Project
Inflammatory response time course, HUVEC (Wada, 2009)
Navigation
Downloads
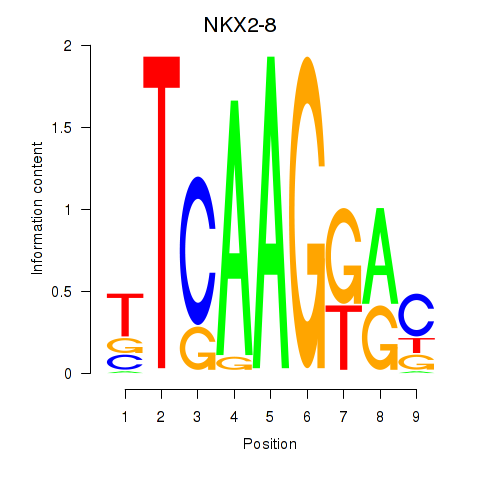
Results for NKX2-8
Z-value: 0.58
Transcription factors associated with NKX2-8
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NKX2-8
|
ENSG00000136327.7 | NKX2-8 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NKX2-8 | hg38_v1_chr14_-_36582593_36582626 | -0.31 | 1.3e-01 | Click! |
Activity profile of NKX2-8 motif
Sorted Z-values of NKX2-8 motif
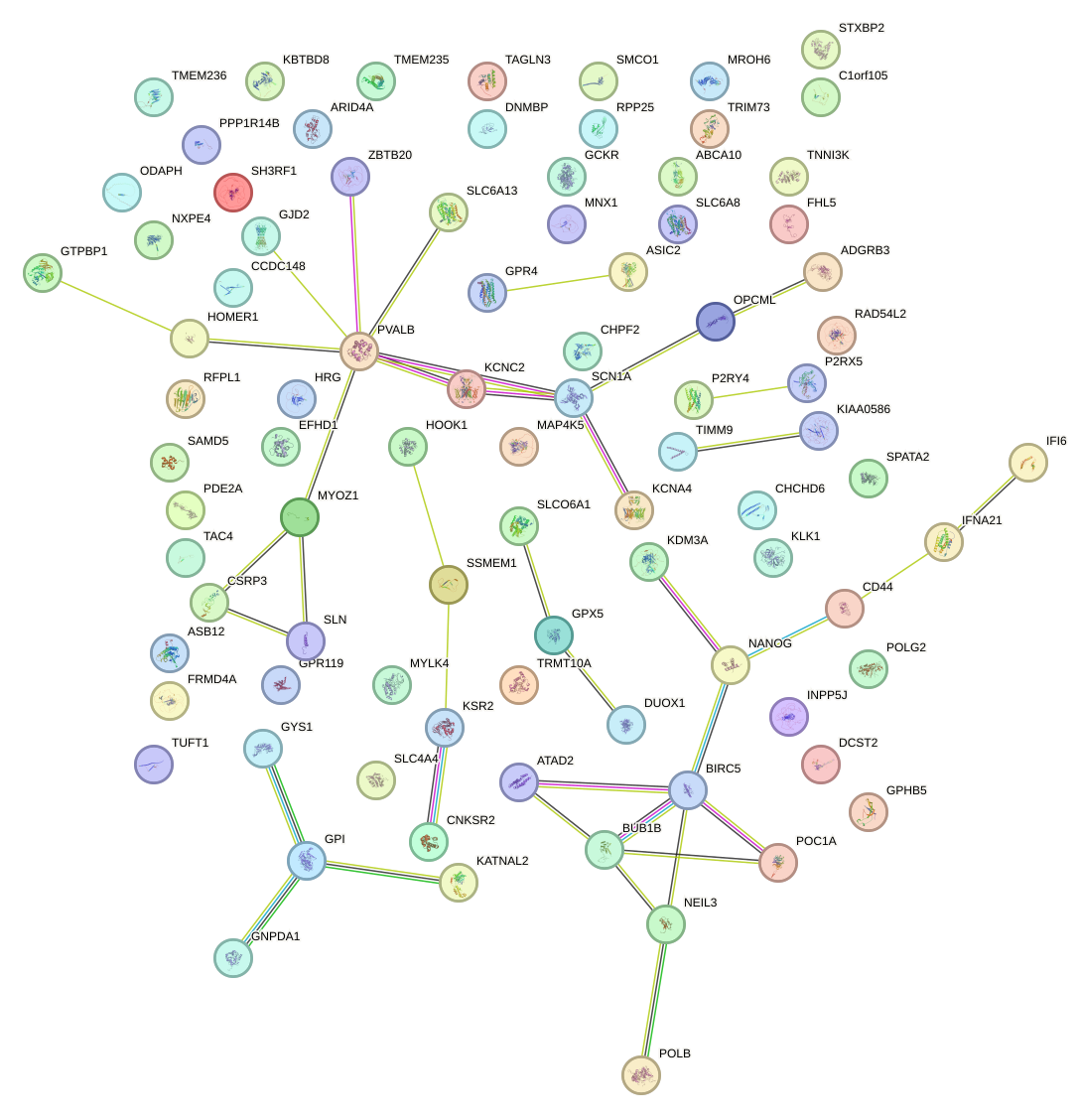
Network of associatons between targets according to the STRING database.
First level regulatory network of NKX2-8
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_-_3691887 | 0.78 |
ENST00000552050.5
|
P2RX5
|
purinergic receptor P2X 5 |
| chr17_-_69150062 | 0.51 |
ENST00000522787.5
ENST00000521538.5 |
ABCA10
|
ATP binding cassette subfamily A member 10 |
| chr11_+_5389377 | 0.46 |
ENST00000328611.5
|
OR51M1
|
olfactory receptor family 51 subfamily M member 1 |
| chr1_+_172420681 | 0.44 |
ENST00000367727.9
|
C1orf105
|
chromosome 1 open reading frame 105 |
| chr1_+_74235377 | 0.44 |
ENST00000326637.8
|
TNNI3K
|
TNNI3 interacting kinase |
| chr14_-_106875069 | 0.41 |
ENST00000390639.2
|
IGHV7-81
|
immunoglobulin heavy variable 7-81 (non-functional) |
| chr3_+_111998739 | 0.40 |
ENST00000393917.6
ENST00000273368.8 |
TAGLN3
|
transgelin 3 |
| chr3_+_111998915 | 0.38 |
ENST00000478951.6
|
TAGLN3
|
transgelin 3 |
| chr16_-_68015911 | 0.36 |
ENST00000574912.1
|
DPEP2NB
|
DPEP2 neighbor |
| chr10_-_73641450 | 0.36 |
ENST00000359322.5
|
MYOZ1
|
myozenin 1 |
| chr2_+_227616998 | 0.32 |
ENST00000641801.1
|
SCYGR4
|
small cysteine and glycine repeat containing 4 |
| chr3_+_126704202 | 0.32 |
ENST00000290913.8
ENST00000508789.5 |
CHCHD6
|
coiled-coil-helix-coiled-coil-helix domain containing 6 |
| chr14_-_106675544 | 0.32 |
ENST00000390632.2
|
IGHV3-66
|
immunoglobulin heavy variable 3-66 |
| chr6_+_29439939 | 0.29 |
ENST00000622521.1
|
OR10C1
|
olfactory receptor family 10 subfamily C member 1 |
| chrX_-_111410420 | 0.29 |
ENST00000371993.7
ENST00000680476.1 |
DCX
|
doublecortin |
| chr22_+_31122923 | 0.28 |
ENST00000620191.4
ENST00000412277.6 ENST00000412985.5 ENST00000331075.10 ENST00000420017.5 ENST00000400294.6 ENST00000405300.5 ENST00000404390.7 |
INPP5J
|
inositol polyphosphate-5-phosphatase J |
| chr14_-_63318933 | 0.28 |
ENST00000621500.2
|
GPHB5
|
glycoprotein hormone subunit beta 5 |
| chr8_-_143568854 | 0.28 |
ENST00000524906.5
ENST00000532862.1 ENST00000534459.5 |
MROH6
|
maestro heat like repeat family member 6 |
| chr19_-_48993300 | 0.28 |
ENST00000323798.8
ENST00000263276.6 |
GYS1
|
glycogen synthase 1 |
| chr11_-_72642407 | 0.28 |
ENST00000376450.7
|
PDE2A
|
phosphodiesterase 2A |
| chr11_-_72642450 | 0.28 |
ENST00000444035.6
ENST00000544570.5 |
PDE2A
|
phosphodiesterase 2A |
| chr1_-_155033778 | 0.27 |
ENST00000368424.4
|
DCST2
|
DC-STAMP domain containing 2 |
| chr4_+_71062642 | 0.26 |
ENST00000649996.1
|
SLC4A4
|
solute carrier family 4 member 4 |
| chr10_-_13972355 | 0.26 |
ENST00000264546.10
|
FRMD4A
|
FERM domain containing 4A |
| chr6_+_28525877 | 0.26 |
ENST00000412168.7
|
GPX5
|
glutathione peroxidase 5 |
| chr11_-_19202004 | 0.25 |
ENST00000648719.1
|
CSRP3
|
cysteine and glycine rich protein 3 |
| chr3_-_52154382 | 0.25 |
ENST00000474012.1
ENST00000296484.7 |
POC1A
|
POC1 centriolar protein A |
| chr11_-_107719657 | 0.25 |
ENST00000525934.1
ENST00000531293.1 |
SLN
|
sarcolipin |
| chr14_-_58427509 | 0.25 |
ENST00000395159.7
|
TIMM9
|
translocase of inner mitochondrial membrane 9 |
| chrX_+_153687918 | 0.25 |
ENST00000253122.10
|
SLC6A8
|
solute carrier family 6 member 8 |
| chr14_-_106593319 | 0.24 |
ENST00000390627.3
|
IGHV3-53
|
immunoglobulin heavy variable 3-53 |
| chr2_+_86441341 | 0.24 |
ENST00000312912.10
ENST00000409064.5 |
KDM3A
|
lysine demethylase 3A |
| chr12_-_262828 | 0.24 |
ENST00000343164.9
ENST00000436453.1 ENST00000445055.6 ENST00000546319.5 |
SLC6A13
|
solute carrier family 6 member 13 |
| chr22_-_36817510 | 0.24 |
ENST00000443735.1
|
PVALB
|
parvalbumin |
| chr12_-_12684490 | 0.23 |
ENST00000540510.1
|
GPR19
|
G protein-coupled receptor 19 |
| chr3_+_66998297 | 0.23 |
ENST00000484414.5
ENST00000460576.5 ENST00000417314.2 |
KBTBD8
|
kelch repeat and BTB domain containing 8 |
| chr7_+_75398915 | 0.23 |
ENST00000437796.1
|
TRIM73
|
tripartite motif containing 73 |
| chr10_+_17752185 | 0.23 |
ENST00000377495.2
|
TMEM236
|
transmembrane protein 236 |
| chr15_+_45129933 | 0.22 |
ENST00000321429.8
ENST00000389037.7 ENST00000558322.5 |
DUOX1
|
dual oxidase 1 |
| chr3_+_186666003 | 0.22 |
ENST00000232003.5
|
HRG
|
histidine rich glycoprotein |
| chrX_-_70260199 | 0.22 |
ENST00000374519.4
|
P2RY4
|
pyrimidinergic receptor P2Y4 |
| chr18_+_46917561 | 0.21 |
ENST00000683218.1
|
KATNAL2
|
katanin catalytic subunit A1 like 2 |
| chr8_+_42338454 | 0.21 |
ENST00000532157.5
ENST00000520008.5 |
POLB
|
DNA polymerase beta |
| chr6_+_69232406 | 0.21 |
ENST00000238918.12
|
ADGRB3
|
adhesion G protein-coupled receptor B3 |
| chr12_-_117968972 | 0.20 |
ENST00000339824.7
|
KSR2
|
kinase suppressor of ras 2 |
| chr6_+_147508645 | 0.19 |
ENST00000367474.2
|
SAMD5
|
sterile alpha motif domain containing 5 |
| chr2_+_232633551 | 0.19 |
ENST00000264059.8
|
EFHD1
|
EF-hand domain family member D1 |
| chr9_+_35909481 | 0.19 |
ENST00000443779.3
|
SPAAR
|
small regulatory polypeptide of amino acid response |
| chr9_+_35910076 | 0.19 |
ENST00000636776.1
|
SPAAR
|
small regulatory polypeptide of amino acid response |
| chr3_+_51538684 | 0.19 |
ENST00000684192.1
|
RAD54L2
|
RAD54 like 2 |
| chr17_+_75476493 | 0.18 |
ENST00000375248.9
|
TMEM94
|
transmembrane protein 94 |
| chr15_+_40161003 | 0.18 |
ENST00000412359.7
ENST00000287598.11 |
BUB1B
|
BUB1 mitotic checkpoint serine/threonine kinase B |
| chr17_+_78214286 | 0.18 |
ENST00000592734.5
ENST00000587746.5 |
BIRC5
|
baculoviral IAP repeat containing 5 |
| chr2_-_158456702 | 0.18 |
ENST00000409889.1
ENST00000283233.10 |
CCDC148
|
coiled-coil domain containing 148 |
| chr17_+_78214186 | 0.18 |
ENST00000301633.8
ENST00000350051.8 ENST00000374948.6 ENST00000590449.1 |
BIRC5
|
baculoviral IAP repeat containing 5 |
| chr10_+_69318831 | 0.18 |
ENST00000359426.7
|
HK1
|
hexokinase 1 |
| chr5_-_79512794 | 0.17 |
ENST00000282260.10
ENST00000508576.5 ENST00000535690.1 |
HOMER1
|
homer scaffold protein 1 |
| chr4_+_75556048 | 0.17 |
ENST00000616557.1
ENST00000435974.2 ENST00000311623.9 |
ODAPH
|
odontogenesis associated phosphoprotein |
| chr11_+_35189869 | 0.17 |
ENST00000525688.5
ENST00000278385.10 ENST00000533222.5 |
CD44
|
CD44 molecule (Indian blood group) |
| chrX_-_130385656 | 0.16 |
ENST00000276218.4
ENST00000682440.1 |
GPR119
|
G protein-coupled receptor 119 |
| chr5_-_102498913 | 0.16 |
ENST00000513675.1
ENST00000379807.7 ENST00000506729.6 |
SLCO6A1
|
solute carrier organic anion transporter family member 6A1 |
| chr19_+_34365173 | 0.16 |
ENST00000589640.5
ENST00000591204.5 ENST00000589399.6 ENST00000356487.11 |
GPI
|
glucose-6-phosphate isomerase |
| chr4_-_169270849 | 0.16 |
ENST00000502315.1
ENST00000284637.14 |
SH3RF1
|
SH3 domain containing ring finger 1 |
| chr3_-_196515315 | 0.16 |
ENST00000397537.3
|
SMCO1
|
single-pass membrane protein with coiled-coil domains 1 |
| chr17_-_33293247 | 0.16 |
ENST00000225823.7
|
ASIC2
|
acid sensing ion channel subunit 2 |
| chr22_+_29438583 | 0.16 |
ENST00000354373.2
|
RFPL1
|
ret finger protein like 1 |
| chr15_-_74956758 | 0.16 |
ENST00000322177.6
|
RPP25
|
ribonuclease P and MRP subunit p25 |
| chr5_-_102499008 | 0.16 |
ENST00000389019.7
|
SLCO6A1
|
solute carrier organic anion transporter family member 6A1 |
| chrX_-_64230600 | 0.16 |
ENST00000362002.3
|
ASB12
|
ankyrin repeat and SOCS box containing 12 |
| chr4_+_177309866 | 0.16 |
ENST00000264596.4
|
NEIL3
|
nei like DNA glycosylase 3 |
| chr2_+_27496830 | 0.15 |
ENST00000264717.7
|
GCKR
|
glucokinase regulator |
| chr20_-_49913693 | 0.15 |
ENST00000422556.1
|
SPATA2
|
spermatogenesis associated 2 |
| chr11_-_19201976 | 0.15 |
ENST00000647990.1
ENST00000649235.1 ENST00000265968.9 ENST00000649842.1 |
CSRP3
|
cysteine and glycine rich protein 3 |
| chr12_+_7789393 | 0.15 |
ENST00000229307.9
|
NANOG
|
Nanog homeobox |
| chr1_-_27672178 | 0.15 |
ENST00000339145.8
ENST00000361157.11 ENST00000362020.4 ENST00000679644.1 |
IFI6
|
interferon alpha inducible protein 6 |
| chr14_-_106211453 | 0.15 |
ENST00000390606.3
|
IGHV3-20
|
immunoglobulin heavy variable 3-20 |
| chr17_+_78231310 | 0.14 |
ENST00000374946.7
ENST00000421688.5 ENST00000586400.5 |
TMEM235
|
transmembrane protein 235 |
| chr22_+_38705922 | 0.14 |
ENST00000216044.10
|
GTPBP1
|
GTP binding protein 1 |
| chr3_-_115071333 | 0.14 |
ENST00000462705.5
|
ZBTB20
|
zinc finger and BTB domain containing 20 |
| chr17_-_8295342 | 0.14 |
ENST00000579192.5
ENST00000577745.2 |
SLC25A35
|
solute carrier family 25 member 35 |
| chr10_-_99913971 | 0.14 |
ENST00000543621.6
|
DNMBP
|
dynamin binding protein |
| chr14_-_106130061 | 0.14 |
ENST00000390602.3
|
IGHV3-13
|
immunoglobulin heavy variable 3-13 |
| chr15_-_34754989 | 0.14 |
ENST00000290374.5
|
GJD2
|
gap junction protein delta 2 |
| chr11_-_114595750 | 0.14 |
ENST00000424261.6
|
NXPE4
|
neurexophilin and PC-esterase domain family member 4 |
| chr4_-_99563984 | 0.13 |
ENST00000394877.7
|
TRMT10A
|
tRNA methyltransferase 10A |
| chr4_-_99564026 | 0.13 |
ENST00000394876.7
|
TRMT10A
|
tRNA methyltransferase 10A |
| chr2_-_29074515 | 0.13 |
ENST00000331664.6
|
PCARE
|
photoreceptor cilium actin regulator |
| chr11_-_64246190 | 0.13 |
ENST00000392210.6
|
PPP1R14B
|
protein phosphatase 1 regulatory inhibitor subunit 14B |
| chr12_-_75209814 | 0.13 |
ENST00000549446.6
|
KCNC2
|
potassium voltage-gated channel subfamily C member 2 |
| chr6_+_125203639 | 0.13 |
ENST00000392482.6
|
TPD52L1
|
TPD52 like 1 |
| chr6_+_96563117 | 0.12 |
ENST00000450218.6
|
FHL5
|
four and a half LIM domains 5 |
| chr14_+_58427385 | 0.12 |
ENST00000354386.10
ENST00000619416.4 |
KIAA0586
|
KIAA0586 |
| chrX_+_48823357 | 0.12 |
ENST00000430858.1
|
HDAC6
|
histone deacetylase 6 |
| chr11_-_132943671 | 0.12 |
ENST00000331898.11
|
OPCML
|
opioid binding protein/cell adhesion molecule like |
| chr9_-_21166660 | 0.12 |
ENST00000380225.1
|
IFNA21
|
interferon alpha 21 |
| chr16_+_33827140 | 0.12 |
ENST00000562905.2
|
IGHV3OR16-13
|
immunoglobulin heavy variable 3/OR16-13 (non-functional) |
| chr14_+_58298497 | 0.12 |
ENST00000348476.7
ENST00000355431.8 ENST00000395168.7 |
ARID4A
|
AT-rich interaction domain 4A |
| chr14_-_58427158 | 0.12 |
ENST00000555097.1
ENST00000556367.6 ENST00000555404.5 |
TIMM9
|
translocase of inner mitochondrial membrane 9 |
| chr12_+_25052732 | 0.12 |
ENST00000547044.5
|
IRAG2
|
inositol 1,4,5-triphosphate receptor associated 2 |
| chr14_-_58427489 | 0.11 |
ENST00000555593.5
|
TIMM9
|
translocase of inner mitochondrial membrane 9 |
| chr6_-_2744126 | 0.11 |
ENST00000647417.1
|
MYLK4
|
myosin light chain kinase family member 4 |
| chr16_+_32995048 | 0.11 |
ENST00000425181.3
|
IGHV3OR16-10
|
immunoglobulin heavy variable 3/OR16-10 (non-functional) |
| chr1_+_151540299 | 0.11 |
ENST00000392712.7
ENST00000368848.6 ENST00000368849.8 ENST00000353024.4 |
TUFT1
|
tuftelin 1 |
| chr8_-_144465648 | 0.11 |
ENST00000424149.6
ENST00000530637.1 |
CYHR1
|
cysteine and histidine rich 1 |
| chr14_+_21887848 | 0.11 |
ENST00000390437.2
|
TRAV12-2
|
T cell receptor alpha variable 12-2 |
| chr8_-_123396412 | 0.11 |
ENST00000287394.10
|
ATAD2
|
ATPase family AAA domain containing 2 |
| chr14_+_58427686 | 0.11 |
ENST00000650904.1
ENST00000652326.2 ENST00000554463.5 ENST00000555833.5 |
KIAA0586
|
KIAA0586 |
| chr2_-_166149120 | 0.11 |
ENST00000641575.1
ENST00000641603.1 |
SCN1A
|
sodium voltage-gated channel alpha subunit 1 |
| chr7_+_151232464 | 0.11 |
ENST00000482173.5
ENST00000495645.5 ENST00000035307.6 |
CHPF2
|
chondroitin polymerizing factor 2 |
| chr5_-_176629943 | 0.11 |
ENST00000510387.5
ENST00000506696.1 |
SNCB
|
synuclein beta |
| chr7_+_130207847 | 0.11 |
ENST00000297819.4
|
SSMEM1
|
serine rich single-pass membrane protein 1 |
| chr12_+_25052512 | 0.11 |
ENST00000557489.5
ENST00000354454.7 ENST00000536173.5 |
IRAG2
|
inositol 1,4,5-triphosphate receptor associated 2 |
| chr11_-_132943092 | 0.11 |
ENST00000612177.4
ENST00000541867.5 |
OPCML
|
opioid binding protein/cell adhesion molecule like |
| chr7_-_157010615 | 0.11 |
ENST00000252971.11
|
MNX1
|
motor neuron and pancreas homeobox 1 |
| chr16_+_58392462 | 0.11 |
ENST00000318129.6
|
GINS3
|
GINS complex subunit 3 |
| chr3_+_63967738 | 0.11 |
ENST00000484332.1
|
ATXN7
|
ataxin 7 |
| chr11_-_40293102 | 0.10 |
ENST00000527150.5
|
LRRC4C
|
leucine rich repeat containing 4C |
| chr14_+_58427970 | 0.10 |
ENST00000261244.9
|
KIAA0586
|
KIAA0586 |
| chr7_-_36724380 | 0.10 |
ENST00000617267.4
|
AOAH
|
acyloxyacyl hydrolase |
| chr2_+_79025696 | 0.10 |
ENST00000272324.10
|
REG3G
|
regenerating family member 3 gamma |
| chr16_-_75267988 | 0.10 |
ENST00000393422.6
|
BCAR1
|
BCAR1 scaffold protein, Cas family member |
| chr2_+_79025678 | 0.10 |
ENST00000393897.6
|
REG3G
|
regenerating family member 3 gamma |
| chr14_+_93927358 | 0.10 |
ENST00000557000.2
|
FAM181A
|
family with sequence similarity 181 member A |
| chr22_+_20858999 | 0.10 |
ENST00000215730.12
|
SNAP29
|
synaptosome associated protein 29 |
| chr1_-_11055820 | 0.10 |
ENST00000490101.1
|
SRM
|
spermidine synthase |
| chr20_+_63738270 | 0.10 |
ENST00000467211.1
|
ENSG00000273047.1
|
novel transcript, LIME1-SLC2A4RG readthrough |
| chr6_-_32763466 | 0.10 |
ENST00000427449.1
ENST00000411527.5 |
HLA-DQB2
|
major histocompatibility complex, class II, DQ beta 2 |
| chr3_-_53130405 | 0.10 |
ENST00000467048.1
ENST00000296292.8 ENST00000394738.7 |
RFT1
|
RFT1 homolog |
| chr14_-_55027045 | 0.10 |
ENST00000455555.1
ENST00000360586.8 ENST00000420358.2 |
WDHD1
|
WD repeat and HMG-box DNA binding protein 1 |
| chrX_-_31178220 | 0.10 |
ENST00000681026.1
|
DMD
|
dystrophin |
| chr14_-_106389858 | 0.10 |
ENST00000390617.2
|
IGHV3-35
|
immunoglobulin heavy variable 3-35 (non-functional) |
| chr14_+_58427425 | 0.10 |
ENST00000619722.5
ENST00000423743.7 |
KIAA0586
|
KIAA0586 |
| chr18_+_31591869 | 0.10 |
ENST00000237014.8
|
TTR
|
transthyretin |
| chr12_+_25052634 | 0.10 |
ENST00000548766.5
|
IRAG2
|
inositol 1,4,5-triphosphate receptor associated 2 |
| chr16_+_78202 | 0.10 |
ENST00000356432.8
ENST00000219431.4 |
MPG
|
N-methylpurine DNA glycosylase |
| chr4_-_99563668 | 0.10 |
ENST00000273962.7
|
TRMT10A
|
tRNA methyltransferase 10A |
| chr3_-_39051935 | 0.10 |
ENST00000668754.1
ENST00000302328.9 |
SCN11A
|
sodium voltage-gated channel alpha subunit 11 |
| chr14_-_106791226 | 0.10 |
ENST00000433072.2
|
IGHV3-72
|
immunoglobulin heavy variable 3-72 |
| chr13_+_108269629 | 0.09 |
ENST00000430559.5
ENST00000375887.9 |
TNFSF13B
|
TNF superfamily member 13b |
| chr12_-_49110840 | 0.09 |
ENST00000550137.5
ENST00000267102.13 ENST00000547382.5 |
LMBR1L
|
limb development membrane protein 1 like |
| chrX_-_31178149 | 0.09 |
ENST00000679437.1
|
DMD
|
dystrophin |
| chr12_+_120978686 | 0.09 |
ENST00000541395.5
ENST00000544413.2 |
HNF1A
|
HNF1 homeobox A |
| chr1_+_180196536 | 0.09 |
ENST00000443059.1
|
QSOX1
|
quiescin sulfhydryl oxidase 1 |
| chr10_-_77637789 | 0.09 |
ENST00000481070.1
ENST00000640969.1 ENST00000286628.14 ENST00000638991.1 ENST00000639913.1 ENST00000480683.2 |
KCNMA1
|
potassium calcium-activated channel subfamily M alpha 1 |
| chr10_-_133357674 | 0.09 |
ENST00000368551.1
|
FUOM
|
fucose mutarotase |
| chr3_+_173584433 | 0.09 |
ENST00000361589.8
|
NLGN1
|
neuroligin 1 |
| chr14_-_106235582 | 0.09 |
ENST00000390607.2
|
IGHV3-21
|
immunoglobulin heavy variable 3-21 |
| chr1_+_149899618 | 0.09 |
ENST00000369150.1
|
BOLA1
|
bolA family member 1 |
| chr2_+_218672027 | 0.09 |
ENST00000392105.7
ENST00000455724.5 ENST00000295709.8 |
STK36
|
serine/threonine kinase 36 |
| chr7_-_36724543 | 0.09 |
ENST00000612871.4
|
AOAH
|
acyloxyacyl hydrolase |
| chr6_+_34466059 | 0.09 |
ENST00000620693.4
ENST00000244458.7 ENST00000374043.6 |
PACSIN1
|
protein kinase C and casein kinase substrate in neurons 1 |
| chr1_+_173824626 | 0.08 |
ENST00000648960.1
ENST00000648807.1 ENST00000649067.1 ENST00000649689.2 |
DARS2
|
aspartyl-tRNA synthetase 2, mitochondrial |
| chr14_-_106269133 | 0.08 |
ENST00000390609.3
|
IGHV3-23
|
immunoglobulin heavy variable 3-23 |
| chr5_+_69369781 | 0.08 |
ENST00000509734.5
ENST00000354868.10 ENST00000521422.5 ENST00000354312.7 ENST00000345306.10 |
RAD17
|
RAD17 checkpoint clamp loader component |
| chr15_-_28174423 | 0.08 |
ENST00000569772.1
|
HERC2
|
HECT and RLD domain containing E3 ubiquitin protein ligase 2 |
| chr19_-_43754901 | 0.08 |
ENST00000270066.11
ENST00000601170.5 |
SMG9
|
SMG9 nonsense mediated mRNA decay factor |
| chr22_+_32357867 | 0.08 |
ENST00000249007.4
|
RFPL3
|
ret finger protein like 3 |
| chr1_-_23424618 | 0.08 |
ENST00000476978.2
|
TCEA3
|
transcription elongation factor A3 |
| chr21_+_32298945 | 0.08 |
ENST00000339944.4
|
MRAP
|
melanocortin 2 receptor accessory protein |
| chr7_-_36724457 | 0.08 |
ENST00000617537.5
ENST00000435386.1 |
AOAH
|
acyloxyacyl hydrolase |
| chr1_+_225825346 | 0.08 |
ENST00000366837.5
|
EPHX1
|
epoxide hydrolase 1 |
| chr6_-_39322968 | 0.08 |
ENST00000507712.5
|
KCNK16
|
potassium two pore domain channel subfamily K member 16 |
| chr10_-_77637721 | 0.08 |
ENST00000638848.1
ENST00000639406.1 ENST00000618048.2 ENST00000639120.1 ENST00000640834.1 ENST00000639601.1 ENST00000638514.1 ENST00000457953.6 ENST00000639090.1 ENST00000639489.1 ENST00000372440.6 ENST00000404771.8 ENST00000638203.1 ENST00000638306.1 ENST00000638351.1 ENST00000638606.1 ENST00000639591.1 ENST00000640182.1 ENST00000640605.1 ENST00000640141.1 |
KCNMA1
|
potassium calcium-activated channel subfamily M alpha 1 |
| chr3_-_16482850 | 0.08 |
ENST00000432519.5
|
RFTN1
|
raftlin, lipid raft linker 1 |
| chr1_-_204166334 | 0.08 |
ENST00000272190.9
|
REN
|
renin |
| chr3_+_109136707 | 0.08 |
ENST00000622536.6
|
C3orf85
|
chromosome 3 open reading frame 85 |
| chr1_+_173824694 | 0.08 |
ENST00000647645.1
|
DARS2
|
aspartyl-tRNA synthetase 2, mitochondrial |
| chr19_+_39342396 | 0.08 |
ENST00000602243.5
ENST00000598913.5 ENST00000314471.10 |
SAMD4B
|
sterile alpha motif domain containing 4B |
| chrX_-_32155462 | 0.07 |
ENST00000359836.5
ENST00000378707.7 ENST00000541735.5 ENST00000684130.1 ENST00000682238.1 ENST00000620040.5 ENST00000474231.5 |
DMD
|
dystrophin |
| chr6_+_116461364 | 0.07 |
ENST00000368606.7
ENST00000368605.3 |
CALHM6
|
calcium homeostasis modulator family member 6 |
| chr14_-_106470788 | 0.07 |
ENST00000434710.1
|
IGHV3-43
|
immunoglobulin heavy variable 3-43 |
| chr20_-_50131125 | 0.07 |
ENST00000371656.3
|
PEDS1
|
plasmanylethanolamine desaturase 1 |
| chr4_+_15339818 | 0.07 |
ENST00000397700.6
ENST00000295297.4 |
C1QTNF7
|
C1q and TNF related 7 |
| chr7_-_95396349 | 0.07 |
ENST00000427422.5
ENST00000451904.5 ENST00000265627.10 |
PON3
|
paraoxonase 3 |
| chr11_+_925840 | 0.06 |
ENST00000448903.7
ENST00000525796.5 ENST00000534328.5 |
AP2A2
|
adaptor related protein complex 2 subunit alpha 2 |
| chr2_-_105438503 | 0.06 |
ENST00000393352.7
ENST00000607522.1 |
FHL2
|
four and a half LIM domains 2 |
| chr3_-_179266971 | 0.06 |
ENST00000349697.2
ENST00000497599.5 |
KCNMB3
|
potassium calcium-activated channel subfamily M regulatory beta subunit 3 |
| chr16_+_58392391 | 0.06 |
ENST00000426538.6
ENST00000328514.11 |
GINS3
|
GINS complex subunit 3 |
| chr21_+_32298849 | 0.06 |
ENST00000303645.10
|
MRAP
|
melanocortin 2 receptor accessory protein |
| chr19_-_4559663 | 0.06 |
ENST00000586582.6
|
SEMA6B
|
semaphorin 6B |
| chr1_+_152985231 | 0.06 |
ENST00000368762.1
|
SPRR1A
|
small proline rich protein 1A |
| chr16_+_176659 | 0.06 |
ENST00000320868.9
ENST00000397797.1 |
HBA1
|
hemoglobin subunit alpha 1 |
| chr10_+_72893734 | 0.06 |
ENST00000334011.10
|
OIT3
|
oncoprotein induced transcript 3 |
| chr14_-_106538331 | 0.06 |
ENST00000390624.3
|
IGHV3-48
|
immunoglobulin heavy variable 3-48 |
| chr20_+_33031648 | 0.06 |
ENST00000349552.1
|
BPIFB6
|
BPI fold containing family B member 6 |
| chr22_-_50085331 | 0.06 |
ENST00000395876.6
|
MLC1
|
modulator of VRAC current 1 |
| chr1_+_16022030 | 0.06 |
ENST00000331433.5
|
CLCNKA
|
chloride voltage-gated channel Ka |
| chr1_-_56966133 | 0.05 |
ENST00000535057.5
ENST00000543257.5 |
C8B
|
complement C8 beta chain |
| chr16_+_172869 | 0.05 |
ENST00000251595.11
ENST00000397806.1 ENST00000484216.1 |
HBA2
|
hemoglobin subunit alpha 2 |
| chr12_-_6493295 | 0.05 |
ENST00000540949.1
|
MRPL51
|
mitochondrial ribosomal protein L51 |
| chr2_-_218671975 | 0.05 |
ENST00000295704.7
|
RNF25
|
ring finger protein 25 |
| chr14_+_63761836 | 0.05 |
ENST00000674003.1
|
SYNE2
|
spectrin repeat containing nuclear envelope protein 2 |
| chr4_+_70334963 | 0.05 |
ENST00000273936.6
|
CABS1
|
calcium binding protein, spermatid associated 1 |
| chr16_+_22490337 | 0.05 |
ENST00000415833.6
|
NPIPB5
|
nuclear pore complex interacting protein family member B5 |
| chr2_-_63588390 | 0.05 |
ENST00000272321.12
ENST00000409562.7 |
WDPCP
|
WD repeat containing planar cell polarity effector |
| chr3_-_114624193 | 0.05 |
ENST00000481632.5
|
ZBTB20
|
zinc finger and BTB domain containing 20 |
| chr14_-_106811131 | 0.05 |
ENST00000424969.2
|
IGHV3-74
|
immunoglobulin heavy variable 3-74 |
| chr17_+_7688427 | 0.05 |
ENST00000396463.7
ENST00000534050.5 |
WRAP53
|
WD repeat containing antisense to TP53 |
| chr8_+_39934955 | 0.05 |
ENST00000502986.2
|
IDO2
|
indoleamine 2,3-dioxygenase 2 |
| chr12_+_49265071 | 0.05 |
ENST00000549183.1
ENST00000639419.1 ENST00000301072.11 |
TUBA1C
|
tubulin alpha 1c |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0033159 | negative regulation of protein import into nucleus, translocation(GO:0033159) |
| 0.1 | 0.3 | GO:1901876 | regulation of calcium ion binding(GO:1901876) negative regulation of calcium ion binding(GO:1901877) |
| 0.1 | 0.2 | GO:0046292 | formaldehyde metabolic process(GO:0046292) |
| 0.1 | 0.2 | GO:0044278 | cell wall disruption in other organism(GO:0044278) |
| 0.1 | 0.3 | GO:0099558 | maintenance of synapse structure(GO:0099558) |
| 0.1 | 0.2 | GO:0006288 | base-excision repair, DNA ligation(GO:0006288) |
| 0.1 | 0.4 | GO:0035995 | detection of muscle stretch(GO:0035995) |
| 0.1 | 0.2 | GO:0042335 | cuticle development(GO:0042335) |
| 0.1 | 0.2 | GO:2001027 | negative regulation of endothelial cell chemotaxis(GO:2001027) |
| 0.1 | 0.2 | GO:0006421 | asparaginyl-tRNA aminoacylation(GO:0006421) |
| 0.1 | 0.5 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.0 | 0.2 | GO:0007206 | phospholipase C-activating G-protein coupled glutamate receptor signaling pathway(GO:0007206) |
| 0.0 | 0.3 | GO:0003431 | growth plate cartilage chondrocyte development(GO:0003431) |
| 0.0 | 0.1 | GO:0070846 | misfolded protein transport(GO:0070843) polyubiquitinated protein transport(GO:0070844) polyubiquitinated misfolded protein transport(GO:0070845) Hsp90 deacetylation(GO:0070846) |
| 0.0 | 0.2 | GO:0050915 | sensory perception of sour taste(GO:0050915) |
| 0.0 | 0.1 | GO:0038060 | nitric oxide-cGMP-mediated signaling pathway(GO:0038060) |
| 0.0 | 0.1 | GO:0035565 | regulation of pronephros size(GO:0035565) |
| 0.0 | 0.2 | GO:0051754 | meiotic sister chromatid cohesion, centromeric(GO:0051754) |
| 0.0 | 0.3 | GO:0002155 | regulation of thyroid hormone mediated signaling pathway(GO:0002155) |
| 0.0 | 1.0 | GO:0035590 | purinergic nucleotide receptor signaling pathway(GO:0035590) |
| 0.0 | 0.4 | GO:0043416 | regulation of skeletal muscle tissue regeneration(GO:0043416) |
| 0.0 | 0.1 | GO:0097368 | establishment of Sertoli cell barrier(GO:0097368) |
| 0.0 | 1.7 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.0 | 0.1 | GO:0046271 | phenylpropanoid catabolic process(GO:0046271) |
| 0.0 | 0.1 | GO:0098923 | retrograde trans-synaptic signaling by soluble gas(GO:0098923) trans-synaptic signaling by soluble gas(GO:0099543) |
| 0.0 | 0.4 | GO:0086069 | bundle of His cell to Purkinje myocyte communication(GO:0086069) |
| 0.0 | 0.3 | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.0 | 0.1 | GO:0016240 | autophagosome docking(GO:0016240) |
| 0.0 | 0.2 | GO:0009438 | methylglyoxal metabolic process(GO:0009438) |
| 0.0 | 0.2 | GO:0072656 | maintenance of protein location in mitochondrion(GO:0072656) |
| 0.0 | 0.2 | GO:0033132 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.0 | 0.3 | GO:0042407 | cristae formation(GO:0042407) |
| 0.0 | 0.4 | GO:1990001 | inhibition of cysteine-type endopeptidase activity involved in apoptotic process(GO:1990001) |
| 0.0 | 0.1 | GO:0007228 | positive regulation of hh target transcription factor activity(GO:0007228) |
| 0.0 | 0.1 | GO:0016476 | regulation of embryonic cell shape(GO:0016476) |
| 0.0 | 0.1 | GO:0055011 | atrial cardiac muscle cell differentiation(GO:0055011) atrial cardiac muscle cell development(GO:0055014) |
| 0.0 | 0.0 | GO:0035674 | tricarboxylic acid transmembrane transport(GO:0035674) |
| 0.0 | 0.1 | GO:0001714 | endodermal cell fate specification(GO:0001714) |
| 0.0 | 0.1 | GO:0051902 | negative regulation of mitochondrial depolarization(GO:0051902) negative regulation of membrane depolarization(GO:1904180) |
| 0.0 | 0.0 | GO:0071676 | negative regulation of mononuclear cell migration(GO:0071676) |
| 0.0 | 0.1 | GO:0008295 | spermidine biosynthetic process(GO:0008295) |
| 0.0 | 0.1 | GO:0045007 | depurination(GO:0045007) |
| 0.0 | 0.2 | GO:1900625 | regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 0.0 | 0.2 | GO:0034465 | response to carbon monoxide(GO:0034465) |
| 0.0 | 0.0 | GO:0003433 | chondrocyte development involved in endochondral bone morphogenesis(GO:0003433) |
| 0.0 | 0.3 | GO:0031115 | negative regulation of microtubule polymerization(GO:0031115) |
| 0.0 | 0.2 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 0.0 | 0.1 | GO:0002457 | T cell antigen processing and presentation(GO:0002457) |
| 0.0 | 0.1 | GO:0046836 | glycolipid transport(GO:0046836) |
| 0.0 | 0.2 | GO:0014029 | neural crest formation(GO:0014029) |
| 0.0 | 0.2 | GO:0015812 | gamma-aminobutyric acid transport(GO:0015812) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:1905103 | integral component of lysosomal membrane(GO:1905103) |
| 0.1 | 0.5 | GO:0042721 | mitochondrial inner membrane protein insertion complex(GO:0042721) |
| 0.0 | 0.3 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.0 | 0.4 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 0.0 | 1.7 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 0.8 | GO:0005639 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) |
| 0.0 | 0.2 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.0 | 0.3 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.0 | 0.3 | GO:0016013 | syntrophin complex(GO:0016013) |
| 0.0 | 0.1 | GO:0020016 | ciliary pocket(GO:0020016) ciliary pocket membrane(GO:0020018) |
| 0.0 | 0.3 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.0 | 0.2 | GO:0035692 | macrophage migration inhibitory factor receptor complex(GO:0035692) |
| 0.0 | 0.1 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.0 | 0.2 | GO:0000778 | condensed nuclear chromosome kinetochore(GO:0000778) |
| 0.0 | 0.4 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.0 | 0.1 | GO:0032021 | NELF complex(GO:0032021) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0050528 | acyloxyacyl hydrolase activity(GO:0050528) |
| 0.1 | 0.3 | GO:0031531 | thyrotropin-releasing hormone receptor binding(GO:0031531) |
| 0.1 | 0.8 | GO:0004931 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.1 | 0.4 | GO:0009019 | tRNA (guanine-N1-)-methyltransferase activity(GO:0009019) |
| 0.1 | 0.8 | GO:0031433 | telethonin binding(GO:0031433) |
| 0.0 | 0.1 | GO:0031780 | corticotropin hormone receptor binding(GO:0031780) type 5 melanocortin receptor binding(GO:0031783) |
| 0.0 | 0.6 | GO:0030911 | cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) TPR domain binding(GO:0030911) |
| 0.0 | 0.2 | GO:0070095 | fructose-6-phosphate binding(GO:0070095) |
| 0.0 | 0.4 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.0 | 0.1 | GO:0004766 | spermidine synthase activity(GO:0004766) |
| 0.0 | 0.2 | GO:0044736 | acid-sensing ion channel activity(GO:0044736) |
| 0.0 | 0.2 | GO:0015065 | uridine nucleotide receptor activity(GO:0015065) G-protein coupled pyrimidinergic nucleotide receptor activity(GO:0071553) |
| 0.0 | 1.7 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 0.2 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.0 | 0.3 | GO:0034485 | phosphatidylinositol-3,4,5-trisphosphate 5-phosphatase activity(GO:0034485) |
| 0.0 | 0.3 | GO:0035251 | UDP-glucosyltransferase activity(GO:0035251) |
| 0.0 | 0.1 | GO:0050510 | N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase activity(GO:0050510) |
| 0.0 | 0.1 | GO:0042903 | tubulin deacetylase activity(GO:0042903) |
| 0.0 | 0.2 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.0 | 0.2 | GO:0008865 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.0 | 0.2 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.0 | 0.1 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
| 0.0 | 0.2 | GO:0060072 | large conductance calcium-activated potassium channel activity(GO:0060072) |
| 0.0 | 0.1 | GO:0046573 | lactonohydrolase activity(GO:0046573) acyl-L-homoserine-lactone lactonohydrolase activity(GO:0102007) |
| 0.0 | 0.0 | GO:0033754 | indoleamine 2,3-dioxygenase activity(GO:0033754) |
| 0.0 | 0.4 | GO:0003906 | DNA-(apurinic or apyrimidinic site) lyase activity(GO:0003906) |
| 0.0 | 0.1 | GO:0042806 | fucose binding(GO:0042806) |
| 0.0 | 0.2 | GO:0042834 | peptidoglycan binding(GO:0042834) oligosaccharide binding(GO:0070492) |
| 0.0 | 0.1 | GO:0030345 | extracellular matrix structural constituent conferring compression resistance(GO:0030021) structural constituent of tooth enamel(GO:0030345) |
| 0.0 | 0.2 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.0 | 0.1 | GO:0004301 | epoxide hydrolase activity(GO:0004301) |
| 0.0 | 0.2 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.0 | 0.2 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.0 | 0.1 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.3 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.0 | GO:0070119 | ciliary neurotrophic factor binding(GO:0070119) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 0.3 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.0 | 0.6 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.2 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |