Project
Inflammatory response time course, HUVEC (Wada, 2009)
Navigation
Downloads
Results for NR2E1
Z-value: 0.74
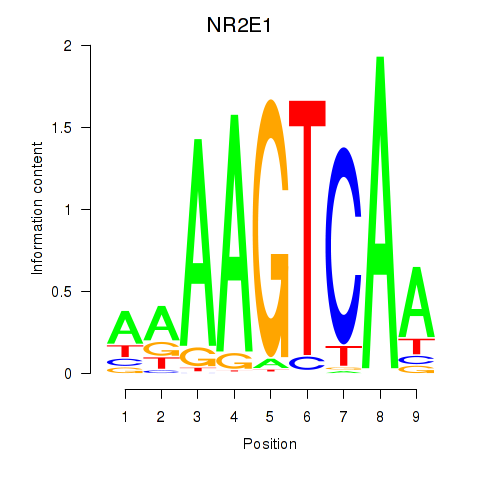
Transcription factors associated with NR2E1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NR2E1
|
ENSG00000112333.12 | NR2E1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NR2E1 | hg38_v1_chr6_+_108166015_108166034 | 0.32 | 1.2e-01 | Click! |
Activity profile of NR2E1 motif
Sorted Z-values of NR2E1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of NR2E1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_169734064 | 3.01 |
ENST00000333360.12
|
SELE
|
selectin E |
| chr3_-_64687613 | 1.95 |
ENST00000295903.8
|
ADAMTS9
|
ADAM metallopeptidase with thrombospondin type 1 motif 9 |
| chr3_-_64687992 | 1.83 |
ENST00000498707.5
|
ADAMTS9
|
ADAM metallopeptidase with thrombospondin type 1 motif 9 |
| chr9_-_16728165 | 1.76 |
ENST00000603713.5
ENST00000603313.5 |
BNC2
|
basonuclin 2 |
| chr16_+_57620077 | 1.57 |
ENST00000567835.5
ENST00000569372.5 ENST00000563548.5 ENST00000562003.5 |
ADGRG1
|
adhesion G protein-coupled receptor G1 |
| chr11_-_8810635 | 1.49 |
ENST00000527510.5
ENST00000528527.5 ENST00000313726.11 ENST00000528523.5 |
DENND2B
|
DENN domain containing 2B |
| chr15_+_67067780 | 1.34 |
ENST00000679624.1
|
SMAD3
|
SMAD family member 3 |
| chr17_-_43546323 | 1.21 |
ENST00000545954.5
ENST00000319349.10 |
ETV4
|
ETS variant transcription factor 4 |
| chr8_+_103372388 | 1.19 |
ENST00000520337.1
|
CTHRC1
|
collagen triple helix repeat containing 1 |
| chr4_-_121164314 | 1.19 |
ENST00000057513.8
|
TNIP3
|
TNFAIP3 interacting protein 3 |
| chr12_+_55681647 | 1.03 |
ENST00000614691.1
|
METTL7B
|
methyltransferase like 7B |
| chr12_+_55681711 | 1.01 |
ENST00000394252.4
|
METTL7B
|
methyltransferase like 7B |
| chr4_+_95091462 | 1.00 |
ENST00000264568.8
|
BMPR1B
|
bone morphogenetic protein receptor type 1B |
| chr1_+_212565334 | 1.00 |
ENST00000366981.8
ENST00000366987.6 |
ATF3
|
activating transcription factor 3 |
| chr11_+_35176611 | 0.99 |
ENST00000279452.10
|
CD44
|
CD44 molecule (Indian blood group) |
| chr12_+_63844758 | 0.98 |
ENST00000631006.2
|
SRGAP1
|
SLIT-ROBO Rho GTPase activating protein 1 |
| chr11_+_35176696 | 0.94 |
ENST00000528455.5
|
CD44
|
CD44 molecule (Indian blood group) |
| chr20_+_54475647 | 0.93 |
ENST00000395939.5
|
DOK5
|
docking protein 5 |
| chr11_+_35176639 | 0.91 |
ENST00000527889.6
|
CD44
|
CD44 molecule (Indian blood group) |
| chr2_+_230759918 | 0.90 |
ENST00000614925.1
|
CAB39
|
calcium binding protein 39 |
| chr11_-_128587551 | 0.90 |
ENST00000392668.8
|
ETS1
|
ETS proto-oncogene 1, transcription factor |
| chr11_+_35176575 | 0.89 |
ENST00000526000.6
|
CD44
|
CD44 molecule (Indian blood group) |
| chr6_+_31948956 | 0.89 |
ENST00000483004.1
|
CFB
|
complement factor B |
| chr1_+_165895564 | 0.77 |
ENST00000469256.6
|
UCK2
|
uridine-cytidine kinase 2 |
| chr20_+_63738270 | 0.76 |
ENST00000467211.1
|
ENSG00000273047.1
|
novel transcript, LIME1-SLC2A4RG readthrough |
| chr8_-_118111806 | 0.75 |
ENST00000378204.7
|
EXT1
|
exostosin glycosyltransferase 1 |
| chr12_+_63844663 | 0.74 |
ENST00000355086.8
|
SRGAP1
|
SLIT-ROBO Rho GTPase activating protein 1 |
| chr2_-_207165923 | 0.72 |
ENST00000309446.11
|
KLF7
|
Kruppel like factor 7 |
| chr19_+_13023958 | 0.70 |
ENST00000587760.5
ENST00000585575.5 |
NFIX
|
nuclear factor I X |
| chr9_-_96302142 | 0.69 |
ENST00000648799.1
|
HSD17B3
|
hydroxysteroid 17-beta dehydrogenase 3 |
| chr2_-_231125032 | 0.68 |
ENST00000258400.4
|
HTR2B
|
5-hydroxytryptamine receptor 2B |
| chr5_-_59216826 | 0.66 |
ENST00000638939.1
|
PDE4D
|
phosphodiesterase 4D |
| chr7_-_13986439 | 0.66 |
ENST00000443608.5
ENST00000438956.5 |
ETV1
|
ETS variant transcription factor 1 |
| chr1_+_158461574 | 0.62 |
ENST00000641432.1
ENST00000641460.1 ENST00000641535.1 ENST00000641971.1 |
OR10K1
|
olfactory receptor family 10 subfamily K member 1 |
| chr7_-_108240049 | 0.61 |
ENST00000379022.8
|
NRCAM
|
neuronal cell adhesion molecule |
| chr17_-_42681840 | 0.58 |
ENST00000332438.4
|
CCR10
|
C-C motif chemokine receptor 10 |
| chr3_-_71132099 | 0.57 |
ENST00000650188.1
ENST00000648121.1 ENST00000648794.1 ENST00000649592.1 |
FOXP1
|
forkhead box P1 |
| chr17_-_3691887 | 0.56 |
ENST00000552050.5
|
P2RX5
|
purinergic receptor P2X 5 |
| chr18_+_80109236 | 0.56 |
ENST00000262198.9
ENST00000560752.5 |
ADNP2
|
ADNP homeobox 2 |
| chr16_-_66918839 | 0.55 |
ENST00000565235.2
ENST00000568632.5 ENST00000565796.5 |
CDH16
|
cadherin 16 |
| chr12_+_111034136 | 0.54 |
ENST00000261726.11
|
CUX2
|
cut like homeobox 2 |
| chr16_-_66918876 | 0.54 |
ENST00000570262.5
ENST00000299752.9 ENST00000394055.7 |
CDH16
|
cadherin 16 |
| chr12_-_55842950 | 0.54 |
ENST00000548629.5
|
MMP19
|
matrix metallopeptidase 19 |
| chr3_+_101827982 | 0.53 |
ENST00000461724.5
ENST00000483180.5 ENST00000394054.6 |
NFKBIZ
|
NFKB inhibitor zeta |
| chr5_-_161546671 | 0.52 |
ENST00000517547.5
|
GABRB2
|
gamma-aminobutyric acid type A receptor subunit beta2 |
| chr19_-_46717076 | 0.52 |
ENST00000601806.5
ENST00000593363.1 ENST00000291281.9 ENST00000598633.1 ENST00000595515.5 ENST00000433867.5 |
PRKD2
|
protein kinase D2 |
| chr14_-_34874887 | 0.52 |
ENST00000382422.6
|
BAZ1A
|
bromodomain adjacent to zinc finger domain 1A |
| chr4_-_156970903 | 0.52 |
ENST00000422544.2
|
PDGFC
|
platelet derived growth factor C |
| chr1_+_147902789 | 0.51 |
ENST00000369235.2
|
GJA8
|
gap junction protein alpha 8 |
| chr6_+_131199979 | 0.51 |
ENST00000537868.5
|
AKAP7
|
A-kinase anchoring protein 7 |
| chr12_+_26195313 | 0.51 |
ENST00000422622.3
|
SSPN
|
sarcospan |
| chr6_+_158649997 | 0.51 |
ENST00000360448.8
ENST00000367081.7 ENST00000611299.5 |
SYTL3
|
synaptotagmin like 3 |
| chr5_-_161546970 | 0.50 |
ENST00000675303.1
|
GABRB2
|
gamma-aminobutyric acid type A receptor subunit beta2 |
| chr19_+_13024573 | 0.50 |
ENST00000358552.7
ENST00000360105.8 ENST00000588228.5 ENST00000676441.1 ENST00000591028.1 |
NFIX
|
nuclear factor I X |
| chr1_+_170664121 | 0.50 |
ENST00000239461.11
|
PRRX1
|
paired related homeobox 1 |
| chr21_-_34888683 | 0.49 |
ENST00000344691.8
ENST00000358356.9 |
RUNX1
|
RUNX family transcription factor 1 |
| chr5_-_160852200 | 0.49 |
ENST00000327245.10
|
ATP10B
|
ATPase phospholipid transporting 10B (putative) |
| chr11_-_63671909 | 0.47 |
ENST00000538786.1
ENST00000540699.1 |
ATL3
|
atlastin GTPase 3 |
| chr5_-_33891941 | 0.47 |
ENST00000352040.7
ENST00000504830.6 |
ADAMTS12
|
ADAM metallopeptidase with thrombospondin type 1 motif 12 |
| chr1_-_158426237 | 0.47 |
ENST00000641042.1
|
OR10K2
|
olfactory receptor family 10 subfamily K member 2 |
| chr20_-_31722533 | 0.46 |
ENST00000677194.1
ENST00000434194.2 ENST00000376062.6 |
BCL2L1
|
BCL2 like 1 |
| chrX_-_66040072 | 0.46 |
ENST00000374737.9
|
VSIG4
|
V-set and immunoglobulin domain containing 4 |
| chr1_+_89821003 | 0.45 |
ENST00000525774.5
ENST00000337338.9 |
LRRC8D
|
leucine rich repeat containing 8 VRAC subunit D |
| chr20_+_44531817 | 0.45 |
ENST00000372889.5
ENST00000372887.5 |
PKIG
|
cAMP-dependent protein kinase inhibitor gamma |
| chr17_+_75979211 | 0.45 |
ENST00000397640.6
ENST00000588202.5 ENST00000590676.1 ENST00000586891.1 |
TEN1
|
TEN1 subunit of CST complex |
| chr11_+_31812307 | 0.45 |
ENST00000643436.1
ENST00000646959.1 ENST00000645942.1 ENST00000530348.5 |
PAUPAR
ENSG00000285283.1
|
PAX6 upstream antisense RNA novel protein |
| chr5_-_179620933 | 0.44 |
ENST00000521173.5
|
HNRNPH1
|
heterogeneous nuclear ribonucleoprotein H1 |
| chrX_-_66040057 | 0.44 |
ENST00000412866.2
|
VSIG4
|
V-set and immunoglobulin domain containing 4 |
| chr5_-_143400716 | 0.43 |
ENST00000424646.6
ENST00000652686.1 |
NR3C1
|
nuclear receptor subfamily 3 group C member 1 |
| chr19_-_633500 | 0.43 |
ENST00000588649.7
|
POLRMT
|
RNA polymerase mitochondrial |
| chr17_-_8087709 | 0.42 |
ENST00000647874.1
|
ALOX12B
|
arachidonate 12-lipoxygenase, 12R type |
| chrX_-_49184789 | 0.42 |
ENST00000453382.5
ENST00000432913.5 |
PRICKLE3
|
prickle planar cell polarity protein 3 |
| chr7_-_106285898 | 0.42 |
ENST00000424768.2
ENST00000681255.1 |
NAMPT
|
nicotinamide phosphoribosyltransferase |
| chrX_-_66040107 | 0.42 |
ENST00000455586.6
|
VSIG4
|
V-set and immunoglobulin domain containing 4 |
| chr7_+_116526277 | 0.42 |
ENST00000393468.1
ENST00000393467.1 |
CAV1
|
caveolin 1 |
| chr3_+_122055355 | 0.41 |
ENST00000330540.7
ENST00000469710.5 ENST00000493101.5 |
CD86
|
CD86 molecule |
| chr4_-_122621011 | 0.41 |
ENST00000611104.2
ENST00000648588.1 |
IL21
|
interleukin 21 |
| chr10_+_5692792 | 0.41 |
ENST00000645567.1
|
TASOR2
|
transcription activation suppressor family member 2 |
| chr4_-_47981535 | 0.41 |
ENST00000402813.9
|
CNGA1
|
cyclic nucleotide gated channel subunit alpha 1 |
| chr14_+_51489112 | 0.41 |
ENST00000356218.8
|
FRMD6
|
FERM domain containing 6 |
| chr12_+_26195647 | 0.40 |
ENST00000535504.1
|
SSPN
|
sarcospan |
| chr14_-_34875348 | 0.40 |
ENST00000360310.6
|
BAZ1A
|
bromodomain adjacent to zinc finger domain 1A |
| chr16_-_69754913 | 0.40 |
ENST00000268802.10
|
NOB1
|
NIN1 (RPN12) binding protein 1 homolog |
| chr12_-_99154867 | 0.39 |
ENST00000549025.6
|
ANKS1B
|
ankyrin repeat and sterile alpha motif domain containing 1B |
| chr6_-_160664270 | 0.39 |
ENST00000316300.10
|
LPA
|
lipoprotein(a) |
| chr12_-_81758641 | 0.39 |
ENST00000552948.5
ENST00000548586.5 |
PPFIA2
|
PTPRF interacting protein alpha 2 |
| chr4_+_80266639 | 0.39 |
ENST00000456523.3
ENST00000312465.12 |
FGF5
|
fibroblast growth factor 5 |
| chr6_+_106098933 | 0.39 |
ENST00000369089.3
|
PRDM1
|
PR/SET domain 1 |
| chr19_+_9087061 | 0.39 |
ENST00000641627.1
|
OR1M1
|
olfactory receptor family 1 subfamily M member 1 |
| chr8_-_121641424 | 0.39 |
ENST00000303924.5
|
HAS2
|
hyaluronan synthase 2 |
| chr10_+_84230660 | 0.39 |
ENST00000652073.1
|
RGR
|
retinal G protein coupled receptor |
| chr3_+_128052390 | 0.38 |
ENST00000481210.5
ENST00000243253.8 |
SEC61A1
|
SEC61 translocon subunit alpha 1 |
| chrX_+_137566119 | 0.38 |
ENST00000287538.10
|
ZIC3
|
Zic family member 3 |
| chr12_+_26195543 | 0.38 |
ENST00000242729.7
|
SSPN
|
sarcospan |
| chr6_-_42722590 | 0.38 |
ENST00000230381.7
|
PRPH2
|
peripherin 2 |
| chr1_+_39955112 | 0.37 |
ENST00000420632.6
ENST00000372811.10 ENST00000434861.5 ENST00000372809.5 |
MFSD2A
|
major facilitator superfamily domain containing 2A |
| chr20_-_31722854 | 0.37 |
ENST00000307677.5
|
BCL2L1
|
BCL2 like 1 |
| chrX_-_115018973 | 0.37 |
ENST00000371936.5
|
IL13RA2
|
interleukin 13 receptor subunit alpha 2 |
| chr5_-_9546066 | 0.36 |
ENST00000382496.10
ENST00000652226.1 |
SEMA5A
|
semaphorin 5A |
| chr1_+_150257247 | 0.35 |
ENST00000647854.1
|
CA14
|
carbonic anhydrase 14 |
| chr17_+_17179527 | 0.35 |
ENST00000579361.1
|
MPRIP
|
myosin phosphatase Rho interacting protein |
| chr1_+_150257764 | 0.35 |
ENST00000369111.9
|
CA14
|
carbonic anhydrase 14 |
| chr12_+_53268289 | 0.35 |
ENST00000257934.9
|
ESPL1
|
extra spindle pole bodies like 1, separase |
| chr3_-_157503574 | 0.35 |
ENST00000494677.5
ENST00000468233.5 |
VEPH1
|
ventricular zone expressed PH domain containing 1 |
| chr3_-_71583683 | 0.34 |
ENST00000649631.1
ENST00000648718.1 |
FOXP1
|
forkhead box P1 |
| chr5_-_161546708 | 0.34 |
ENST00000393959.6
|
GABRB2
|
gamma-aminobutyric acid type A receptor subunit beta2 |
| chr1_+_67685342 | 0.33 |
ENST00000617962.2
|
GADD45A
|
growth arrest and DNA damage inducible alpha |
| chr3_-_116445458 | 0.33 |
ENST00000490035.7
|
LSAMP
|
limbic system associated membrane protein |
| chr5_-_16936231 | 0.33 |
ENST00000507288.1
ENST00000274203.13 ENST00000513610.6 |
MYO10
|
myosin X |
| chr6_+_45422564 | 0.32 |
ENST00000625924.1
|
RUNX2
|
RUNX family transcription factor 2 |
| chrX_-_53684119 | 0.32 |
ENST00000342160.7
ENST00000446750.1 |
HUWE1
|
HECT, UBA and WWE domain containing E3 ubiquitin protein ligase 1 |
| chr3_+_12351493 | 0.32 |
ENST00000683699.1
|
PPARG
|
peroxisome proliferator activated receptor gamma |
| chr9_-_21994345 | 0.31 |
ENST00000579755.2
ENST00000530628.2 |
CDKN2A
|
cyclin dependent kinase inhibitor 2A |
| chr16_+_7304219 | 0.31 |
ENST00000675562.1
|
RBFOX1
|
RNA binding fox-1 homolog 1 |
| chr12_-_99154492 | 0.31 |
ENST00000546568.5
ENST00000546960.5 |
ANKS1B
|
ankyrin repeat and sterile alpha motif domain containing 1B |
| chr14_-_56810448 | 0.30 |
ENST00000339475.10
ENST00000555006.5 ENST00000672264.2 ENST00000554559.5 ENST00000555804.1 |
OTX2
|
orthodenticle homeobox 2 |
| chr17_-_8295342 | 0.30 |
ENST00000579192.5
ENST00000577745.2 |
SLC25A35
|
solute carrier family 25 member 35 |
| chr17_-_31297231 | 0.30 |
ENST00000247271.5
|
OMG
|
oligodendrocyte myelin glycoprotein |
| chr14_+_36661852 | 0.30 |
ENST00000361487.7
|
PAX9
|
paired box 9 |
| chr12_-_55842927 | 0.30 |
ENST00000322569.9
ENST00000409200.7 |
MMP19
|
matrix metallopeptidase 19 |
| chr6_+_42564060 | 0.30 |
ENST00000372903.6
|
UBR2
|
ubiquitin protein ligase E3 component n-recognin 2 |
| chr12_-_99154746 | 0.30 |
ENST00000549558.6
ENST00000550693.6 ENST00000549493.6 |
ANKS1B
|
ankyrin repeat and sterile alpha motif domain containing 1B |
| chr1_+_170663917 | 0.29 |
ENST00000497230.2
|
PRRX1
|
paired related homeobox 1 |
| chr6_+_13013513 | 0.29 |
ENST00000675203.1
|
PHACTR1
|
phosphatase and actin regulator 1 |
| chr18_+_74597850 | 0.29 |
ENST00000582337.5
ENST00000299687.10 |
ZNF407
|
zinc finger protein 407 |
| chr5_+_177600104 | 0.29 |
ENST00000029410.10
ENST00000510761.1 ENST00000505468.1 |
B4GALT7
|
beta-1,4-galactosyltransferase 7 |
| chr6_+_45422485 | 0.28 |
ENST00000359524.7
|
RUNX2
|
RUNX family transcription factor 2 |
| chr17_-_75262344 | 0.28 |
ENST00000579743.2
ENST00000578348.5 ENST00000582486.5 ENST00000582717.5 |
GGA3
|
golgi associated, gamma adaptin ear containing, ARF binding protein 3 |
| chr7_-_51316754 | 0.28 |
ENST00000632460.1
ENST00000441453.5 ENST00000648294.1 ENST00000265136.12 ENST00000395542.6 ENST00000395540.6 |
COBL
|
cordon-bleu WH2 repeat protein |
| chr1_-_94927079 | 0.28 |
ENST00000370206.9
ENST00000394202.8 |
CNN3
|
calponin 3 |
| chr9_-_111330224 | 0.28 |
ENST00000302681.3
|
OR2K2
|
olfactory receptor family 2 subfamily K member 2 |
| chr4_+_87650277 | 0.28 |
ENST00000339673.11
ENST00000282479.8 |
DMP1
|
dentin matrix acidic phosphoprotein 1 |
| chr9_-_92482350 | 0.27 |
ENST00000375543.2
|
ASPN
|
asporin |
| chr5_+_32711723 | 0.27 |
ENST00000415167.2
|
NPR3
|
natriuretic peptide receptor 3 |
| chr20_+_18813777 | 0.27 |
ENST00000377428.4
|
SCP2D1
|
SCP2 sterol binding domain containing 1 |
| chr17_-_75393656 | 0.27 |
ENST00000392563.5
|
GRB2
|
growth factor receptor bound protein 2 |
| chr1_+_110873135 | 0.27 |
ENST00000271324.6
|
CD53
|
CD53 molecule |
| chr14_+_22508602 | 0.26 |
ENST00000390504.1
|
TRAJ33
|
T cell receptor alpha joining 33 |
| chr17_-_676348 | 0.26 |
ENST00000681510.1
ENST00000679680.1 |
VPS53
|
VPS53 subunit of GARP complex |
| chr11_-_5269933 | 0.26 |
ENST00000396895.3
|
HBE1
|
hemoglobin subunit epsilon 1 |
| chr17_+_2056073 | 0.26 |
ENST00000576444.1
ENST00000322941.3 |
HIC1
|
HIC ZBTB transcriptional repressor 1 |
| chr3_-_71583592 | 0.26 |
ENST00000650156.1
ENST00000649596.1 |
FOXP1
|
forkhead box P1 |
| chr5_+_119629552 | 0.26 |
ENST00000613773.4
ENST00000620555.4 ENST00000515256.5 ENST00000509264.1 |
FAM170A
|
family with sequence similarity 170 member A |
| chr3_-_116444983 | 0.26 |
ENST00000333617.8
|
LSAMP
|
limbic system associated membrane protein |
| chr8_+_85209213 | 0.25 |
ENST00000520225.1
|
E2F5
|
E2F transcription factor 5 |
| chr20_-_31722949 | 0.25 |
ENST00000376055.9
|
BCL2L1
|
BCL2 like 1 |
| chr16_-_3018170 | 0.25 |
ENST00000572154.1
ENST00000328796.5 |
CLDN6
|
claudin 6 |
| chr19_+_58544045 | 0.25 |
ENST00000253024.10
ENST00000593582.5 |
TRIM28
|
tripartite motif containing 28 |
| chr1_-_243850216 | 0.24 |
ENST00000673466.1
|
AKT3
|
AKT serine/threonine kinase 3 |
| chr17_-_75393741 | 0.24 |
ENST00000578961.5
ENST00000392564.5 |
GRB2
|
growth factor receptor bound protein 2 |
| chr1_+_213989691 | 0.24 |
ENST00000607425.1
|
PROX1
|
prospero homeobox 1 |
| chr1_+_152878312 | 0.24 |
ENST00000368765.4
|
SMCP
|
sperm mitochondria associated cysteine rich protein |
| chr18_+_34710307 | 0.23 |
ENST00000679796.1
|
DTNA
|
dystrobrevin alpha |
| chr17_+_42609641 | 0.23 |
ENST00000681413.1
ENST00000251413.8 ENST00000591509.5 |
TUBG1
|
tubulin gamma 1 |
| chr16_+_30664334 | 0.23 |
ENST00000287468.5
|
FBRS
|
fibrosin |
| chr5_+_137867852 | 0.23 |
ENST00000421631.6
ENST00000239926.9 |
MYOT
|
myotilin |
| chr17_-_28368012 | 0.23 |
ENST00000555059.2
|
ENSG00000273171.1
|
novel protein, readthrough between VTN and SEBOX |
| chr1_-_115768702 | 0.23 |
ENST00000261448.6
|
CASQ2
|
calsequestrin 2 |
| chr4_-_108168950 | 0.23 |
ENST00000379951.6
|
LEF1
|
lymphoid enhancer binding factor 1 |
| chr8_-_673547 | 0.23 |
ENST00000522893.1
|
ERICH1
|
glutamate rich 1 |
| chr4_-_40630588 | 0.23 |
ENST00000514014.1
|
RBM47
|
RNA binding motif protein 47 |
| chr4_-_108168919 | 0.23 |
ENST00000265165.6
|
LEF1
|
lymphoid enhancer binding factor 1 |
| chrX_+_47218670 | 0.23 |
ENST00000357227.9
ENST00000519758.5 ENST00000520893.5 ENST00000622098.4 ENST00000517426.5 |
CDK16
|
cyclin dependent kinase 16 |
| chr7_+_123601815 | 0.23 |
ENST00000451215.6
|
ASB15
|
ankyrin repeat and SOCS box containing 15 |
| chr17_+_7630094 | 0.22 |
ENST00000441599.6
ENST00000380450.9 ENST00000416273.7 ENST00000575903.5 ENST00000571153.5 ENST00000575618.5 ENST00000576152.1 ENST00000576830.5 |
SHBG
|
sex hormone binding globulin |
| chr5_-_102296260 | 0.22 |
ENST00000310954.7
|
SLCO4C1
|
solute carrier organic anion transporter family member 4C1 |
| chr22_-_30299482 | 0.22 |
ENST00000434291.5
|
ENSG00000248751.6
|
novel protein |
| chr5_+_32711313 | 0.22 |
ENST00000265074.13
|
NPR3
|
natriuretic peptide receptor 3 |
| chrX_+_47218232 | 0.22 |
ENST00000457458.6
ENST00000522883.1 |
CDK16
|
cyclin dependent kinase 16 |
| chr12_-_119803383 | 0.21 |
ENST00000392520.2
ENST00000678677.1 ENST00000679249.1 ENST00000676849.1 |
CIT
|
citron rho-interacting serine/threonine kinase |
| chrX_+_136532205 | 0.21 |
ENST00000370634.8
|
VGLL1
|
vestigial like family member 1 |
| chr7_+_123601836 | 0.21 |
ENST00000434204.5
|
ASB15
|
ankyrin repeat and SOCS box containing 15 |
| chr6_-_138499487 | 0.21 |
ENST00000343505.9
|
NHSL1
|
NHS like 1 |
| chr7_+_114414997 | 0.21 |
ENST00000462331.5
ENST00000393491.7 ENST00000403559.8 ENST00000408937.7 ENST00000393498.6 ENST00000393495.7 ENST00000378237.7 |
FOXP2
|
forkhead box P2 |
| chr3_+_138621225 | 0.21 |
ENST00000479848.1
|
FAIM
|
Fas apoptotic inhibitory molecule |
| chr16_+_21678514 | 0.21 |
ENST00000286149.8
ENST00000388958.8 |
OTOA
|
otoancorin |
| chr6_-_130222813 | 0.20 |
ENST00000437477.6
ENST00000439090.7 |
SAMD3
|
sterile alpha motif domain containing 3 |
| chr11_-_31807617 | 0.20 |
ENST00000639920.1
ENST00000640460.1 |
PAX6
|
paired box 6 |
| chr14_+_22168387 | 0.20 |
ENST00000557168.1
|
TRAV30
|
T cell receptor alpha variable 30 |
| chr3_+_189631373 | 0.20 |
ENST00000264731.8
ENST00000418709.6 ENST00000320472.9 ENST00000392460.7 ENST00000440651.6 |
TP63
|
tumor protein p63 |
| chr14_-_56810380 | 0.20 |
ENST00000672125.1
ENST00000673481.1 |
OTX2
|
orthodenticle homeobox 2 |
| chr16_+_30075967 | 0.20 |
ENST00000279387.12
ENST00000562664.5 ENST00000627746.2 ENST00000562222.5 |
PPP4C
|
protein phosphatase 4 catalytic subunit |
| chr3_-_51499950 | 0.19 |
ENST00000423656.5
ENST00000504652.5 ENST00000684031.1 |
DCAF1
|
DDB1 and CUL4 associated factor 1 |
| chr6_-_87702221 | 0.19 |
ENST00000257787.6
|
AKIRIN2
|
akirin 2 |
| chr10_-_59753444 | 0.19 |
ENST00000594536.5
ENST00000414264.6 |
MRLN
|
myoregulin |
| chr7_-_107803215 | 0.19 |
ENST00000340010.10
ENST00000453332.1 |
SLC26A3
|
solute carrier family 26 member 3 |
| chr8_-_115492221 | 0.19 |
ENST00000518018.1
|
TRPS1
|
transcriptional repressor GATA binding 1 |
| chr17_+_45061296 | 0.19 |
ENST00000590310.2
ENST00000677949.1 ENST00000258960.7 |
NMT1
|
N-myristoyltransferase 1 |
| chr6_+_31547560 | 0.19 |
ENST00000376148.9
ENST00000376145.8 |
NFKBIL1
|
NFKB inhibitor like 1 |
| chrX_-_153971169 | 0.19 |
ENST00000369984.4
|
HCFC1
|
host cell factor C1 |
| chr22_-_41446777 | 0.19 |
ENST00000434408.1
ENST00000327492.4 |
TOB2
|
transducer of ERBB2, 2 |
| chr10_-_24721866 | 0.18 |
ENST00000416305.1
ENST00000320481.10 |
ARHGAP21
|
Rho GTPase activating protein 21 |
| chr3_-_197183806 | 0.18 |
ENST00000671246.1
ENST00000660553.1 |
DLG1
|
discs large MAGUK scaffold protein 1 |
| chr8_+_79611036 | 0.18 |
ENST00000220876.12
ENST00000518111.5 |
STMN2
|
stathmin 2 |
| chr8_+_76681208 | 0.18 |
ENST00000651372.2
|
ZFHX4
|
zinc finger homeobox 4 |
| chrX_-_124963768 | 0.18 |
ENST00000371130.7
ENST00000422452.2 |
TENM1
|
teneurin transmembrane protein 1 |
| chr10_-_72088972 | 0.18 |
ENST00000317376.8
ENST00000412663.5 |
SPOCK2
|
SPARC (osteonectin), cwcv and kazal like domains proteoglycan 2 |
| chr11_-_70717994 | 0.18 |
ENST00000659264.1
|
SHANK2
|
SH3 and multiple ankyrin repeat domains 2 |
| chr10_+_84173793 | 0.18 |
ENST00000372126.4
|
C10orf99
|
chromosome 10 open reading frame 99 |
| chr12_+_80716906 | 0.18 |
ENST00000228644.4
|
MYF5
|
myogenic factor 5 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:0061394 | regulation of transcription from RNA polymerase II promoter in response to arsenic-containing substance(GO:0061394) |
| 0.3 | 4.1 | GO:1900625 | regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 0.2 | 3.8 | GO:0045636 | positive regulation of melanocyte differentiation(GO:0045636) |
| 0.2 | 0.9 | GO:2000681 | negative regulation of rubidium ion transport(GO:2000681) negative regulation of rubidium ion transmembrane transporter activity(GO:2000687) |
| 0.2 | 1.3 | GO:0061767 | negative regulation of lung blood pressure(GO:0061767) |
| 0.2 | 0.8 | GO:0046035 | CMP salvage(GO:0006238) CMP biosynthetic process(GO:0009224) CMP metabolic process(GO:0046035) |
| 0.2 | 1.2 | GO:0043932 | ossification involved in bone remodeling(GO:0043932) |
| 0.2 | 1.0 | GO:1900383 | regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.1 | 1.0 | GO:0048165 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) negative regulation of chondrocyte proliferation(GO:1902731) |
| 0.1 | 0.4 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.1 | 0.4 | GO:0002665 | negative regulation of T cell tolerance induction(GO:0002665) negative regulation of T cell anergy(GO:0002668) negative regulation of lymphocyte anergy(GO:0002912) regulation of lymphotoxin A production(GO:0032681) positive regulation of lymphotoxin A production(GO:0032761) regulation of lymphotoxin A biosynthetic process(GO:0043016) positive regulation of lymphotoxin A biosynthetic process(GO:0043017) |
| 0.1 | 1.1 | GO:0060154 | cellular process regulating host cell cycle in response to virus(GO:0060154) |
| 0.1 | 0.7 | GO:0010513 | positive regulation of phosphatidylinositol biosynthetic process(GO:0010513) |
| 0.1 | 0.4 | GO:1990654 | regulation of extrathymic T cell differentiation(GO:0033082) sebum secreting cell proliferation(GO:1990654) |
| 0.1 | 2.3 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.1 | 0.4 | GO:1900085 | negative regulation of peptidyl-tyrosine autophosphorylation(GO:1900085) negative regulation of inward rectifier potassium channel activity(GO:1903609) |
| 0.1 | 3.9 | GO:0061756 | leukocyte adhesion to vascular endothelial cell(GO:0061756) |
| 0.1 | 1.6 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.1 | 0.5 | GO:1902202 | proteoglycan catabolic process(GO:0030167) regulation of hepatocyte growth factor receptor signaling pathway(GO:1902202) |
| 0.1 | 0.8 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.1 | 0.5 | GO:0061152 | trachea submucosa development(GO:0061152) trachea gland development(GO:0061153) |
| 0.1 | 0.4 | GO:0031117 | positive regulation of microtubule depolymerization(GO:0031117) |
| 0.1 | 0.8 | GO:0001554 | luteolysis(GO:0001554) |
| 0.1 | 0.4 | GO:0003383 | apical constriction(GO:0003383) |
| 0.1 | 0.2 | GO:0043049 | hepatocyte cell migration(GO:0002194) otic placode formation(GO:0043049) branching involved in pancreas morphogenesis(GO:0061114) acinar cell differentiation(GO:0090425) positive regulation of forebrain neuron differentiation(GO:2000979) |
| 0.1 | 0.3 | GO:0019355 | nicotinamide nucleotide biosynthetic process from aspartate(GO:0019355) 'de novo' NAD biosynthetic process from aspartate(GO:0034628) |
| 0.1 | 0.3 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.1 | 0.2 | GO:0021757 | caudate nucleus development(GO:0021757) putamen development(GO:0021758) |
| 0.1 | 0.3 | GO:0045875 | negative regulation of sister chromatid cohesion(GO:0045875) |
| 0.1 | 0.6 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 0.1 | 0.7 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.1 | 0.4 | GO:0070171 | negative regulation of tooth mineralization(GO:0070171) |
| 0.1 | 0.2 | GO:1990164 | histone H2A phosphorylation(GO:1990164) |
| 0.1 | 0.4 | GO:0039019 | pronephric nephron development(GO:0039019) |
| 0.1 | 0.3 | GO:1902080 | regulation of calcium ion import into sarcoplasmic reticulum(GO:1902080) negative regulation of calcium ion import into sarcoplasmic reticulum(GO:1902081) |
| 0.1 | 0.2 | GO:0019046 | release from viral latency(GO:0019046) |
| 0.1 | 1.1 | GO:0032625 | interleukin-21 production(GO:0032625) interleukin-21 secretion(GO:0072619) |
| 0.1 | 0.2 | GO:0090290 | positive regulation of osteoclast proliferation(GO:0090290) |
| 0.1 | 0.5 | GO:0089700 | protein kinase D signaling(GO:0089700) |
| 0.1 | 0.2 | GO:0001897 | cytolysis by symbiont of host cells(GO:0001897) |
| 0.1 | 1.0 | GO:0051386 | regulation of neurotrophin TRK receptor signaling pathway(GO:0051386) |
| 0.1 | 0.8 | GO:0048664 | neuron fate determination(GO:0048664) |
| 0.1 | 0.3 | GO:0001757 | somite specification(GO:0001757) |
| 0.1 | 0.1 | GO:0060648 | mammary gland bud morphogenesis(GO:0060648) |
| 0.1 | 0.4 | GO:0043402 | glucocorticoid mediated signaling pathway(GO:0043402) |
| 0.1 | 1.4 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.1 | 0.2 | GO:0007499 | ectoderm and mesoderm interaction(GO:0007499) |
| 0.0 | 0.2 | GO:1901536 | positive regulation of methylation-dependent chromatin silencing(GO:0090309) regulation of DNA demethylation(GO:1901535) negative regulation of DNA demethylation(GO:1901536) regulation of genetic imprinting(GO:2000653) |
| 0.0 | 0.2 | GO:0018008 | N-terminal peptidyl-glycine N-myristoylation(GO:0018008) |
| 0.0 | 0.4 | GO:0035469 | determination of pancreatic left/right asymmetry(GO:0035469) |
| 0.0 | 0.4 | GO:0051122 | hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.0 | 1.5 | GO:0043586 | tongue development(GO:0043586) |
| 0.0 | 0.1 | GO:1903033 | regulation of microtubule plus-end binding(GO:1903031) positive regulation of microtubule plus-end binding(GO:1903033) |
| 0.0 | 0.4 | GO:0048842 | positive regulation of axon extension involved in axon guidance(GO:0048842) |
| 0.0 | 0.5 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.0 | 0.6 | GO:0019227 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.0 | 0.2 | GO:0015722 | canalicular bile acid transport(GO:0015722) |
| 0.0 | 0.3 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.0 | 0.2 | GO:1902775 | mitochondrial large ribosomal subunit assembly(GO:1902775) |
| 0.0 | 0.2 | GO:0007341 | penetration of zona pellucida(GO:0007341) |
| 0.0 | 0.3 | GO:2000230 | negative regulation of pancreatic stellate cell proliferation(GO:2000230) |
| 0.0 | 0.5 | GO:0002158 | osteoclast proliferation(GO:0002158) |
| 0.0 | 0.1 | GO:2000863 | negative regulation of macrophage chemotaxis(GO:0010760) positive regulation of estrogen secretion(GO:2000863) positive regulation of estradiol secretion(GO:2000866) |
| 0.0 | 0.4 | GO:0043305 | negative regulation of mast cell degranulation(GO:0043305) |
| 0.0 | 0.1 | GO:0060024 | rhythmic synaptic transmission(GO:0060024) |
| 0.0 | 0.1 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.0 | 0.2 | GO:0097475 | motor neuron migration(GO:0097475) |
| 0.0 | 0.3 | GO:0070170 | regulation of tooth mineralization(GO:0070170) |
| 0.0 | 0.3 | GO:0035986 | senescence-associated heterochromatin focus assembly(GO:0035986) |
| 0.0 | 0.5 | GO:2000543 | positive regulation of gastrulation(GO:2000543) |
| 0.0 | 0.2 | GO:0086029 | Purkinje myocyte to ventricular cardiac muscle cell signaling(GO:0086029) Purkinje myocyte to ventricular cardiac muscle cell communication(GO:0086068) |
| 0.0 | 0.1 | GO:0061055 | myotome development(GO:0061055) |
| 0.0 | 0.3 | GO:0035989 | tendon development(GO:0035989) |
| 0.0 | 0.1 | GO:0033634 | positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) negative regulation of endothelial cell chemotaxis(GO:2001027) |
| 0.0 | 0.1 | GO:0072268 | pattern specification involved in metanephros development(GO:0072268) |
| 0.0 | 0.6 | GO:0060670 | branching involved in labyrinthine layer morphogenesis(GO:0060670) |
| 0.0 | 0.1 | GO:0035054 | atrial septum secundum morphogenesis(GO:0003290) embryonic heart tube anterior/posterior pattern specification(GO:0035054) |
| 0.0 | 0.1 | GO:0021847 | ventricular zone neuroblast division(GO:0021847) |
| 0.0 | 0.1 | GO:1904732 | regulation of electron carrier activity(GO:1904732) |
| 0.0 | 0.2 | GO:0030950 | establishment or maintenance of actin cytoskeleton polarity(GO:0030950) |
| 0.0 | 1.1 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 0.2 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.0 | 0.1 | GO:0071802 | negative regulation of podosome assembly(GO:0071802) |
| 0.0 | 0.4 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.0 | 0.5 | GO:0007614 | short-term memory(GO:0007614) |
| 0.0 | 0.2 | GO:0048743 | positive regulation of skeletal muscle fiber development(GO:0048743) |
| 0.0 | 0.1 | GO:0040030 | regulation of molecular function, epigenetic(GO:0040030) positive regulation of melanin biosynthetic process(GO:0048023) positive regulation of secondary metabolite biosynthetic process(GO:1900378) |
| 0.0 | 0.1 | GO:0032416 | negative regulation of sodium:proton antiporter activity(GO:0032416) musculoskeletal movement, spinal reflex action(GO:0050883) |
| 0.0 | 0.2 | GO:2000660 | negative regulation of interleukin-1-mediated signaling pathway(GO:2000660) |
| 0.0 | 0.4 | GO:0023019 | signal transduction involved in regulation of gene expression(GO:0023019) |
| 0.0 | 0.4 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 0.4 | GO:0048934 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.0 | 0.1 | GO:0035610 | protein side chain deglutamylation(GO:0035610) |
| 0.0 | 0.3 | GO:0042481 | regulation of odontogenesis(GO:0042481) |
| 0.0 | 0.1 | GO:0042663 | regulation of endodermal cell fate specification(GO:0042663) |
| 0.0 | 0.1 | GO:0046116 | queuosine biosynthetic process(GO:0008616) queuosine metabolic process(GO:0046116) |
| 0.0 | 0.1 | GO:0060054 | positive regulation of epithelial cell proliferation involved in wound healing(GO:0060054) |
| 0.0 | 0.4 | GO:0034356 | NAD biosynthesis via nicotinamide riboside salvage pathway(GO:0034356) |
| 0.0 | 0.1 | GO:0001807 | type IV hypersensitivity(GO:0001806) regulation of type IV hypersensitivity(GO:0001807) negative regulation of type IV hypersensitivity(GO:0001808) |
| 0.0 | 0.5 | GO:0007171 | activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) |
| 0.0 | 0.2 | GO:0045852 | pH elevation(GO:0045852) intracellular pH elevation(GO:0051454) |
| 0.0 | 0.5 | GO:0035641 | locomotory exploration behavior(GO:0035641) |
| 0.0 | 0.4 | GO:0060856 | establishment of blood-brain barrier(GO:0060856) |
| 0.0 | 0.1 | GO:0000720 | pyrimidine dimer repair by nucleotide-excision repair(GO:0000720) |
| 0.0 | 0.1 | GO:0007060 | male meiosis chromosome segregation(GO:0007060) |
| 0.0 | 0.6 | GO:0035590 | purinergic nucleotide receptor signaling pathway(GO:0035590) |
| 0.0 | 0.3 | GO:0045078 | positive regulation of interferon-gamma biosynthetic process(GO:0045078) |
| 0.0 | 0.5 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.0 | 0.4 | GO:0000469 | cleavage involved in rRNA processing(GO:0000469) |
| 0.0 | 0.3 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.0 | 0.1 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.0 | 0.3 | GO:0034374 | low-density lipoprotein particle remodeling(GO:0034374) |
| 0.0 | 0.1 | GO:0055059 | asymmetric neuroblast division(GO:0055059) |
| 0.0 | 0.2 | GO:0000212 | meiotic spindle organization(GO:0000212) |
| 0.0 | 0.2 | GO:0031665 | negative regulation of lipopolysaccharide-mediated signaling pathway(GO:0031665) |
| 0.0 | 0.1 | GO:0090158 | endoplasmic reticulum membrane organization(GO:0090158) |
| 0.0 | 0.2 | GO:0003322 | pancreatic A cell development(GO:0003322) |
| 0.0 | 0.1 | GO:0090292 | nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.0 | 0.1 | GO:0010961 | cellular magnesium ion homeostasis(GO:0010961) |
| 0.0 | 0.1 | GO:0071350 | interleukin-15-mediated signaling pathway(GO:0035723) cellular response to interleukin-15(GO:0071350) |
| 0.0 | 0.3 | GO:0043517 | positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) |
| 0.0 | 0.4 | GO:0022400 | regulation of rhodopsin mediated signaling pathway(GO:0022400) |
| 0.0 | 0.5 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.0 | 0.6 | GO:0034142 | toll-like receptor 4 signaling pathway(GO:0034142) |
| 0.0 | 0.3 | GO:0071850 | mitotic cell cycle arrest(GO:0071850) |
| 0.0 | 0.2 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.0 | 0.3 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.0 | 0.2 | GO:1903764 | regulation of voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization(GO:1903760) regulation of potassium ion export across plasma membrane(GO:1903764) |
| 0.0 | 0.1 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.0 | 0.4 | GO:0032211 | negative regulation of telomere maintenance via telomerase(GO:0032211) |
| 0.0 | 0.7 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 0.3 | GO:1901741 | positive regulation of myoblast fusion(GO:1901741) |
| 0.0 | 0.1 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.0 | 0.1 | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.0 | 0.2 | GO:1900103 | positive regulation of endoplasmic reticulum unfolded protein response(GO:1900103) |
| 0.0 | 0.1 | GO:2000402 | negative regulation of lymphocyte migration(GO:2000402) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 3.7 | GO:0035692 | macrophage migration inhibitory factor receptor complex(GO:0035692) |
| 0.3 | 1.0 | GO:1990622 | CHOP-ATF3 complex(GO:1990622) |
| 0.3 | 0.9 | GO:0008623 | CHRAC(GO:0008623) |
| 0.2 | 1.6 | GO:0097451 | glial limiting end-foot(GO:0097451) |
| 0.1 | 1.3 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.1 | 0.4 | GO:1990879 | CST complex(GO:1990879) |
| 0.1 | 0.3 | GO:0005592 | collagen type XI trimer(GO:0005592) |
| 0.1 | 1.1 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.1 | 0.5 | GO:0070436 | Grb2-EGFR complex(GO:0070436) |
| 0.1 | 1.0 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.0 | 0.2 | GO:0030289 | protein phosphatase 4 complex(GO:0030289) |
| 0.0 | 0.2 | GO:1990578 | perinuclear endoplasmic reticulum membrane(GO:1990578) |
| 0.0 | 0.4 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 0.3 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.0 | 0.3 | GO:1990745 | EARP complex(GO:1990745) |
| 0.0 | 1.4 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.0 | 0.3 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.0 | 0.2 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.0 | 0.1 | GO:0030981 | cortical microtubule cytoskeleton(GO:0030981) |
| 0.0 | 0.5 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.0 | 3.3 | GO:0005901 | caveola(GO:0005901) |
| 0.0 | 0.1 | GO:0002139 | stereocilia coupling link(GO:0002139) periciliary membrane compartment(GO:1990075) |
| 0.0 | 0.2 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.0 | 0.6 | GO:0005639 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) |
| 0.0 | 0.5 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 0.1 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 0.0 | 0.4 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.0 | 0.3 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.0 | 0.3 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.0 | 0.6 | GO:0090665 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 0.3 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.0 | 0.5 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.0 | 0.1 | GO:0000110 | nucleotide-excision repair factor 1 complex(GO:0000110) |
| 0.0 | 0.2 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.0 | 1.0 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 0.1 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.1 | GO:0035061 | perichromatin fibrils(GO:0005726) interchromatin granule(GO:0035061) |
| 0.0 | 0.2 | GO:0005883 | neurofilament(GO:0005883) |
| 0.0 | 0.2 | GO:0046581 | intercellular canaliculus(GO:0046581) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 3.0 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.3 | 0.8 | GO:0050509 | N-acetylglucosaminyl-proteoglycan 4-beta-glucuronosyltransferase activity(GO:0050509) |
| 0.2 | 1.3 | GO:0031962 | mineralocorticoid receptor binding(GO:0031962) |
| 0.1 | 1.4 | GO:0005237 | inhibitory extracellular ligand-gated ion channel activity(GO:0005237) |
| 0.1 | 0.4 | GO:0070320 | inward rectifier potassium channel inhibitor activity(GO:0070320) |
| 0.1 | 0.5 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.1 | 3.7 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.1 | 0.4 | GO:0035373 | chondroitin sulfate proteoglycan binding(GO:0035373) |
| 0.1 | 0.4 | GO:0004883 | glucocorticoid receptor activity(GO:0004883) glucocorticoid-activated RNA polymerase II transcription factor binding transcription factor activity(GO:0038051) |
| 0.1 | 0.4 | GO:0004052 | arachidonate 12-lipoxygenase activity(GO:0004052) |
| 0.1 | 0.4 | GO:0047280 | nicotinamide phosphoribosyltransferase activity(GO:0047280) |
| 0.1 | 1.1 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.1 | 1.0 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.1 | 0.4 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.1 | 0.3 | GO:0000309 | nicotinamide-nucleotide adenylyltransferase activity(GO:0000309) |
| 0.1 | 0.8 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.1 | 0.4 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.1 | 0.2 | GO:0005150 | interleukin-1, Type I receptor binding(GO:0005150) |
| 0.1 | 0.7 | GO:0051378 | amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 0.1 | 0.7 | GO:0005168 | neurotrophin TRKA receptor binding(GO:0005168) |
| 0.1 | 0.6 | GO:0035381 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.0 | 0.2 | GO:0019107 | glycylpeptide N-tetradecanoyltransferase activity(GO:0004379) myristoyltransferase activity(GO:0019107) |
| 0.0 | 0.5 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.0 | 0.3 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.0 | 0.7 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.3 | GO:0060002 | plus-end directed microfilament motor activity(GO:0060002) |
| 0.0 | 0.5 | GO:0030284 | estrogen receptor activity(GO:0030284) |
| 0.0 | 0.4 | GO:0005221 | intracellular cyclic nucleotide activated cation channel activity(GO:0005221) cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.0 | 0.6 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.0 | 0.3 | GO:0003831 | beta-N-acetylglucosaminylglycopeptide beta-1,4-galactosyltransferase activity(GO:0003831) |
| 0.0 | 0.1 | GO:0002046 | opsin binding(GO:0002046) |
| 0.0 | 4.9 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.6 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.0 | 0.5 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.0 | 0.4 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 0.7 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 1.2 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.1 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.0 | 0.2 | GO:0035184 | histone threonine kinase activity(GO:0035184) |
| 0.0 | 0.9 | GO:0001848 | complement binding(GO:0001848) |
| 0.0 | 0.2 | GO:0070728 | leucine binding(GO:0070728) |
| 0.0 | 0.5 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.0 | 0.1 | GO:0004167 | dopachrome isomerase activity(GO:0004167) |
| 0.0 | 0.7 | GO:0031698 | beta-2 adrenergic receptor binding(GO:0031698) |
| 0.0 | 0.2 | GO:0004704 | NF-kappaB-inducing kinase activity(GO:0004704) |
| 0.0 | 0.1 | GO:0030060 | L-malate dehydrogenase activity(GO:0030060) |
| 0.0 | 1.8 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.0 | 0.2 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.0 | 0.5 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 0.2 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.0 | 0.4 | GO:0015245 | fatty acid transporter activity(GO:0015245) |
| 0.0 | 0.9 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.0 | 0.7 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.4 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.0 | 1.5 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 0.8 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.1 | GO:0098821 | BMP receptor activity(GO:0098821) |
| 0.0 | 0.2 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.0 | 0.3 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.0 | 1.0 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.0 | 0.5 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 1.2 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.0 | 0.4 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.0 | 0.1 | GO:0016531 | copper chaperone activity(GO:0016531) |
| 0.0 | 0.2 | GO:0043995 | histone acetyltransferase activity (H4-K5 specific)(GO:0043995) histone acetyltransferase activity (H4-K8 specific)(GO:0043996) histone acetyltransferase activity (H4-K16 specific)(GO:0046972) |
| 0.0 | 0.2 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.0 | 0.3 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.0 | 0.2 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.0 | 0.2 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.0 | 0.1 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.0 | 0.2 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 0.3 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.0 | 0.2 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.0 | 0.5 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.0 | 1.3 | GO:0050681 | androgen receptor binding(GO:0050681) |
| 0.0 | 0.1 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 0.0 | 0.1 | GO:0001595 | angiotensin receptor activity(GO:0001595) angiotensin type II receptor activity(GO:0004945) |
| 0.0 | 0.1 | GO:0031781 | melanocortin receptor binding(GO:0031779) type 3 melanocortin receptor binding(GO:0031781) type 4 melanocortin receptor binding(GO:0031782) |
| 0.0 | 0.5 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.1 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.0 | 0.8 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.0 | 0.9 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.0 | 0.1 | GO:1990599 | 3' overhang single-stranded DNA endodeoxyribonuclease activity(GO:1990599) |
| 0.0 | 0.4 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 0.4 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 0.1 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 0.0 | 0.4 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.0 | 0.1 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.0 | 0.6 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 0.3 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.0 | 0.2 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.0 | 0.5 | GO:0043425 | bHLH transcription factor binding(GO:0043425) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.7 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.1 | 5.5 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.0 | 1.2 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 1.2 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 1.1 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.0 | 0.4 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.0 | 2.1 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 0.4 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 0.9 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.0 | 1.1 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 0.3 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 0.9 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.0 | 1.9 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 0.9 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 5.0 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.4 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 0.7 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 0.4 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.7 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.1 | 0.4 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.0 | 0.7 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.0 | 0.9 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 0.0 | 0.7 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.0 | 0.9 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.0 | 1.3 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 1.1 | REACTOME INFLAMMASOMES | Genes involved in Inflammasomes |
| 0.0 | 1.4 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 0.9 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.0 | 0.4 | REACTOME OPSINS | Genes involved in Opsins |
| 0.0 | 0.5 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 1.0 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 1.0 | REACTOME ACTIVATION OF GENES BY ATF4 | Genes involved in Activation of Genes by ATF4 |
| 0.0 | 1.2 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 0.5 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.0 | 0.8 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 0.7 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.0 | 0.2 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 0.4 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.0 | 0.6 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.2 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.0 | 0.4 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.1 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.0 | 0.2 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.0 | 0.7 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.4 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |
| 0.0 | 0.7 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |