Project
Inflammatory response time course, HUVEC (Wada, 2009)
Navigation
Downloads
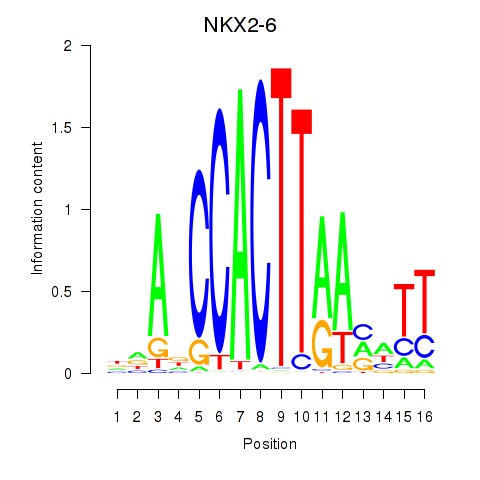
Results for NKX2-6
Z-value: 0.88
Transcription factors associated with NKX2-6
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NKX2-6
|
ENSG00000180053.8 | NKX2-6 |
Activity profile of NKX2-6 motif
Sorted Z-values of NKX2-6 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of NKX2-6
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_-_5978022 | 3.30 |
ENST00000525219.6
|
IL15RA
|
interleukin 15 receptor subunit alpha |
| chr10_-_5977589 | 3.25 |
ENST00000620345.4
ENST00000397251.7 ENST00000397248.6 ENST00000622442.4 ENST00000620865.4 |
IL15RA
|
interleukin 15 receptor subunit alpha |
| chr10_-_5977535 | 3.24 |
ENST00000379977.8
|
IL15RA
|
interleukin 15 receptor subunit alpha |
| chr3_-_123620496 | 2.96 |
ENST00000578202.1
|
MYLK
|
myosin light chain kinase |
| chr3_-_123620571 | 2.89 |
ENST00000583087.5
|
MYLK
|
myosin light chain kinase |
| chr10_-_5977492 | 2.31 |
ENST00000530685.5
ENST00000397255.7 ENST00000379971.5 ENST00000528354.5 ENST00000397250.6 ENST00000429135.2 |
IL15RA
|
interleukin 15 receptor subunit alpha |
| chr12_+_55681711 | 2.26 |
ENST00000394252.4
|
METTL7B
|
methyltransferase like 7B |
| chr12_+_55681647 | 2.22 |
ENST00000614691.1
|
METTL7B
|
methyltransferase like 7B |
| chr2_+_102311502 | 2.20 |
ENST00000404917.6
ENST00000410040.5 |
IL1RL1
IL18R1
|
interleukin 1 receptor like 1 interleukin 18 receptor 1 |
| chr1_+_205256189 | 2.05 |
ENST00000329800.7
|
TMCC2
|
transmembrane and coiled-coil domain family 2 |
| chr15_+_67067780 | 2.01 |
ENST00000679624.1
|
SMAD3
|
SMAD family member 3 |
| chr1_-_120051714 | 1.80 |
ENST00000579475.7
|
NOTCH2
|
notch receptor 2 |
| chr2_-_100104530 | 1.67 |
ENST00000432037.5
ENST00000673232.1 ENST00000423966.6 ENST00000409236.6 |
AFF3
|
AF4/FMR2 family member 3 |
| chr9_+_94084458 | 1.46 |
ENST00000620992.5
ENST00000288976.3 |
PTPDC1
|
protein tyrosine phosphatase domain containing 1 |
| chr3_-_121660892 | 1.38 |
ENST00000428394.6
ENST00000314583.8 |
HCLS1
|
hematopoietic cell-specific Lyn substrate 1 |
| chr8_+_27771942 | 1.30 |
ENST00000523566.5
|
ESCO2
|
establishment of sister chromatid cohesion N-acetyltransferase 2 |
| chr12_-_89526011 | 1.16 |
ENST00000313546.8
|
POC1B
|
POC1 centriolar protein B |
| chr12_-_89526164 | 1.10 |
ENST00000548729.5
|
POC1B-GALNT4
|
POC1B-GALNT4 readthrough |
| chr15_-_55270874 | 1.09 |
ENST00000567380.5
ENST00000565972.5 ENST00000569493.5 |
RAB27A
|
RAB27A, member RAS oncogene family |
| chr12_-_89526253 | 1.07 |
ENST00000547474.1
|
POC1B-GALNT4
|
POC1B-GALNT4 readthrough |
| chr19_-_43670153 | 1.04 |
ENST00000601723.5
ENST00000339082.7 ENST00000340093.8 |
PLAUR
|
plasminogen activator, urokinase receptor |
| chr12_-_70788914 | 1.04 |
ENST00000342084.8
|
PTPRR
|
protein tyrosine phosphatase receptor type R |
| chr16_+_78022538 | 0.99 |
ENST00000651443.1
ENST00000299642.10 |
CLEC3A
|
C-type lectin domain family 3 member A |
| chr5_+_52787899 | 0.90 |
ENST00000274311.3
ENST00000282588.7 |
PELO
ITGA1
|
pelota mRNA surveillance and ribosome rescue factor integrin subunit alpha 1 |
| chr9_-_83063159 | 0.90 |
ENST00000340717.4
|
RASEF
|
RAS and EF-hand domain containing |
| chr3_+_171843337 | 0.87 |
ENST00000334567.9
ENST00000619900.4 ENST00000450693.1 |
TMEM212
|
transmembrane protein 212 |
| chr2_+_69915100 | 0.86 |
ENST00000264444.7
|
MXD1
|
MAX dimerization protein 1 |
| chr3_+_100709424 | 0.84 |
ENST00000675246.1
ENST00000675692.1 ENST00000675890.1 ENST00000675586.1 ENST00000674645.1 ENST00000675958.1 ENST00000479672.6 ENST00000675420.1 ENST00000674615.1 ENST00000676431.1 ENST00000674758.1 |
TFG
|
trafficking from ER to golgi regulator |
| chr3_+_100709382 | 0.80 |
ENST00000620299.5
|
TFG
|
trafficking from ER to golgi regulator |
| chr7_+_75915148 | 0.76 |
ENST00000461988.6
ENST00000421059.1 ENST00000394893.5 ENST00000412521.5 ENST00000414186.5 |
POR
|
cytochrome p450 oxidoreductase |
| chr3_+_100709344 | 0.76 |
ENST00000418917.7
ENST00000675553.1 |
TFG
|
trafficking from ER to golgi regulator |
| chr1_+_19251786 | 0.75 |
ENST00000330263.5
|
MRTO4
|
MRT4 homolog, ribosome maturation factor |
| chr2_+_69915041 | 0.75 |
ENST00000540449.5
|
MXD1
|
MAX dimerization protein 1 |
| chr3_+_100709473 | 0.75 |
ENST00000240851.9
ENST00000675243.1 ENST00000676111.1 ENST00000476228.5 ENST00000463568.6 ENST00000676395.1 |
TFG
|
trafficking from ER to golgi regulator |
| chr8_+_49911801 | 0.74 |
ENST00000643809.1
|
SNTG1
|
syntrophin gamma 1 |
| chr14_+_23185316 | 0.73 |
ENST00000399910.5
ENST00000492621.5 |
RNF212B
|
ring finger protein 212B |
| chr14_-_106875069 | 0.72 |
ENST00000390639.2
|
IGHV7-81
|
immunoglobulin heavy variable 7-81 (non-functional) |
| chr4_-_78939352 | 0.70 |
ENST00000512733.5
|
PAQR3
|
progestin and adipoQ receptor family member 3 |
| chr1_+_156369202 | 0.70 |
ENST00000537040.6
|
RHBG
|
Rh family B glycoprotein |
| chr3_+_100709290 | 0.69 |
ENST00000675047.1
ENST00000490574.6 ENST00000675543.1 ENST00000676308.1 ENST00000675499.1 |
TFG
|
trafficking from ER to golgi regulator |
| chrY_-_23694579 | 0.65 |
ENST00000343584.10
|
PRYP3
|
PTPN13 like Y-linked pseudogene 3 |
| chr10_+_119892692 | 0.64 |
ENST00000369075.8
|
SEC23IP
|
SEC23 interacting protein |
| chr12_-_55842927 | 0.63 |
ENST00000322569.9
ENST00000409200.7 |
MMP19
|
matrix metallopeptidase 19 |
| chr19_+_51573171 | 0.62 |
ENST00000436511.2
|
ZNF175
|
zinc finger protein 175 |
| chr13_+_43879895 | 0.62 |
ENST00000325686.7
|
LACC1
|
laccase domain containing 1 |
| chr14_+_64715677 | 0.61 |
ENST00000634379.2
|
PLEKHG3
|
pleckstrin homology and RhoGEF domain containing G3 |
| chr11_+_111255982 | 0.58 |
ENST00000637637.1
|
C11orf53
|
chromosome 11 open reading frame 53 |
| chr7_+_123601815 | 0.58 |
ENST00000451215.6
|
ASB15
|
ankyrin repeat and SOCS box containing 15 |
| chr9_+_72114595 | 0.57 |
ENST00000545168.5
|
GDA
|
guanine deaminase |
| chr7_+_123601836 | 0.56 |
ENST00000434204.5
|
ASB15
|
ankyrin repeat and SOCS box containing 15 |
| chr6_+_125781108 | 0.55 |
ENST00000368357.7
|
NCOA7
|
nuclear receptor coactivator 7 |
| chr7_+_142791635 | 0.55 |
ENST00000633705.1
|
TRBC1
|
T cell receptor beta constant 1 |
| chr19_-_50476725 | 0.55 |
ENST00000595790.5
|
FAM71E1
|
family with sequence similarity 71 member E1 |
| chr6_+_63563448 | 0.53 |
ENST00000673199.1
|
PTP4A1
|
protein tyrosine phosphatase 4A1 |
| chr19_-_50476838 | 0.50 |
ENST00000600100.6
|
FAM71E1
|
family with sequence similarity 71 member E1 |
| chr3_-_48089203 | 0.50 |
ENST00000468075.2
ENST00000360240.10 |
MAP4
|
microtubule associated protein 4 |
| chr4_+_69280472 | 0.50 |
ENST00000335568.10
ENST00000511240.1 |
UGT2B28
|
UDP glucuronosyltransferase family 2 member B28 |
| chr4_-_48080172 | 0.49 |
ENST00000507351.1
|
TXK
|
TXK tyrosine kinase |
| chrX_-_133097095 | 0.48 |
ENST00000511190.5
|
USP26
|
ubiquitin specific peptidase 26 |
| chr15_-_40828699 | 0.45 |
ENST00000299174.10
ENST00000427255.2 |
PPP1R14D
|
protein phosphatase 1 regulatory inhibitor subunit 14D |
| chr11_-_64759967 | 0.44 |
ENST00000377432.7
ENST00000164139.4 |
PYGM
|
glycogen phosphorylase, muscle associated |
| chrX_+_30243715 | 0.44 |
ENST00000378981.8
ENST00000397550.6 |
MAGEB1
|
MAGE family member B1 |
| chr22_-_23754376 | 0.44 |
ENST00000398465.3
ENST00000248948.4 |
VPREB3
|
V-set pre-B cell surrogate light chain 3 |
| chr18_+_58362467 | 0.44 |
ENST00000675101.1
|
NEDD4L
|
NEDD4 like E3 ubiquitin protein ligase |
| chr11_+_65860394 | 0.44 |
ENST00000308110.9
|
MUS81
|
MUS81 structure-specific endonuclease subunit |
| chr12_-_110583305 | 0.43 |
ENST00000354300.5
|
PPTC7
|
protein phosphatase targeting COQ7 |
| chr2_-_178702479 | 0.43 |
ENST00000414766.5
|
TTN
|
titin |
| chr17_+_35587478 | 0.43 |
ENST00000618940.4
|
AP2B1
|
adaptor related protein complex 2 subunit beta 1 |
| chr1_+_16440700 | 0.43 |
ENST00000504551.6
ENST00000457722.6 ENST00000337132.10 ENST00000443980.6 |
NECAP2
|
NECAP endocytosis associated 2 |
| chr2_-_162243375 | 0.42 |
ENST00000188790.9
ENST00000443424.5 |
FAP
|
fibroblast activation protein alpha |
| chr1_+_158289916 | 0.40 |
ENST00000368170.8
|
CD1C
|
CD1c molecule |
| chr4_-_170003738 | 0.39 |
ENST00000502832.1
ENST00000393704.3 |
MFAP3L
|
microfibril associated protein 3 like |
| chr3_+_122077776 | 0.38 |
ENST00000264468.9
|
CD86
|
CD86 molecule |
| chr4_-_155953851 | 0.38 |
ENST00000679136.1
ENST00000433477.4 ENST00000679996.1 ENST00000680741.1 ENST00000680553.1 ENST00000679942.1 |
CTSO
|
cathepsin O |
| chr11_-_66593031 | 0.38 |
ENST00000333861.5
|
CCDC87
|
coiled-coil domain containing 87 |
| chr1_+_171512032 | 0.38 |
ENST00000426496.6
|
PRRC2C
|
proline rich coiled-coil 2C |
| chr12_-_49900250 | 0.38 |
ENST00000552669.5
|
FAIM2
|
Fas apoptotic inhibitory molecule 2 |
| chr19_+_496454 | 0.37 |
ENST00000346144.8
ENST00000215637.8 ENST00000382683.8 |
MADCAM1
|
mucosal vascular addressin cell adhesion molecule 1 |
| chr17_-_36534841 | 0.37 |
ENST00000614623.5
ENST00000621344.4 |
MYO19
|
myosin XIX |
| chr19_-_51108365 | 0.36 |
ENST00000421832.3
|
CTU1
|
cytosolic thiouridylase subunit 1 |
| chr1_+_202462730 | 0.36 |
ENST00000290419.9
ENST00000491336.5 |
PPP1R12B
|
protein phosphatase 1 regulatory subunit 12B |
| chr2_-_174598206 | 0.36 |
ENST00000392546.6
ENST00000436221.1 |
WIPF1
|
WAS/WASL interacting protein family member 1 |
| chr22_-_50085331 | 0.35 |
ENST00000395876.6
|
MLC1
|
modulator of VRAC current 1 |
| chr17_+_4932248 | 0.35 |
ENST00000329125.6
|
GP1BA
|
glycoprotein Ib platelet subunit alpha |
| chr7_+_141764097 | 0.35 |
ENST00000247879.2
|
TAS2R3
|
taste 2 receptor member 3 |
| chr17_-_42964437 | 0.35 |
ENST00000427569.7
ENST00000430739.5 |
AARSD1
|
alanyl-tRNA synthetase domain containing 1 |
| chr22_+_29205877 | 0.34 |
ENST00000334018.11
ENST00000429226.5 ENST00000404755.7 ENST00000404820.7 ENST00000430127.1 |
EMID1
|
EMI domain containing 1 |
| chr17_+_4932285 | 0.33 |
ENST00000611961.1
|
GP1BA
|
glycoprotein Ib platelet subunit alpha |
| chr6_-_89315291 | 0.33 |
ENST00000402938.4
|
GABRR2
|
gamma-aminobutyric acid type A receptor subunit rho2 |
| chr11_-_61920267 | 0.33 |
ENST00000531922.2
ENST00000301773.9 |
RAB3IL1
|
RAB3A interacting protein like 1 |
| chr3_+_122055355 | 0.32 |
ENST00000330540.7
ENST00000469710.5 ENST00000493101.5 |
CD86
|
CD86 molecule |
| chr7_+_111091006 | 0.32 |
ENST00000451085.5
ENST00000422987.3 ENST00000421101.1 |
LRRN3
|
leucine rich repeat neuronal 3 |
| chr2_-_68952880 | 0.32 |
ENST00000481498.1
ENST00000328895.9 |
GKN2
|
gastrokine 2 |
| chr1_-_173824322 | 0.31 |
ENST00000356198.6
|
CENPL
|
centromere protein L |
| chr5_-_140557414 | 0.31 |
ENST00000336283.9
|
SRA1
|
steroid receptor RNA activator 1 |
| chr22_-_50085414 | 0.31 |
ENST00000311597.10
|
MLC1
|
modulator of VRAC current 1 |
| chr7_+_111091119 | 0.31 |
ENST00000308478.10
|
LRRN3
|
leucine rich repeat neuronal 3 |
| chr3_+_186996444 | 0.31 |
ENST00000676633.1
|
ST6GAL1
|
ST6 beta-galactoside alpha-2,6-sialyltransferase 1 |
| chr4_+_168497044 | 0.30 |
ENST00000505667.6
|
PALLD
|
palladin, cytoskeletal associated protein |
| chr4_+_165378998 | 0.30 |
ENST00000402744.9
|
CPE
|
carboxypeptidase E |
| chr11_+_65040895 | 0.29 |
ENST00000531072.1
ENST00000398846.6 |
SAC3D1
|
SAC3 domain containing 1 |
| chr20_-_1393045 | 0.29 |
ENST00000400137.9
ENST00000381715.4 |
FKBP1A
|
FKBP prolyl isomerase 1A |
| chr7_+_142800957 | 0.29 |
ENST00000466254.1
|
TRBC2
|
T cell receptor beta constant 2 |
| chr6_+_29396555 | 0.29 |
ENST00000623183.1
|
OR12D2
|
olfactory receptor family 12 subfamily D member 2 |
| chr10_+_18340821 | 0.29 |
ENST00000612743.1
ENST00000612134.4 |
CACNB2
|
calcium voltage-gated channel auxiliary subunit beta 2 |
| chr11_+_63369779 | 0.28 |
ENST00000279178.4
|
SLC22A9
|
solute carrier family 22 member 9 |
| chr11_-_106022209 | 0.28 |
ENST00000301919.9
ENST00000534458.1 ENST00000530108.1 ENST00000530788.1 |
MSANTD4
|
Myb/SANT DNA binding domain containing 4 with coiled-coils |
| chr4_+_168497066 | 0.28 |
ENST00000261509.10
|
PALLD
|
palladin, cytoskeletal associated protein |
| chr12_+_119593758 | 0.27 |
ENST00000426426.3
|
TMEM233
|
transmembrane protein 233 |
| chr12_+_69825221 | 0.27 |
ENST00000552032.7
|
MYRFL
|
myelin regulatory factor like |
| chr5_+_140691427 | 0.27 |
ENST00000643996.1
ENST00000509299.6 ENST00000645065.1 ENST00000642752.1 ENST00000503873.6 ENST00000642970.1 ENST00000230771.9 ENST00000646468.1 ENST00000645749.1 |
HARS2
|
histidyl-tRNA synthetase 2, mitochondrial |
| chr12_+_69825273 | 0.26 |
ENST00000547771.6
|
MYRFL
|
myelin regulatory factor like |
| chr7_+_31687208 | 0.26 |
ENST00000409146.3
ENST00000342032.8 |
PPP1R17
|
protein phosphatase 1 regulatory subunit 17 |
| chrX_+_47193796 | 0.26 |
ENST00000442035.5
ENST00000335972.11 ENST00000457753.5 |
UBA1
|
ubiquitin like modifier activating enzyme 1 |
| chr19_+_55600277 | 0.26 |
ENST00000301073.4
|
ZNF524
|
zinc finger protein 524 |
| chr2_-_178651500 | 0.26 |
ENST00000446966.1
ENST00000426232.5 |
TTN
|
titin |
| chr4_+_119027335 | 0.26 |
ENST00000627783.2
|
SYNPO2
|
synaptopodin 2 |
| chr8_+_49911604 | 0.26 |
ENST00000642164.1
ENST00000644093.1 ENST00000643999.1 ENST00000647073.1 ENST00000646880.1 |
SNTG1
|
syntrophin gamma 1 |
| chr7_+_121873152 | 0.25 |
ENST00000650826.1
ENST00000650728.1 ENST00000393386.7 ENST00000651390.1 ENST00000651842.1 ENST00000650681.1 |
PTPRZ1
|
protein tyrosine phosphatase receptor type Z1 |
| chr19_+_51571249 | 0.25 |
ENST00000262259.7
ENST00000545217.5 ENST00000596504.1 |
ZNF175
|
zinc finger protein 175 |
| chr15_+_75347431 | 0.25 |
ENST00000567657.5
|
NEIL1
|
nei like DNA glycosylase 1 |
| chr1_-_173824107 | 0.25 |
ENST00000345664.10
ENST00000367710.7 |
CENPL
|
centromere protein L |
| chr1_-_28058087 | 0.24 |
ENST00000373864.5
|
EYA3
|
EYA transcriptional coactivator and phosphatase 3 |
| chr7_-_91880672 | 0.24 |
ENST00000456229.1
ENST00000442961.1 ENST00000406735.6 ENST00000419292.1 ENST00000351870.8 |
MTERF1
|
mitochondrial transcription termination factor 1 |
| chr1_+_158254414 | 0.24 |
ENST00000289429.6
|
CD1A
|
CD1a molecule |
| chr3_+_122077850 | 0.24 |
ENST00000482356.5
ENST00000393627.6 |
CD86
|
CD86 molecule |
| chr6_+_31571957 | 0.24 |
ENST00000454783.5
|
LTA
|
lymphotoxin alpha |
| chr6_+_32439866 | 0.24 |
ENST00000374982.5
ENST00000395388.7 |
HLA-DRA
|
major histocompatibility complex, class II, DR alpha |
| chr1_-_247536440 | 0.23 |
ENST00000366487.4
ENST00000641802.1 |
OR2C3
|
olfactory receptor family 2 subfamily C member 3 |
| chr3_+_114294020 | 0.23 |
ENST00000383671.8
|
TIGIT
|
T cell immunoreceptor with Ig and ITIM domains |
| chr10_+_32446102 | 0.23 |
ENST00000277657.12
ENST00000362006.11 |
CCDC7
|
coiled-coil domain containing 7 |
| chr22_-_43143339 | 0.23 |
ENST00000327555.5
ENST00000290429.11 |
MCAT
|
malonyl-CoA-acyl carrier protein transacylase |
| chr1_+_171248471 | 0.23 |
ENST00000402921.6
ENST00000617670.6 ENST00000367750.7 |
FMO1
|
flavin containing dimethylaniline monoxygenase 1 |
| chr17_-_76053639 | 0.23 |
ENST00000602720.5
|
SRP68
|
signal recognition particle 68 |
| chr17_-_36534883 | 0.22 |
ENST00000620640.4
|
MYO19
|
myosin XIX |
| chr1_-_200669708 | 0.22 |
ENST00000436897.1
ENST00000447706.6 ENST00000331314.11 |
DDX59
|
DEAD-box helicase 59 |
| chr1_-_173824856 | 0.22 |
ENST00000682279.1
|
CENPL
|
centromere protein L |
| chr7_+_150801695 | 0.22 |
ENST00000475536.5
ENST00000468689.2 |
TMEM176A
|
transmembrane protein 176A |
| chr12_-_49897056 | 0.22 |
ENST00000552863.5
|
FAIM2
|
Fas apoptotic inhibitory molecule 2 |
| chr6_+_10528326 | 0.22 |
ENST00000379597.7
|
GCNT2
|
glucosaminyl (N-acetyl) transferase 2 (I blood group) |
| chr9_+_122371036 | 0.22 |
ENST00000619306.5
ENST00000426608.6 ENST00000223423.8 |
PTGS1
|
prostaglandin-endoperoxide synthase 1 |
| chr3_-_39192584 | 0.22 |
ENST00000340369.4
ENST00000421646.1 ENST00000396251.1 |
XIRP1
|
xin actin binding repeat containing 1 |
| chr6_-_166587594 | 0.22 |
ENST00000444122.2
|
RAMACL
|
RNA guanine-7 methyltransferase activating subunit like (pseudogene) |
| chr12_-_56645955 | 0.21 |
ENST00000552959.5
ENST00000551020.5 ENST00000553007.2 ENST00000262030.8 |
ATP5F1B
|
ATP synthase F1 subunit beta |
| chr17_-_36534927 | 0.21 |
ENST00000610930.4
|
MYO19
|
myosin XIX |
| chr12_+_4720395 | 0.21 |
ENST00000252318.7
|
GALNT8
|
polypeptide N-acetylgalactosaminyltransferase 8 |
| chr5_+_140691591 | 0.20 |
ENST00000508522.5
ENST00000448069.2 |
HARS2
|
histidyl-tRNA synthetase 2, mitochondrial |
| chr4_+_90127555 | 0.20 |
ENST00000509176.5
|
CCSER1
|
coiled-coil serine rich protein 1 |
| chr11_-_65856944 | 0.20 |
ENST00000524553.5
|
CFL1
|
cofilin 1 |
| chr2_-_174597728 | 0.19 |
ENST00000409891.5
ENST00000410117.5 |
WIPF1
|
WAS/WASL interacting protein family member 1 |
| chr3_-_39154558 | 0.19 |
ENST00000514182.1
|
CSRNP1
|
cysteine and serine rich nuclear protein 1 |
| chr1_-_77979054 | 0.19 |
ENST00000370768.7
ENST00000370767.5 ENST00000421641.1 |
FUBP1
|
far upstream element binding protein 1 |
| chr11_-_65857007 | 0.19 |
ENST00000527344.5
|
CFL1
|
cofilin 1 |
| chr8_+_55102012 | 0.18 |
ENST00000327381.7
|
XKR4
|
XK related 4 |
| chr7_+_121873089 | 0.18 |
ENST00000651065.1
|
PTPRZ1
|
protein tyrosine phosphatase receptor type Z1 |
| chr6_-_34696733 | 0.18 |
ENST00000374023.8
|
ILRUN
|
inflammation and lipid regulator with UBA-like and NBR1-like domains |
| chr17_-_28576882 | 0.18 |
ENST00000395319.7
ENST00000581807.5 ENST00000226253.9 ENST00000584086.5 ENST00000395321.6 |
ALDOC
|
aldolase, fructose-bisphosphate C |
| chr6_-_34696839 | 0.17 |
ENST00000374026.7
|
ILRUN
|
inflammation and lipid regulator with UBA-like and NBR1-like domains |
| chr1_+_109620578 | 0.17 |
ENST00000531203.6
ENST00000256578.8 |
AMPD2
|
adenosine monophosphate deaminase 2 |
| chr2_+_17541157 | 0.17 |
ENST00000406397.1
|
VSNL1
|
visinin like 1 |
| chr2_+_70978380 | 0.17 |
ENST00000272421.10
ENST00000441349.5 ENST00000457410.5 |
ANKRD53
|
ankyrin repeat domain 53 |
| chr14_-_63318933 | 0.17 |
ENST00000621500.2
|
GPHB5
|
glycoprotein hormone subunit beta 5 |
| chr16_+_715092 | 0.16 |
ENST00000568223.7
|
METRN
|
meteorin, glial cell differentiation regulator |
| chr20_-_1393074 | 0.16 |
ENST00000614856.2
ENST00000678408.1 ENST00000618612.5 ENST00000439640.5 ENST00000381719.8 ENST00000677533.1 |
FKBP1A
|
FKBP prolyl isomerase 1A |
| chr19_-_42242526 | 0.16 |
ENST00000222330.8
ENST00000676537.1 |
GSK3A
|
glycogen synthase kinase 3 alpha |
| chr1_-_107688492 | 0.15 |
ENST00000415432.6
|
VAV3
|
vav guanine nucleotide exchange factor 3 |
| chr1_-_153312919 | 0.15 |
ENST00000683862.1
|
PGLYRP3
|
peptidoglycan recognition protein 3 |
| chr1_+_169367934 | 0.15 |
ENST00000367807.7
ENST00000329281.6 ENST00000420531.1 |
BLZF1
|
basic leucine zipper nuclear factor 1 |
| chr7_+_23680130 | 0.15 |
ENST00000409192.7
ENST00000409653.5 ENST00000409994.3 ENST00000344962.9 |
FAM221A
|
family with sequence similarity 221 member A |
| chr12_-_262828 | 0.15 |
ENST00000343164.9
ENST00000436453.1 ENST00000445055.6 ENST00000546319.5 |
SLC6A13
|
solute carrier family 6 member 13 |
| chr15_+_81182579 | 0.14 |
ENST00000302987.9
|
IL16
|
interleukin 16 |
| chr2_+_112911159 | 0.14 |
ENST00000263326.8
|
IL37
|
interleukin 37 |
| chr7_+_16661182 | 0.14 |
ENST00000446596.5
ENST00000452975.6 ENST00000438834.5 |
BZW2
|
basic leucine zipper and W2 domains 2 |
| chr11_-_111379268 | 0.13 |
ENST00000393067.8
|
POU2AF1
|
POU class 2 homeobox associating factor 1 |
| chr6_-_39322540 | 0.13 |
ENST00000425054.6
ENST00000373227.8 ENST00000373229.9 |
KCNK16
|
potassium two pore domain channel subfamily K member 16 |
| chr7_+_100602344 | 0.13 |
ENST00000223061.6
|
PCOLCE
|
procollagen C-endopeptidase enhancer |
| chr12_+_54280842 | 0.13 |
ENST00000678077.1
ENST00000548688.5 |
HNRNPA1
|
heterogeneous nuclear ribonucleoprotein A1 |
| chr17_+_62423891 | 0.13 |
ENST00000616852.1
ENST00000311506.10 |
METTL2A
|
methyltransferase like 2A |
| chr15_-_28099293 | 0.13 |
ENST00000431101.1
ENST00000445578.5 ENST00000353809.9 ENST00000354638.8 |
OCA2
|
OCA2 melanosomal transmembrane protein |
| chr2_-_174597795 | 0.12 |
ENST00000679041.1
|
WIPF1
|
WAS/WASL interacting protein family member 1 |
| chr19_+_10452891 | 0.12 |
ENST00000344979.7
|
PDE4A
|
phosphodiesterase 4A |
| chr11_+_1870252 | 0.12 |
ENST00000612798.4
|
LSP1
|
lymphocyte specific protein 1 |
| chr10_-_27240505 | 0.12 |
ENST00000375888.5
ENST00000676732.1 |
ACBD5
|
acyl-CoA binding domain containing 5 |
| chr11_-_28108109 | 0.12 |
ENST00000263181.7
|
KIF18A
|
kinesin family member 18A |
| chr7_+_128476748 | 0.11 |
ENST00000262432.13
ENST00000480046.5 |
METTL2B
|
methyltransferase like 2B |
| chr1_+_173824626 | 0.11 |
ENST00000648960.1
ENST00000648807.1 ENST00000649067.1 ENST00000649689.2 |
DARS2
|
aspartyl-tRNA synthetase 2, mitochondrial |
| chr4_-_46124046 | 0.11 |
ENST00000295452.5
|
GABRG1
|
gamma-aminobutyric acid type A receptor subunit gamma1 |
| chr1_+_203128279 | 0.11 |
ENST00000367235.1
ENST00000618295.1 |
ADORA1
|
adenosine A1 receptor |
| chr3_-_46208304 | 0.11 |
ENST00000296140.4
|
CCR1
|
C-C motif chemokine receptor 1 |
| chr8_-_100950549 | 0.11 |
ENST00000395951.7
|
YWHAZ
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein zeta |
| chr9_-_101738640 | 0.10 |
ENST00000361820.6
|
GRIN3A
|
glutamate ionotropic receptor NMDA type subunit 3A |
| chr9_+_4839761 | 0.10 |
ENST00000448872.6
ENST00000441844.2 |
RCL1
|
RNA terminal phosphate cyclase like 1 |
| chr1_+_20186076 | 0.10 |
ENST00000375099.4
|
UBXN10
|
UBX domain protein 10 |
| chr5_-_140691312 | 0.10 |
ENST00000438307.6
ENST00000415192.6 ENST00000509087.2 ENST00000675698.1 ENST00000504156.7 ENST00000457527.6 ENST00000307633.7 ENST00000676327.1 ENST00000675851.1 ENST00000675366.1 ENST00000675204.1 ENST00000507746.7 ENST00000431330.7 |
HARS1
|
histidyl-tRNA synthetase 1 |
| chr22_+_41621977 | 0.09 |
ENST00000405506.2
|
XRCC6
|
X-ray repair cross complementing 6 |
| chr1_+_173824694 | 0.09 |
ENST00000647645.1
|
DARS2
|
aspartyl-tRNA synthetase 2, mitochondrial |
| chr6_+_28267107 | 0.09 |
ENST00000621053.1
ENST00000617168.4 ENST00000421553.7 ENST00000611552.2 ENST00000623276.3 |
ENSG00000276302.1
ZSCAN26
|
novel protein zinc finger and SCAN domain containing 26 |
| chr4_+_94489030 | 0.09 |
ENST00000510099.5
|
PDLIM5
|
PDZ and LIM domain 5 |
| chr3_-_48595267 | 0.09 |
ENST00000328333.12
ENST00000681320.1 |
COL7A1
|
collagen type VII alpha 1 chain |
| chr22_+_22395005 | 0.09 |
ENST00000390298.2
|
IGLV7-43
|
immunoglobulin lambda variable 7-43 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 5.9 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.3 | 1.0 | GO:0038195 | urokinase plasminogen activator signaling pathway(GO:0038195) |
| 0.3 | 2.0 | GO:0061767 | negative regulation of lung blood pressure(GO:0061767) |
| 0.3 | 0.9 | GO:0002665 | negative regulation of T cell tolerance induction(GO:0002665) negative regulation of T cell anergy(GO:0002668) negative regulation of lymphocyte anergy(GO:0002912) regulation of lymphotoxin A production(GO:0032681) positive regulation of lymphotoxin A production(GO:0032761) regulation of lymphotoxin A biosynthetic process(GO:0043016) positive regulation of lymphotoxin A biosynthetic process(GO:0043017) |
| 0.3 | 2.0 | GO:0038172 | interleukin-33-mediated signaling pathway(GO:0038172) |
| 0.3 | 1.3 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.3 | 0.8 | GO:0090031 | regulation of growth plate cartilage chondrocyte proliferation(GO:0003420) positive regulation of steroid hormone biosynthetic process(GO:0090031) |
| 0.2 | 0.7 | GO:0070634 | transepithelial ammonium transport(GO:0070634) |
| 0.2 | 0.6 | GO:0006147 | guanine catabolic process(GO:0006147) |
| 0.2 | 1.1 | GO:1903435 | positive regulation of constitutive secretory pathway(GO:1903435) |
| 0.2 | 0.9 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.2 | 1.0 | GO:1903385 | regulation of homophilic cell adhesion(GO:1903385) |
| 0.1 | 0.4 | GO:0072429 | response to intra-S DNA damage checkpoint signaling(GO:0072429) |
| 0.1 | 0.7 | GO:0030241 | skeletal muscle myosin thick filament assembly(GO:0030241) |
| 0.1 | 0.4 | GO:0097325 | melanocyte proliferation(GO:0097325) |
| 0.1 | 1.8 | GO:0061314 | Notch signaling involved in heart development(GO:0061314) |
| 0.1 | 1.4 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.1 | 0.5 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.1 | 0.5 | GO:0060335 | positive regulation of response to interferon-gamma(GO:0060332) positive regulation of interferon-gamma-mediated signaling pathway(GO:0060335) |
| 0.1 | 0.2 | GO:0006393 | termination of mitochondrial transcription(GO:0006393) |
| 0.1 | 0.2 | GO:0002876 | positive regulation of chronic inflammatory response to antigenic stimulus(GO:0002876) |
| 0.1 | 0.2 | GO:0002503 | peptide antigen assembly with MHC class II protein complex(GO:0002503) |
| 0.1 | 0.4 | GO:0099590 | neurotransmitter receptor internalization(GO:0099590) |
| 0.1 | 0.2 | GO:0006421 | asparaginyl-tRNA aminoacylation(GO:0006421) |
| 0.1 | 0.6 | GO:0001554 | luteolysis(GO:0001554) |
| 0.1 | 0.4 | GO:0034227 | tRNA thio-modification(GO:0034227) |
| 0.1 | 0.6 | GO:0048007 | antigen processing and presentation of lipid antigen via MHC class Ib(GO:0048003) antigen processing and presentation, exogenous lipid antigen via MHC class Ib(GO:0048007) |
| 0.1 | 0.3 | GO:0030070 | insulin processing(GO:0030070) |
| 0.1 | 0.2 | GO:0032826 | natural killer cell differentiation involved in immune response(GO:0002325) negative regulation of natural killer cell differentiation(GO:0032824) regulation of natural killer cell differentiation involved in immune response(GO:0032826) negative regulation of natural killer cell differentiation involved in immune response(GO:0032827) positive regulation of cytolysis in other organism(GO:0051714) |
| 0.0 | 0.1 | GO:0070256 | negative regulation of circadian sleep/wake cycle, non-REM sleep(GO:0042323) negative regulation of mucus secretion(GO:0070256) |
| 0.0 | 4.6 | GO:0048208 | vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) |
| 0.0 | 0.1 | GO:0006726 | eye pigment biosynthetic process(GO:0006726) eye pigment metabolic process(GO:0042441) pigment metabolic process involved in developmental pigmentation(GO:0043324) pigment metabolic process involved in pigmentation(GO:0043474) |
| 0.0 | 0.7 | GO:0047484 | regulation of response to osmotic stress(GO:0047484) |
| 0.0 | 0.4 | GO:1904879 | positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1904879) |
| 0.0 | 0.2 | GO:2000077 | UDP-glucose catabolic process(GO:0006258) negative regulation of type B pancreatic cell development(GO:2000077) |
| 0.0 | 0.4 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.0 | 0.2 | GO:2000784 | regulation of unidimensional cell growth(GO:0051510) negative regulation of unidimensional cell growth(GO:0051511) establishment of cell polarity regulating cell shape(GO:0071964) regulation of establishment or maintenance of cell polarity regulating cell shape(GO:2000769) positive regulation of establishment or maintenance of cell polarity regulating cell shape(GO:2000771) regulation of establishment of cell polarity regulating cell shape(GO:2000782) positive regulation of establishment of cell polarity regulating cell shape(GO:2000784) positive regulation of barbed-end actin filament capping(GO:2000814) |
| 0.0 | 0.4 | GO:0022417 | protein maturation by protein folding(GO:0022417) |
| 0.0 | 0.1 | GO:0002314 | germinal center B cell differentiation(GO:0002314) |
| 0.0 | 0.2 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.0 | 0.6 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.0 | 0.7 | GO:0070493 | thrombin receptor signaling pathway(GO:0070493) |
| 0.0 | 0.2 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.0 | 0.4 | GO:0070445 | oligodendrocyte progenitor proliferation(GO:0070444) regulation of oligodendrocyte progenitor proliferation(GO:0070445) |
| 0.0 | 0.4 | GO:0021702 | cerebellar Purkinje cell layer formation(GO:0021694) cerebellar Purkinje cell differentiation(GO:0021702) |
| 0.0 | 0.6 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.0 | 1.6 | GO:0035116 | embryonic hindlimb morphogenesis(GO:0035116) |
| 0.0 | 0.3 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.0 | 0.3 | GO:0015747 | urate transport(GO:0015747) |
| 0.0 | 1.8 | GO:0042982 | amyloid precursor protein metabolic process(GO:0042982) |
| 0.0 | 1.6 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.0 | 0.8 | GO:0006418 | tRNA aminoacylation for protein translation(GO:0006418) |
| 0.0 | 0.8 | GO:0061641 | CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.0 | 0.2 | GO:1902412 | regulation of mitotic cytokinesis(GO:1902412) |
| 0.0 | 0.8 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.0 | 0.8 | GO:0090140 | regulation of mitochondrial fission(GO:0090140) |
| 0.0 | 0.2 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.0 | 0.3 | GO:0006054 | N-acetylneuraminate metabolic process(GO:0006054) |
| 0.0 | 0.1 | GO:1903936 | cellular response to sodium arsenite(GO:1903936) |
| 0.0 | 0.2 | GO:2001199 | negative regulation of dendritic cell differentiation(GO:2001199) |
| 0.0 | 0.2 | GO:0002155 | thyroid hormone mediated signaling pathway(GO:0002154) regulation of thyroid hormone mediated signaling pathway(GO:0002155) |
| 0.0 | 0.7 | GO:0034067 | protein localization to Golgi apparatus(GO:0034067) |
| 0.0 | 0.1 | GO:1903251 | multi-ciliated epithelial cell differentiation(GO:1903251) |
| 0.0 | 0.1 | GO:0097680 | cellular hyperosmotic salinity response(GO:0071475) double-strand break repair via classical nonhomologous end joining(GO:0097680) |
| 0.0 | 0.1 | GO:0000480 | endonucleolytic cleavage in 5'-ETS of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000480) |
| 0.0 | 1.2 | GO:0001895 | retina homeostasis(GO:0001895) |
| 0.0 | 0.1 | GO:0035973 | aggrephagy(GO:0035973) |
| 0.0 | 0.1 | GO:0000393 | spliceosomal conformational changes to generate catalytic conformation(GO:0000393) |
| 0.0 | 0.4 | GO:0005980 | glycogen catabolic process(GO:0005980) |
| 0.0 | 0.2 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.9 | GO:0034665 | integrin alpha1-beta1 complex(GO:0034665) |
| 0.2 | 2.0 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.1 | 3.8 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.1 | 1.5 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.1 | 0.4 | GO:1990425 | ryanodine receptor complex(GO:1990425) |
| 0.1 | 0.4 | GO:0048476 | Holliday junction resolvase complex(GO:0048476) |
| 0.1 | 0.4 | GO:0072534 | perineuronal net(GO:0072534) |
| 0.1 | 1.3 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.1 | 1.1 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.1 | 1.0 | GO:0016013 | syntrophin complex(GO:0016013) |
| 0.1 | 5.9 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 1.6 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 0.7 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.0 | 0.7 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.0 | 0.2 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) |
| 0.0 | 0.1 | GO:0043564 | Ku70:Ku80 complex(GO:0043564) |
| 0.0 | 0.4 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.0 | 0.8 | GO:0030125 | clathrin vesicle coat(GO:0030125) |
| 0.0 | 9.3 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.0 | 0.1 | GO:0030934 | anchoring collagen complex(GO:0030934) |
| 0.0 | 0.9 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.0 | 0.5 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.0 | 0.1 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.0 | 0.8 | GO:0016459 | myosin complex(GO:0016459) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.2 | GO:0042008 | interleukin-18 receptor activity(GO:0042008) |
| 0.7 | 5.9 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.3 | 2.0 | GO:0031962 | mineralocorticoid receptor binding(GO:0031962) |
| 0.2 | 1.8 | GO:0038049 | transcription factor activity, ligand-activated RNA polymerase II transcription factor binding(GO:0038049) |
| 0.2 | 0.6 | GO:0008892 | guanine deaminase activity(GO:0008892) |
| 0.2 | 1.3 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.2 | 0.8 | GO:0008941 | nitric oxide dioxygenase activity(GO:0008941) oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of two atoms of oxygen into one donor(GO:0016708) |
| 0.1 | 0.4 | GO:0098640 | integrin binding involved in cell-matrix adhesion(GO:0098640) |
| 0.1 | 0.9 | GO:0098634 | protein binding involved in cell-matrix adhesion(GO:0098634) |
| 0.1 | 0.4 | GO:0008184 | glycogen phosphorylase activity(GO:0008184) |
| 0.1 | 0.7 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.1 | 0.8 | GO:0060002 | plus-end directed microfilament motor activity(GO:0060002) |
| 0.1 | 0.2 | GO:0004314 | [acyl-carrier-protein] S-malonyltransferase activity(GO:0004314) S-malonyltransferase activity(GO:0016419) malonyltransferase activity(GO:0016420) |
| 0.1 | 0.6 | GO:0030884 | lipid antigen binding(GO:0030882) endogenous lipid antigen binding(GO:0030883) exogenous lipid antigen binding(GO:0030884) |
| 0.1 | 11.0 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.1 | 0.3 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.0 | 0.4 | GO:0048256 | flap endonuclease activity(GO:0048256) |
| 0.0 | 0.7 | GO:0031433 | telethonin binding(GO:0031433) |
| 0.0 | 0.3 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.0 | 0.1 | GO:0015173 | aromatic amino acid transmembrane transporter activity(GO:0015173) |
| 0.0 | 0.2 | GO:0031531 | thyrotropin-releasing hormone receptor binding(GO:0031531) |
| 0.0 | 0.2 | GO:0008745 | N-acetylmuramoyl-L-alanine amidase activity(GO:0008745) |
| 0.0 | 1.1 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 0.2 | GO:0016427 | tRNA (cytosine) methyltransferase activity(GO:0016427) |
| 0.0 | 0.3 | GO:0002161 | aminoacyl-tRNA editing activity(GO:0002161) |
| 0.0 | 0.2 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.0 | 0.2 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.0 | 0.2 | GO:0004666 | prostaglandin-endoperoxide synthase activity(GO:0004666) |
| 0.0 | 0.1 | GO:0032795 | heterotrimeric G-protein binding(GO:0032795) |
| 0.0 | 0.3 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 1.6 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 0.2 | GO:0030942 | signal recognition particle binding(GO:0005047) endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.0 | 0.4 | GO:0034713 | type I transforming growth factor beta receptor binding(GO:0034713) |
| 0.0 | 0.9 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 0.6 | GO:0016876 | aminoacyl-tRNA ligase activity(GO:0004812) ligase activity, forming carbon-oxygen bonds(GO:0016875) ligase activity, forming aminoacyl-tRNA and related compounds(GO:0016876) |
| 0.0 | 0.1 | GO:0003963 | RNA-3'-phosphate cyclase activity(GO:0003963) |
| 0.0 | 0.5 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 2.0 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 0.7 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.0 | 0.1 | GO:0061752 | telomeric repeat-containing RNA binding(GO:0061752) |
| 0.0 | 0.4 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.0 | 0.3 | GO:0015143 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.0 | 0.4 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.0 | 0.3 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.0 | 0.1 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.0 | 0.9 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.0 | 0.7 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.0 | 0.3 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.0 | 0.2 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.0 | 0.1 | GO:0015056 | corticotrophin-releasing factor receptor activity(GO:0015056) |
| 0.0 | 0.4 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.0 | 4.4 | GO:0008168 | methyltransferase activity(GO:0008168) |
| 0.0 | 0.4 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.0 | 0.4 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 0.1 | GO:0035673 | oligopeptide transmembrane transporter activity(GO:0035673) |
| 0.0 | 0.4 | GO:0019789 | SUMO transferase activity(GO:0019789) |
| 0.0 | 0.5 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 0.1 | GO:0004974 | leukotriene receptor activity(GO:0004974) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 5.9 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.1 | 3.4 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.1 | 1.4 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 1.5 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 1.2 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 0.4 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 1.9 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.0 | 1.7 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 1.8 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 0.6 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 0.2 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 6.8 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.1 | 1.8 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.1 | 2.0 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 1.4 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.9 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.0 | 0.7 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.0 | 0.7 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 0.6 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 1.1 | REACTOME POST TRANSLATIONAL MODIFICATION SYNTHESIS OF GPI ANCHORED PROTEINS | Genes involved in Post-translational modification: synthesis of GPI-anchored proteins |
| 0.0 | 0.4 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 0.5 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.0 | 0.4 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 0.4 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.0 | 0.3 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.3 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |