 0.8 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.2 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.0 | 0.2 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.6 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 0.2 | GO:1990239 | steroid hormone binding(GO:1990239) |
| 0.0 | 0.6 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.5 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.0 | 0.4 | GO:0019870 | potassium channel inhibitor activity(GO:0019870) |
| 0.0 | 1.0 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.0 | 0.1 | GO:0016427 | tRNA (cytosine) methyltransferase activity(GO:0016427) |
| 0.0 | 0.5 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.0 | 0.4 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.4 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.5 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.0 | 0.1 | GO:0043141 | ATP-dependent 5'-3' DNA helicase activity(GO:0043141) |
| 0.0 | 0.4 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.0 | 0.1 | GO:0050051 | alkane 1-monooxygenase activity(GO:0018685) leukotriene-B4 20-monooxygenase activity(GO:0050051) |
| 0.0 | 0.2 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 0.3 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.0 | 0.1 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.0 | 1.0 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 0.1 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.0 | 1.8 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.0 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.0 | 0.1 | GO:0009019 | tRNA (guanine-N1-)-methyltransferase activity(GO:0009019) |
| 0.0 | 0.4 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 0.3 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 0.2 | GO:0022820 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.0 | 1.3 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.0 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 1.3 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 1.6 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.0 | 1.2 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.7 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 1.6 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.0 | 1.8 | PID FGF PATHWAY | FGF signaling pathway |
| 0.0 | 1.4 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.0 | 0.7 | PID BMP PATHWAY | BMP receptor signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.1 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.1 | 0.9 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.1 | 1.0 | REACTOME INTEGRATION OF PROVIRUS | Genes involved in Integration of provirus |
| 0.1 | 2.0 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 1.3 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 0.2 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.0 | 1.5 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 2.2 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 1.4 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 1.2 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 0.7 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.8 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 1.1 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.0 | 0.5 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.0 | 0.5 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.0 | 0.6 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.3 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.0 | 1.6 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 0.7 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 0.3 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 0.6 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 1.3 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 0.7 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.2 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 0.3 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 0.3 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.0 | 0.7 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.6 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.4 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.5 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |