Project
Inflammatory response time course, HUVEC (Wada, 2009)
Navigation
Downloads
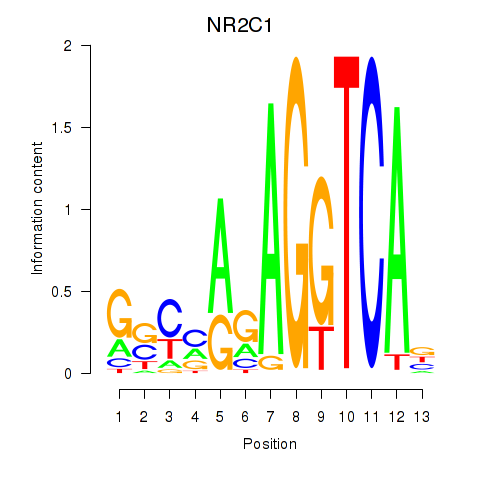
Results for NR2C1
Z-value: 0.97
Transcription factors associated with NR2C1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NR2C1
|
ENSG00000120798.17 | NR2C1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NR2C1 | hg38_v1_chr12_-_95073490_95073574 | 0.61 | 1.4e-03 | Click! |
Activity profile of NR2C1 motif
Sorted Z-values of NR2C1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of NR2C1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_-_159727324 | 6.44 |
ENST00000401980.3
ENST00000545162.5 |
SOD2
|
superoxide dismutase 2 |
| chr6_-_159726871 | 6.05 |
ENST00000535561.5
|
SOD2
|
superoxide dismutase 2 |
| chr2_+_109129199 | 3.11 |
ENST00000309415.8
|
SH3RF3
|
SH3 domain containing ring finger 3 |
| chr2_+_218568865 | 2.75 |
ENST00000295701.9
|
CNOT9
|
CCR4-NOT transcription complex subunit 9 |
| chr6_+_159727561 | 2.68 |
ENST00000631126.2
ENST00000337387.4 |
WTAP
|
WT1 associated protein |
| chr19_+_10086787 | 2.62 |
ENST00000590378.5
ENST00000397881.7 |
SHFL
|
shiftless antiviral inhibitor of ribosomal frameshifting |
| chr6_+_159726998 | 2.55 |
ENST00000614346.4
|
WTAP
|
WT1 associated protein |
| chr13_-_33185994 | 2.11 |
ENST00000255486.8
|
STARD13
|
StAR related lipid transfer domain containing 13 |
| chr19_+_10086305 | 1.85 |
ENST00000253110.16
ENST00000591813.5 |
SHFL
|
shiftless antiviral inhibitor of ribosomal frameshifting |
| chr2_+_74529923 | 1.81 |
ENST00000258080.8
ENST00000352222.7 |
HTRA2
|
HtrA serine peptidase 2 |
| chr3_+_155083523 | 1.75 |
ENST00000680057.1
|
MME
|
membrane metalloendopeptidase |
| chr2_+_74530018 | 1.69 |
ENST00000437202.1
|
HTRA2
|
HtrA serine peptidase 2 |
| chr3_+_155083889 | 1.67 |
ENST00000680282.1
|
MME
|
membrane metalloendopeptidase |
| chr19_+_676385 | 1.46 |
ENST00000166139.9
|
FSTL3
|
follistatin like 3 |
| chr2_+_218568558 | 1.37 |
ENST00000627282.2
ENST00000542068.5 |
CNOT9
|
CCR4-NOT transcription complex subunit 9 |
| chr19_+_48325323 | 1.37 |
ENST00000596315.5
|
EMP3
|
epithelial membrane protein 3 |
| chr19_+_48325522 | 1.36 |
ENST00000594198.1
ENST00000270221.11 ENST00000597279.5 ENST00000593437.1 |
EMP3
|
epithelial membrane protein 3 |
| chr3_-_116444983 | 1.26 |
ENST00000333617.8
|
LSAMP
|
limbic system associated membrane protein |
| chr20_+_45857607 | 1.15 |
ENST00000255152.3
|
ZSWIM3
|
zinc finger SWIM-type containing 3 |
| chr2_+_218568809 | 1.14 |
ENST00000273064.11
|
CNOT9
|
CCR4-NOT transcription complex subunit 9 |
| chr19_+_50415799 | 1.12 |
ENST00000599632.1
|
ENSG00000142539.9
|
novel protein |
| chr11_-_65662931 | 1.02 |
ENST00000615805.4
ENST00000612991.4 |
RELA
|
RELA proto-oncogene, NF-kB subunit |
| chr3_-_58627596 | 1.02 |
ENST00000474531.5
ENST00000465970.1 |
FAM107A
|
family with sequence similarity 107 member A |
| chr10_+_24466487 | 1.00 |
ENST00000396446.5
ENST00000396445.5 ENST00000376451.4 |
KIAA1217
|
KIAA1217 |
| chr17_-_37745018 | 0.96 |
ENST00000613727.4
ENST00000614313.4 ENST00000617811.5 ENST00000621123.4 |
HNF1B
|
HNF1 homeobox B |
| chr14_-_72894091 | 0.92 |
ENST00000556509.6
|
DPF3
|
double PHD fingers 3 |
| chr12_+_131929259 | 0.91 |
ENST00000542167.2
ENST00000376649.8 ENST00000538037.5 ENST00000456665.6 |
PUS1
|
pseudouridine synthase 1 |
| chr11_-_65663083 | 0.91 |
ENST00000308639.13
|
RELA
|
RELA proto-oncogene, NF-kB subunit |
| chr7_+_39623886 | 0.90 |
ENST00000436179.1
|
RALA
|
RAS like proto-oncogene A |
| chr3_-_58627567 | 0.88 |
ENST00000649301.1
|
FAM107A
|
family with sequence similarity 107 member A |
| chr19_+_39125769 | 0.87 |
ENST00000602004.1
ENST00000599470.5 ENST00000321944.8 ENST00000593480.5 ENST00000358301.7 ENST00000593690.5 ENST00000599386.5 |
PAK4
|
p21 (RAC1) activated kinase 4 |
| chr10_+_26697653 | 0.85 |
ENST00000376215.10
|
PDSS1
|
decaprenyl diphosphate synthase subunit 1 |
| chr19_-_48868454 | 0.82 |
ENST00000355496.9
|
PLEKHA4
|
pleckstrin homology domain containing A4 |
| chr3_-_113746218 | 0.81 |
ENST00000497255.1
ENST00000240922.8 ENST00000478020.1 ENST00000493900.5 |
NAA50
|
N-alpha-acetyltransferase 50, NatE catalytic subunit |
| chr12_+_131929219 | 0.80 |
ENST00000322060.9
|
PUS1
|
pseudouridine synthase 1 |
| chr11_-_45918014 | 0.78 |
ENST00000525192.5
|
PEX16
|
peroxisomal biogenesis factor 16 |
| chr19_-_48868590 | 0.75 |
ENST00000263265.11
|
PLEKHA4
|
pleckstrin homology domain containing A4 |
| chr1_+_78490966 | 0.75 |
ENST00000370757.8
ENST00000370756.3 |
PTGFR
|
prostaglandin F receptor |
| chr2_+_209771972 | 0.75 |
ENST00000439458.5
ENST00000272845.10 |
UNC80
|
unc-80 homolog, NALCN channel complex subunit |
| chr11_+_65530639 | 0.74 |
ENST00000279270.10
|
SCYL1
|
SCY1 like pseudokinase 1 |
| chr3_-_50567646 | 0.72 |
ENST00000426034.5
ENST00000441239.5 |
C3orf18
|
chromosome 3 open reading frame 18 |
| chr5_-_1801284 | 0.71 |
ENST00000505818.1
|
MRPL36
|
mitochondrial ribosomal protein L36 |
| chr11_-_45917867 | 0.69 |
ENST00000378750.10
|
PEX16
|
peroxisomal biogenesis factor 16 |
| chr6_+_43770202 | 0.68 |
ENST00000372067.8
ENST00000672860.2 |
VEGFA
|
vascular endothelial growth factor A |
| chr6_+_159727492 | 0.66 |
ENST00000621533.5
|
WTAP
|
WT1 associated protein |
| chr17_-_2025289 | 0.66 |
ENST00000331238.7
|
RTN4RL1
|
reticulon 4 receptor like 1 |
| chr11_-_45917823 | 0.66 |
ENST00000533151.5
ENST00000241041.7 |
PEX16
|
peroxisomal biogenesis factor 16 |
| chr3_+_184186023 | 0.65 |
ENST00000429586.6
ENST00000292808.5 |
ABCF3
|
ATP binding cassette subfamily F member 3 |
| chr2_-_162242998 | 0.64 |
ENST00000627638.2
ENST00000447386.5 |
FAP
|
fibroblast activation protein alpha |
| chr15_-_82262660 | 0.64 |
ENST00000557844.1
ENST00000359445.7 ENST00000268206.12 |
EFL1
|
elongation factor like GTPase 1 |
| chrX_+_22136552 | 0.62 |
ENST00000682888.1
ENST00000684356.1 |
PHEX
|
phosphate regulating endopeptidase homolog X-linked |
| chr3_-_116445458 | 0.61 |
ENST00000490035.7
|
LSAMP
|
limbic system associated membrane protein |
| chr6_+_31547560 | 0.61 |
ENST00000376148.9
ENST00000376145.8 |
NFKBIL1
|
NFKB inhibitor like 1 |
| chr3_-_126517764 | 0.60 |
ENST00000290868.7
|
UROC1
|
urocanate hydratase 1 |
| chrX_-_50470818 | 0.59 |
ENST00000611977.2
|
DGKK
|
diacylglycerol kinase kappa |
| chr1_-_229434086 | 0.58 |
ENST00000366684.7
ENST00000684723.1 ENST00000366683.4 |
ACTA1
|
actin alpha 1, skeletal muscle |
| chr21_+_44012296 | 0.57 |
ENST00000291574.9
ENST00000380221.7 |
TRAPPC10
|
trafficking protein particle complex 10 |
| chr3_+_37243333 | 0.56 |
ENST00000431105.1
|
GOLGA4
|
golgin A4 |
| chr3_+_42856021 | 0.56 |
ENST00000493193.1
|
ACKR2
|
atypical chemokine receptor 2 |
| chr1_+_22052627 | 0.56 |
ENST00000400259.5
ENST00000344548.7 ENST00000315554.13 ENST00000656825.1 ENST00000651171.1 ENST00000652582.1 ENST00000667384.1 |
CDC42
|
cell division cycle 42 |
| chr3_+_184337591 | 0.56 |
ENST00000383847.7
|
FAM131A
|
family with sequence similarity 131 member A |
| chr19_+_16661121 | 0.55 |
ENST00000187762.7
ENST00000599479.1 |
TMEM38A
|
transmembrane protein 38A |
| chr8_+_12951583 | 0.53 |
ENST00000528753.2
|
TRMT9B
|
tRNA methyltransferase 9B (putative) |
| chr16_-_11915991 | 0.52 |
ENST00000420576.6
|
GSPT1
|
G1 to S phase transition 1 |
| chr16_+_30699155 | 0.52 |
ENST00000262518.9
|
SRCAP
|
Snf2 related CREBBP activator protein |
| chr11_-_67508091 | 0.51 |
ENST00000531506.1
|
CDK2AP2
|
cyclin dependent kinase 2 associated protein 2 |
| chr2_-_218270099 | 0.51 |
ENST00000248450.9
ENST00000444053.5 |
AAMP
|
angio associated migratory cell protein |
| chr2_-_162243375 | 0.50 |
ENST00000188790.9
ENST00000443424.5 |
FAP
|
fibroblast activation protein alpha |
| chr21_-_14546351 | 0.50 |
ENST00000619120.4
|
SAMSN1
|
SAM domain, SH3 domain and nuclear localization signals 1 |
| chr7_+_39623547 | 0.49 |
ENST00000005257.7
|
RALA
|
RAS like proto-oncogene A |
| chr21_-_14546297 | 0.49 |
ENST00000400566.6
ENST00000400564.5 |
SAMSN1
|
SAM domain, SH3 domain and nuclear localization signals 1 |
| chr17_+_73193029 | 0.49 |
ENST00000299886.9
ENST00000438720.7 |
COG1
|
component of oligomeric golgi complex 1 |
| chr11_-_67508152 | 0.48 |
ENST00000301488.8
|
CDK2AP2
|
cyclin dependent kinase 2 associated protein 2 |
| chr19_+_6464229 | 0.47 |
ENST00000600229.6
ENST00000356762.7 |
CRB3
|
crumbs cell polarity complex component 3 |
| chr12_-_48105808 | 0.46 |
ENST00000448372.5
|
SENP1
|
SUMO specific peptidase 1 |
| chr11_-_134411854 | 0.45 |
ENST00000392580.5
|
B3GAT1
|
beta-1,3-glucuronyltransferase 1 |
| chr16_-_11915878 | 0.45 |
ENST00000439887.6
ENST00000434724.7 |
GSPT1
|
G1 to S phase transition 1 |
| chr11_-_116792386 | 0.44 |
ENST00000433069.2
ENST00000542499.5 |
APOA5
|
apolipoprotein A5 |
| chr20_+_48921701 | 0.43 |
ENST00000371917.5
|
ARFGEF2
|
ADP ribosylation factor guanine nucleotide exchange factor 2 |
| chr3_+_101827982 | 0.43 |
ENST00000461724.5
ENST00000483180.5 ENST00000394054.6 |
NFKBIZ
|
NFKB inhibitor zeta |
| chr12_-_42237727 | 0.43 |
ENST00000548917.1
|
YAF2
|
YY1 associated factor 2 |
| chr12_-_48106042 | 0.42 |
ENST00000551798.1
ENST00000549518.6 |
SENP1
|
SUMO specific peptidase 1 |
| chr14_-_21098570 | 0.42 |
ENST00000360947.8
|
ZNF219
|
zinc finger protein 219 |
| chr2_+_119366924 | 0.41 |
ENST00000535757.5
ENST00000535617.5 ENST00000627093.2 ENST00000355857.8 ENST00000409094.5 ENST00000542275.5 ENST00000311521.8 |
DBI
|
diazepam binding inhibitor, acyl-CoA binding protein |
| chr2_-_88128049 | 0.41 |
ENST00000393750.3
ENST00000295834.8 |
FABP1
|
fatty acid binding protein 1 |
| chrX_+_16786421 | 0.41 |
ENST00000398155.4
ENST00000380122.10 |
TXLNG
|
taxilin gamma |
| chr2_-_96035974 | 0.41 |
ENST00000434632.5
|
GPAT2
|
glycerol-3-phosphate acyltransferase 2, mitochondrial |
| chr2_-_201451446 | 0.40 |
ENST00000332624.8
ENST00000430254.1 |
TRAK2
|
trafficking kinesin protein 2 |
| chr20_+_38962299 | 0.39 |
ENST00000373325.6
ENST00000373323.8 ENST00000252011.8 ENST00000615559.1 |
DHX35
ENSG00000277581.1
|
DEAH-box helicase 35 novel transcript, sense intronic to DHX35 |
| chr8_+_12951325 | 0.39 |
ENST00000400069.7
|
TRMT9B
|
tRNA methyltransferase 9B (putative) |
| chr4_-_110623051 | 0.39 |
ENST00000557119.2
ENST00000644743.1 |
PITX2
|
paired like homeodomain 2 |
| chr14_+_73569115 | 0.39 |
ENST00000622407.4
ENST00000238651.10 |
ACOT2
|
acyl-CoA thioesterase 2 |
| chr3_-_50567711 | 0.39 |
ENST00000357203.8
|
C3orf18
|
chromosome 3 open reading frame 18 |
| chr22_-_50527689 | 0.39 |
ENST00000652401.1
|
TYMP
|
thymidine phosphorylase |
| chr2_-_42493970 | 0.39 |
ENST00000394973.4
ENST00000306078.2 |
KCNG3
|
potassium voltage-gated channel modifier subfamily G member 3 |
| chr12_-_42238203 | 0.38 |
ENST00000327791.8
ENST00000534854.7 |
YAF2
|
YY1 associated factor 2 |
| chr2_+_119367669 | 0.37 |
ENST00000393103.2
|
DBI
|
diazepam binding inhibitor, acyl-CoA binding protein |
| chr3_+_37243177 | 0.36 |
ENST00000361924.6
ENST00000444882.5 ENST00000356847.8 ENST00000617480.4 ENST00000450863.6 ENST00000429018.5 |
GOLGA4
|
golgin A4 |
| chr17_+_4950147 | 0.35 |
ENST00000522301.5
|
ENO3
|
enolase 3 |
| chr22_-_31140494 | 0.34 |
ENST00000215885.4
|
PLA2G3
|
phospholipase A2 group III |
| chr2_-_218269619 | 0.33 |
ENST00000447885.1
ENST00000420660.5 |
AAMP
|
angio associated migratory cell protein |
| chr16_-_88686453 | 0.33 |
ENST00000332281.6
|
SNAI3
|
snail family transcriptional repressor 3 |
| chr21_-_31558977 | 0.32 |
ENST00000286827.7
ENST00000541036.5 |
TIAM1
|
TIAM Rac1 associated GEF 1 |
| chr4_-_75673112 | 0.32 |
ENST00000395719.7
ENST00000677489.1 |
G3BP2
|
G3BP stress granule assembly factor 2 |
| chr15_+_90184912 | 0.30 |
ENST00000561085.1
ENST00000332496.10 |
SEMA4B
|
semaphorin 4B |
| chr1_-_166975407 | 0.30 |
ENST00000528703.5
ENST00000525740.5 ENST00000529387.5 ENST00000469934.6 ENST00000529071.5 ENST00000526687.1 ENST00000271417.8 |
ILDR2
|
immunoglobulin like domain containing receptor 2 |
| chr3_-_150546403 | 0.29 |
ENST00000239944.7
ENST00000491660.1 ENST00000487153.1 |
SERP1
|
stress associated endoplasmic reticulum protein 1 |
| chr10_+_125973373 | 0.29 |
ENST00000417114.5
ENST00000445510.5 ENST00000368691.5 |
FANK1
|
fibronectin type III and ankyrin repeat domains 1 |
| chr11_-_64246190 | 0.29 |
ENST00000392210.6
|
PPP1R14B
|
protein phosphatase 1 regulatory inhibitor subunit 14B |
| chr14_-_21098848 | 0.28 |
ENST00000556174.5
ENST00000554478.5 ENST00000553980.1 ENST00000421093.6 |
ZNF219
|
zinc finger protein 219 |
| chr11_+_7987314 | 0.28 |
ENST00000531572.2
ENST00000651655.1 |
EIF3F
|
eukaryotic translation initiation factor 3 subunit F |
| chr11_-_66438788 | 0.27 |
ENST00000329819.4
ENST00000310999.11 ENST00000430466.6 |
MRPL11
|
mitochondrial ribosomal protein L11 |
| chr12_+_95858928 | 0.26 |
ENST00000266735.9
ENST00000553192.5 ENST00000552085.1 |
SNRPF
|
small nuclear ribonucleoprotein polypeptide F |
| chr7_+_121873152 | 0.26 |
ENST00000650826.1
ENST00000650728.1 ENST00000393386.7 ENST00000651390.1 ENST00000651842.1 ENST00000650681.1 |
PTPRZ1
|
protein tyrosine phosphatase receptor type Z1 |
| chr19_-_4066892 | 0.25 |
ENST00000322357.9
|
ZBTB7A
|
zinc finger and BTB domain containing 7A |
| chr2_-_218568291 | 0.25 |
ENST00000418019.5
ENST00000454775.5 ENST00000338465.5 ENST00000415516.5 ENST00000258399.8 |
USP37
|
ubiquitin specific peptidase 37 |
| chr2_+_201451711 | 0.25 |
ENST00000194530.8
ENST00000392249.6 |
STRADB
|
STE20 related adaptor beta |
| chr17_+_14069489 | 0.24 |
ENST00000429152.6
ENST00000261643.8 |
COX10
|
cytochrome c oxidase assembly factor heme A:farnesyltransferase COX10 |
| chr16_+_2205708 | 0.24 |
ENST00000397124.5
ENST00000565250.1 |
MLST8
|
MTOR associated protein, LST8 homolog |
| chr4_-_75672868 | 0.23 |
ENST00000678123.1
ENST00000678578.1 ENST00000677876.1 ENST00000676839.1 ENST00000678265.1 |
G3BP2
|
G3BP stress granule assembly factor 2 |
| chr11_+_94768331 | 0.23 |
ENST00000317829.12
ENST00000433060.3 |
AMOTL1
|
angiomotin like 1 |
| chr14_+_73537135 | 0.23 |
ENST00000311148.9
|
ACOT1
|
acyl-CoA thioesterase 1 |
| chr4_-_75673139 | 0.23 |
ENST00000677566.1
ENST00000503660.5 ENST00000677060.1 ENST00000678552.1 |
G3BP2
|
G3BP stress granule assembly factor 2 |
| chr11_-_64245816 | 0.22 |
ENST00000542235.1
|
PPP1R14B
|
protein phosphatase 1 regulatory inhibitor subunit 14B |
| chr1_+_160127672 | 0.22 |
ENST00000447527.1
|
ATP1A2
|
ATPase Na+/K+ transporting subunit alpha 2 |
| chr12_-_22334683 | 0.22 |
ENST00000404299.3
ENST00000396037.9 |
ST8SIA1
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 1 |
| chr9_-_35685462 | 0.22 |
ENST00000607559.1
|
TPM2
|
tropomyosin 2 |
| chr3_-_113746059 | 0.21 |
ENST00000477813.5
|
NAA50
|
N-alpha-acetyltransferase 50, NatE catalytic subunit |
| chr10_+_62804710 | 0.21 |
ENST00000373783.3
|
ADO
|
2-aminoethanethiol dioxygenase |
| chr1_-_226186673 | 0.21 |
ENST00000366812.6
|
ACBD3
|
acyl-CoA binding domain containing 3 |
| chr11_+_65615755 | 0.21 |
ENST00000355703.4
|
PCNX3
|
pecanex 3 |
| chr7_+_121873089 | 0.20 |
ENST00000651065.1
|
PTPRZ1
|
protein tyrosine phosphatase receptor type Z1 |
| chr3_-_113746185 | 0.20 |
ENST00000616174.1
|
NAA50
|
N-alpha-acetyltransferase 50, NatE catalytic subunit |
| chr3_+_113747022 | 0.20 |
ENST00000273398.8
ENST00000496747.5 ENST00000475322.1 |
ATP6V1A
|
ATPase H+ transporting V1 subunit A |
| chr14_+_21768482 | 0.20 |
ENST00000390428.3
|
TRAV6
|
T cell receptor alpha variable 6 |
| chr2_-_213150236 | 0.20 |
ENST00000442445.1
ENST00000342002.6 |
IKZF2
|
IKAROS family zinc finger 2 |
| chr4_+_26319636 | 0.20 |
ENST00000342295.6
ENST00000506956.5 ENST00000512671.6 ENST00000345843.8 |
RBPJ
|
recombination signal binding protein for immunoglobulin kappa J region |
| chr9_-_34397800 | 0.18 |
ENST00000297623.7
|
C9orf24
|
chromosome 9 open reading frame 24 |
| chr14_-_77320741 | 0.18 |
ENST00000682795.1
ENST00000682247.1 |
POMT2
|
protein O-mannosyltransferase 2 |
| chr20_+_32358303 | 0.18 |
ENST00000651418.1
ENST00000375687.10 ENST00000542461.5 ENST00000613218.4 ENST00000646367.1 ENST00000620121.4 |
ASXL1
|
ASXL transcriptional regulator 1 |
| chr6_+_33204645 | 0.18 |
ENST00000374662.4
|
HSD17B8
|
hydroxysteroid 17-beta dehydrogenase 8 |
| chr11_-_123885627 | 0.17 |
ENST00000528595.1
ENST00000375026.7 |
TMEM225
|
transmembrane protein 225 |
| chrX_-_153944621 | 0.17 |
ENST00000393700.8
|
RENBP
|
renin binding protein |
| chr1_+_44674688 | 0.17 |
ENST00000418644.5
ENST00000458657.6 ENST00000535358.6 ENST00000441519.5 ENST00000445071.5 |
ARMH1
|
armadillo like helical domain containing 1 |
| chr15_+_82262781 | 0.16 |
ENST00000566205.1
ENST00000339465.5 ENST00000569120.1 ENST00000682753.1 ENST00000566861.5 ENST00000565432.1 |
SAXO2
|
stabilizer of axonemal microtubules 2 |
| chr12_-_24562438 | 0.15 |
ENST00000646273.1
ENST00000659413.1 ENST00000446891.7 |
SOX5
|
SRY-box transcription factor 5 |
| chr1_+_110034607 | 0.15 |
ENST00000369795.8
|
STRIP1
|
striatin interacting protein 1 |
| chrX_+_38561530 | 0.14 |
ENST00000378482.7
ENST00000286824.6 |
TSPAN7
|
tetraspanin 7 |
| chr2_-_74529670 | 0.14 |
ENST00000377526.4
|
AUP1
|
AUP1 lipid droplet regulating VLDL assembly factor |
| chr12_-_53335737 | 0.13 |
ENST00000303846.3
|
SP7
|
Sp7 transcription factor |
| chr13_+_21140774 | 0.13 |
ENST00000450573.5
ENST00000467636.1 |
SAP18
|
Sin3A associated protein 18 |
| chr8_-_80030232 | 0.13 |
ENST00000518271.1
ENST00000276585.9 ENST00000521605.1 |
MRPS28
|
mitochondrial ribosomal protein S28 |
| chr2_-_38076076 | 0.13 |
ENST00000614273.1
ENST00000610745.5 ENST00000490576.1 |
CYP1B1
|
cytochrome P450 family 1 subfamily B member 1 |
| chr1_-_29181809 | 0.12 |
ENST00000466448.4
ENST00000373795.7 |
SRSF4
|
serine and arginine rich splicing factor 4 |
| chr2_+_46698909 | 0.12 |
ENST00000650611.1
ENST00000306503.5 |
LINC01118
SOCS5
|
long intergenic non-protein coding RNA 1118 suppressor of cytokine signaling 5 |
| chr5_-_88268801 | 0.12 |
ENST00000506536.5
ENST00000512429.5 ENST00000514135.5 ENST00000296595.11 ENST00000509387.5 |
TMEM161B
|
transmembrane protein 161B |
| chr11_-_26722051 | 0.10 |
ENST00000396005.8
|
SLC5A12
|
solute carrier family 5 member 12 |
| chr19_+_11239602 | 0.10 |
ENST00000252453.12
|
ANGPTL8
|
angiopoietin like 8 |
| chr10_-_121596117 | 0.10 |
ENST00000351936.11
|
FGFR2
|
fibroblast growth factor receptor 2 |
| chr3_+_150546671 | 0.10 |
ENST00000487799.5
|
EIF2A
|
eukaryotic translation initiation factor 2A |
| chr1_-_155911340 | 0.10 |
ENST00000368323.8
|
RIT1
|
Ras like without CAAX 1 |
| chr15_+_45129933 | 0.10 |
ENST00000321429.8
ENST00000389037.7 ENST00000558322.5 |
DUOX1
|
dual oxidase 1 |
| chr1_+_40247926 | 0.09 |
ENST00000372766.4
|
TMCO2
|
transmembrane and coiled-coil domains 2 |
| chr2_-_42361198 | 0.09 |
ENST00000234301.3
|
COX7A2L
|
cytochrome c oxidase subunit 7A2 like |
| chr22_+_41381923 | 0.08 |
ENST00000266304.9
|
TEF
|
TEF transcription factor, PAR bZIP family member |
| chr14_+_41608344 | 0.08 |
ENST00000554120.5
|
LRFN5
|
leucine rich repeat and fibronectin type III domain containing 5 |
| chr1_-_982086 | 0.07 |
ENST00000341290.6
|
PERM1
|
PPARGC1 and ESRR induced regulator, muscle 1 |
| chr1_-_113905020 | 0.07 |
ENST00000432415.5
ENST00000369571.2 ENST00000256658.8 ENST00000369564.5 |
AP4B1
|
adaptor related protein complex 4 subunit beta 1 |
| chr5_+_79612410 | 0.07 |
ENST00000423041.6
ENST00000504233.5 ENST00000453514.6 ENST00000428308.6 |
TENT2
|
terminal nucleotidyltransferase 2 |
| chr19_+_43596388 | 0.06 |
ENST00000391965.6
ENST00000525771.1 |
ZNF576
|
zinc finger protein 576 |
| chr3_+_150546765 | 0.06 |
ENST00000406576.7
ENST00000460851.6 ENST00000482093.5 ENST00000273435.9 |
EIF2A
|
eukaryotic translation initiation factor 2A |
| chr1_-_19923617 | 0.06 |
ENST00000375116.3
|
PLA2G2E
|
phospholipase A2 group IIE |
| chr7_+_151048526 | 0.06 |
ENST00000349064.10
|
ASIC3
|
acid sensing ion channel subunit 3 |
| chr5_+_171419635 | 0.06 |
ENST00000274625.6
|
FGF18
|
fibroblast growth factor 18 |
| chr17_-_41586887 | 0.06 |
ENST00000167586.7
|
KRT14
|
keratin 14 |
| chr19_-_43198079 | 0.05 |
ENST00000597374.5
ENST00000599371.1 |
PSG4
|
pregnancy specific beta-1-glycoprotein 4 |
| chrX_-_40177213 | 0.05 |
ENST00000672922.2
ENST00000378455.8 ENST00000342274.8 |
BCOR
|
BCL6 corepressor |
| chr22_-_40862070 | 0.05 |
ENST00000307221.5
|
DNAJB7
|
DnaJ heat shock protein family (Hsp40) member B7 |
| chr1_+_212624472 | 0.04 |
ENST00000294829.5
|
FAM71A
|
family with sequence similarity 71 member A |
| chr16_-_4242068 | 0.04 |
ENST00000399609.7
|
SRL
|
sarcalumenin |
| chr12_+_109139397 | 0.04 |
ENST00000377854.9
ENST00000377848.7 |
ACACB
|
acetyl-CoA carboxylase beta |
| chr1_-_30908681 | 0.03 |
ENST00000339394.7
|
SDC3
|
syndecan 3 |
| chr14_-_21023954 | 0.03 |
ENST00000554094.5
|
NDRG2
|
NDRG family member 2 |
| chr17_-_41047267 | 0.03 |
ENST00000542137.1
ENST00000391419.3 |
KRTAP2-1
|
keratin associated protein 2-1 |
| chr3_+_46497970 | 0.03 |
ENST00000296142.4
|
RTP3
|
receptor transporter protein 3 |
| chr6_+_10585748 | 0.03 |
ENST00000265012.5
|
GCNT2
|
glucosaminyl (N-acetyl) transferase 2 (I blood group) |
| chr6_+_54307856 | 0.02 |
ENST00000370869.7
|
TINAG
|
tubulointerstitial nephritis antigen |
| chr22_+_24423597 | 0.02 |
ENST00000424232.5
ENST00000611543.4 ENST00000486108.1 |
ADORA2A
|
adenosine A2a receptor |
| chr12_-_118372883 | 0.02 |
ENST00000542532.5
ENST00000392533.8 |
TAOK3
|
TAO kinase 3 |
| chrX_-_43882411 | 0.02 |
ENST00000378069.5
|
MAOB
|
monoamine oxidase B |
| chr14_-_77320855 | 0.02 |
ENST00000556394.2
ENST00000261534.9 |
POMT2
|
protein O-mannosyltransferase 2 |
| chr14_-_77320813 | 0.02 |
ENST00000682467.1
|
POMT2
|
protein O-mannosyltransferase 2 |
| chr7_-_143647646 | 0.00 |
ENST00000636941.1
|
TCAF2C
|
TRPM8 channel associated factor 2C |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.5 | 12.5 | GO:0001315 | age-dependent response to oxidative stress(GO:0001306) age-dependent response to reactive oxygen species(GO:0001315) regulation of systemic arterial blood pressure by acetylcholine(GO:0003068) vasodilation by acetylcholine involved in regulation of systemic arterial blood pressure(GO:0003069) regulation of systemic arterial blood pressure by neurotransmitter(GO:0003070) age-dependent general metabolic decline(GO:0007571) |
| 1.1 | 5.3 | GO:2000325 | regulation of ligand-dependent nuclear receptor transcription coactivator activity(GO:2000325) positive regulation of ligand-dependent nuclear receptor transcription coactivator activity(GO:2000327) |
| 0.9 | 3.5 | GO:1904924 | negative regulation of mitophagy in response to mitochondrial depolarization(GO:1904924) |
| 0.7 | 5.9 | GO:0080009 | mRNA methylation(GO:0080009) |
| 0.6 | 1.9 | GO:0006117 | acetaldehyde metabolic process(GO:0006117) |
| 0.6 | 1.7 | GO:0070902 | mitochondrial tRNA pseudouridine synthesis(GO:0070902) |
| 0.5 | 2.1 | GO:0032581 | ER-dependent peroxisome organization(GO:0032581) |
| 0.4 | 1.2 | GO:0071962 | mitotic sister chromatid cohesion, centromeric(GO:0071962) |
| 0.4 | 2.1 | GO:0097498 | endothelial tube lumen extension(GO:0097498) |
| 0.3 | 3.4 | GO:0071492 | cellular response to UV-A(GO:0071492) |
| 0.3 | 1.0 | GO:0002184 | cytoplasmic translational termination(GO:0002184) |
| 0.3 | 1.0 | GO:0035565 | regulation of pronephros size(GO:0035565) |
| 0.3 | 1.1 | GO:0097325 | melanocyte proliferation(GO:0097325) |
| 0.2 | 0.7 | GO:1903572 | regulation of protein kinase D signaling(GO:1903570) positive regulation of protein kinase D signaling(GO:1903572) |
| 0.2 | 0.6 | GO:0071338 | submandibular salivary gland formation(GO:0060661) hair follicle cell proliferation(GO:0071335) regulation of hair follicle cell proliferation(GO:0071336) positive regulation of hair follicle cell proliferation(GO:0071338) |
| 0.2 | 0.6 | GO:1904383 | response to sodium phosphate(GO:1904383) |
| 0.1 | 1.0 | GO:2000035 | regulation of stem cell division(GO:2000035) |
| 0.1 | 0.4 | GO:0021763 | subthalamic nucleus development(GO:0021763) prolactin secreting cell differentiation(GO:0060127) superior vena cava morphogenesis(GO:0060578) |
| 0.1 | 0.6 | GO:0019556 | histidine catabolic process to glutamate and formamide(GO:0019556) histidine catabolic process to glutamate and formate(GO:0019557) formamide metabolic process(GO:0043606) |
| 0.1 | 0.6 | GO:0090131 | mesenchyme migration(GO:0090131) |
| 0.1 | 0.9 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.1 | 0.4 | GO:0010902 | positive regulation of very-low-density lipoprotein particle remodeling(GO:0010902) |
| 0.1 | 2.7 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.1 | 0.4 | GO:0098937 | dendritic transport(GO:0098935) anterograde dendritic transport(GO:0098937) |
| 0.1 | 1.4 | GO:0051665 | membrane raft localization(GO:0051665) |
| 0.1 | 1.5 | GO:0032926 | negative regulation of activin receptor signaling pathway(GO:0032926) |
| 0.1 | 0.3 | GO:0036135 | Schwann cell migration(GO:0036135) regulation of Schwann cell migration(GO:1900147) |
| 0.1 | 0.8 | GO:0071798 | response to prostaglandin D(GO:0071798) cellular response to prostaglandin D stimulus(GO:0071799) |
| 0.1 | 0.2 | GO:1900920 | regulation of amino acid uptake involved in synaptic transmission(GO:0051941) regulation of glutamate uptake involved in transmission of nerve impulse(GO:0051946) regulation of L-glutamate import(GO:1900920) |
| 0.1 | 1.9 | GO:0035641 | locomotory exploration behavior(GO:0035641) |
| 0.1 | 0.2 | GO:0035359 | negative regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035359) |
| 0.1 | 0.6 | GO:0042256 | mature ribosome assembly(GO:0042256) |
| 0.0 | 0.4 | GO:0006072 | glycerol-3-phosphate metabolic process(GO:0006072) |
| 0.0 | 0.8 | GO:0007253 | cytoplasmic sequestering of NF-kappaB(GO:0007253) |
| 0.0 | 0.1 | GO:0071603 | endothelial cell-cell adhesion(GO:0071603) |
| 0.0 | 0.4 | GO:0032000 | positive regulation of fatty acid beta-oxidation(GO:0032000) |
| 0.0 | 0.7 | GO:0048681 | negative regulation of axon regeneration(GO:0048681) |
| 0.0 | 1.2 | GO:0036151 | phosphatidylcholine acyl-chain remodeling(GO:0036151) |
| 0.0 | 1.0 | GO:0051290 | protein heterotetramerization(GO:0051290) |
| 0.0 | 0.2 | GO:1990928 | response to amino acid starvation(GO:1990928) |
| 0.0 | 0.6 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.0 | 0.9 | GO:0000042 | protein targeting to Golgi(GO:0000042) |
| 0.0 | 0.6 | GO:0042994 | cytoplasmic sequestering of transcription factor(GO:0042994) |
| 0.0 | 0.2 | GO:1904098 | regulation of protein O-linked glycosylation(GO:1904098) positive regulation of protein O-linked glycosylation(GO:1904100) |
| 0.0 | 0.1 | GO:0045629 | negative regulation of T-helper 2 cell differentiation(GO:0045629) |
| 0.0 | 0.5 | GO:0070445 | oligodendrocyte progenitor proliferation(GO:0070444) regulation of oligodendrocyte progenitor proliferation(GO:0070445) |
| 0.0 | 0.2 | GO:0006051 | mannosamine metabolic process(GO:0006050) N-acetylmannosamine metabolic process(GO:0006051) |
| 0.0 | 0.2 | GO:1905068 | positive regulation of ephrin receptor signaling pathway(GO:1901189) positive regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901297) positive regulation of canonical Wnt signaling pathway involved in heart development(GO:1905068) |
| 0.0 | 0.2 | GO:0035871 | protein K11-linked deubiquitination(GO:0035871) |
| 0.0 | 0.1 | GO:0042335 | cuticle development(GO:0042335) |
| 0.0 | 0.6 | GO:0046339 | diacylglycerol metabolic process(GO:0046339) |
| 0.0 | 1.0 | GO:0050869 | negative regulation of B cell activation(GO:0050869) |
| 0.0 | 0.2 | GO:0055059 | asymmetric neuroblast division(GO:0055059) |
| 0.0 | 0.1 | GO:0035602 | fibroblast growth factor receptor signaling pathway involved in negative regulation of apoptotic process in bone marrow(GO:0035602) fibroblast growth factor receptor signaling pathway involved in hemopoiesis(GO:0035603) fibroblast growth factor receptor signaling pathway involved in positive regulation of cell proliferation in bone marrow(GO:0035604) fibroblast growth factor receptor signaling pathway involved in mammary gland specification(GO:0060595) mammary gland bud formation(GO:0060615) branch elongation involved in salivary gland morphogenesis(GO:0060667) mesenchymal cell differentiation involved in lung development(GO:0060915) |
| 0.0 | 0.3 | GO:0060124 | positive regulation of growth hormone secretion(GO:0060124) |
| 0.0 | 0.4 | GO:0008655 | pyrimidine-containing compound salvage(GO:0008655) pyrimidine nucleoside salvage(GO:0043097) |
| 0.0 | 0.3 | GO:0075522 | IRES-dependent viral translational initiation(GO:0075522) |
| 0.0 | 0.2 | GO:0000098 | sulfur amino acid catabolic process(GO:0000098) |
| 0.0 | 0.5 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.0 | 0.4 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.6 | GO:0000038 | very long-chain fatty acid metabolic process(GO:0000038) |
| 0.0 | 0.1 | GO:0000415 | negative regulation of histone H3-K36 methylation(GO:0000415) |
| 0.0 | 0.3 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.0 | 0.1 | GO:0050915 | iron chelate transport(GO:0015688) siderophore transport(GO:0015891) sensory perception of sour taste(GO:0050915) |
| 0.0 | 1.1 | GO:0042273 | ribosomal large subunit biogenesis(GO:0042273) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 5.9 | GO:0036396 | MIS complex(GO:0036396) mRNA editing complex(GO:0045293) |
| 0.6 | 1.9 | GO:0035525 | NF-kappaB p50/p65 complex(GO:0035525) |
| 0.4 | 5.3 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.3 | 1.0 | GO:0018444 | translation release factor complex(GO:0018444) |
| 0.2 | 3.5 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.2 | 1.2 | GO:0031415 | NatA complex(GO:0031415) |
| 0.1 | 0.5 | GO:0072534 | perineuronal net(GO:0072534) |
| 0.1 | 1.1 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.1 | 2.1 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.1 | 1.0 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.1 | 0.4 | GO:0032280 | symmetric synapse(GO:0032280) |
| 0.0 | 0.7 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 0.2 | GO:0035517 | PR-DUB complex(GO:0035517) |
| 0.0 | 0.3 | GO:0034715 | pICln-Sm protein complex(GO:0034715) |
| 0.0 | 0.6 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 0.3 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.0 | 0.1 | GO:0000839 | Hrd1p ubiquitin ligase ERAD-L complex(GO:0000839) |
| 0.0 | 0.2 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.0 | 0.4 | GO:0045179 | apical cortex(GO:0045179) |
| 0.0 | 0.3 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.0 | 0.8 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 0.9 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.0 | 0.2 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.0 | 14.3 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 0.2 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.0 | 2.1 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 3.2 | GO:0005903 | brush border(GO:0005903) |
| 0.0 | 0.8 | GO:0005865 | striated muscle thin filament(GO:0005865) |
| 0.0 | 0.4 | GO:0042627 | chylomicron(GO:0042627) |
| 0.0 | 0.2 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.0 | 0.7 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 0.3 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 1.9 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.2 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 12.5 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.6 | 1.7 | GO:0004730 | pseudouridylate synthase activity(GO:0004730) |
| 0.5 | 1.4 | GO:0031755 | endothelial differentiation G-protein coupled receptor binding(GO:0031753) Edg-2 lysophosphatidic acid receptor binding(GO:0031755) |
| 0.4 | 1.2 | GO:0052858 | peptidyl-lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0052858) |
| 0.3 | 0.8 | GO:0030156 | benzodiazepine receptor binding(GO:0030156) |
| 0.3 | 0.8 | GO:0004958 | prostaglandin F receptor activity(GO:0004958) |
| 0.2 | 1.9 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.2 | 0.9 | GO:0070137 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 0.2 | 0.6 | GO:0032427 | GBD domain binding(GO:0032427) |
| 0.1 | 0.7 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.1 | 0.4 | GO:0009032 | thymidine phosphorylase activity(GO:0009032) pyrimidine-nucleoside phosphorylase activity(GO:0016154) |
| 0.1 | 1.0 | GO:0003747 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.1 | 0.5 | GO:0015018 | galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity(GO:0015018) |
| 0.1 | 5.4 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.1 | 0.4 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.1 | 0.7 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.1 | 1.4 | GO:0048185 | activin binding(GO:0048185) |
| 0.1 | 0.4 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.1 | 0.4 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.1 | 0.2 | GO:0047025 | 3-oxoacyl-[acyl-carrier-protein] reductase (NADH) activity(GO:0047025) |
| 0.1 | 0.7 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.0 | 0.6 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.0 | 1.1 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 0.3 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.0 | 0.5 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.0 | 0.8 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.2 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.0 | 1.6 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 0.6 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.9 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.0 | 0.3 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.0 | 0.2 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.0 | 4.0 | GO:0008238 | exopeptidase activity(GO:0008238) |
| 0.0 | 4.3 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 0.2 | GO:0004311 | farnesyltranstransferase activity(GO:0004311) |
| 0.0 | 0.6 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 1.0 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.0 | 0.2 | GO:1990239 | steroid hormone binding(GO:1990239) |
| 0.0 | 0.8 | GO:0030159 | receptor signaling complex scaffold activity(GO:0030159) |
| 0.0 | 0.5 | GO:0019198 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 0.1 | GO:0005111 | type 2 fibroblast growth factor receptor binding(GO:0005111) |
| 0.0 | 0.5 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.0 | 0.2 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.0 | 0.3 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 0.1 | GO:0044736 | acid-sensing ion channel activity(GO:0044736) |
| 0.0 | 0.2 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.0 | 0.4 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 0.1 | GO:0097027 | ubiquitin-protein transferase activator activity(GO:0097027) |
| 0.0 | 0.1 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 13.9 | PID FOXO PATHWAY | FoxO family signaling |
| 0.1 | 1.9 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.0 | 0.7 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 0.6 | ST GAQ PATHWAY | G alpha q Pathway |
| 0.0 | 1.8 | SIG CHEMOTAXIS | Genes related to chemotaxis |
| 0.0 | 0.9 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.9 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 0.3 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.0 | 0.6 | PID RHOA PATHWAY | RhoA signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.9 | REACTOME REGULATED PROTEOLYSIS OF P75NTR | Genes involved in Regulated proteolysis of p75NTR |
| 0.1 | 0.8 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.1 | 1.4 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.0 | 0.7 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.9 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 0.6 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.0 | 0.4 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 0.4 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 0.4 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.0 | 0.3 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.0 | 1.0 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.0 | 0.6 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.5 | REACTOME A TETRASACCHARIDE LINKER SEQUENCE IS REQUIRED FOR GAG SYNTHESIS | Genes involved in A tetrasaccharide linker sequence is required for GAG synthesis |
| 0.0 | 0.2 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.0 | 0.5 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |